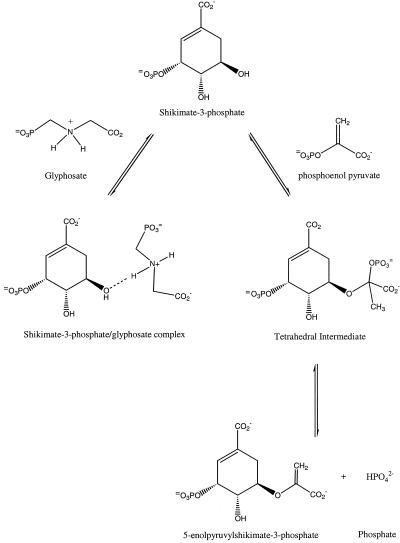
Among enzyme inhibitors used in agriculture, glyphosate (N-phosphomethyl glycine) is remarkable. A nonselective herbicide discovered in 1970 by a group of scientists at Monsanto led by Dr. John Franz (1), glyphosate, since first commercialization under the trade name Roundup, has been used globally as a safe and effective means of weed control. The discovery of glyphosate's herbicidal activity was not quite serendipity, but instead resulted from a synthetic strategy based on the hypothesis that the weak herbicidal activities of related compounds derived from the possibility of their similar metabolic fate (2). Nevertheless, the initial discovery by greenhouse screening has been followed by intensive biochemical studies that have now led to nearly complete understanding of glyphosate's mode of action. In 1972, scientists at Monsanto led by Dr. E. Jaworski observed (3) that application of glyphosate resulted in the inhibition of aromatic amino acid biosynthesis in plants. In 1980, Professor N. Amrhein and coworkers (4) identified its target enzyme from the shikimate pathway (4): 5-enolpyruvoylshikimate-3-phosphate synthase (EPSPS; EC 2.5.1.19). EPSPS is a key enzyme involved in aromatic amino acid biosynthesis (5). The enzyme catalyzes an unusual reaction, wherein the enolpyruvoyl group from phosphoenol pyruvate (PEP) is transferred to the 5-hydroxyl of shikimate-3-phosphate (S3P) to form the products 5-enolpyruvylshikimate-3-phosphate (EPSP) and inorganic phosphate (Pi). The only other enzyme known to catalyze carboxyvinyl transfer by using PEP is UDP-N-acetylglucosamine enolpyruvyl transferase (MurA), which catalyzes the first committed step in the biosynthesis of the peptidoglycan layer of the bacterial cell. In the case of EPSPS, the reaction proceeds through a tetrahedral intermediate (Scheme S1) formed from S3P and PEP (6). Inhibition of EPSPS by glyphosate has been shown to proceed through the formation of an EPSPS-S3P-glyphosate ternary complex and the binding is ordered with glyphosate binding to the enzyme only after the formation of a binary EPSPS-S3P complex. Binding of glyphosate to EPSPS has been shown to be competitive with PEP and uncompetitive with respect to S3P (7). What makes glyphosate a remarkable inhibitor and herbicide? Glyphosate is a relatively simple molecule—an N-methyl phosphonate derivative of glycine with a chemical structure not unlike that of the universal high energy phosphoryl-transfer agent PEP. Despite this, glyphosate retains exquisite specificity for EPSPS and is not known to appreciatively inhibit any other enzyme, even MurA (8).
Scheme 1.
The EPSPS reaction is the penultimate step in the shikimic acid pathway for the biosynthesis of aromatic amino acids (Phe, Tyr, and Trp) and many secondary metabolites, including tetrahydrofolate, ubiquinone, and vitamin K (9). The shikimic acid pathway, present in plants and microorganisms, is completely absent in mammals, fish, birds, reptiles, and insects. The importance of the shikimate pathway in plants is further substantiated by the estimation that up to 35% or more of the ultimate plant mass in dry weight is represented by aromatic molecules derived from the shikimate pathway (2). On the basis of this information, it is readily apparent why EPSPS is a good target for novel antibiotics and herbicides.
The crystal structure of the ternary EPSPS-S3P-glyphosate complex was reported in PNAS at a stunning resolution of 1.5 Å and allows detailed visualization of the complete set of interactions of the ligands with their target (10). Using formate and phosphate as part of their crystallization protocol, the authors also report the high-resolution structure of the quaternary EPSPS-S3P-Pi-formate complex and suggest that the structure of this complex is representative of the true binary EPSPS-S3P complex. The new structures advance our understanding of the interaction of the widely used, broad spectrum herbicide glyphosate with EPSPS and provide an important opportunity for designing new and unique inhibitors for use as herbicides as well as antibiotic and antiparasitic drugs.
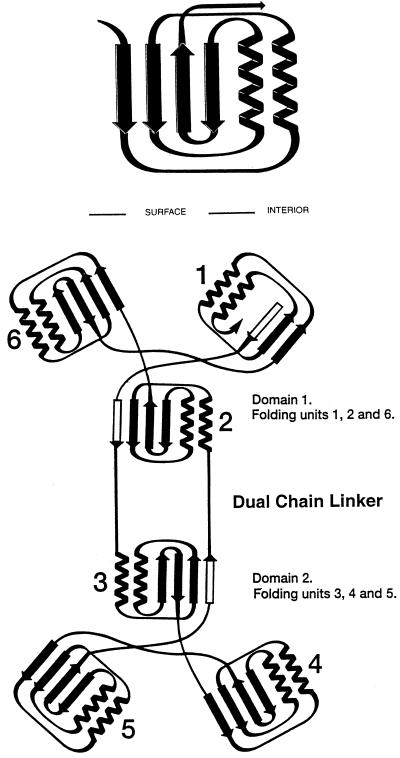
In recent years significant progress has been made toward understanding how glyphosate inhibits EPSPS. The first three-dimensional x-ray crystal structure of native Escherichia coli EPSPS (Fig. 1) showed that the enzyme consists of two domains of nearly equal size and that each domain contains six parallel α-helices aligned in a way that their macrodipoles create a significant electropositive attraction for the anionic ligands at the interface between the two domains (11). The EPSPS structure was found to be remarkably similar to the x-ray crystal structure of MurA (12), the closely related enolpyruvyl transferase whose structure was reported by some of the same investigators who now describe the new EPSPS structures. Binding of substrate (UDP-N-acetylglucosamine) and inhibitor (fosfomycin) to MurA was shown in these studies to induce a dramatic conformational change. On ligand binding, the MurA tertiary structure changes from the “open” form of the enzyme to the “closed” form, in which the two domains rearrange substantially into a tightly packed conformation (13). Using limited tryptic digestion and matrix-assisted laser desorption ionization–time-of-flight mass spectroscopy, Krekel et al. (14) have suggested that binding of S3P in combination with glyphosate also stabilizes EPSPS in a closed conformation. In addition, fluorescence spectroscopy studies imply that the association of glyphosate with the EPSPS-S3P binary complex induces a substantial conformational change in the enzyme (15). In a separate study using radiolabeled plant PreEPSPS (mature enzyme with a short chloroplast transit peptide), Della-Cioppa and Kishore (16) have shown that the native precursor protein is transported into chloroplasts, whereas a PreEPSPS-S3P-glyphosate ternary complex was not readily transported into the chloroplast stroma, suggesting that the ternary complex in its closed form is probably hindered in temporarily undergoing a conformational change to facilitate the process of importation across the chloroplast membrane. Collectively, these results have suggested that EPSPS undergoes a macroconformational change on the binding of S3P and glyphosate.
Figure 1.
Folding and topological symmetry of EPSPS (adapted from ref. 11). The two domain structure is formed by 6-fold replication of a protein folding unit (Upper) comprising two parallel helices and a four-stranded β-sheet. Each domain is formed from three of these folding units, which are related by an approximate 3-fold topological symmetry axis. In the open form of EPSPS, these axes are not collinear, but are presumably more so in the closed formed of the enzyme reported by Schönbrunn et al. (10). In each domain, three of the helices are buried and the surface of the molecule formed from the three β-sheets and the solvent-accessible faces of the other three helices. The N and C termini are located in Domain 1 with two crossover polypeptide segments creating a double hinge that links the two domains (Lower). Among the six folding units, three are folded from continuous segments of polypeptide chain. The other three contain the same arrangement of secondary structural features, but the sequences are not from a continuous chain. The arrangement positions the N termini of all 12 helices near the interface that results when the enzyme closes to form the active site. The positive macrodipoles from these helices presumably contribute to binding of substrates, products, and inhibitors, which are all multiply charged anions.
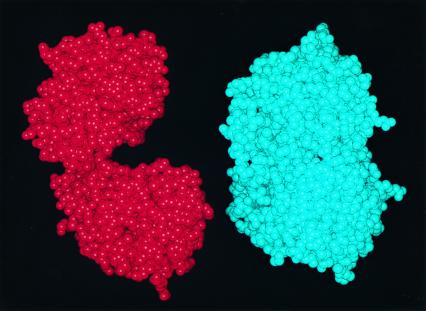
The results presented in this issue of PNAS clearly show that the two domains of EPSPS converge on ligand binding (Fig. 2). The x-ray structures also indicate that residues required for ligand binding are found in both globular domains and this effective interaction likely helps exclude bulk solvent from the active site during catalysis. The results further demonstrate that the x-ray structure of EPSPS bound with S3P, but not glyphosate, is virtually identical to that of EPSPS-S3P-glyphosate complex. Because S3P is necessary for crystal formation, the authors suggest that it is S3P alone that triggers the conformational change of the enzyme from the open to the closed state. It should be noted that there are potential alternative explanations for this observation. Studies using fluorescence spectroscopy have shown that there is no fluorescence change on the binding of S3P or glyphosate alone, but that formation of the ternary enzyme complex is signaled by a fluorescence decrease (15). Studies using native, open-form E. coli enzyme crystals soaked in buffered 100 mM S3P solution have shown that EPSPS-S3P binary complex, like the native enzyme, contains two widely separated globular domains (2). Another point to note would be that, if indeed the enzyme is in a closed state after S3P binding and formation of the binary complex, how then does glyphosate enter the solvent-inaccessible active site? Channels or interactions that might mediate this process are not identified by the authors in their analysis of the structures. The possibility that the phosphate and formate ions occupying the glyphosate binding site in the glyphosate-free structure are partly responsible for inducing the macroconformational change from the open to the closed form of the ligand-bound enzyme is also not considered.
Figure 2.
(Left) Space-filling model of E. coli EPSPS in the open form. (Right) Model of an EPSPS molecule in the closed, ligand-bound form. Comparisons of the figures illustrate the dramatic conformational change that attends ligand binding.
By using the x-ray crystal structure of MurA bound with a fluorinated analog of its tetrahedral intermediate, the authors have identified residues involved in the interaction with the PEP moiety. Comparisons of this structure with that of the newly observed binding mode for glyphosate to EPSPS reveal that homologous residues stabilize the binding of glyphosate in the ternary complex. Based on this and the observation that the formate and phosphate ions in the inhibitor free structure bind in the same anionic sites as the corresponding groups in glyphosate, the authors suggest that PEP occupies the glyphosate binding site in the EPSPS enzyme. This suggestion is at odds with kinetic and direct binding data that have shown that glyphosate can bind to an EPSPS-EPSP binary complex forming an EPSPS-EPSP-glyphosate ternary complex (17). Furthermore, glyphosate was found to be an uncompetitive inhibitor versus EPSP in the reverse reaction (17). These results would suggest that the glyphosate and PEP binding sites are not completely superimposable in their ternary EPSPS complexes. More studies will be required to fully detail the interactions of PEP and possible binding differences between PEP and glyphosate in the EPSPS active site.
Regardless of whether the true binary complex with S3P is open or closed, or whether the PEP and glyphosate sites partly overlap, the new EPSPS structures are significant new tools for the rational design and discovery of novel inhibitors. Quite recently, the growth of several pathogenic parasites such as Plasmodium falciparum, Taxoplasma gondii, and Cryptosporidium parvum was shown to be inhibited by glyphosate (18). Thus, the new EPSPS structures come at an especially opportune time for impact on new infectious diseases programs. Studies on the binding of the tetrahedral reaction intermediate to EPSPS have demonstrated that tapping into the structural determinants involved in S3P and glyphosate recognition could lead to inhibitors of picomolar affinity (19). However, though the detailed description of the active site can be used for virtual screening or even de novo ligand design, the catalytic site has been designed by nature to bind multiply charged anionic ligands like S3P, PEP, glyphosate, and products, EPSP and Pi. Given that interactions with these anionic ligands are stabilized by an array of basic EPSPS side chains and helix macrodipoles that, on ligand binding, converge by a massive conformationally induced fit mechanism, new ligands complementary to this site would also be expected to be highly anionic. Such molecules might be unlikely drugable leads unless facilitated transport mechanisms were available for membrane passage.
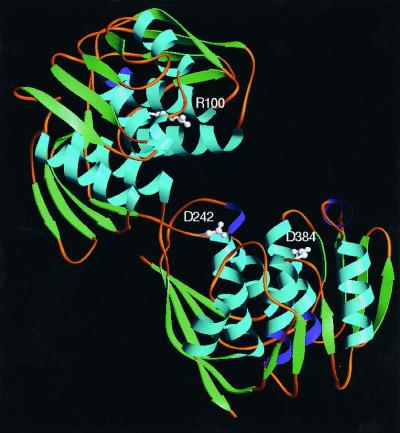
With some wisdom, the authors therefore propose an alternate strategy for structure-based inhibitor design. The results presented in the paper clearly demonstrate what other studies have predicted: the binding of ligands to EPSPS causes a macroconformational change from an open form to a closed form of the enzyme. Now, using studies that spatially identify residues responsible for the conformational change in MurA and mapping them in the EPSPS crystal structure (20–22), the authors have identified a new array of residues common to both enzymes (Fig. 3) that might be important for the conformational change and thereby provide new templates for future efforts that target the design of novel antimicrobial and herbicidal agents that block closure of the enzymes and formation of the catalytic sites.
Figure 3.
Crystal structure of the open form of EPSPS. Schönbrunn et al. (10) identify residues in MurA and their homologs in EPSPS that are determinants in the control of domain closure, and suggest that inhibitors that bind to these residues will interfere with closure of the enzymes and the formation of their active sites. Arg-100 (Upper, or Domain 2) is relatively close to the active site. Asp-384 is in the Lower domain (Domain 1). Asp-242 is near the two-stranded hinge that links the two domains.
Footnotes
See companion article on page 1376 in issue 4 of volume 98.
References
- 1.Baird D D, Upchurch R P, Homesley W B, Franz J E. Proc North Cent Weed Control Conf. 1971;26:64–68. [Google Scholar]
- 2.Franz J E, Mao M K, Sikorski J A. Glyphosate: A Unique Global Herbicide. Washington, DC: Am. Chem. Soc.; 1997. [Google Scholar]
- 3.Jaworski E G. J Agric Food Chem. 1972;20:1195–1198. [Google Scholar]
- 4.Amrhein N, Deus B, Gehrke P, Steinrücken H C. Plant Physiol. 1980;66:830–834. doi: 10.1104/pp.66.5.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haslam E. Shikimic Acid: Metabolism and Metabolites. New York: Wiley; 1993. [Google Scholar]
- 6.Anderson K S, Johnson K A. Chem Rev (Washington, DC) 1990;90:1131–1149. [Google Scholar]
- 7.Kishore G M, Shah D M. Annu Rev Biochem. 1988;57:627–663. doi: 10.1146/annurev.bi.57.070188.003211. [DOI] [PubMed] [Google Scholar]
- 8.Gruys K J, Sikorski J A. In: Comprehensive Biological Catalysis. Sinnott M, editor. London: Academic; 1997. pp. 273–291. [Google Scholar]
- 9.Gruys K J, Sikorski J A. Inhibitors of Tryptophan, Phenylalanine and Tyrosine Biosynthesis as Herbicides. New York: Dekker; 1999. [Google Scholar]
- 10.Schönbrunn E, Eschenburg S, Shuttleworth W A, Schloss J V, Amrhein N, Evans J N S, Kabsch W. Proc Natl Acad Sci USA. 2001;98:1376–1380. doi: 10.1073/pnas.98.4.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stallings W C, Meguid-Abdel S S, Lim L W, Shieh H-S, Dayringer H E, Leimgruber N K, Stegeman R A, Anderson K S, Sikorski J A, Padgette S R, et al. Proc Natl Acad Sci USA. 1991;88:5046–5050. doi: 10.1073/pnas.88.11.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schönbrunn E, Sack S, Eschenburg S, Perrakis A, Krekel F, Amrhein N, Mandelkow E. Structure. 1996;4:1065–1075. doi: 10.1016/s0969-2126(96)00113-x. [DOI] [PubMed] [Google Scholar]
- 13.Skarzynski T, Mistry A, Wonacott A, Hutchinson S E, Kelly V A, Duncan K. Structure. 1996;4:1465–1474. doi: 10.1016/s0969-2126(96)00153-0. [DOI] [PubMed] [Google Scholar]
- 14.Krekel F, Oecking C, Amrhein N, Macheroux P. Biochemistry. 1999;38:8864–8878. doi: 10.1021/bi990412o. [DOI] [PubMed] [Google Scholar]
- 15.Anderson K S, Sikorski J A, Johnson K A. Biochemistry. 1988;27:1604–1610. doi: 10.1021/bi00405a032. [DOI] [PubMed] [Google Scholar]
- 16.Della-Cioppa G, Kishore G M. EMBO J. 1988;7:1299–1305. doi: 10.1002/j.1460-2075.1988.tb02944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sammons R D, Gruys K J, Anderson K S, Johnson K A, Sikorski J A. Biochemistry. 1995;34:6433–6439. doi: 10.1021/bi00019a024. [DOI] [PubMed] [Google Scholar]
- 18.Roberts F, Roberts C W, Johnson J J, Kyle D E, Krell T, Coggins J R, Coombs G H, Milhous W K, Tzipori S, Ferguson D J P, et al. Nature (London) 1998;393:801–805. doi: 10.1038/31723. [DOI] [PubMed] [Google Scholar]
- 19.Anderson K S, Johnson K A. J Biol Chem. 1990;265:5567–5572. [PubMed] [Google Scholar]
- 20.Schönbrunn E, Eschenburg S, Krekel F, Luger K, Amrhein N. Biochemistry. 2000;39:2164–2173. doi: 10.1021/bi991091j. [DOI] [PubMed] [Google Scholar]
- 21.Schönbrunn E, Eschenburg S, Luger K, Kabsch W, Amrhein N. Proc Natl Acad Sci USA. 2000;97:6345–6349. doi: 10.1073/pnas.120120397. . (First Published May 23, 2000; 10.1073/pnas.120120397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eschenburg S, Schönbrunn E. Proteins Struct Funct Genet. 2000;40:290–298. doi: 10.1002/(sici)1097-0134(20000801)40:2<290::aid-prot90>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]