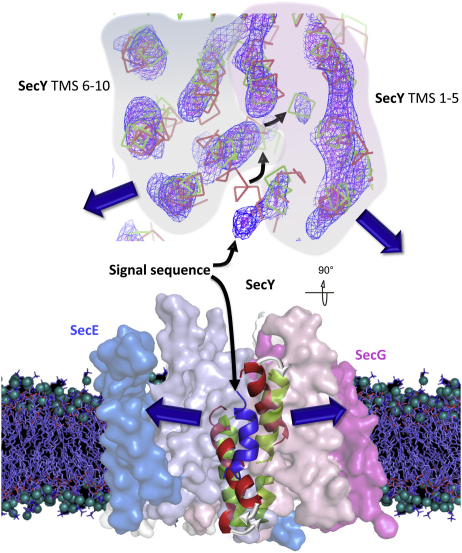
Summary
The Sec complex forms the core of a conserved machinery coordinating the passage of proteins across or into biological membranes. The bacterial complex SecYEG interacts with the ATPase SecA or translating ribosomes to translocate secretory and membrane proteins accordingly. A truncated preprotein competes with the physiological full-length substrate and primes the protein-channel complex for transport. We have employed electron cryomicroscopy of two-dimensional crystals to determine the structure of the complex unlocked by the preprotein. Its visualization in the native environment of the membrane preserves the active arrangement of SecYEG dimers, in which only one of the two channels is occupied by the polypeptide substrate. The signal sequence could be identified along with the corresponding conformational changes in SecY, including relocation of transmembrane segments 2b and 7 as well as the plug, which presumably then promote channel opening. Therefore, we propose that the structure describes the translocon unlocked by preprotein and poised for protein translocation.
Graphical abstract
Highlights
► Structure determination of the membrane-bound SecYEG complex unlocked by preprotein ► The SecYEG dimer binds only one substrate polypeptide ► Signal sequence binds at the interface between SecYEG and the lipid bilayer ► The association transmits a conformational change enabling channel opening
A major outstanding question in the protein translocation field lies in the nature of the active protein channel upon engagement with its substrate. Hizlan et al. describe the structure of the ubiquitous Sec complex associated with a bone fide mimic of a presecretory protein in the native environment of the membrane. The results reveal how the binding of the signal sequence unlocks the Sec complex prior to channel opening and preprotein transport.
Introduction
A prerequisite of the signal sequence hypothesis is the availability of a membrane-bound machinery for recognition and transport of preproteins (Blobel and Dobberstein, 1975). Protein secretion in bacteria generally relies on the peripheral association of the ATPase SecA with the protein-channel complex SecYEG (Brundage et al., 1990). Both factors interact with the preprotein signal sequence (Gelis et al., 2007; Plath et al., 1998; Van den Berg et al., 2004), which is transferred from SecA to SecYEG prior to protein translocation across the membrane through the center of SecY (Cannon et al., 2005; Van den Berg et al., 2004). Membrane proteins require the signal recognition particle (SRP) and its receptor for nascent chain targeting to the Sec complex prior to translocation, which is driven by their synthesis. A lateral gate for insertion is formed between transmembrane segment (TMS) 2b and TMS 7 of SecY, which is also the binding site for the signal sequence (Plath et al., 1998; Van den Berg et al., 2004).
The structure of the protein-channel complex has been resolved by electron and X-ray crystallography. The former was determined in the membrane and revealed SecYEG dimers in a back-to-back configuration (Breyton et al., 2002). This dimeric arrangement is required for translocation (Deville et al., 2011), even though translocation proceeds through a single SecYEG complex (Osborne and Rapoport, 2007). Together, the two copies provide a productive high-affinity binding site for SecA, to secure the interaction during transport. Notably, the ADP-associated state of SecA has a lower affinity for SecYEG, compared to the ATP-bound state (Robson et al., 2007, 2009b). Therefore, the 10-fold higher affinity of SecA for the dimer over the monomer (Deville et al., 2011) helps prevent the dissociation and abolition of translocation at the ADP-associated stage.
The X-ray structure, determined using solubilizing detergent, identified monomers with the central channel held closed by an annulus of hydrophobic residues at the center of SecY and a short helix (2a) or plug (Park and Rapoport, 2011; Van den Berg et al., 2004). The structure of the SecYEG-SecA complex (also in detergent solution) contains one copy of each and shows that the association opens a “window” at the lateral gate by the separation of TMS 2b and 7, in preparation for signal sequence binding and protein translocation (Zimmer et al., 2008). The plug and the TMS lining the channel and lateral gate need to move further in order to accommodate and transport secretory and membrane proteins. The nature of this conformational change holds the key to understanding the molecular mechanism of transport.
Recently, the Escherichia coli SecYEG complex has been visualized in the membrane environment associated with a ribosome nascent chain complex (RNC) (Frauenfeld et al., 2011). The nascent chain contains the N-terminal signal anchor (SA) of FtsQ, a classical substrate of the cotranslational insertion pathway. In this structure, the SecY conformation is apparently very similar to the posttranslational complex of SecYEG-SecA determined without any substrate (Zimmer et al., 2008). A density on the outside of the complex at the lateral gate was attributed to the SA.
In order to learn more about the active process we set out to determine the structure of SecYEG, using electron microscopy of two-dimensional (2D) crystals, in association with a substrate polypeptide. Our approach involved the analysis of membrane-bound SecYEG in complex with a 40 amino acid (aa) polypeptide of the N terminus of the precursor of LamB. This protein is a β-barrel outer membrane porin and a typical substrate of the SecA-dependent posttranslational secretory pathway. A number of mutations and deletions of the signal sequence, mostly around the central hydrophobic core, render the substrate defective in translocation (Emr et al., 1980). Suppressors of these mutations, the prl alleles, have been instrumental in the identification and analysis of SecY (prlA), SecE, SecG, and SecA (Emr et al., 1981; Smith et al., 2005).
The interaction of the signal sequence with SecA and SecYEG is retained by representative synthetic peptides of LamB (denoted SS). The association with SecA results in the competitive inhibition of protein translocation; the structural basis of the recognition was characterized by NMR spectroscopy (Gelis et al., 2007). The peptide also opens channels in membranes containing SecYEG (Simon and Blobel, 1992). In addition, signal sequence peptides, including of LamB, act as allosteric activators of the translocation machinery, allowing the efficient transport of secretory proteins without signal sequences (Gouridis et al., 2009).
We further explored the ability of the peptide to bind and activate the SecYEG complex. A peptide containing the signal sequence and a short stretch of the mature protein was useful in this respect and allowed us to stabilize and crystallize its complex with SecYEG for structure determination by electron cryomicroscopy. The map of the membrane-bound translocon engaged and activated by this preprotein mimic provides a clear view of the signal sequence and the TMS of the SecYEG dimer. The conformational changes induced by the association help to explain some of the prlA phenotypes, and suggest a mechanism for the initiation of preprotein transport and channel opening.
Results
A Peptide Mimic of the Preprotein Acts on the Physiological Translocation Site
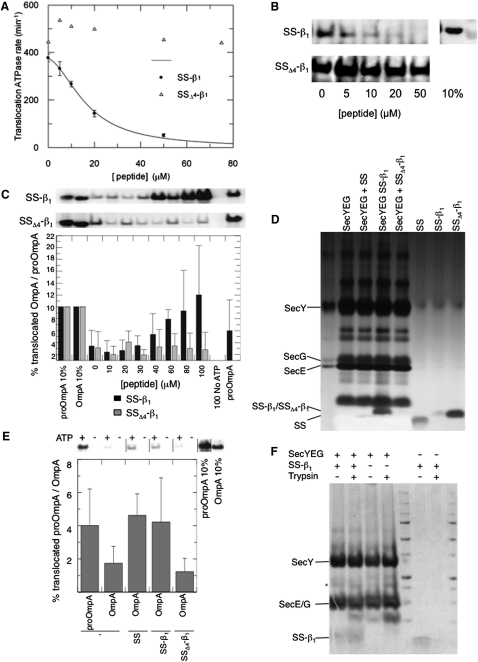
An extended 40 aa peptide of the LamB signal sequence (SS) containing an additional 15 aa β strand of the mature protein (SS-β1) quantitatively inhibits assays reconstituting SecA/ATP-driven translocation of the preprotein proOmpA into proteoliposomes containing SecYEG (Figures 1A and 1B). Conversely, a similar peptide with a 4 aa deletion (SSΔ4-β1), corresponding to a classic defective signal sequence (Emr et al., 1980), did not.
Figure 1.
A 40 aa Peptide of LamB SS-β1 Competes for Full-Length Preprotein and Activates the SecY Complex
(A) Addition of LamB SS-β1, but not SSΔ4-β1, inhibits translocation-associated ATPase activity of SecA. ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase linked assay in the presence of 50 nM SecA, 1 mM ATP, 1 μM SecYEG proteoliposomes, and 0.7 μM proOmpA after preincubation with increasing concentrations of the signal sequence peptides.
(B) Samples used in the ATPase assays shown in (A) were tested for translocation efficiency according to protease protection of proOmpA. Successfully translocated, protease-protected proOmpA was visualized by western blot. The top right-hand lane was loaded as a measure of 10% of the total proOmpA present in the samples.
(C) Translocation assays (as in B) using OmpA instead of proOmpA. The upper two panels show representative western blots for successfully translocated substrate in the presence of wild-type and mutant peptides; each lane corresponds to the bars in the quantification below, for increasing concentrations (0–100 μM) of peptides: SS-β1 (black bars) or SSΔ4-β1 (gray bars); n = 4–6. The translocation efficiency was calibrated against a 10% standard. Negative (no ATP with 100 μM peptide) and positive (proOmpA without peptide) controls are shown on the right. All error bars denote SD.
(D) SecYEG vesicles reconstituted in the presence of SS, SS-β1, or SSΔ4-β1 were loaded onto an SDS-PAGE gel and the polypeptides visualized by silver staining.
(E) Translocation assays for OmpA (as in B) using vesicles incorporating SecYEG with or without peptide. The upper panel shows a representative western, quantified (as in C) in the lower panel (n = 8). The peptide present in the initial reconstitution is indicated below. proOmpA was used as a positive control testing the competence of vesicles reconstituted without preprotein peptides (far left). All error bars denote SD.
(F) Coomassie-stained SDS-PAGE gel of 2D crystals of SecYEG grown in the presence or absence of SS-β1, before and after exposure to 1:100 (w/w) trypsin for 20 min at room temperature. ∗Well-known breakdown product of SecY (Collinson et al., 2001; Robson et al., 2007).
The SS-β1 Peptide Transactivates SecYEG Conferring the Ability to Translocate a Signal Sequence-less Secretory Protein
To confirm the peptide was acting physiologically on SecY, we exploited the allosteric transactivation of translocation by signal sequence peptides (Gouridis et al., 2009). SS-β1 and SSΔ4-β1 were titrated into translocation reactions of OmpA without signal sequence (Figure 1C). As expected, the wild-type peptide conferred the ability to promote translocation of OmpA much more effectively than the mutant.
Next, the SecYEG complex was reconstituted into phospholipid vesicles in the absence or presence of SS, SS-β1, or SSΔ4-β1. The vesicles were then reisolated from the excess unbound peptide and analyzed for incorporated peptide and their ability to promote the translocation of OmpA. SS and SS-β1, but not SSΔ4-β1, were retained following coreconstitution with SecYEG (Figure 1D), and permitted the translocation of OmpA (Figure 1E). The induced increase in OmpA transport was not as high as observed upon direct addition to translocation assays (Figure 1C). This was due to the inevitable dissociation of some of the SecYEG-bound peptide during vesicle reisolation from the unbound excess peptide (the apparent Kd ∼10 μM; Figure 1A). Therefore, the resulting partially loaded Sec complex, as expected, exhibited a lower level of activation. Nevertheless, the results do show the preprotein mimic must be acting directly on the SecYEG complex. In this membrane- and substrate-bound state the complex is unlocked and primed to allow the passage of the mature part of the secretory protein.
Growth and Analysis of 2D Crystals of SecYEG with and without SS-β1
Electron microscopy of 2D crystals enables visualization of membrane proteins reconstituted into lipid bilayers. The structure of SecYEG without bound peptide previously determined in this way revealed the complex was a dimer in the membrane (Breyton et al., 2002), whereas all structures of SecY complexes determined in the presence of detergent showed monomers (Egea and Stroud, 2010; Van den Berg et al., 2004; Zimmer et al., 2008). As this particular dimeric arrangement of SecYEG is obligatory for the productive engagement of substrate (Deville et al., 2011), we used 2D crystallography again for the complex with preprotein peptide. The membrane-embedded crystals were prepared in much the same way as the vesicles for activity measurements.
Crystals were grown with or without SS-β1 and then isolated from the crystallization liquor and analyzed by SDS-PAGE. Those grown in the presence of the peptide retained it (Figure 1F). Its resistance to proteolysis suggested that the peptide was occluded within the crystalline membranes, rather than at the surface. For the purposes of data collection the sample was taken directly from the crystallization liquor, where the peptide was maintained in solution at all times to prevent the dissociation observed above.
The Architecture of the SecYEG Complex Bound to the Preprotein Peptide
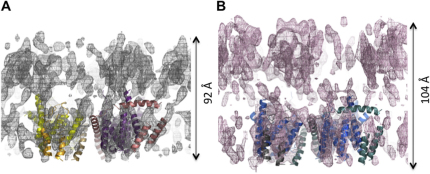
The unit cell dimensions (112 × 58 Å) of the 2D crystals with or without the peptide were the same, and similar to those obtained previously (104 × 57 Å) (Breyton et al., 2002). Images of SecYEG-SS-β1 crystals recorded at various tilt angles were used for 3D reconstruction (Table S1 available online). As before, the 3D map showed that the crystals consisted of two stacked membranes connected by facing cytosolic loops of SecY (Figure S1). The two layers were more tightly packed than in the former study (Breyton et al., 2002), reducing the thickness of the crystals. This slight difference in the crystal packing was independent of the peptide and most likely the result of subtle variations in the growth conditions.
Figure S1.
Comparison of Double Membrane Crystals of SecYEG with and without Preprotein
Side view of the SecYEG map with (A) and without (B) bound preprotein (determined previously (Breyton et al., 2002), without the imposed non-crystallographic symmetry) and with one dimer fitted into the maps (color coding is as Figure 2). Both maps are at 8 Å in-plane resolution and contoured at 1.5 SD. The double-membrane crystals of SecYEG prepared in this study (A) are around 12 Å less thick compared to those determined previously (B), and the unit cell is slightly larger. This effect is independent of the presence of the peptide and is due to variation in the crystal packing.
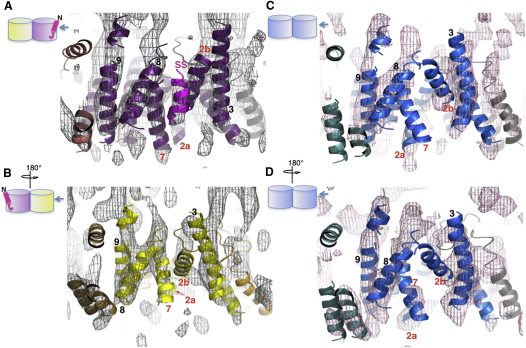
Previously, the structure of SecYEG alone was of sufficient quality to fit all 15 TMS of SecY (10), SecE (3), and SecG (2) (Breyton et al., 2002). This fitting was subsequently verified by the X-ray structure (Bostina et al., 2005; Van den Berg et al., 2004). The new map was of very similar resolution and quality (Figures 2A and 2B; Table S1). Clear rod-like densities of transmembrane helices enabled accurate docking of an atomic homology model of E. coli SecYEG (Bostina et al., 2005; Deville et al., 2011; Figures 2A and 2C–2E). All TMS could be accurately fitted, except the highly tilted TMS 3 of SecE, due to the inherently lower resolution perpendicular to the membrane plane (Table S1; Breyton et al., 2002).
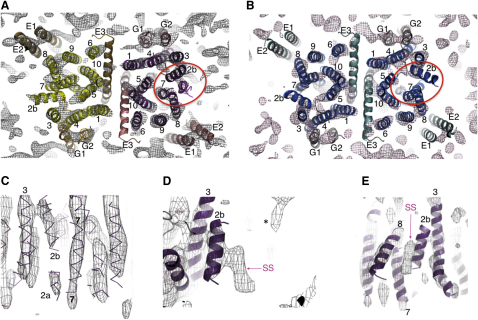
Figure 2.
Structure of SecYEG Bound to the Preprotein Peptide SS-β1
The TMS are labeled for SecY (1-10), SecE (E1-3), SecG (G1 and G2), and the signal sequence (SS). Maps are contoured at 1.5 SD.
(A and B) Top views from the cytoplasmic side of one crystalline membrane of SecYEG showing map density and super-imposed E. coli models (Experimental Procedures). The lateral gate of the substrate-occupied complex and its equivalent in the complex visualized without peptide are circled in red. (A) Structure of the SecYEG dimer bound to the preprotein mimic. The occupied complex with bound preprotein peptide is on the right-hand side of the dimer, with SecY, E, and G shown, respectively, in purple, salmon, and dark pink. The unoccupied complex is on the left-hand side of the dimer shown with SecY, E, and G in yellow, sand, and orange. (B) SecYEG, without bound preprotein peptide and without the applied 2-fold symmetry (Breyton et al., 2002), is shown for comparison with SecY, E, and G in blue, light teal, and gray.
(C–E) Detailed side views of map density of the SecYEG complex bound to the preprotein mimic (as in (A), right hand complex), with corresponding fitted E. coli homology model (purple lines). (C) view from the center of one SecYEG complex out through the lateral gate. (D) Side view toward TMS 2b and 3. ∗Denotes the density from the second crystal layer, which is not part of the structure being viewed. (E) looking into the lateral gate from the outside.
The Asymmetric Association of the Preprotein Peptide to the SecYEG Dimer
The overall architecture of the complex associated with the peptide was largely unchanged from that without it (Breyton et al., 2002), showing the SecYEG dimers in the functional back-to-back conformation (Deville et al., 2011; Figures 2A and 2B). However, upon closer inspection the maps revealed significant differences. In the absence of substrate the two SecYEG complexes had the same structure, apart from random noise (correlation coefficient [CC] = 0.557) (Figure 2B). Previously, the noise contribution was reduced by applying noncrystallographic 2-fold symmetry (Breyton et al., 2002). In the presence of peptide, however, the structure of one SecYEG complex appeared to be different from its partner (CC = 0.467), and from the two copies of the complex determined without peptide prior to symmetry imposition (Figure 2B; CC = 0.417 and 0.406). This is consistent with substrate binding and induced conformational change in only one of the SecYEG complexes.
The TMS of the E. coli homology model were fitted individually to each monomer complex of the equivalent unsymmetrized maps determined with and without the preprotein peptide. One of the monomer complexes of the peptide-bound dimer possesses an extra prominent density just outside the lateral gate, adjacent to TMS 2b, 7, and 8 (Figures 2A and 2B, red circle). This was the only significant density within the membrane not accounted for by the SecYEG model (Figures 2D and 2E), and was therefore assigned to the bound substrate. The extra feature was clearly absent from the other monomer complex in the peptide-bound SecYEG dimer, as well as from both complexes of the dimer determined without bound preprotein (Breyton et al., 2002; Figures 2A, 2B, and 3). Further additional densities unaccounted for by adjacent SecYEG dimers (Figure 2A) are due to the cytosolic loops of complexes in the other membrane layer (Figure S1), which do not penetrate the membrane (e.g., asterisk in Figure 2D).
Figure 3.
Comparison of the Lateral Gates of SecYEG Complexes Determined with and without the Preprotein Peptide
(A–D) Side view detail from outside the lateral gate, as indicated in the respective scheme of the SecYEG dimer (side view, cytoplasm uppermost). The experimental map density is shown (contoured at 1.5 SD) along with the docked TMS (ribbon representation) of the E. coli atomic homology model (Experimental Procedures), colored as in Figure 2. Selected helices of SecY are labeled by their corresponding number (red numbers denote key TMS). The SecYEG dimer associated with SS-β1, with the substrate-occupied (A) and unoccupied (B) monomer complexes; the fitted core α helix of the signal sequence peptide (Gelis et al., 2007) is shown in magenta in (A). (C and D) The SecYEG complex determined without peptide, prior to symmetry imposition of the noncrystallographic dimer (Breyton et al., 2002).
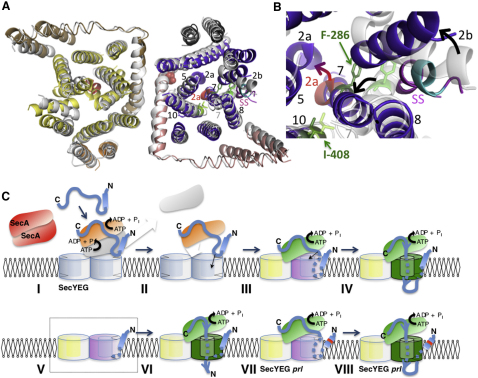
Signal-Sequence-Induced Conformational Changes in SecYEG
The structure of the hydrophobic α-helical core of the LamB signal sequence determined in complex with SecA (Gelis et al., 2007) fits into the extra density outside the lateral gate (Figures 2A and 3A). The mutations resulting in defective signal sequences, including the SSΔ4 deletion used above, all fall within this helical core (Emr et al., 1980; cyan in Figures 4A and 4B). The signal sequence location in the map does not agree exactly with the proposed position intercalated between TMS 2b and 7 (Van den Berg et al., 2004), but is consistent with defined crosslinks between the signal sequence and phospholipids, TMS 2b, 7, and 8 (Plath et al., 1998). The hydrophobic core of the helix also correlates well with its central position in the lipid bilayer.
Figure 4.
Mechanism of Preprotein-Induced Activation of the SecY Complex
(A) Cytosolic view of the fitted models of SecYEG determined previously (Bostina et al., 2005) overlaid with the one bound by the preprotein peptide; color coding as in Figure 2, except that the SecYEG formerly determined without peptide is shown in white with the plug (2a) in red. The 4 residues -LAVA- of the signal sequence, which when deleted ablate preprotein transport and peptide activity (Figures 1A–1E), are shown in cyan. The sites associated with the signal sequence suppressor allele prlA4 (SecY-F286Y in TMS 7 and SecY-I408N in TMS 10) are shown in SecY determined without added peptide (lime green sticks on white ribbon) and in SecY bound to the preprotein mimic (dark green sticks on purple ribbon). SecE-S120 at the C-terminal periplasmic region of TMS 3, known to interact with the plug (2a; SecY-F67) (Flower et al., 1995; Tam et al., 2005), is represented by red spheres.
(B) Detail of the protein channel and lateral gate (as in A). The arrows describe the movement of TMS 2b, 7, and the plug (2a) associated with the binding of signal sequence (magenta, SS).
(C) Model for activation, channel opening and translocation. Preprotein (blue) with an N-terminal signal sequence (cylinder) is engaged by SecA (red inactive dimer) and targeted to the translocation machinery SecYEG (symmetrical light blue dimer). (I). The initiation of translocation involves ATP hydrolysis and the dissociation of SecA dimers (orange and white monomers) (Duong, 2003; Or et al., 2002; Robson et al., 2007), which exposes the signal sequence (II) to facilitate binding at the protein-lipid interface of SecYEG. (III). The association activates one monomer in the SecYEG dimer, breaking the 2-fold symmetry. The activated complex (as in A and B) is primed for translocation (purple), while the passive complex (yellow) becomes tightly closed and assists in the binding of SecA, now fully active (green) (Deville et al., 2011). (IV). ATP hydrolysis results in the intercalation of preprotein, channel opening (green) and translocation. (V). The activated asymmetric conformation can also be promoted by a trans-acting signal sequence peptide (dashed box) (Figures 1C–1E) and is visualized by the structure described here. (VI). In this bound state it is capable of transporting signal sequence-less substrates. (VII and VIII). The prlA mutants are predisposed to the activated form of SecYEG (purple) and capable of translocating proteins with defective signal sequences (red band on blue cylinder).
The signal sequence helix is in close contact with TMS 2b, which has consequently tilted away from the lateral gate (compare Figure 3A with Figures 3C and 3D; Figures 4A and 4B). Another change triggered upon contact with the preprotein peptide is a major relocation of TMS 7: a straightening of 40° toward center of the channel brings its periplasmic end 15 Å toward TMS 5 and 10 (Figure 4B). This, in turn, results in the displacement of the channel plug (helix 2a) 10 Å toward TMS 3 of SecE (Figures 4A and 4B). Our confidence in the positioning of the signal sequence and the description of the conformational change is reflected by the fits to well-resolved and clear densities (Figures 2C–2E and 3A; see helices denoted 2a, 2b, 7, and SS).
The local differences in TMS 2b and 7 between the two monomeric complexes in the substrate-bound dimer are more pronounced than those between the peptide-containing monomer in this dimer and the complex crystallized without peptide (compare Figure 3A with Figure 3B, and Figure 3A with Figures 3C and 3D). The lateral gate in the unoccupied complex of the substrate-bound dimer appears to be more tightly closed than in both copies of the inactive complex.
Discussion
The Sec complex, like most membrane proteins, is prone to detergent exposure and the depletion of lipids (Bessonneau et al., 2002; Deville et al., 2011; Gold et al., 2010). In this study, these known destabilizing effects on the active arrangement of the translocon have been avoided by crystallizing the complex within the membrane. Its reconstitution with a bona fide preprotein mimetic at the physiological site has provided a detailed view of an activated translocon, showing just one substrate bound per SecYEG dimer.
This substrate-induced asymmetry is consistent with the requirement for two distinct copies of the channel, with only one of them being active (Deville et al., 2011; Osborne and Rapoport, 2007) and evidently, is inherent to SecYEG. The binding of substrate to one SecYEG appears to induce an inaccessible state in the other, possibly accounting for the preference of the dimer to bind only one copy of the preprotein.
In the active monomer the substrate binding site and induced conformational changes in the SecYEG protein channel are consistent with the critical role played by TMS 2b, 7, and the plug in substrate recognition and transport (Van den Berg et al., 2004). The genetic analyses of LamB secretion also support this interpretation (Emr et al., 1980, 1981). The prl suppressors of defective signal sequences do not directly complement substrate binding. Instead they are thought to stabilize the open form of the complex (or destabilize the closed form) (Bondar et al., 2010; Smith et al., 2005), a state normally achieved in the wild-type upon signal sequence binding. Hence, the prl mutants readily adopt an active conformation, allowing the transit of substrates with defective signal sequences. These mutations in SecY (prlA) map mostly to the plug (helix 2a), TMS 7, and TMS 10 (Osborne and Silhavy, 1993; Smith et al., 2005; Figure 6 in Van den Berg et al., 2004) in the right position to promote the conformational changes observed here. Therefore, we suggest that the structure represents an activated form of the complex favored by the prl mutants. For example, the potent suppressor prlA4 (SecYF286Y;I408N) may achieve this by promoting the displacement of TMS 7 (by SecYF286Y) and stabilizing an interaction between TMS 7 and 10 (by SecYI408N), predisposing it to the conformation observed here in complex with the signal peptide (Figure 4B). This particular suppressor also promotes the displacement of the plug helix toward TMS 3 of SecE (Tam et al., 2005), as described here. This displacement is also consistent with a known interaction between the prlA3 site (SecYF67C in helix 2a) and SecES120C (TMS 3, red sphere in Figure 4A) in the active complex (Harris and Silhavy, 1999; Tam et al., 2005). This relocation also closely matches a prediction of the plug position in the open state (Robson et al., 2009a).
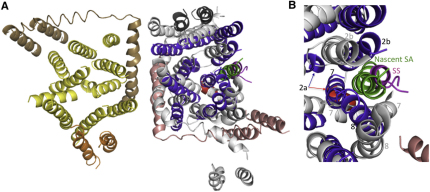
The SecYEG-ribosome complex in the act of cotranslational transport of a membrane protein has also been visualized in a lipid bilayer, in this case encapsulated by nanodiscs (Frauenfeld et al., 2011). The structure determined by electron cryomicroscopy of single particles does not report the movements we observe, but does show the nascent signal anchor in a very similar position to the signal sequence (Figure S2).
Figure S2.
Comparison of SecYEG Structure Bound to the Presecretory Signal Sequence and to a Nascent Signal Anchor
(A) Cytosolic view of the fitted model of the SecYEG dimer bound by preprotein (colored as in Figure 2) aligned and overlaid with the SecYEG monomer bound by the ribosome nascent chain (RNC) complex (Frauenfeld et al., 2011). For the SecYEG-RNC complex the ribosome has been omitted for the purposes of clarity and the complex is colored as follows: SecY, SecE, and SecG (white), nascent signal anchor (SA, green) and plug (2a, red).
(B) Detail showing the lateral gate (same color coding). Key TMS are labeled in black (SecYEG bound to SSβ1) or gray (SecYEG bound to RNC).
The experiments described here profit from the ability of signal sequence peptides to transactivate the SecYEG complex (Figures 1C–1E; Gouridis et al., 2009). The preprotein mimics are too short to fully engage the translocon, instead they act to unlock or prime the complex. Therefore, the results reveal part of the preprotein binding site and the architecture of the activated complex in the early stages of the cycle prior to translocation. The signal sequence binding site on the outside of the complex is compatible with a mechanism for translocation described in Figure 4C and explains why the activation mechanism is allosteric.
The conformational changes in the substrate-bound state, particularly involving TMS 7 of SecY, were not apparent in the complex activated by SecA (Zimmer et al., 2008) or any other structure. Therefore, they must be specific to the preprotein. These conformational changes would undoubtedly affect the lining of the protein channel, including the hydrophobic seal in the center of SecY (Park and Rapoport, 2011; Van den Berg et al., 2004), and thereby, together with the displacement of the plug, facilitate channel opening and intercalation of the translocating polypeptide (Figure 4C). This putative activation step may promote the dislocation of the two halves of SecY about the hinge region between TMS 5 and 6, to allow the passage of proteins through or into membrane (Van den Berg et al., 2004), possibly in the manner described by the structure of the slightly more open state (Egea and Stroud, 2010). The next challenge is to determine the structure of the fully open complex engaged in secretion or with a membrane protein trapped during the insertion process.
Experimental Procedures
Chromatography media was from GE Healthcare. Detergents were obtained from Glycon and lipids from Avanti. SilverQuest silver staining kit and NuPAGE gels were purchased from Invitrogen. Bio-Beads SM2 were from Bio-Rad. All other materials were supplied by Sigma Aldrich.
Peptide and Protein Production
The LamB signal sequence MMITLRKLPLAVAVAAGVMSAQAYA (SS), LamB signal sequence plus the first β strand MMITLRKLPLAVAVAAGVMSAQAYA-VDFHGYARSGIGWTG (SS-β1) and the mutant version (Emr et al., 1980) MMITLRKLP-(ΔLAVA)-VAAGVMSAQAYA-VDFHGYARSGIGWTG (SSΔ4-β1) were synthesized by Dr Graham Bloomberg (University of Bristol).
Protein samples were purified according to published protocols (Robson et al., 2009b).
ATPase and Translocation Assays
In vitro ATPase and translocation assays involving proOmpA and OmpA were performed essentially as described previously (Robson et al., 2009b); see Extended Experimental Procedures as well for further details.
Extended Experimental Procedures.
ATPase and Translocation Assays
In vitro ATPase and translocation assays using proOmpA and OmpA were performed at 25°C in 50 mM triethanolamine, 50 mM KCl and 2 mM MgCl2 at pH 7.5. Pyruvate kinase (7 U/ml), lactate dehydrogenase (10 U/ml), phosphoenol pyruvate (2 mM) and NADH (0.2 mM) were mixed with SecA (50 nM) and SecYEG (1 μM) reconstituted into proteoliposomes. The reaction was initiated with 1 mM ATP, followed by addition of the peptides in 6 M urea, which had no effect on the ATPase rate in the absence of preprotein. Subsequent addition of proOmpA or OmpA (0.7 μM) initiated translocation-associated ATPase activity, which was measured by the change in absorbance at 340 nm using a Lambda 25 Spectrophotometer (Perkin Elmer). The inclusion of urea at the concentrations used had no effect on the ATPase rates or translocation efficiency.
Translocation efficiency was assayed by taking samples from the ATPase reactions and subjecting them to proteinase K digestion (0.2 mg/ml for 45 min on ice). After precipitation by trichloroacetic acid (20%, plus 1 mg/ml BSA, for 30 min on ice), protease-protected substrate was detected by western blot using an antibody raised against proOmpA.
Electron Microscopy and Image Processing
Specimens for electron cryomicroscopy were prepared by the back-injection method in 4% trehalose (Breyton et al., 2002). Images were recorded at liquid-helium temperature in a JEOL 3000 SSF electron microscope at specimen tilts of 0°, 10°, 20°, 30° and 45° at 300 kV and at a magnification of 53,000X in spot-scan mode. Crystal quality was evaluated by optical diffraction and well-ordered areas of 4,000 × 4,000 pixels were digitized on a ZEISS SCAI scanner at 7 μm step size. The lattices were processed by 2dx_image (Gipson et al., 2007) and the image data were merged in plane group p121b with the ORIGTILTK program in the MRC package (Crowther et al., 1996; Henderson et al., 1986). Image amplitudes were scaled with an average temperature factor of −600 Å to compensate for the resolution-dependent degradation of image amplitudes as described previously (Breyton et al., 2002). The 3D map was calculated with the CCP4 program suite (1994).
Model Building
The E. coli molecular model of SecYEG (Bostina et al., 2005; Deville et al., 2011) was modified to include the missing helices of SecE and SecG. This combined model was then used as a template for the version of SecYEG (Breyton et al., 2002) and for SecYEG-SS-β1, fitted to the electron density using COOT (Emsley and Cowtan, 2004). The models were prepared in cartoon representation using PyMOL (The PyMOL Molecular Graphics System, Version 1.3.x, Schrödinger, LLC). The maps were contoured at 1.5 SD.
Coreconstitution of SecYEG with Preprotein Peptides
Proteoliposomes containing SecYEG were reconstituted in the absence or presence of SS, SS-β1, or SSΔ4-β1. The reconstitution mixture contained 1.65 μM SecYEG, 3.2 mg/ml (∼4.6 mM) E. coli polar lipids, ±10 μM peptide; for further details see (Robson et al., 2009b) and Extended Experimental Procedures. Following detergent removal by dialysis and Bio-Bead adsorption, the proteoliposomes were separated from excess unbound peptide by centrifugation and resuspended to give a final SecYEG concentration of 4.6 μM (8.9 mg/ml lipid). The protein composition was then evaluated by SDS-PAGE. High concentrations of lipid disturbed the migration of the peptides at the lower regions of the gel, therefore, reduced quantities (18 pmol ∼1.4 μg SecYEG and 35 μg lipid) were loaded and the peptide content was evaluated by silver staining. The vesicles were then challenged in assays monitoring the translocation of proOmpA and OmpA (as above).
Growth and Analysis of 2D Crystals Containing SecYEG and SS-β1
Two-dimensional crystals were grown of SecYEG (3.4 μM) as before (Breyton et al., 2002), in the absence or presence of 10 μM SS-β1 in the sample and 5 μM in the outside dialysis buffer (sufficient to saturate the sites on SecYEG as the concentrations employed stipulate tight-binding conditions). The crystals (10 μg with respect to the protein, and ∼2 μg lipid) were then subjected to SDS-PAGE before and after exposure to trypsin (0.1 μg for 20 min at room temperature).
Electron Microscopy, Image Processing, and Model Building
Electron cryomicroscopy, structure determination and model fitting was carried out in the manner already described for SecYEG alone (Breyton et al., 2002); see Extended Experimental Procedures as well for further details. Files describing the models fitted to the experimental map density of the structure are available upon request.
Determination of Correlation Coefficients
The map density covering one monomer in each map was masked out and overlapped to the other according to the noncrystallographic 2-fold symmetry (Breyton et al., 2002) by MAPROT. The correlation coefficients were then calculated by OVERLAPMAP.
Acknowledgments
Our appreciation goes to Dr. Mirko Lotz for his contributions during the early stages of the project. We thank Dr Richard Sessions for advice on model building and Dr Ryan Schulze for critical reading of the manuscript. We would also like to thank Henning Stahlberg and Anchi Cheng for computer programs, and Dr. Özkan Yildiz for his continual advice on computation. The work was supported by the BBSRC (Project Grants to I.C., BB/F007248/1 and BB/F002343/1), the Wellcome Trust (equipment grant to I.C., 082140), the Deutsche Forschungsgemeinschaft (SFB 807) and the Max-Planck-Gesellschaft. None of the authors of this work have a financial interest related to this work.
Published online: January 26, 2012
Footnotes
Supplemental Information includes Extended Experimental Procedures, two figures, one table, and one movie and can be found with this article online at doi:10.1016/j.celrep.2011.11.003.
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 Unported License (CC-BY-NC-ND; http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode).
Supplemental Information
The movie depicts the dynamic activation of the SecY complex (cyan; viewed in ribbon representation, from the side with the cytosolic face uppermost). The association of the signal sequence (appears in blue) at the lateral gate coincides with a conformational change in trans-membrane segments 2b (TM2b), 7 (TM7) and the plug (TM2a) of SecY (red to green).
References
- Bessonneau P., Besson V., Collinson I., Duong F. The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure. EMBO J. 2002;21:995–1003. doi: 10.1093/emboj/21.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 1975;67:835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondar A.N., del Val C., Freites J.A., Tobias D.J., White S.H. Dynamics of SecY translocons with translocation-defective mutations. Structure. 2010;18:847–857. doi: 10.1016/j.str.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostina M., Mohsin B., Kühlbrandt W., Collinson I. Atomic model of the E. coli membrane-bound protein translocation complex SecYEG. J. Mol. Biol. 2005;352:1035–1043. doi: 10.1016/j.jmb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Breyton C., Haase W., Rapoport T.A., Kühlbrandt W., Collinson I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature. 2002;418:662–665. doi: 10.1038/nature00827. [DOI] [PubMed] [Google Scholar]
- Brundage L., Hendrick J.P., Schiebel E., Driessen A.J., Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- Cannon K.S., Or E., Clemons W.M., Jr., Shibata Y., Rapoport T.A. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J. Cell Biol. 2005;169:219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson I., Breyton C., Duong F., Tziatzios C., Schubert D., Or E., Rapoport T., Kühlbrandt W. Projection structure and oligomeric properties of a bacterial core protein translocase. EMBO J. 2001;20:2462–2471. doi: 10.1093/emboj/20.10.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deville K., Gold V.A., Robson A., Whitehouse S., Sessions R.B., Baldwin S.A., Radford S.E., Collinson I. The oligomeric state and arrangement of the active bacterial translocon. J. Biol. Chem. 2011;286:4659–4669. doi: 10.1074/jbc.M110.175638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong F. Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J. 2003;22:4375–4384. doi: 10.1093/emboj/cdg418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea P.F., Stroud R.M. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc. Natl. Acad. Sci. USA. 2010;107:17182–17187. doi: 10.1073/pnas.1012556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emr S.D., Hedgpeth J., Clément J.-M., Silhavy T.J., Hofnung M. Sequence analysis of mutations that prevent export of lambda receptor, an Escherichia coli outer membrane protein. Nature. 1980;285:82–85. doi: 10.1038/285082a0. [DOI] [PubMed] [Google Scholar]
- Emr S.D., Hanley-Way S., Silhavy T.J. Suppressor mutations that restore export of a protein with a defective signal sequence. Cell. 1981;23:79–88. doi: 10.1016/0092-8674(81)90272-5. [DOI] [PubMed] [Google Scholar]
- Flower A.M., Osborne R.S., Silhavy T.J. The allele-specific synthetic lethality of prlA-prlG double mutants predicts interactive domains of SecY and SecE. EMBO J. 1995;14:884–893. doi: 10.1002/j.1460-2075.1995.tb07070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfeld J., Gumbart J., Sluis E.O., Funes S., Gartmann M., Beatrix B., Mielke T., Berninghausen O., Becker T., Schulten K., Beckmann R. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 2011;18:614–621. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelis I., Bonvin A.M., Keramisanou D., Koukaki M., Gouridis G., Karamanou S., Economou A., Kalodimos C.G. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131:756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold V.A., Robson A., Bao H., Romantsov T., Duong F., Collinson I. The action of cardiolipin on the bacterial translocon. Proc. Natl. Acad. Sci. USA. 2010;107:10044–10049. doi: 10.1073/pnas.0914680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouridis G., Karamanou S., Gelis I., Kalodimos C.G., Economou A. Signal peptides are allosteric activators of the protein translocase. Nature. 2009;462:363–367. doi: 10.1038/nature08559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris C.R., Silhavy T.J. Mapping an interface of SecY (PrlA) and SecE (PrlG) by using synthetic phenotypes and in vivo cross-linking. J. Bacteriol. 1999;181:3438–3444. doi: 10.1128/jb.181.11.3438-3444.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Or E., Navon A., Rapoport T.A. Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J. 2002;21:4470–4479. doi: 10.1093/emboj/cdf471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne R.S., Silhavy T.J. PrlA suppressor mutations cluster in regions corresponding to three distinct topological domains. EMBO J. 1993;12:3391–3398. doi: 10.1002/j.1460-2075.1993.tb06013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne A.R., Rapoport T.A. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Park E., Rapoport T.A. Preserving the membrane barrier for small molecules during bacterial protein translocation. Nature. 2011;473:239–242. doi: 10.1038/nature10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K., Mothes W., Wilkinson B.M., Stirling C.J., Rapoport T.A. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- Robson A., Booth A.E., Gold V.A., Clarke A.R., Collinson I. A large conformational change couples the ATP binding site of SecA to the SecY protein channel. J. Mol. Biol. 2007;374:965–976. doi: 10.1016/j.jmb.2007.09.086. [DOI] [PubMed] [Google Scholar]
- Robson A., Carr B., Sessions R.B., Collinson I. Synthetic peptides identify a second periplasmic site for the plug of the SecYEG protein translocation complex. FEBS Lett. 2009;583:207–212. doi: 10.1016/j.febslet.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Robson A., Gold V.A., Hodson S., Clarke A.R., Collinson I. Energy transduction in protein transport and the ATP hydrolytic cycle of SecA. Proc. Natl. Acad. Sci. USA. 2009;106:5111–5116. doi: 10.1073/pnas.0809592106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S.M., Blobel G. Signal peptides open protein-conducting channels in E. coli. Cell. 1992;69:677–684. doi: 10.1016/0092-8674(92)90231-z. [DOI] [PubMed] [Google Scholar]
- Smith M.A., Clemons W.M., Jr., DeMars C.J., Flower A.M. Modeling the effects of prl mutations on the Escherichia coli SecY complex. J. Bacteriol. 2005;187:6454–6465. doi: 10.1128/JB.187.18.6454-6465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P.C., Maillard A.P., Chan K.K., Duong F. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 2005;24:3380–3388. doi: 10.1038/sj.emboj.7600804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg B., Clemons W.M., Jr., Collinson I., Modis Y., Hartmann E., Harrison S.C., Rapoport T.A. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- Zimmer J., Nam Y., Rapoport T.A. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- Collaborative Computational Project, Number 4. (1994). The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763. [DOI] [PubMed]
- Bostina, M., Mohsin, B., Kühlbrandt, W., and Collinson, I. (2005). Atomic model of the E. coli membrane-bound protein translocation complex SecYEG. J. Mol. Biol. 352, 1035–1043. [DOI] [PubMed]
- Breyton, C., Haase, W., Rapoport, T.A., Kühlbrandt, W., and Collinson, I. (2002). Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature 418, 662–665. [DOI] [PubMed]
- Crowther, R.A., Henderson, R., and Smith, J.M. (1996). MRC image processing programs. J. Struct. Biol. 116, 9–16. [DOI] [PubMed]
- Deville, K., Gold, V.A., Robson, A., Whitehouse, S., Sessions, R.B., Baldwin, S.A., Radford, S.E., and Collinson, I. (2011). The oligomeric state and arrangement of the active bacterial translocon. J. Biol. Chem. 286, 4659–4669. [DOI] [PMC free article] [PubMed]
- Emsley, P., and Cowtan, K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132. [DOI] [PubMed]
- Frauenfeld, J., Gumbart, J., Sluis, E.O., Funes, S., Gartmann, M., Beatrix, B., Mielke, T., Berninghausen, O., Becker, T., Schulten, K., and Beckmann, R. (2011). Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 18, 614–621. [DOI] [PMC free article] [PubMed]
- Gipson, B., Zeng, X., Zhang, Z.Y., and Stahlberg, H. (2007). 2dx—user-friendly image processing for 2D crystals. J. Struct. Biol. 157, 64–72. [DOI] [PubMed]
- Henderson, R., Baldwin, J., Downing, K., and Zemlin, F. (1986). Structure of purple membrane from Halobacterium halobium. Recording, measurement and evaluation of electron micrographs at 3.5 Å resolution. Ultramicroscopy 19, 147–178.
- Unger, V.M., and Schertler, G.F. (1995). Low resolution structure of bovine rhodopsin determined by electron cryo-microscopy. Biophys. J. 68, 1776–1786. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The movie depicts the dynamic activation of the SecY complex (cyan; viewed in ribbon representation, from the side with the cytosolic face uppermost). The association of the signal sequence (appears in blue) at the lateral gate coincides with a conformational change in trans-membrane segments 2b (TM2b), 7 (TM7) and the plug (TM2a) of SecY (red to green).