Abstract
Background
Red man syndrome (RMS) is a well known adverse reaction that occurs in pediatric patients receiving vancomycin, yet reported prevalence is varied, and characteristics and risk factors, are not well understood. Our objective was to determine the prevalence, characteristics, and risk factors for red man syndrome in pediatric patients receiving vancomycin, including contributing genetic factors.
Methods
A multi-center retrospective study of 546 subjects (0.5– 21 years) who received at least one dose of intravenous vancomycin was conducted. Demographic and symptom data were collected through chart review and parent/nurse report. Genotype analysis included ten single nucleotide polymorphisms (SNPs) in the histamine pathway.
Results
RMS was observed in 77 (14%) subjects receiving vancomycin. 40% of subjects with RMS symptoms developed rash, pruritis and flushing, without hypotension. Antecedent antihistamine use was identified as a risk factor for RMS (p<0.001). Multivariate regression analysis identified age >2 years (p=0.008), previous RMS (p<0.001), VC dose (p=0.02), and VC concentration (p=0.017) as RMS risk factors, while African-American race was protective (p=0.011). We observed an apparent association between RMS and a SNP in the diamine oxidase gene (p=0.044); however, no associations were revealed by multifactor dimensionality reduction analysis.
Conclusions
RMS is a common adverse event in children receiving vancomycin. Identified risk factors are Caucasian ethnicity, age ≥ 2 years, previous RMS history, vancomycin dose ≥10 mg/kg, vancomycin concentration ≥5 mg/ml and antecedent antihistamine use. Known genetic variants in histamine metabolism or receptors do not appear to be substantial contributors to risk of RMS.
Keywords: Vancomycin, Histamine, Red Man Syndrome, Single nucleotide polymorphisms
Introduction
Red man syndrome (RMS) is the most common adverse drug reaction (ADR) that occurs with vancomycin with estimated rates of 5– 50% in hospitalized subjects, and up to 90% in healthy control subjects.1–5 However, its true incidence remains unknown and, previous studies of RMS in children have described widely varying clinical features.6–10 RMS encompasses a spectrum of symptoms that ranges from a mild reaction such as flushing, urticarial rash, and/or pruritis, to a severe reaction that includes generalized erythema, intense pruritis, and even hypotension.1, 3, 10 Although there is general consensus on clinical features included in the symptom complex of RMS, there is no standard definition based on a required constellation of features that constitutes this ADR. The lack of a precise phenotype may account for widely reported estimates of incidence in the literature.
Vancomycin is widely used in children for treatment of serious gram positive infections. Recently published methicillin-resistant S. aureus (MRSA) guidelines recommend vancomycin as a first line agent in the setting of serious or invasive MRSA infections.11 Therefore, characterization of this ADR is important for optimizing the therapeutic benefit of vancomycin while employing methods to prevent occurrence of RMS.
RMS is believed to be an anaphylactoid type of reaction due to vancomycin-induced direct mast cell degranulation. It has been shown to be associated with a rise in blood histamine level in some studies; however, conflicting data exist.3–5, 12–14 Increasing evidence suggests that altered histamine metabolism may contribute to the pathogenesis of hypersensitivity reactions, including RMS.15–17 Histamine is synthesized from L-histidine and primarily metabolized by histamine N-methyltransferase (HNMT) and diamine oxidase (DAO) (Supplemental Digital Content 1, Figure).18–20 Both of these enzymes are polymorphically expressed. Several single nucleotide polymorphisms (SNPs) in the H1 and H4 histamine receptors also have been described. It is known that certain SNPs in the H4 receptor, which is expressed on mast cells, are associated with atopic dermatitis and pruritus. It is possible that one or more of these SNPs may contribute to altered function of these receptors. 21–23
The purpose of this study was to precisely describe the clinical syndrome, further characterize the epidemiology of RMS, identify risk factors for RMS in pediatric subjects, and explore associations between RMS and known SNPs in genes involved in histamine production, response, and degradation.
Methods
Study Design and Participants
Hospitalized subjects between 6 months and 21 years of age who received at least one dose of intravenous vancomycin during a hospitalization between April of 2007 and October of 2009 were enrolled. Subjects who continued to receive vancomycin after enrollment were followed prospectively until vancomycin was stopped to monitor for development of RMS, whereas subjects with RMS at the time of enrollment were not followed further.
Initial screening for RMS was based on presence of one or more of the following signs or symptoms: macular or papular rash, flushing, itching, and/or a documented decrease of either systolic or diastolic blood pressure (BP) by > 10 mm/hg in association with a dose of vancomycin. Confirmation of RMS required the presence of at least two of these signs/symptoms. Reactions were then further characterized by extent: local rash, pruritis, and flushing were defined as affecting only one body part (ex: face, neck, or torso); generalized rash included a combination of ≥ 3 body parts; and generalized flushing or itch included ≥ 2 body parts. Involvement of ≥2 extremities was considered generalized regardless of association with other body parts. Presence of generalized symptoms, such as a combination of rash on at least 3 body parts and flushing or itch of at least 2 body parts in any of the above categories was defined as a severe reaction.
Immune deficiency was classified as primary or secondary, and defined by either presence of an underlying diagnosis of an immune system disorder or receipt of treatment with an agent intended to result in immune suppression. Immune suppressive treatment was defined as: 1) therapy of at least 2 weeks duration with corticosteroids (e.g. prednisone); 2) at least one round of chemotherapy with a myelosuppressive agent; 3) or chronic immune modulator therapy for a rheumatic disease or solid organ transplant (e.g. tacrolimus). Medications known to modify histamine responses also were recorded.
The protocol was approved by the institutional review boards of the respective participating centers; Children’s Mercy Hospitals and Clinics (CMHC), Arkansas Children’s Hospital, Kosair Children’s Hospital, and Texas Children’s Hospital. Parental permission was obtained for all subjects and assent was obtained for subjects ≥7 years that were deemed capable by their parent or legal guardian. This trial was registered at clinical trials.gov (# NCT00824122) and with the Pediatric Pharmacology Research Units #10914.
Clinical methods
Diagnosis of RMS, age, gender, ethnicity, RMS signs and symptoms, previous vancomycin receipt, and previous RMS symptomatology were identified through medical record review and parent/nurse report. Cases required confirmation by more than one source (e.g. parent and nurse report). Clinical diagnosis for which vancomycin therapy was initiated; chronic co-morbid conditions, use of agents with antihistamine properties, leukotriene antagonists, immune suppression, treatment with immunosuppressive agents, and treatment for RMS symptoms were obtained from the medical records. Chronic comorbid conditions were categorized as previously described.24
Specific characteristics of vancomycin administration included drug concentration (mg/ml), interruption or slowing of infusion, dosing interval, and dose (mg/kg/dose). Intermittent doses of corticosteroids and receipt of blood transfusion within 24 hours prior to vancomycin initiation was recorded as a possible confounder for the determination of RMS symptoms. Standard practice at each institution included an initial infusion rate of 60 minutes. However, Vancomycin infusion rate was not included in the analysis as some patients with previous RMS had initial infusion rates that were prolonged for their course of therapy and some patients with first time RMS symptomatology had a prolonged infusion time at enrollment.
Genotyping methods
Genomic DNA was isolated from whole blood or saliva using a QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA). The following SNPs were genotyped: HNMT (rs6430764, -1639 C>T; rs2071048, -464 C>T; rs11558538, 314 C>T, Thr105Ile; rs1050900, 3’ UTR A>T), DAO (rs10156191, 47 C>T, Thr16Met; rs1049742, 995 C>T, Ser332Phe; rs1049793, 4107 C>G, His645Asp), HDC (rs17740607, 92 C>T, Met31Thr), H1 (rs901865, -17 C>T) and H4 (rs11665084, 413C>T, Ala138Val). A restriction fragment length polymorphism assay was used to detect H1 rs901865. All other SNPs were detected using predesigned TaqMan assays (Applied Biosystems, Carlsbad, CA).
Further testing of possible gene-gene interactions was performed with multifactor dimensionality reduction (MDR) using previously described techniques.25 MDR in conjunction with other data mining and machine learning techniques offers a strategy to detect possible nonlinear complex gene-gene interactions that is not explained by traditional categorical analysis.19, 26 One-locus model characterizes the main effect of each SNP while multi-locus model investigates the interactions among relevant polymorphisms. The validity and significance of the selected models were assessed by 1000-count permutation testing. MDR was performed using the open source software mdr2.0 and model goodness-of- fit and significance were assessed by software mdrpt1.0.26
Statistical analysis
Chi square analysis was used to compare categorical variables; Student’s t test and ANOVA were used to compare means. Odds ratios (OR) and 95% confidence intervals (CI) were reported along with p-values. Wilcoxon rank sum was used for non-parametric testing and logistic regression with backward selection was employed to identify the predictive factors related to the development of RMS. Age, ethnicity, vancomycin per kilogram dosing, vancomycin concentration in mg/dl, previous vancomycin receipt, presence of a chronic co-morbid condition, previous RMS, and antihistamine use prior to first dose of vancomycin were included in the multivariate logistic regression model. Antihistamine use during vancomycin treatment, and slowed infusion were not included in the model, as these two events were more likely to be initiated by the presence of RMS symptomatology. Co-variates that were statistically insignificant were removed from the model by stepwise elimination.
A multifactor dimensionality reduction (MDR) analysis was performed to evaluate the DNA sequence variations and gene-gene interactions associated with the incidence of RMS. Ten SNPs were included as risk factors and the outcome variable was the development of RMS during vancomycin.
Results
Demographics
546 subjects were enrolled and had clinical data available (391 CMHC, 116 Arkansas, 24 Kosair, and 15 Texas). The RMS rate was 14.1% (77/546) (95% CI 12%, 17%), with equal gender distribution (Table 1). The majority of subjects were Caucasian or African American, 79.1% and 12.5% respectively. Caucasians and other non-African American ethnic groups were more likely to be diagnosed as having RMS symptoms than African American subjects (OR =12.5, 95%CI (1.7, 90.1), p=0.013, OR =16.3, 95%CI (2.0, 133.7), p=0.009) (Table 1). The median age for all subjects in the study was 6.7 years (range 0.5, 21.8). Subjects with RMS were significantly older at 8.7 years (IQR 4.3–13.3) than subjects without RMS at 6.1 years (IQR 2–13) (p= 0.009). Further, children older than 2 years of age were 4.6 times more likely to develop RMS than children under 2 years of age. There was no difference found for development of RMS in those 2–16 years vs. >16–21 years of age, or in severity of symptomatology between age groups.
Table 1.
Comparison of demographic features, age, reason for VC, and CCC in patients with and without RMS.
| RMS (+) (%) | RMS (−) (%) | P-value | ||
|---|---|---|---|---|
| N=546 | 77 (14.1) | 469 (85.9) | ||
| Male | 40 (13) | 263 (86.8) | NS | |
| Ethnicity | ||||
| Non-Hispanic White | 67 (15.5) | 365 (84.5) | <0.0001 | |
| Non-Hispanic Black | 1 (0.1) | 67 (99.9) | ||
| Other (Incl Hispanic) | 9 (19.6) | 37 (80.4) | ||
| Age | 0.0013 | |||
| ≤ 2 years | 5 (4.2) | 112 (95.7) | ||
| > 2 years | 72 (16.8) | 357 (83.2) | ||
| Reason for VC use | NS | |||
| Bacteremia | 15 (8.9) | 154 (91.1) | ||
| Pneumonia | 12 (12.1) | 87 (87.9) | ||
| SSTI/Osteomyelitis | 19 (19.6) | 78 (80.4) | ||
| CNS | 11 (15.9) | 58 (84.1) | ||
| Fever & Neutropenia | 10 (17.2) | 48 (82.8) | ||
| Other infection† | 10 (22.7) | 44 (81.5) | ||
| Chronic Co-morbid Condition (CCC) |
||||
| No CCC | 13 (8.8) | 134 (91.2) | 0.0321** | |
| Presence of CCC | 64 (16) | 335 (84) | ||
Other infection includes tracheitis, intra-abdominal, UTI/pyelonephritis, mastoiditis, surgical prophylaxis, not-specified
Healthy subjects were less likely to develop RMS as compared to patients with co-morbidities (9% vs. 16%, p=0.0321). But no specific diseases are linked with a higher risk of RMS (p=0.2315).
73.1% (399/546) of children had a chronic co-morbid condition. The most common chronic co-morbid condition was an oncologic process (183/399; 45.9%). Subjects with an underlying chronic co-morbid conditions were significantly more likely to develop RMS compared to healthy subjects (OR=1.8, 95% CI (1.1, 3.7), p=0.032). However, no association was found with any specific chronic co-morbid conditions and development of RMS. The most frequent diagnoses for which vancomycin was given were suspected or proven bacteremia 169/546 (31%) and pneumonia 99/546 (18.1%).
Clinical Features
Subjects who had received vancomycin in the past without a history of RMS were not found to be at higher risk of developing RMS than those who had never been exposed. However, subjects with a previous history of RMS from exposure to vancomycin were more likely to develop RMS with their current vancomycin therapeutic course (43% vs. 11%, OR=6.1, 95%CI (2.0, 133.7), p<0.001).
RMS developed more frequently in subjects who received anti-histamines for any reason (e.g. anti-emetic or sleep aid) prior to the first dose of vancomycin (22%; 40/185 vs 11%; 37/361, OR=2.3, 95% CI (1.5, 3.9), p<0.001) than those who had not received prior anti-histamines. In subjects with a history of RMS, those who received an antihistamine prior to the first vancomycin dose were as likely to develop RMS as those who did not (42.9%; 15/35 vs. 44.4%; 8/18, p>0.05). However, in subjects without prior history of RMS, those who received an antihistamine prior to the first vancomycin dose were more likely to develop RMS than those who did not (16.7%; 25/150 vs 8.8%; 29/343, OR=2.2, 95% CI (1.2, 3.8), p<0.01).
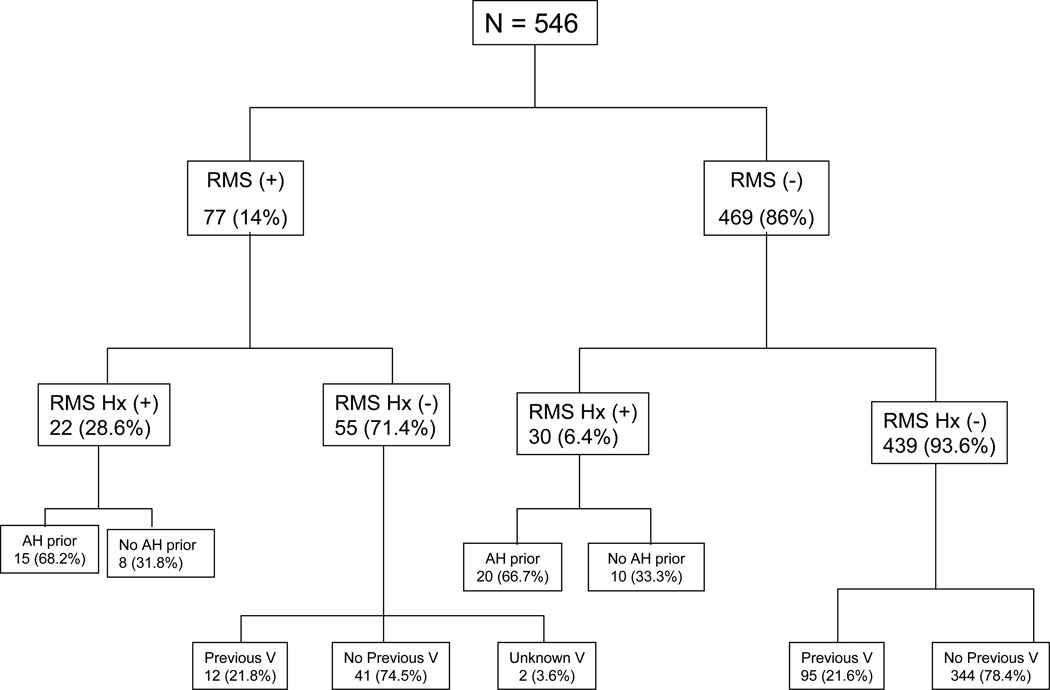
Twenty-two of 52 subjects who had a history of previous RMS (42.3%) developed recurrent RMS. The majority of subjects with a history of RMS received prophylactic antihistamines before the first dose of vancomycin whether they had recurrent (68.2%) symptomatology or not (66.7%) (Figure 1). There was no difference in the presence or severity of rash (p=0.81, p=0.85) and pruritis (p=0.13, p=0.23) between subjects who received a dose of antihistamine prior to vancomycin and those who did not. However, presence and severity of flushing appeared to be modified by antecedent antihistamine use (p=0.006, p=0.04) respectively.
Figure 1.
Flow diagram of patients with and without current RMS and history of RMS, as well as antihistamine and previous vancomycin exposure.
Hx= History
RMS Hx (+) = RMS history positive
RMS Hx (−) = RMS history negative
AH = Antihistamine
V = Vancomycin
Pruritis, rash and flushing were common in all subjects who developed RMS. Hypotension was not observed in any subject (Table 2). When evaluating combined symptomatology, 59.7% (46/77) developed itch and rash, 49.4% (38/77) developed rash and flushing, 61% (47/77) developed flushing and itch, and 40.3% (31/77) of subjects developed flushing, rash, and itching simultaneously. Subjects with recurrent RMS developed similar symptoms of equal severity as subjects with new RMS (Table 2). However, subjects with recurrent RMS were more likely to develop the combination of itch and rash together (Table 2).
Table 2.
Phenotype Characteristics of Patients with current RMS without and with a history of RMS
| New | Recurrent | Total | P-value | ||
|---|---|---|---|---|---|
| N=55 (71.4) | N=22 (28.6) | N=77 (%) | |||
| Pruritis | 43 (78.2) | 21 (95.5) | 64 (82) | 0.09 | |
| Local | 11 (25.6) | 7 (33.3) | 18 (28.1) | ||
| General | 32 (74.4) | 14 (66.7) | 46 (71.9) | 0.13 | |
| Rash | 34 (61.8) | 19 (86.4) | 53 (67.9) | 0.055 | |
| Local | 7 (20.6) | 5 (26.3) | 12 (22.6) | ||
| General | 27 (79.4) | 14 (73.7) | 41 (77.4) | 0.067 | |
| Flushing | 43 (78.2) | 17 (77.3) | 60 (76.9) | 1 | |
| Local | 19 (44.2) | 9 (52.9) | 28 (46.7) | ||
| General | 24 (55.8) | 8 (47.1) | 32 (53.3) | 0.96 | |
| Pruritis + Rash | 28 (50.9) | 18 (81.8) | 46 (59.7) | 0.03 | |
| Pruritis + Rash + Flush | NS | ||||
Drug Administration
We also determined the timing of RMS symptom onset, which was available for 61/77 (79%) cases. The majority (62%) of subjects with RMS developed symptoms within 30 minutes of infusion initiation, 21% had symptoms between 30 and 60 minutes and 16% had symptoms more than one hour into their infusion. Subjects with earlier detection of symptoms were more likely to have their infusion slowed (86% for 0–30 min, 54% for 30–60 min and 20% for >60 min, OR=5.1, 95% CI (2.2, 12.3), p<0.001).
The incidence of RMS was vancomycin dose (mg/kg) dependent; with 8% in subjects who received 10mg/kg, 16% in subjects who received 15mg/kg, and 27% in subjects who received 20mg/kg dosing (p=0.003). Risk of RMS was also associated with the concentration of vancomycin preparations; with the 5 mg/ml preparation conferring lower risk of RMS than other preparations including 10 mg/ml, 50 mg/ml, 100 mg/ml (p<0.001).
There were no differences among subjects who received intermittent steroid dosing, blood product transfusion prior to first dose of vancomycin, immune suppressive therapy, or leukotriene inhibitors with regard to risk of developing RMS symptomatology.
Co-variates that remained significant after multivariate analysis included age >2 years (p=0.008), vancomycin dose (p=0.002), previous RMS (p< 0.0001), and vancomycin concentration (p=0.0169). African American ethnicity conferred decreased risk (p=0.011). All other univariate factors (previous vancomycin, chronic co-morbid conditions, and prior antihistamine) did not remain significant in the multivariate analysis, partly due to the correlation among risk factors. For instance, previous RMS was correlated with prior antihistamine; therefore prior antihistamine became insignificant in the multivariate analysis.
Genotype analysis
Genotype results for all 10 SNPs were obtained on a total of 523 (95.8%) subjects. Twenty-one subjects had no sample available and 2 samples were inferior quality. The 995 C>T SNP in the DAO gene was significantly associated with RMS symptomatology in the Caucasian and Other non-African American ethnicity categories. Among subjects with RMS symptomatology, 44% were found to have this sequence variation on at least one allele compared to 15% who were homozygous for the wild-type allele (p=0.044). There were no significant associations with any of the other SNPs.
MDR analysis was performed to investigate potential gene-gene interactions associated with the development of RMS. The optimal model at one locus, two-loci and three-loci were developed and significance of these selected models was evaluated by Testing Accuracy and Crossing Validation Counts. The most promising gene-gene interaction was rs1049742 (995 C>T, Ser332Phe) +HDC with testing accuracy at 57% (p=0.083). However, there was no statistically significant association between development of RMS symptomatology and any sequence variation combinations, partly due to the relatively small sample size and the complex RMS phenotype.
Discussion
RMS associated with vancomycin use has long been recognized as an adverse drug reaction. However, few systematic investigations have been conducted in pediatric subjects to date. One previous retrospective study evaluated RMS in 650 children who had been exposed to vancomycin, and found a low rate (1.6%) of RMS, which limited their ability to determine risk factors. Other studies included only small numbers of children and revealed wide variations in clinical symptoms of RMS, including hypotension as a common finding, forming the basis for which slowing the infusion has become a common ameliorating practice. 7–9 Therefore, the present study is the first to characterize RMS features and risk factors across a large population and to evaluate for a potential SNP association.
The prevalence of RMS found in this study is consistent with the range noted in previous studies,3–5 and our data indicate that vancomycin dose, vancomycin concentration, previous RMS history, age, and ethnicity are risk factors for RMS. It is unclear why patients of African-American ethnicity are less likely to develop RMS, although it is possible that rash and flushing are harder to determine in this patient population.
While antihistamines were commonly employed before receipt of the initial vancomycin dose, they were not found to protect subjects from developing RMS or ameliorate rash and pruritis. However, antecedent antihistamine use was associated with statistically significant less flushing in subjects with RMS. Not only were antihistamines not protective for RMS, administration of an antihistamine for any reason prior to receiving vancomycin was associated with increased risk of RMS. This observation is somewhat counter-intuitive and a biologically plausible explanation is not readily apparent. However, these findings underscore the complex nature of the RMS reaction, and suggest that although the histaminergic pathway is likely involved, alternative inflammatory pathways that may or may not involve histamine release (e.g. complement receptor activation on the mast cell) may play a more dominant role.
Genotype analysis for sequence variations in the histamine biotransformation enzymes 15–17 and H1 and H4 receptors 21–23 revealed one SNP with an apparent association of risk for RMS. However, further MDR analysis did not detect a significant association in any combination of subject symptoms and SNPs. This indicates that these particular gene polymorphisms are unlikely to be important determinants of RMS risk.
This study has several limitations. First, the study had both retrospective and prospective components. Clinical data for those patients who already had developed RMS at the time of enrollment were collected retrospectively, whereas subjects who had not developed RMS at enrollment were followed prospectively, thus we were unable to collect the timing of development of RMS symptoms on all patients. Potential ascertainment bias was mitigated by enrolling subjects solely based on age and current receipt of vancomycin, without regard to previous vancomycin receipt or RMS reaction. Second, our criteria for determining whether a subject developed RMS were stricter than previously published clinical criteria, raising the possibility that we excluded subjects with milder symptomatology. However, our goal was to rigorously define the population of subjects with definite RMS and the prevalence of RMS in our study was consistent with previous reports. Furthermore, the use of stricter criteria facilitated more rigorous identification of risk factors for developing RMS. Third, while we evaluated all commonly used H1 and H2 antihistamines, as well as other agents with antihistamine and immune suppressant effects in our study,27, 28 it is possible that medications, other than the ones we evaluated, could play a role in ameliorating RMS symptomatology (e.g. cromolyn sodium). Finally, given the short half-life of these medications, data was not collected regarding the length of antihistamine use. It is possible that longer term use could play a role in development of RMS or amelioration of symptomatology.
Using a rigorous and specific definition, the prevalence of RMS in a large sample of hospitalized children receiving vancomycin was 14%. Our study suggests that dose (mg/kg) and concentration (mg/ml) of infused vancomycin play a key role in the development of RMS syndrome. To our knowledge this is the first study to report this finding; although previous studies have also shown the development of RMS in the setting of standard one hour infusion times, dose and concentration have not been independently evaluated. 7, 29 Additional risk factors for RMS include RMS history, non-African American ethnicity, and age ≥ 2 years. Known genetic polymorphisms of histamine pathway and histamine receptor genes have little influence on risk of RMS. Unexpectedly, pre-treatment with an antihistamine did not prevent RMS from developing, and did not ameliorate most symptoms once present. However, duration of antihistamine treatment was not collected. Nevertheless these observations merit confirmation in a prospective controlled study.
Supplementary Material
Supplemental Digital Content 1, Figure Precursor, actions and metabolism of histamine. Histidine Decarboxylase (HDC), Histamine N-methyltransferase (HNMT), Diamine Oxidase (DAO), Histamine 1,2,3,4 receptors (H1R, H2R, H3R, H4R)
Acknowledgements
Source of Funding
This study was funded in part by the Katherine Berry Richardson Foundation, and also supported by U 10 HD31313-17 (GLK), Pediatric Pharmacology Research Unit Network, the Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, MD.
We would like to acknowledge Katie Klockau and Jackie Riley, both recipients of Summer Scholarships, for their contribution with genotype analysis. We would also like to thank Mr. Greyson Twist and Mrs. Liliane Njountché for technical assistance. In addition, we are grateful to all research coordinators (University of Arkansas and Arkansas Children’s Hospital, Lee Howard and Leah Dawson; the University of Louisville, Carrie Schanie and Nitya Narayan; Baylor College of Medicine and Texas Children’s Hospital, Christina Lopez and Cynthia Bourdreaux). We would also like to thank Janice Sullivan, MD at the University of Louisville, and Lisa Bomgaars, MD at Baylor College of Medicine and Texas Children’s Hospital. Finally, we would like to acknowledge Dr. Ralph Kauffman for helpful editing of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors state they have no conflicts of interest to declare.
References
- 1.Levine DP. Vancomycin: a history. Clin Infect Dis. 2006;42(Suppl 1):S5–S12. doi: 10.1086/491709. [DOI] [PubMed] [Google Scholar]
- 2.Wang G, Hindler JF, Ward KW, Bruckner DA. Increased vancomycin MICs for Staphylococcus aureus clinical isolates from a university hospital during a 5-year period. J Clin Microbiol. 2006;44:3883–3886. doi: 10.1128/JCM.01388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace MR, Mascola JR, Oldfield EC., 3rd Red man syndrome: incidence, etiology, and prophylaxis. J Infect Dis. 1991;164:1180–1185. doi: 10.1093/infdis/164.6.1180. [DOI] [PubMed] [Google Scholar]
- 4.O'Sullivan TL, Ruffing MJ, Lamp KC, Warbasse LH, Rybak MJ. Prospective evaluation of red man syndrome in patients receiving vancomycin. J Infect Dis. 1993;168:773–776. doi: 10.1093/infdis/168.3.773. [DOI] [PubMed] [Google Scholar]
- 5.Rybak MJ, Bailey EM, Warbasse LH. Absence of "red man syndrome" in patients being treated with vancomycin or high-dose teicoplanin. Antimicrob Agents Chemother. 1992;36:1204–1207. doi: 10.1128/aac.36.6.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy M, Koren G, Dupuis L, Read SE. Vancomycin-induced red man syndrome. Pediatrics. 1990;86:572–580. [PubMed] [Google Scholar]
- 7.Odio C, Mohs E, Sklar FH, Nelson JD, McCracken GH., Jr Adverse reactions to vancomycin used as prophylaxis for CSF shunt procedures. Am J Dis Child. 1984;138:17–19. doi: 10.1001/archpedi.1984.02140390009004. [DOI] [PubMed] [Google Scholar]
- 8.Schaad UB, McCracken GH, Jr, Nelson JD. Clinical pharmacology and efficacy of vancomycin in pediatric patients. J Pediatr. 1980;96:119–126. doi: 10.1016/s0022-3476(80)80347-7. [DOI] [PubMed] [Google Scholar]
- 9.Fujita K, Murono K, Yoshikawa M, Miyamoto K. Intravenous vancomycin treatment in children; its clinical usefulness and serum concentration monitoring. Jpn J Antibiot. 1993;46:505–510. [PubMed] [Google Scholar]
- 10.Le J, Nguyen T, Law AV, Hodding J. Adverse drug reactions among children over a 10-year period. Pediatrics. 2006;118:555–562. doi: 10.1542/peds.2005-2429. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, Bayer A, Cosgrove SE, et al. Clinical practice guidelines by the infectious diseases society of america for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: executive summary. Clin Infect Dis. 52:285–292. doi: 10.1093/cid/cir034. [DOI] [PubMed] [Google Scholar]
- 12.Toyoguchi T, Ebihara M, Ojima F, et al. Histamine release induced by antimicrobial agents and effects of antimicrobial agents on vancomycin-induced histamine release from rat peritoneal mast cells. J Pharm Pharmacol. 2000;52:327–331. doi: 10.1211/0022357001773878. [DOI] [PubMed] [Google Scholar]
- 13.Sahai J, Healy DP, Shelton MJ, et al. Comparison of vancomycin- and teicoplanin-induced histamine release and "red man syndrome". Antimicrob Agents Chemother. 1990;34:765–769. doi: 10.1128/aac.34.5.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polk RE, Healy DP, Schwartz LB, et al. Vancomycin and the red-man syndrome: pharmacodynamics of histamine release. J Infect Dis. 1988;157:502–507. doi: 10.1093/infdis/157.3.502. [DOI] [PubMed] [Google Scholar]
- 15.Petersen J, Raithel M, Schwelberger HG. Characterisation of functional polymorphisms of the human diamine oxidase gene. Inflamm Res. 2005;54(Suppl 1):S58–S59. doi: 10.1007/s00011-004-0426-6. [DOI] [PubMed] [Google Scholar]
- 16.Raithel M, Matek M, Baenkler HW, Jorde W, Hahn EG. Mucosal histamine content and histamine secretion in Crohn's disease, ulcerative colitis and allergic enteropathy. Int Arch Allergy Immunol. 1995;108:127–133. doi: 10.1159/000237129. [DOI] [PubMed] [Google Scholar]
- 17.Yan L, Galinsky RE, Bernstein JA, Liggett SB, Weinshilboum RM. Histamine N-methyltransferase pharmacogenetics: association of a common functional polymorphism with asthma. Pharmacogenetics. 2000;10:261–266. doi: 10.1097/00008571-200004000-00007. [DOI] [PubMed] [Google Scholar]
- 18.Barnes PJ, Chung KF, Page CP. Inflammatory mediators and asthma. Pharmacol Rev. 1988;40:49–84. [PubMed] [Google Scholar]
- 19.Maslinski C. Histamine and its metabolism in mammals. Part II: Catabolism of histamine and histamine liberation. Agents Actions. 1975;5:183–225. doi: 10.1007/BF02026434. [DOI] [PubMed] [Google Scholar]
- 20.Chen GL, Wang H, Wang W, et al. Histamine N-methyltransferase gene polymorphisms in Chinese and their relationship with enzyme activity in erythrocytes. Pharmacogenetics. 2003;13:389–397. doi: 10.1097/00008571-200307000-00004. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Martin E, Ayuso P, Martinez C, Blanca M, Agundez JA. Histamine pharmacogenomics. Pharmacogenomics. 2009;10:867–883. doi: 10.2217/pgs.09.26. [DOI] [PubMed] [Google Scholar]
- 22.Choi JH, Kim SH, Suh CH, Nahm DH, Park HS. Polymorphisms of high-affinity IgE receptor and histamine-related genes in patients with ASA-induced urticaria/angioedema. J Korean Med Sci. 2005;20:367–372. doi: 10.3346/jkms.2005.20.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thurmond RL, Gelfand EW, Dunford PJ. The role of histamine H1 and H4 receptors in allergic inflammation: the search for new antihistamines. Nat Rev Drug Discov. 2008;7:41–53. doi: 10.1038/nrd2465. [DOI] [PubMed] [Google Scholar]
- 24.Feudtner C, Hays RM, Haynes G, et al. Deaths attributed to pediatric complex chronic conditions: national trends and implications for supportive care services. Pediatrics. 2001;107:E99. doi: 10.1542/peds.107.6.e99. [DOI] [PubMed] [Google Scholar]
- 25.Kononenko MR-SaI. Theoretical and Empirical Analysis of ReliefF and RReliefF. Machine Learning Journal. 2003;53:23–69. [Google Scholar]
- 26.Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–382. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- 27.Scadding GW, Scadding GK. Recent advances in antileukotriene therapy. Curr Opin Allergy Clin Immunol. 2010;10:370–376. doi: 10.1097/ACI.0b013e32833bfa20. [DOI] [PubMed] [Google Scholar]
- 28.Gupta MA, Guptat AK. The use of antidepressant drugs in dermatology. J Eur Acad Dermatol Venereol. 2001;15:512–518. doi: 10.1046/j.1468-3083.2001.00278.x. [DOI] [PubMed] [Google Scholar]
- 29.Healy DP, Sahai JV, Fuller SH, Polk RE. Vancomycin-induced histamine release and "red man syndrome": comparison of 1- and 2-hour infusions. Antimicrob Agents Chemother. 1990;34:550–554. doi: 10.1128/aac.34.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Digital Content 1, Figure Precursor, actions and metabolism of histamine. Histidine Decarboxylase (HDC), Histamine N-methyltransferase (HNMT), Diamine Oxidase (DAO), Histamine 1,2,3,4 receptors (H1R, H2R, H3R, H4R)