Abstract
We employ a combination of 13C/15N magic angle spinning (MAS) NMR and 2H NMR to study the structural and functional consequences of different membrane environments on VDAC1, and, conversely, the effect of VDAC1 on the structure of the lipid bilayer. MAS spectra reveal a well structured VDAC1 in 2D crystals of dimyristoylphosphatidylcholine (DMPC) and diphytanoylphosphatidylcholine (DPhPC) and their temperature dependence suggests that VDAC structure does not change conformation above and below the lipid phase transition temperature. The same data show that the N-terminus remains structured at both low and high temperatures. Importantly, functional studies based on electrophysiological measurements on these same samples show fully functional channels, even without the presence of Triton X-100 that has been found necessary for in-vitro-refolded channels. 2H solid state NMR and differential scanning calorimetry were used to investigate the dynamics and phase behavior of the lipids within the VDAC1 2D crystals. 2H NMR spectra indicate that the presence of protein in DMPC results in a broad lipid phase transition that is shifted from 19°C to ~27°C and show the existence of different lipid populations, consistent with the presence of both annular and bulk lipids in the functionally and structurally homogeneous samples.
Keywords: MAS NMR, 2H NMR, ion channels, VDAC, membrane proteins, electrophysiology, lipid bilayers, 2D crystals, annular lipids, protein-lipid interaction
1. Introduction
Interactions between integral membrane proteins and the membrane lipid environment influence the conformation and function of membrane proteins1. Lipid phase, composition, and membrane thickness are known to affect the structure and activity of membrane proteins. A key example has been observed in cells where most bilayers are reported to be in a liquid-crystalline (Lα) phase2 in which membrane proteins are known to be functional; changes from Lα to the gel phase (Lβ) have been correlated with a loss of activity3–5. Simultaneously, the presence of membrane proteins in the bilayer affects the properties of the surrounding lipids, which can be directly observed through changes in phase transition temperatures, enthalpies, and 2H NMR spectra that probe lipid order parameters. Protein-lipid and protein-protein interactions in the bilayer mediate biological functions including membrane fusion6 and signal transduction7. Furthermore, some studies of cell membranes suggest that the formation of lipid rafts are correlated with high protein concentration8 although this relationship is still regarded to be controversial and has yet to be fully understood9.
The effects of protein-lipid and protein-protein interactions can be particularly prominent when the protein concentration is high and, in certain cases, ordered unilamellar or multilamellar sheet-like arrays, known as 2-dimensional (2D) crystals, can form. 2D membrane protein crystals are known to be present in vivo for several proteins, including bacteriorhodopsin10, water channels11, and the nicotinic acetylcholine receptor12. Many additional examples exist of membrane proteins that form 2D crystals when isolated and reconstituted into single-component lipid bilayers13.
2D crystals are also of great interest within the structural biology community as the inherent order facilitates studies with electron microscopy (EM)14 and atomic force microscopy15. Not surprisingly, a number of 3D membrane protein structures have been determined at various resolutions with cryo-EM, including bacteriorhodopsin10, light harvesting complex II16, gap junction channels17, and bacterial porins18. Another approach to examine 2D crystals of membrane proteins is magic angle spinning NMR (MAS NMR). In particular, the microscopic order in 2D crystals results in high resolution spectra that yield atomic level details of membrane protein structure and mechanistic information as demonstrated by several investigations19–30. Thus, 2D crystal preparations are a promising alternative to 3D crystallization since: (1) membrane proteins can be reconstituted into a more native-like lipid bilayer15, (2) membrane proteins in 2D crystals can retain full functionality31, and (3) 2D crystals are possibly easier to obtain. Furthermore, unlike 3D crystals, which contain fewer lipid molecules per protein, membrane proteins in 2D crystals are surrounded by a continuous lipid bilayer32.
Despite the extensive use of 2D crystals in structural studies, little is known about the lipid dynamics and phase behavior of the lipid bilayers in these systems. Specifically, what is the nature of the lipid environment in 2D crystals and how does it compares to lipid environments established for well-studied pure lipids 33–35 or with lower protein concentration36? Lipid-protein interactions in a 2D crystal were previously studied by EM for the water channel AQP011,32,37 where Gonen et al. found that most annular lipids are tightly packed between adjacent tetramers of AQP0, mediating lattice interactions11. This suggested that channel mobility and conformational flexibility within the bilayer were very restricted. However, while EM can provide a picture of lipid arrangement and adaptation to the membrane protein, it does not directly provide information about the dynamics of the lipid bilayers, including changes in transition temperatures and lipid order parameters.
Solid state 2H NMR is probably the most useful technique to investigate lipid dynamics on an atomic level and in a noninvasive manner. For a CD bond in a polymethylene chain the 2H quadrupole coupling constant is ~167 kHz. This allows one to examine molecular motion on time scale of ~10−3–10−8 s based on quadrupole echo lineshapes38–41. Such 2H spectra have been used extensively to study lipid dynamics and changes in lipid phase behavior due to heterogeneous lipid composition, lipid-cholesterol interactions 42–46, and hydrophobic mismatch47,48. Despite its extensive use, 2H NMR has not been utilized to examine changes in lipid dynamics for 2D crystals and, in particular, it has not been used to study the effect of β-barrel insertion on lipid dynamics. In addition, with a 13C/15N labeled protein and 2H labeled lipids it is possible to observe static 2H NMR and MAS NMR spectra under identical conditions to obtain paired measurements of lipid dynamics and atomic level protein structural information. MAS NMR has yielded high resolution spectra on a growing number of membrane proteins, including bR24,28,29,,49, KcsA50, phospholamban51, DsbB52, and OmpG30. By combining both 2H static NMR and MAS NMR, we can probe the influence of membrane proteins on surrounding lipids, and, at the same time the influence of the lipid environment on the integral membrane proteins under similar conditions.
The voltage-dependent anion channel isoform 1 (VDAC1) is a 32 kDa integral membrane protein that controls transport of metabolites between the outer mitochondrial space and the cytosol53–56. VDAC1 is a typical beta barrel ion channel with structures known from detergent based solution NMR and crystallographic studies57–59. Its function has been extensively studied54,56,60–62, and 2D crystals of VDAC1 have been previously characterized by EM63–66. In analogy to various other membrane proteins, several studies suggest a significant impact of membrane lipid composition and protein lipid-interactions on VDAC activity. For example, (1) VDAC gating may be regulated by characteristic mitochondrial lipids67, VDAC channels isolated from the seeds of Phaseolus coccineus are sensitive to cholesterol and phytosterols68, (3) VDAC has been reported to associate with detergent resistant microdomains isolated from mitochondria69, and (4) interaction of VDAC with proteins such as Bcl-xL are suggested to depend on membrane composition70. Thus, it is pertinent to understand the effect of these varying lipid environments on the structure of the embedded protein.
Here we apply a combination of functional assays, static 2H NMR, 13C,15N MAS NMR, and differential scanning calorimetry (DSC) to investigate VDAC1 2D crystals containing different lipids or lipid mixtures. Importantly, functional assays performed on VDAC1 reconstituted directly from such 2D crystals show they insert into planar lipid membranes and that resulting channels gate properly. Furthermore, insertion and gating occurs in the absence of Triton X-100 in the sample buffer, in contrast to the situation for detergent solubilized samples where it is required for insertion into the lipid bilayer57. We employ DSC and temperature dependent solid state 2H NMR to examine the phase equilibria and dynamic behavior of DMPC lipids in the 2D crystals over a temperature range from 14 °C to 45 °C. Our results reveal a phase transition for lipids in 2D crystals shifted to a higher TM (~27 °C) and suggest the coexistence of both gel and liquid crystalline phases across this temperature range. Finally, MAS NMR spectra of 13C/15N labeled VDAC1 in 2D crystals has permitted a detailed residue-specific structural inspection of the protein under conditions of varying lipid phase, lipid composition and hydrophobic mismatch, facilitated by the high spectral resolution and sensitivity permitted by our preparation protocol. Earlier MAS NMR studies on VDAC1 were performed under significantly higher lipid-to-protein ratios that are more consistent with the formation of liposomes rather than 2D crystals71. Under those conditions the protein was found to feature a rigid N-terminus, which was unaffected by changes in lipid composition and temperature explored in the study71. Our study shows that the protein behaves similarly in 2D crystals. For 2D crystals comprised of DMPC, the 13C/15N spectra show that neither the N-terminus nor the β-barrel of the protein show structural changes between 4–30°C, despite the change in lipid phase that we observe by DSC and 2H NMR. In addition, the VDAC1 structure is very similar in pure DMPC, DMPC/dioleylphosphatidylglycerol (DOPG) mixtures, and pure diphytanoylphosphatidylcholine (DPhPC), where the latter is of particular interest given its use in VDAC1 functional studies. Our data provide insights into the consequences of 2D crystal formation for membrane protein studies, permit us to postulate a model for the number of annular and ‘bulk’ lipid molecules in the 2D crystal and explore the extent to which the protein and lipids undergo conformational changes to minimize hydrophobic mismatch.
2. Experimental Section
Materials
1,2-dimyristoyl(d54)-sn-glycero-3-phosphocholine (d54-DMPC), 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DOPG), and 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC) were obtained from Avanti Polar Lipids (Alabaster, AL). Deuterium depleted H2O was obtained from Cambridge Isotope Laboratory (Andover, MA). Octyl-POE (polyoxyethelene) was obtained from Bachem (King of Prussia, PA). All other reagents were obtained from Fischer.
Preparation of VDAC1/DMPC 2D crystals
Protocols for expression, refolding, and purification of VDAC1 were based on the work of Malia and Wagner,72 and Hiller et al73. Human VDAC1 was expressed in BL21 (DE3) cells transformed with VDAC1 plasmid containing a 6x C-terminal histidine tag. Cells were grown in either natural abundance LB media or M9 media containing 13C glucose and 15N ammonium chloride (Cambridge Isotope Labs, Andover MA) at 37 °C until reaching an OD600 of 0.8. Overexpression was induced with 1 mM IPTG for 3–5 hours. After lysis of cells and isolation of inclusion bodies, VDAC1 was purified under denaturing conditions (8 M urea, 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 20 mM imidazole) over Ni-agarose resin and eluted with the same buffer containing 250 mM imidazole. Fractions containing VDAC1 were dialyzed against 4 L of buffer (50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM EDTA, 20 mM BME). Precipitated VDAC1 was then centrifuged and dissolved in 8 M urea buffer or 6 M GuHCl buffer. VDAC1 was refolded at 4 °C by dropwise dilution into 10x volume of refolding buffer (25 mM NaPO4, pH 7.0, 100 mM NaCl, 1 mM EDTA, 20 mM BME, 43 mM lauryldimethylamine oxide (LDAO)). Refolded VDAC1 samples were dialyzed against 20x volume of buffer (25 mM NaPO4, pH 7.0, 2–3 mM DTT, 1mM EDTA). Final purification was done by cation exchange chromatography, and fractions containing folded VDAC1 were concentrated using Centricon MWCO 10,000 centrifugal filters.
2D crystals were prepared according to the procedure published by Dolder et al.63 with some modifications. Purified, refolded VDAC1 in 4.3 mM LDAO was dialyzed for 24 hr against 2–4 L of buffer (0.6% (w/v) octyl-POE, 50 mM Tris-HCl, pH 8.0, 4–5 mM dithiothreitol (DTT). Dry DMPC (d54-DMPC for 2H NMR studies) was solubilized in 1% octyl-POE and added to VDAC1 at a lipid/protein ratio of 1:2 (w/w) or as indicated. The mixture was then dialyzed against buffer containing 150 mM NaCl, 20 mM MgCl2, 10 mM MES, and 4 mM DTT at pH 6.5 at room temperature for several days, with twice per day 4 L buffer changes in a dialysis cassette with 10 kDa MWCO. Dialysis was allowed to continue for at least an additional 4–5 days after crystals appeared fully formed. Harvested crystals were washed with 25 mM NaPO4 at pH 7 to remove any remaining detergent and salt for NMR studies. Samples used for 2H NMR studies were washed with buffer made from 2H depleted H2O. Samples are refrigerated at 4 °C when not in use.
Differential scanning calorimetry
DSC measurements were performed using a MicroCal VP-DSC (Piscataway, NJ). Pure d54-DMPC and VDAC1/d54-DMPC 2D crystals were each mixed with excess 25mM phosphate buffer at pH 7.0 at room temperature. All buffers and samples were degassed for 10 minutes under vacuum prior to the experiments. The scan rate was 1 °C/min from 1–40 °C with 30 minutes between each scan to allow temperature re-equilibration. The buffer itself was scanned before the samples to obtain a reproducible baseline. Each experiment was allowed to run overnight to ensure reproducibility. Data analysis was performed using the Origin DSC software included with the calorimeter.
NMR Spectroscopy
Solid state 2H NMR experiments were performed on a custom-built spectrometer (Courtesy of Dr. D. Ruben) operating at 60.8 MHz for 2H using a single-channel probe with 4.0 mm coil. Spectra were obtained with a quadrupolar echo sequence38 with a π/2 pulse of 2.5 μs and a delay of 30 μs between the two pulses. Oriented 2H NMR spectra (θ = 0°) were calculated by de-Pake-ing method described by Sternin et al.74
13C-13C correlation spectra were acquired with RFDR75,76 mixing at 12.5 kHz MAS and 750 MHz 1H field strength on a custom built spectrometer (courtesy of Dr. D. Ruben) equipped with a 3.2 mm E-Free MAS probe (Bruker BioSpin, Billerica MA) and at 20 kHz MAS and 900 MHz 1H field strength on an Avance 2 spectrometer equipped with a 3.2 mm E-Free MAS probe (Bruker Biopsin, Billerica MA). 83 kHz TPPM decoupling77 was used during evolution and acquisition periods (5.8 us pulse with relative phases of 0° and 13° spinning at 12.5 kHz and 0° and 18° spinning at 20 kHz). Each spectrum averaged 32 scans per point unless otherwise noted. The temperature was calibrated at a given spinning frequency with a specified amount of cooling gas according to the chemical shift of 79Br 78. This provided a lower bound on the actual sample temperature, as it did not account for RF heating. Above the 2D crystal phase transition temperature, the sample temperature was calibrated at approximately 30 °C, and below the phase transition the temperature was calibrated to 0 °C without RF heating.
VDAC reconstitution and conductance measurements
Planar lipid membranes were formed on a 70–90 μm diameter orifice in the 15-μm thick Teflon partition that separated two chambers as previously described67. Lipid bilayers used for membrane formation were made from a 5 mg/ml solution of DPhPC (purchased from Avanti Polar Lipids, Inc. Alabaster, AL) in pentane. The chambers were filled with of 1M KCl buffered with 5 mM HEPES at pH 7.4. Channel insertion was achieved by adding protein solutions into the 1.2 ml aqueous phase in the cis compartment while stirring. The protein added was either a 5 μL suspension of VDAC1/DMPC 2D crystals (undiluted), or 0.5 μL of VDAC1/DMPC 2D crystals that had been diluted 1/100 (v/v) in a buffer “A” containing 10 mM Tris, 50 mM KCl, 1mM EDTA, 15% (v/v) DMSO, 2.5% (v/v) Triton X-100, pH 7.0. The potential was defined as positive when it was greater at the side of VDAC addition (cis-side). Current recordings were performed as described previously67 using an Axopatch 200B amplifier (Axon Instruments, Inc., Foster City, CA) in the voltage clamp mode. Single-channel data were filtered by a low-pass 8-pole Butterworth filter (Model LPF-8, Warner Instrument Corp.) at 15 kHz and directly saved into the computer memory with a sampling frequency of 50 kHz.
The voltage-dependent properties of a VDAC-containing membranes were assessed following the protocol devised by Colombini and colleagues60,79 in which gating is inferred from the channel response to a slowly changing periodic transmembrane voltage. A symmetrical 5 mHz triangular voltage wave with ± 60 mV amplitude from a Function Waveform Generator 33120A (Hewlett Packard) was used. Data were acquired with Digidata 1322A board (Axon Instruments Inc.) at a sampling frequency of 1 Hz and analyzed using the pClamp 10.2 software (Axon Instruments, Inc.). Analysis of VDAC voltage-gating was performed following published protocols67. In each experiment, current records were collected in response to 5–8 periods of triangular voltage waves. Only the part of the wave during which the channels were reopening was used for the subsequent analysis. Given the variable number of channels per experiment, the average conductance (G) was normalized to the maximum conductance (Gmax).
3. Results
Electron Microscopy
The preparation of VDAC1 2D crystals from purified recombinantly expressed VDAC1 was described previously by Dolder et al.63. Since our preparations differed somewhat from the protocol used in Dolder et al., the first step in this work was to verify the formation of proper 2D crystals. Dialysis of approximately 20 mg VDAC1/10 mg DMPC dissolved in octyl-POE in a total volume of 40 mL showed signs of precipitation within 10–12 hours at room temperature, and mature 2D crystals formed after 1–2 days. Dialysis proceeded for at least an additional 4–5 days after crystals were fully formed to remove lingering detergent. The rate of 2D crystal formation depended to a large extent on the purity of refolded VDAC1 and to some extent on the amount of DTT present in the dialysis buffer; with more DTT 2D crystals formed more quickly. Preliminary samples not used in this study contained mixed populations of properly folded and unfolded VDAC and took at least twice as long to observe 2D crystal formation. 2D crystal samples were characterized by electron microscopy. An example image of VDAC1 2D crystals is shown in Figure S1 with overlapping sheets of VDAC1 2D crystals readily visible. It was difficult to isolate unilamellar sheets, similar to the reports by Dolder et al.63
Functional assays
VDAC voltage gating in planar lipid membranes
Functional assays were performed by reconstitution of the VDAC1-DMPC 2D crystals directly into planar membranes made of DPhPC and testing VDAC’s gating properties. Non-ionic detergents, such as Triton X-100, are routinely used to stimulate VDAC insertion into the planar membranes57. However, the presence of detergent might affect VDAC folding in the planar membrane,80 and consequently change channel properties. In order to rule out this possibility, we compared the effect of the presence or absence of detergent on VDAC’s properties. VDAC was reconstituted into the planar membranes using two protocols: (1) directly from VDAC1/DMPC or (2) after dilution in buffer “A” (see Experimental Section) containing Triton X-100. Channel reconstitution from VDAC1 2D crystals without Triton X-100 required 103 fold more protein compared to channels reconstituted in the presence of the detergent. This confirms that Triton X-100 stimulates channel insertion. However, VDAC reconstituted with or without detergent showed typical VDAC basic properties, such as single channel conductance of 4.2 nS in 1 M KCl56 and voltage gating at high, > 40 mV, applied voltages.
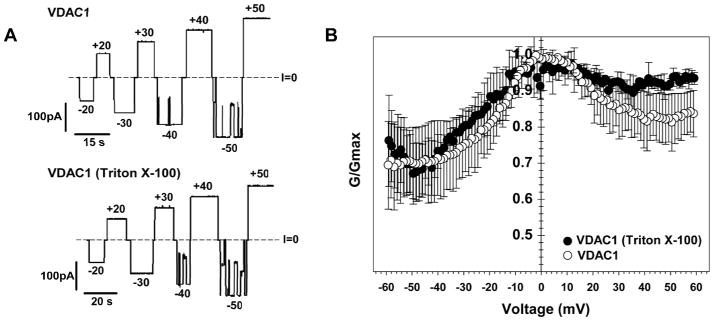
Figure 1A shows representative traces of ion currents through single channels reconstituted from VDAC-DMPC 2D crystals without (top) or with (bottom) Triton X-100 detergent. Both traces show the typical VDAC gating when 40 – 50 mV of applied voltage induced channel conductance transitions from the highly conducting “open” state to the low conducting “closed states”. Voltage gating is a characteristic property of VDAC56, and the most reliable method to study VDAC voltage gating was designed by Colombini and co-workers.81,79 In this method, a slow symmetrical triangular voltage wave is applied to the membrane containing multiple channels allowing collection of statistics on many channels.
Figure 1.
(A) Representative single channel current traces of VDAC1 reconstituted into planar lipid membranes from 2D crystals without (top) or with (bottom) the presence of Triton X-100 in the sample buffer used for functional studies. Applied voltages are shown. Dashed lines indicate zero current levels. (B) Characteristic bell-shape plots of the normalized average conductance vs. applied voltage. Each data point is a mean of three experiments ±STD.
Representative current traces in response to these voltage waves are shown in Figure S2 without (top) and with (bottom) Triton X-100 in the buffer used for channel reconstitution. The analysis of multi-channel currents (15 to 340 channels) as shown in Figure S2 gave the calculation of the conductance at different voltages applied, resulting in characteristic bell-shaped plots regardless of the presence of detergent (Figure 1B). VDAC closure was more pronounced at negative voltages in single and multi-channel (both with and without Triton X-100). This is a manifestation of an intrinsic asymmetry of VDAC56 where VDAC has been shown to exhibit two different closed states depending on a sign of applied voltage.
Differential scanning calorimetry
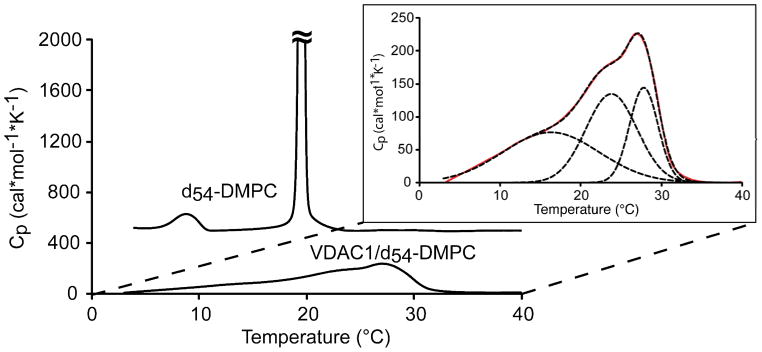
Figure 2 shows the endotherm of both pure d54-DMPC and VDAC1/d54-DMPC 2D crystals (VDAC:d54-DMPC wt ratio of 2:1, molar ratio ~1:25). Pure d54-DMPC exhibits a sharp transition at 19 °C (rippled gel phase Pβ′ to lamellar liquid crystal phase Lα), as expected, that has large transition enthalpy and is highly cooperative, with a pretransition at 8 °C (Lamellar gel phase Lβ′ to rippled gel phase Pβ′) that is broader with smaller transition enthalpy.
Figure 2.
DSC thermograms of pure d54-DMPC and VDAC1/d54-DMPC 2D crystal. The largest peak at 19 °C for pure DMPC is cutoff in this figure so that the 2D crystals can be visualized on the same scale. Inset: Expansion of the DSC thermogram of VDAC1/d54-DMPC 2D crystals and simulations using two narrow and one broad component. Experimental data are graphed by the solid red line, and the simulated values are shown by the black dashed lines.
The effect of VDAC1 on the lipid phase transition is immediately apparent in the DSC of the 2D crystals. VDAC1/d54-DMPC 2D crystals show a much broader phase transition spanning 20 degrees with a maximum at the transition temperature, TM, near 27 °C. The magnitude of the maximum observed transition enthalpy, ΔH, of VDAC1/d54-DMPC is only 6% of pure d54-DMPC, indicating the transition is almost completely eliminated or severely broadened. These observations are reminiscent of lipid samples containing a large amount of protein or cholesterol showing a large disruption in lipid acyl chain packing.46,82 The reduction of transition enthalpy and broadening of transition temperatures show the transition is less cooperative in the 2D crystal. The transition profile is asymmetric and can be decomposed into two narrow components and a broader component as shown in Figure 2. The broad component is centered at 16 °C with a half-width of 8 degrees. The two narrower transitions are centered at 24 and 28 °C, with half width of 2.5 and 4.5 degrees, respectively.
2H NMR Spectroscopy
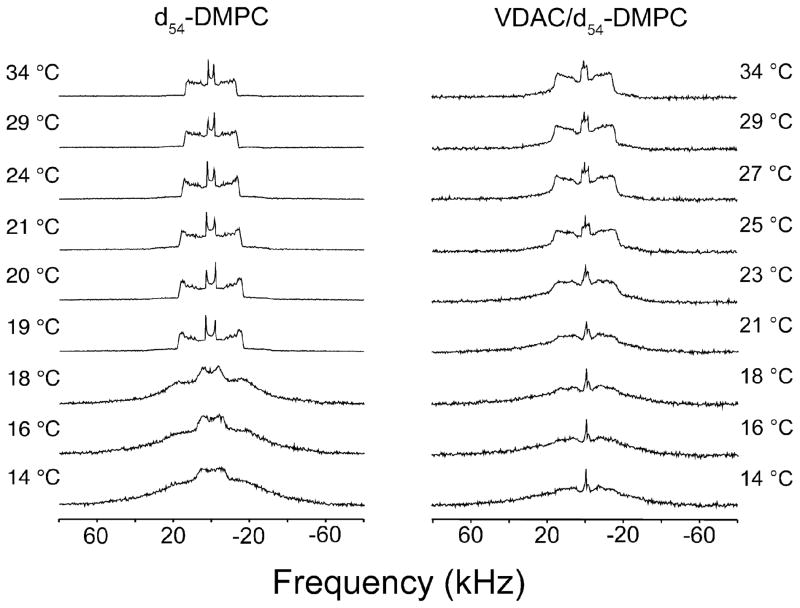
The 2H NMR spectra of pure d54-DMPC (mixed with 2H depleted buffer at a ratio of 1:1 (w/w)) and that of VDAC1/d54-DMPC 2D crystals (protein-to-lipid molar ratio of ~1:25) are presented in Figure 3. Lipid liquid crystal and gel phases have distinct 2H NMR lineshapes, therefore the phase boundary is apparent by examining 2H NMR spectra as a function of temperature. In the liquid crystal phase, perpendicular edge quadrupolar splittings, ΔνQ⊥ of various 2H’s in the hydrocarbon chain are between 5 to 30 kHz depending on their location in the lipid bilayer and the degree of acyl chain flexibility. The terminal methyl group of the acyl chain, located in the middle of the bilayer, has the smallest ΔνQ⊥ that is also the easiest to quantify. The width of the , is determined by splittings of the least mobile methylenes that are near the middle of the acyl chain and closer to the phosphate group. Due to extensive overlap, the splittings of individual methylene groups are not easily determined. The line shapes of d54-DMPC/VDAC 2D crystals share some features with the pure gel phase lipid 83 where the gauche-trans isomerization and axial diffusion slows and the spectrum broadens.
Figure 3.
2H NMR spectra of d54-DMPC (left) and VDAC1/d54-DMPC 2D crystals (right) as a function of temperature. For pure DMPC, a sharp transition between the liquid crystalline to gel phase is observed between 18–19 °C. For the 2D crystals, the transition is more gradual over a larger temperature range. All spectra are plotted on a normalized intensity scale.
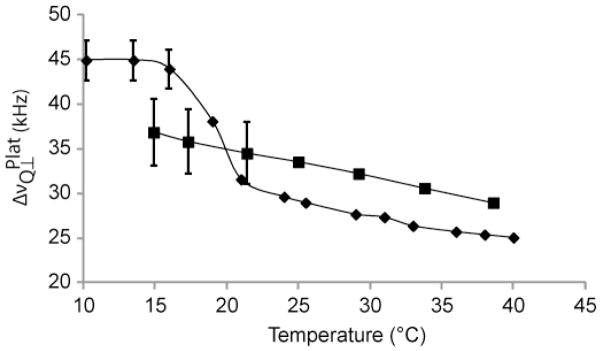
As shown in Figure 3, d54-DMPC undergoes a sharp phase transition at 19 °C, with little evidence of coexistence between liquid crystal and gel phase, as expected. However, for VDAC1/d54-DMPC 2D crystals, the transition is more gradual. Compared to the spectra of pure lipid, the spectra of 2D crystals appear to be a superposition of liquid crystal and gel phase spectra, suggesting coexistence of both liquid and gel phase over a range of at least 10 °C. The sharp peak in the middle of the spectra is most likely due to residual deuterated water. The phase transition can be visualized by examining as a function of temperature. For pure d54-DMPC, the phase transition is apparent by a break in linearity of versus temperature below 19 °C, the phase transition temperature, as shown in Figure 4. For the 2D crystals, is larger at temperatures above and smaller below 20 °C compared to the pure lipid (see also Figure S3). Although there is no obvious break in linearity, the fact that the 2H NMR lineshape at 39 °C resembles that of liquid crystal phase, but at 14 °C that of gel phase, indicates that a phase transition must exist, albeit very gradual and hard to quantify. Interestingly, the terminal methyl group appears to show a decreased splitting in the 2D crystals (Figure S3) suggesting that the rigidification of the acyl chains is not necessarily homogeneous across the lipid bilayer.
Figure 4.
Perpendicular quadrupolar splitting, ΔνQ⊥, as a function of temperature. (◆) d54-DMPC, (■) VDAC1/d54-DMPC 2D crystal. ΔνQ⊥ were difficult to determine with precision at lower temperatures due to poorer signal-to-noise ratio caused by spectral broadening in the gel phase. ΔνQ⊥ were reproducible to within 5% (d54-DMPC) and 10% (VDAC1/d54-DMPC) for these spectra.
Thus, consistent with the DSC experiments, the 2H NMR experiments show a significant effect of the protein on the lipid phase behavior, causing a broadening of transition temperatures. This suggests that the transition is less cooperative in the 2D crystal. In addition, both NMR and DSC indicate the coexistence of different domains of lipids in distinct phases, with at least part of the lipids eventually transitioning to a liquid crystalline state that is reminiscent of the bulk phase of fluid DMPC bilayers.
In addition to the temperature dependent data presented above, we also acquired 2H NMR temperature-dependent data using a higher protein-to-lipid molar ratio of 1:50, which is the same amount used in a previous MAS NMR study71. This protein-to-lipid ratio is outside the range reported by Dolder et al. for forming 2D crystals63 and under these conditions samples were likely liposomes. See Figure S5. Even with a higher lipid content we see significant effects of the protein on the lipids. There appears to be no indication of significant amounts of bulk lipids, nor does there appear to be separate phases (2D VDAC1 crystals surrounded by macroscopically separated lipids) since we see no indications of a second component with the same phase behavior as bulk DMPC. This is consistent with the idea that this protein-to-lipid ratio forms homogenous samples where VDAC is distributed evenly in the membrane and not forming 2D crystals.
Structural comparison of VDAC1 in liquid crystalline and gel phases
Generally most membrane proteins are most active in the Lα phase of lipid bilayers. The underlying reasons for this have been studied in detail for several membrane proteins84,85, and it has been reported that decrease in protein function is correlated with changes in membrane protein structure as the lipids transition from the Lα to an Lβ phase. In bilayers containing both Lβ and Lα phases, membrane proteins preferentially partition into Lα domains86–88. Here we examine the possibility of conformational changes observed for VDAC based on MAS NMR of 13C,15N isotopically labeled VDAC1 in 2D DMPC crystals. Comparison of chemical shifts for 13C correlation spectra at temperatures above and below the lipid phase transition temperature of 27 °C permit us to look for conformational changes as a function of lipid phase, which also probes the effect of changes in lipid bilayer thickness. Our experiments also allow us to detect possible aggregation or exclusion of VDAC1 as a function of lipid phase by comparison the resolution and chemical shift dispersion of the 13C NMR spectra.
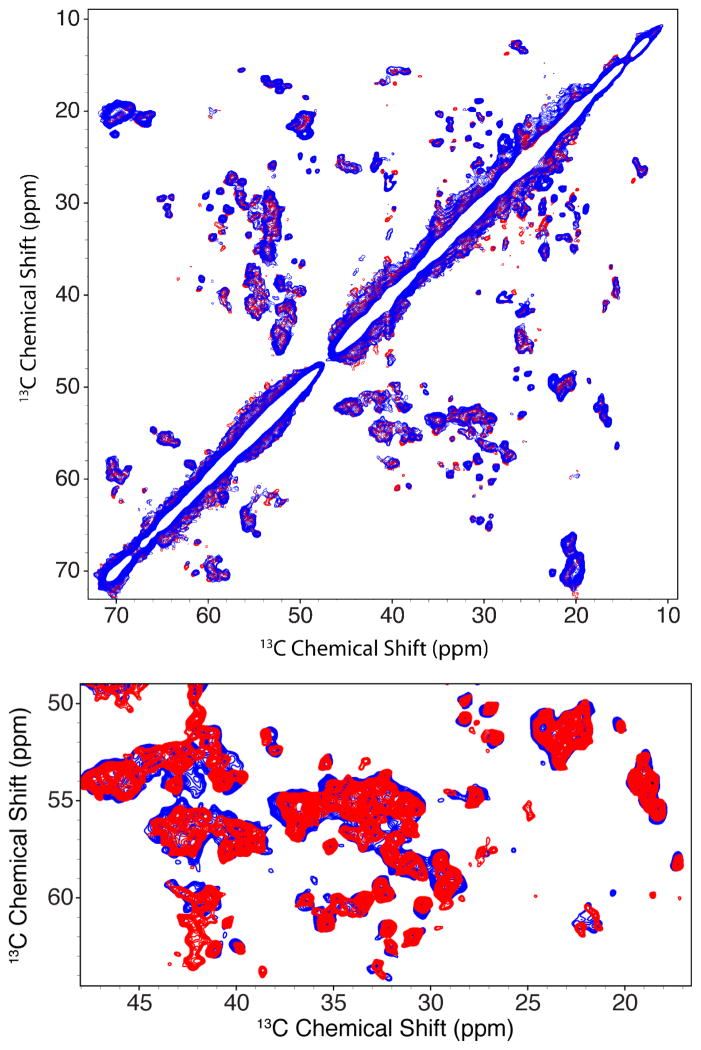
Figure 5 shows 13C-13C correlation spectra of VDAC1 in 2D crystals collected near 0 °C and 30 °C, above and below the elevated DMPC TM. Assignment of peaks arising from the N-terminal region of VDAC1 was achieved by 2D and 3D 13C-13C and 15N-13C-13C correlation experiments that will be described in a future publication. Chemical shifts for peaks arising from the N-terminal region of VDAC1 as well as the β-barrel are nearly identical, indicating that the phase of the lipid bilayer does not perturb the channel structure. Furthermore, the resolution and chemical shift dispersion for data collected above and below TM are also very similar, indicating that VDAC1 remains well structured and is not aggregated when the lipid bilayer is in the Lβ phase. Furthermore, since RFDR relies on homonuclear dipolar couplings, cross peak intensities are sensitive to local motion on the NMR time scale. Cross peak intensities and peak linewidths for data collected above and below the phase transition are nearly identical, indicating little difference in local dynamics. Spectra acquired below the TM exhibit line widths that appear only marginally broader, <5% or less than spectra acquired above TM. Furthermore, the MAS spectra show that the N-terminal region is rigid above and below the phase transition. In these samples, the overall structure of VDAC1 appears to be robust over a range of temperatures and relatively unaffected by the DMPC lipid phase transition.
Figure 5.
Comparison of 13C-13C RFDR correlation experiments performed on VDAC1 DMPC 2D crystals at 4°C (blue) and above the phase transition temperature (>30°C, red). Comparison of chemical shifts show the structure of VDAC1 is not perturbed by changes in the DMPC phase. The spectra were recorded with a 1.3 ms RFDR mixing period, at ωr/2π= 12.5 kHz, ω0I/2π=750 MHz and with 83 kHz TPPM decoupling during evolution and acquisition. Spectra were plotted and processed in nmrPipe with the same parameters and plotted on the same scale in Sparky. In the top spectrum, the low temperature spectrum was overlaid on top of the high temperature spectrum. For the bottom spectrum, the high temperature data was overlaid on top of the low temperature data and contour levels for both spectra were reduced to emphasize potential differences.
Comparison between VDAC1 in DMPC and DPhPC
The VDAC1 functional studies in this paper, as well as many others, were performed in DPhPC lipids. DPhPC lipids do not exhibit a phase transition over a wide temperature range and have an ether linkage that protects them from hydrolysis. These samples permit us to probe the structure of VDAC1 in 2D crystals in the same lipids used to reconstitute VDAC1 for the functional studies. Since the NMR experiments are performed under conditions where no voltage is applied across the membrane, it is likely that VDAC is in the “open” conformation for the NMR experiments. We reconstituted VDAC1 in DPhPC lipids at a molar ratio of 1:20 (1:2 w/w, protein:lipid) under two different dialysis conditions. In the first sample the dialysis buffer was the same as the one used to prepare DMPC 2D crystals, including Mg2+. In the second dialysis buffer, only 50 mM sodium phosphate was used. The second preparation was done in order to probe the effect of Mg2+ in the dialysis buffer, which has been reported to be required for the formation of DMPC 2D crystals63.
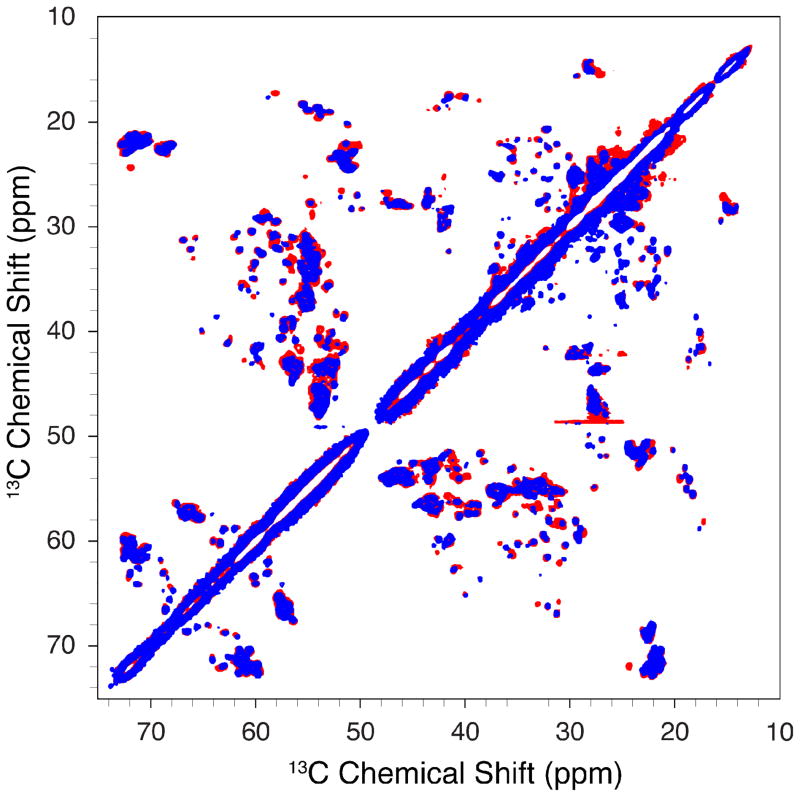
13C MAS NMR correlation spectra comparing VDAC1 in DMPC to VDAC1 in DPhPC are shown in Figures 6 and 7. The spectrum of VDAC1 in DPhPC shows a high dispersion of sharp peaks, similar to VDAC1 in DMPC. This demonstrates that VDAC1 in DPhPC under our reconstitution conditions yields samples with a high degree of microscopic order. Clearly some differences are present, but the overall dispersion of peaks is essentially identical in both spectra and all of the chemical shifts that can be observed in both spectra completely overlap, including for the N-terminal residues. This demonstrates that the backbone structure of VDAC1 reconstituted in DMPC and that of the protein reconstituted in DPhPC are nearly identical. Interestingly, linewidths in the DPhPC data are about 10–15% narrower when compared with VDAC1 in DMPC collected under identical conditions. Furthermore, data in DPhPC show a number of peaks for Cα-Cβ backbone sites that were missing compared to data in DMPC and that fall within the beta barrel region of the protein. This suggests differences in dynamics for the β-barrel backbone between DMPC and DPhPC and is under further investigation. Furthermore, DPhPC samples prepared with and without the presence of Mg2+ yielded identical spectra, indicating that magnesium was not required for the formation of a high degree of microscopic order in DPhPC lipids.
Figure 6.
Comparison between DMPC and DPhPC lipids. Both samples were prepared in the presence of Mg2+ U-13C 15N-VDAC1 1.6 ms RFDR, 20 kHz MAS, 900 MHz 1H field, T=4°C. The horizontal line at approximately 48 ppm is an artifact. Experimental times for each spectrum were about 16 hours.
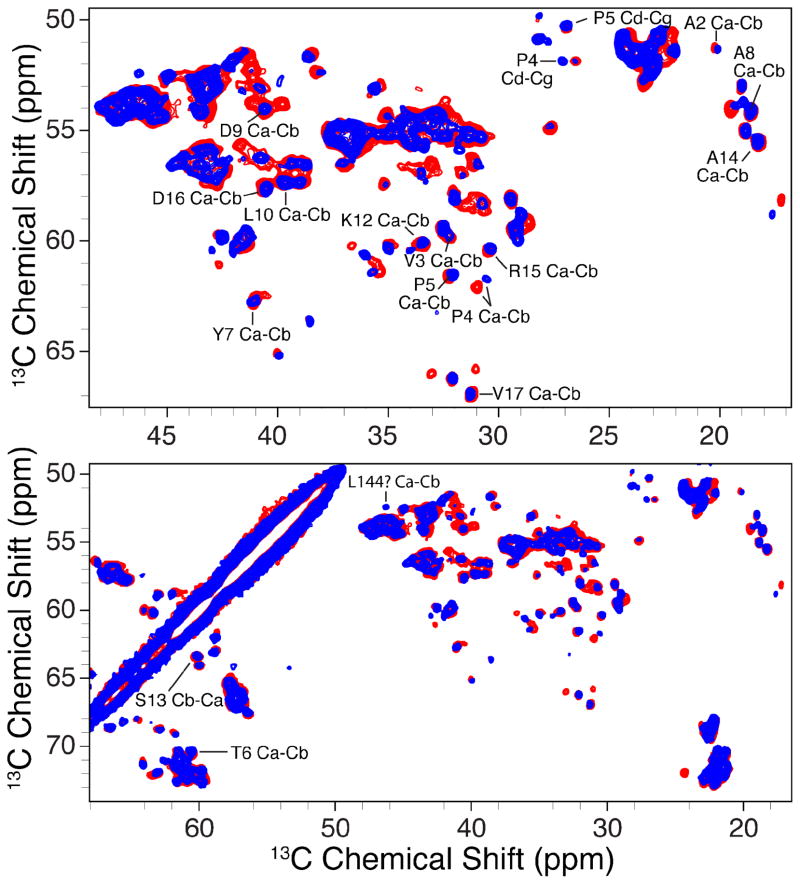
Figure 7.
Expansion of Figure 6. Selective assignments are pointed out for several residues in the N-terminal region. Chemical shifts for these residues are consistent between 2D crystals prepared with DMPC, DPhPC, and assignments reported for VDAC1 in liposomes71. One exception is L144, which was not independently confirmed here due to the lack of signal for this crosspeak for VDAC1 samples in DMPC.
4. Discussion
Electron Microscopy
Electron microscopy was used to validate our preparation of VDAC-DMPC 2D crystals and confirm that we were studying the same system as reported in Dolder et al. Furthermore EM images of DMPC/DOPG preparations at a protein-to-lipid molar ratio of 1:25 showed 2D crystals that appeared to be qualitatively very similar to 2D crystals obtained with only DMPC (see Figure S6), although a detailed comparison of the lattice parameters was not undertaken. VDAC1 reconstituted in DPhPC also appeared to yield 2D crystals (see Figure S7) at a protein-to-lipid ratio of 2:1 (w/w), 1:20 molar ratio.
Functional characterization of VDAC
Triton X-100 and cholesterol were previously proposed to be required for recombinantly expressed, refolded, and reconstituted VDAC1 to form properly functioning channels57,89. Here we demonstrate that refolded recombinant VDAC1 reconstituted into planar bilayers directly from VDAC1-DMPC 2D crystals forms functional channels with characteristic single-channel conductance and voltage-gating behavior (Figures 1 and S2) without the requirement of either cholesterol or detergent. These properties are similar to those of VDAC1 isolated from mitochondria of different sources and reconstituted into planar membranes at similar experimental conditions81,90–93. The presence of detergent could also affect VDAC ability to refold from the detergent solution into the planar membrane.80 However, circular dichroism analysis suggests that the conformation of fungal VDAC in Triton X-100 micelles is very similar to its pore-forming conformation in lipid membranes.94 Our data also show that presence of Triton-X-100 in VDAC-DMPC lipid crystals does not affect channel properties. Triton-X-100 for channel reconstitution from 2D crystals is not required. These channels gate properly regardless of the presence of Triton X-100.
Experiments with reconstituted channels, including those described here, require stable planar membranes that must last for up to several hours. Such stability is very difficult if not impossible to obtain with DMPC bilayers, most likely because the phase transition of C14 acyl chains lipids is near room temperature. Therefore, VDAC was reconstituted into membranes made of DPhPC, a lipid widely used in ion channels reconstitution experiments95–98. Insertion of VDAC1 from DMPC 2D crystals into DPhPC bilayers may involve membrane fusion, and it is possible that protein might undergo significant structural rearrangements upon transitioning from detergent micelles or lipid 2D crystals into the lipid planar bilayer as was suggested earlier91. We cannot entirely discard the possibility of a conformational change upon transition of VDAC from 2D crystal to the planar membrane in our experimental design to measure VDAC gating; however, MAS NMR results of VDAC1 reconstituted into DPhPC suggest that VDAC1 adopts a very similar if not the same conformation between DMPC and DPhPC.
DSC thermogram of VDAC 2D crystals and estimation of amounts of bulk and annular lipids
DSC and 2H NMR measurements show a broadened, multicomponent phase transition with a maximum at 27° C for lipids in the VDAC1/DMPC 2D crystals. This observation suggests the existence of bulk and bound lipids in the 2D crystals that contribute to the phase transition. However, considerations of the area of the VDAC surface and protein-to-lipid ratio do not appear consistent with this simple interpretation. The formation of 2D crystals occurs over a specific molar ratio of protein to lipids and permits us to measure of the minimum number of lipids required to prevent protein aggregation and properly “solvate” the protein molecules. These solvating lipid molecules could occupy an annulus, and we can estimate the number of annular versus bulk lipids by considering the size of the pore as determined from diffraction and NMR structures for VDAC1. These dimensions permit us to estimate the number of lipid molecules required to cover the surface area.
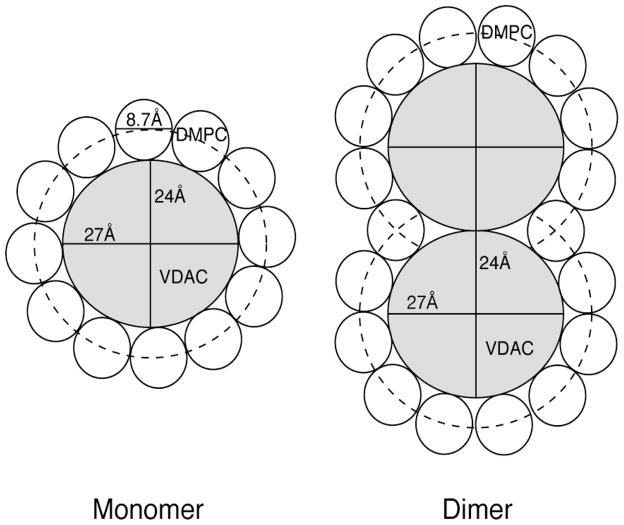
We first consider the case where the 2D crystals could be composed entirely of VDAC1 monomers. The pore of VDAC1 is elliptical with dimensions of 27 Å and 24 Å for the longer and shorter axes58. With an effective lipid diameter of 8.7 Å99, approximately 24 lipid molecules are required to form the first shell of the bilayer around each channel. This is illustrated in Figure 8, which shows a projection of VDAC1 surrounded by a single shell of 12 DMPC molecules per monolayer. The optimal molar ratio used to form homogenous samples of 2D crystals was approximately 1:25 (protein:lipid) as determined by optimization of the quality of NMR spectra. For the case of VDAC1 monomers, this ratio would be approximately the minimal amount required for each protein molecule to have one annulus, and there would be no bulk lipid present in the 2D crystals. This result contrasts the idea that at least some bulk lipids contribute to the phase transition. Also this contrasts the observation of regular occurring depressions observed on the surface of the 2D crystals which are attributed to areas of lipid density63. Since samples with DMPC were prepared using the same dialysis conditions as Dolder et al. it is more likely that VDAC1 forms dimers in our 2D crystal samples as was previously reported63. For the case of dimers, if two protein molecules share annular lipids along one face, the number of lipids required to occupy a shell around both protein molecules is 20 to 21; if protein-protein interactions occlude the presence of lipids entirely around both molecules, the number of required lipids reduces to 18. This case is illustrated in Figure 8. For the case of dimers, approximately 20% to 28% of lipid molecules could be available as bulk. It is interesting to note that Dolder et al. reported the formation of 2D crystals between relative weight ratios of 2:1 (protein:lipid) and 5:1 (protein:lipid), corresponding to a molar range between 1:25 and 1:12.5 (protein:lipid)63. In our hands we also observed formation of 2D crystals over this range. However, a protein-to-lipid molar ratio of less than 1:25 produced samples that appeared to be heterogeneous mixtures of 2D crystals and amorphous precipitates, and the quality of resulting NMR spectra was compromised, especially for the case where a protein-to-lipid molar ratio of 1:12.5 was used. Given our estimation of the number of annular and bulk lipids present for VDAC dimers, this suggests that at protein:lipid ratios lower than 1:20 to 1:18 there are insufficient number of lipids for the formation of a complete annulus, leading to possible VDAC1 aggregation in order to minimize exposed hydrophobic regions for some fraction of the protein:lipid mixture. It also suggests that some bulk lipids are required to form homogenous populations of purely 2D crystals. However, a detailed molecular interpretation of the DSC curve and its connection to the 2H spectral lineshapes requires additional experiments.
Figure 8.
Schematic illustrations of a projection of the VDAC1 monomer and dimer and the surrounding ~12 and ~18 DMPC molecules in the one half of the bilayer. The VDAC1 pore forms an ellipse with approximate dimensions of 24 and 27 Angstroms, while the diameter of a DMPC molecule was reported to be 8.7 Angstroms. Left: 12 DMPC molecules in the two halves of the bilayer are required to form a shell around a monomer of the protein. Right: VDAC1 dimers would require 18–20 DMPC molecules to form a shell if some lipids are shared between molecules or overlapping protein-protein interactions preclude the need for a complete annulus between molecules.
A second interesting feature is the increase in TM from 19 to 27 °C This is also a different result from consideration of two-component systems. The increase in TM was predicted by Marcelja100 in the context of lipid-mediated protein-protein interactions present in bilayers with high membrane protein concentration. According to Marcelja’s model, attractive forces between protein molecules promote clustering, which decreases the total free energy of the membrane by decreasing the total amount of boundary lipid. Furthermore, stronger protein-lipid interactions shift the bulk phase transition temperature higher than for pure lipid bilayers. These general theoretical results agree with our experimental data. The idea of strong protein-lipid interactions was also supported in the observation of very small variations in lattice parameters for VDAC 2D crystals63.
2H NMR spectra and acyl chain packing in the lipid hydrophobic core
In this paper, we have shown the DSC and 2H NMR spectra observed are reminiscent of previous studies of lipid reconstituted with large amount of cholesterol42 or peptides82 relative to phospholipid, consistent with the fact that formation of the 2D crystal can be induced by decreasing lipid content,65 thereby increasing the protein-to-lipid ratio. In this lattice, the liquid crystal phase of the bilayer becomes more ordered and takes on gel phase characteristics with slower dynamics, while the gel phase becomes disordered and more fluid, leading to a smoother, gradual phase transition as observed by the width of in Figure 4, determined by the section of acyl chain closer to the lipid headgroup. This finding is actually in contrast with previous studies of lipid-protein systems, which showed disordering of the gel phase, but not ordering of the liquid crystal phase above Tc101–103. At first glance, the 2D crystals studied here appeared to be more cholesterol-lipid like, rather than protein-lipid. Nevertheless, this ordering effect is predicted for a system with strong protein-lipid interaction according to the theoretical model by Marèelja.100 Therefore, this observation might be a unique feature of 2D crystals compared to other model and biological membranes.
In contrast with the ordering effect observed at the top of the acyl chain, the behavior observed at the hydrophobic core of the bilayer, formed by the lower part of the acyl chain, is not the same. While the plateau region is broadened for the 2D crystals, the methyl splittings are however not broadened and even appear narrower at lower temperatures in the crystals up to 29 °C, where the splittings roughly match to those in pure DMPC (see Figure 4 and Figure S3). This suggests that the presence of VDAC1 perturbs the hydrophobic core of the bilayer and disrupts acyl chain packing. VDAC1 insertion appears to make the bilayer as a whole more rigid while the terminal methyl groups are more mobile. This could be due to the shape of VDAC1, since the channel pore has a slightly concave shape59 and may allow for greater motion for the terminal methyl groups. Similar disordering at the terminal methyl group was previously observed in the study of DMPC with cytochrome oxidase, showing that boundary lipid is disordered.101,102 This behavior would also explain the large decrease in transition enthalpy and increase in transition temperature range measured by DSC, as the creation of a protein perturbed region in the bilayer would lead to loss of cooperativity as lipid-lipid contacts are disrupted.
Since the de-Paked spectrum shows only one lipid environment exists in the liquid crystal phase, perturbation of the lipid bilayer by VDAC1 is universal. This shows that there exists very little conformational flexibility for lipids in the space between VDAC1, similar to a previous study104 involving cholesteryl-β-cyclodextrin (βCC) derivatives as well as the electron crystallography study11 with AQP0. The tight packing of proteins in the lattice is expected since 2D crystals are known to be rigid and stable, thus is possible that no lipid is left unexposed to VDAC1. Tight lipid packing can also explain the increase of the main phase transition temperature, TM, observed in DSC as electrostatic attraction between VDAC1 and the lipid headgroups increases. However, the presence of three distinct transitions in the deconvoluted DSC thermogram eludes a complete explanation. The asymmetric DSC profile suggests there may very well be a second lipid environment. Potentially the two phases could be similar to each other, and line broadening in the 2D crystal 2H spectrum makes them indistinguishable. Alternatively, the transition might be more complicated than the simple two-state transition model assumed by DSC deconvolution, as discussed by Huang et al.46
MAS spectra of VDAC/DMPC 2D crystals
As shown in Figures 5–7, the 2D crystals of VDAC yield NMR spectra with superb resolution. Typically 13C linewidths are between 0.25 and 0.5 ppm for uniformly labeled VDAC and 15N linewidths are less than 0.5 ppm at 750 MHz 1H frequency. This resolution appears to be better than that observed in VDAC1 preparations in liposomes described elsewhere71. The sensitivity afforded by 2D crystal preparations allows acquisition of 2D spectra in 8 hours or less and 3D experiments require 5 to 7 days. 13C and 15N chemical shifts assignments for the N-terminus, which consists of approximately the first 20 residues, for VDAC1 in 2D crystals could be identified this way. For example, cross peaks due to A2, V3, P4, etc. and extending up to V17 are illustrated in Figure 7. The lack of change in these resonances between different 2D crystal preparations suggests that the conformation of the N-terminus, which is proposed to be involved in channel gating, is not sensitive to temperature and choice of lipid matrix (see Figure S8). Moreover, the N-terminus shifts are also consistent with those reported for preparations in liposomes, where it was also reported that resonances from the N-terminus remained unaffected by changes in sample temperature and liposome composition71. It is worth noting that, aside from an overall gain in sensitivity, many additional peaks are detected in the 13C-13C and 15N-13C spectra of 2D crystals that are not observed with VDAC1 in liposomes. For example, a significantly greater number of peaks were observed in the 15N-13C one-bond TEDOR spectrum shown in Figure S9 compared to 15N-13C correlation data reported for VDAC1 in liposomes. These differences are likely due to different dynamic processes present in the samples and will be discussed in future publications. We note that the cross peaks in Figures 5–7 are narrow and single and we have not seen any doubling that would be indicative of multiple, rather than a single, conformation in 2D crystals. Although the resolution and sensitivity of the MAS NMR spectra did not vary strongly with the choice of the lipid matrix, our results do indicate that it is crucial to optimize the protein-to-lipid ratio and remove any improperly folded molecules prior to reconstitution in lipids.
Temperature dependent experiments
Changes in lipid phase could induce changes in the structure of VDAC, changes in protein dynamics, or possibly induce protein aggregation. Spectra recorded above and below TM are nearly identical, indicating that there are likely no global changes in protein structure. In addition, crosspeak intensities in both spectra are also comparable within experimental error, indicating little to no changes in protein dynamics. Additional peaks are possibly observed in overlapped regions of the spectra at lower temperatures but this effect is not dramatic. In part due to its different secondary structure, we could unequivocally assign the resonances of the N-terminus and show that the conformation and dynamics of the N-terminus remain unchanged over a range of temperatures and lipid compositions. These observations are consistent with previous MAS NMR studies of VDAC in liposomes, which showed that N-terminal residues did not change their structure or dynamics in response to changes in the temperature or liposome lipid composition71. Thus, it appears that the N-terminal rigidity and apparent overall structural robustness of VDAC in these 2D crystals is not simply due to stabilizing effects induced by 2D crystal formation. A detailed comparison between MAS NMR data of VDAC1 DMPC 2D crystals acquired near 30 °C and the previously published data of VDAC1 DOPE liposomes acquired near 25 °C in Figure S5 of Reference 71 appear to reveal some differences. For example, A2 appears to be shifted or possibly assigned differently between DOPE liposomes at 25 °C and DMPC liposomes at 5 °C and DMPC 2D crystals at 30 °C. Furthermore, crosspeaks L10 Ca-Cb, V3 Ca-Cb, and L144 Ca-Cb appear to be diminished or highly asymmetric in the spectrum collected at 25 °C in DOPE compared DMPC 2D crystals at 30 °C. As these data were acquired on different samples under somewhat different experimental conditions, it is not clear if these differences can be attributed directly to differences in dynamics between the liposomes and 2D crystals and further studies are needed to delineate possible differences. Overall, the overall structure of VDAC1 in 2D crystals appears to be robust over a range of temperatures and relatively unaffected by the DMPC lipid phase transition. This is in contrast to some α-helical membrane proteins, which have been shown to exhibit structural changes as a function of temperature and lipid phase, which consequently perturb their function3,101,105,106.
Estimation of hydrophobic thickness and comparison of MAS spectra for lipids with different acyl chain lengths
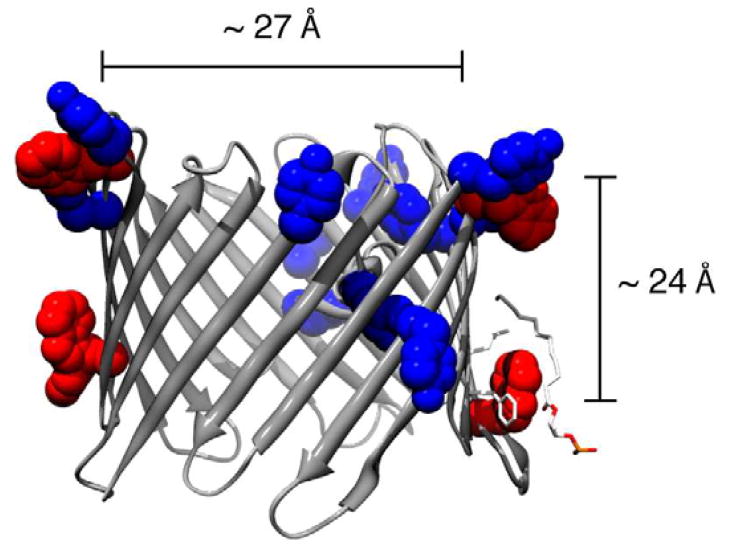
The hydrophobic region of integral membrane proteins and length of surrounding fatty acyl chains is often matched as closely as possible to minimize the cost of unfavorably exposing the acyl chains or protein transmembrane region to an aqueous environment. Differences between the protein hydrophobic thickness and acyl chain length can be minimized by protein conformational changes, protein aggregation, or stretching/compressing of fatty acyl chains. Stretching of the acyl chains leads to apparent ordering47, which can be observed as a decrease in order parameters and is consistent with our measurement of order parameters for VDAC1-DMPC 2D crystals. To probe the effect of possible hydrophobic mismatch in VDAC1 2D crystals, we estimate the hydrophobic thickness and compare it to values of acyl chain length for lipids used in this study. Since tryptophan and tyrosine residues frequently flank bilayer spanning regions of integral membrane proteins, we can estimate the hydrophobic thickness of VDAC1 from the distance between these residues, as observed in Figure 9. From the positions of these residues in the crystal structure of murine VDAC1 (PDB 3EMN), we estimate the hydrophobic thickness to be 24 Å, which is in line with the average hydrophobic thickness of many β-barrel bacterial porins. The thickness of DMPC bilayers above the phase transition is approximately 23 Å, slightly shorter than the effective hydrophobic thickness of VDAC. For DPhPC, the bilayer thickness is approximately 25.5 Å, slightly longer than the estimated hydrophobic thickness of VDAC1.
Figure 9.
The crystal structure of VDAC1 shown with the approximate pore diameter and estimation of the hydrophobic thickness. The average distance between tryptophan (red) and tyrosine (blue) residues flanking the barrel is approximately 24 Å. The crystal structure is from the PDB file 3EMN and the molecular graphics image was produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR001081).107
Hydrophobic mismatch can induce changes in lipid bilayer structure, membrane protein structure, or both. Figures 6 and 7 permit structural comparisons of VDAC1 in 2D crystals for lipids that are slightly shorter and slightly longer than the estimated hydrophobic thickness. To assess the affect of possible hydrophobic mismatch on the structure of the open channel, we compare 13C NMR spectra of VDAC1 in DMPC and DPhPC. The chemical shifts for almost all Cα-Cβ cross peaks in Figures 6 and 7 are identical between DMPC and DPhPC spectra, indicating that the VDAC1 backbone structures are nearly identical in both DMPC and DPhPC samples. Secondary chemical shifts of missing peaks indicate that almost all peaks missing in the DPhPC spectra occur for sites in the β-strands of the protein. Peaks missing from β-strand regions in the DPhPC spectra may be broadened beyond detection by protein dynamics interfering with cross polarization or 1H decoupling during the NMR experiments108,109. Peaks arising from residues in the N-terminal domain strongly appear in both DPhPC and DMPC, suggesting this region is very rigid in both lipids. Interestingly, the crosspeak assigned as L144 Ca-Cb for VDAC1 in liposomes (Ca at 52.2 and Cb at 46.2 ppm)71 is not present in spectra for VDAC1 in DMPC 2D crystals but does appear in spectra for VDAC1 in DPhPC 2D crystals (Figures 6–7). The reason for this difference is under further investigation and may be correlated to changes in dynamics for some regions of the protein between DMPC and DPhPC.
5. Concluding Remarks
In this study we have shown that there is sufficient bulk lipid in VDAC1 2D crystals to observe a phase transition and that TM is increased by approximately 8 degrees and the transition broaden relative to that of pure DMPC. Experimental results agree, at least qualitatively, with the extended molecular field model proposed by Marcelja, whereby lipid-mediated protein-protein interactions and lipid-protein interactions are necessary to explain the broadened phase transition and elevated TM found in 2D crystals. A minimum number of lipids is required to form homogenous samples of 2D crystals. This can be understood by examining the number of lipids required to occupy the VDAC1 annulus, which appears to be roughly 2/3 of the total amount of lipids required to form 2D crystals.
The structure of VDAC1 appears to be unaffected by lipid phase and small differences in acyl chain length. No conformational changes were detected between experiments collected above and below the lipid phase transition, and likewise no conformational changes were detected between lipids with acyl chain lengths slightly shorter and slightly longer than the estimated VDAC1 hydrophobic thickness. The N-terminal region is rigid and well structured over the range of temperatures probed in this study, including temperatures similar to those used in solution NMR studies. The MAS NMR data of VDAC in 2D crystals, along with the functional studies, indicate that the formation of 2D crystals do not cause a measurable structure change, nor cause the channel function to be different. Indeed, VDAC1 reconstituted into planar membranes from 2D DMPC crystals actually does not require the presence of Triton X-100 nor the presence of cholesterol to function properly, in contrast to electrophysiological measurements performed on VDAC1 in LDAO micelles. Our observations also suggest that 2D crystal formation is a useful tool for maximizing MAS SSNMR sensitivity, without necessarily sacrificing biological relevance.
Supplementary Material
Acknowledgments
The authors wish to thank Nicki Watson from the Whitehead Institute and Maria Ericsson from Harvard Medical School for acquiring EM images and Dr. David Ruben for help with de-Pake-ing. The Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems (NSF-0070319 and NIH GM68762) is gratefully acknowledged for help acquiring the DSC data. This research was supported by NIH grants EB001960, EB002026 and GM075879. O.T. and T.K.R. wish to thank Sergey Bezrukov for fruitful discussions and Kely Sheldon for help with experiments and acknowledge support by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH.
References
- 1.Marsh D. FEBS Lett. 1990;268:371–5. doi: 10.1016/0014-5793(90)81288-y. [DOI] [PubMed] [Google Scholar]
- 2.Singer SJ, Nicolson GL. Science. 1972;175:720–31. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 3.Oldfield E. Science. 1973;180:982–983. doi: 10.1126/science.180.4089.982. [DOI] [PubMed] [Google Scholar]
- 4.Hesketh TR, Smith GA, Houslay MD, Mcgill KA, Birdsall NJM, Metcalfe JC, Warren GB. Biochemistry. 1976;15:4145–4151. doi: 10.1021/bi00664a002. [DOI] [PubMed] [Google Scholar]
- 5.Starling AP, East JM, Lee AG. Biochemistry. 1995;34:3084–3091. doi: 10.1021/bi00009a040. [DOI] [PubMed] [Google Scholar]
- 6.Lorin A, Charloteaux B, Fridmann-Sirkis Y, Thomas A, Shai Y, Brasseur R. Journal of Biological Chemistry. 2007;282:18388–18396. doi: 10.1074/jbc.M700099200. [DOI] [PubMed] [Google Scholar]
- 7.Escribá PV, Wedegaertner PB, Goñi FM, Vögler O. Biochim Biophys Acta. 2007;1768:836–52. doi: 10.1016/j.bbamem.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Brown DA, London E. Annu Rev Cell Dev Biol. 1998;14:111–36. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 9.Simons K, Vaz WLC. Annu Rev Biophys Biomol Struct. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 10.Henderson R, Unwin PN. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- 11.Gonen T, Cheng YF, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Unwin N. J Struct Biol. 1998;121:181–90. doi: 10.1006/jsbi.1997.3949. [DOI] [PubMed] [Google Scholar]
- 13.Dolder M, Engel A, Zulauf M. FEBS Lett. 1996;382:203–8. doi: 10.1016/0014-5793(96)00180-9. [DOI] [PubMed] [Google Scholar]
- 14.Walz T, Grigorieff N. J Struct Biol. 1998;121:142–61. doi: 10.1006/jsbi.1998.3945. [DOI] [PubMed] [Google Scholar]
- 15.Stahlberg H, Fotiadis D, Scheuring S, Rémigy H, Braun T, Mitsuoka K, Fujiyoshi Y, Engel A. FEBS Lett. 2001;504:166–72. doi: 10.1016/s0014-5793(01)02746-6. [DOI] [PubMed] [Google Scholar]
- 16.Kühlbrandt W, Wang DN. Nature. 1991;350:130–4. doi: 10.1038/350130a0. [DOI] [PubMed] [Google Scholar]
- 17.Unger VM, Kumar NM, Gilula NB, Yeager M. Nat Struct Biol. 1997;4:39–43. doi: 10.1038/nsb0197-39. [DOI] [PubMed] [Google Scholar]
- 18.Jap BK, Downing KH, Walian PJ. J Struct Biol. 1990;103:57–63. doi: 10.1016/1047-8477(90)90086-r. [DOI] [PubMed] [Google Scholar]
- 19.Harbison GS, Herzfeld J, Griffin RG. Biochemistry. 1983;22:1–5. doi: 10.1021/bi00270a600. [DOI] [PubMed] [Google Scholar]
- 20.Harbison GS, Mulder PPJ, Pardoen H, Lugtenburg J, Herzfeld J, Griffin RG. J Am Chem Soc. 1985;107:4810–4816. [Google Scholar]
- 21.Harbison GS, Smith SO, Pardoen JA, Courtin JML, Lugtenburg J, Herzfeld J, Mathies RA, Griffin RG. Biochemistry. 1985;24:6955–6962. doi: 10.1021/bi00345a031. [DOI] [PubMed] [Google Scholar]
- 22.Degroot HJM, Harbison GS, Herzfeld J, Griffin RG. Biochemistry. 1989;28:3346–3353. doi: 10.1021/bi00434a033. [DOI] [PubMed] [Google Scholar]
- 23.Hu JG, Griffin RG, Herzfeld J. Proc Natl Acad Sci U S A. 1994;91:8880–8884. doi: 10.1073/pnas.91.19.8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffiths JM, Lakshmi KV, Bennett AE, Raap J, Vanderwielen CM, Lugtenburg J, Herzfeld J, Griffin RG. J Am Chem Soc. 1994;116:10178–10181. [Google Scholar]
- 25.Jaroniec CP, Lansing JC, Tounge BA, Belenky M, Herzfeld J, Griffin RG. J Am Chem Soc. 2001;123:12929–12930. doi: 10.1021/ja016923r. [DOI] [PubMed] [Google Scholar]
- 26.Shastri S, Vonck J, Pfleger N, Haase W, Kuehlbrandt W, Glaubitz C. Biochim Biophys Acta. 2007;1768:3012–9. doi: 10.1016/j.bbamem.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Shi L, Ahmed MAM, Zhang W, Whited G, Brown LS, Ladizhansky V. J Mol Bio. 2009:1–9. doi: 10.1016/j.jmb.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 28.Bajaj VS, Mak-Jurkauskas ML, Belenky M, Herzfeld J, Griffin RG. Proc Nat’l Acad Sci. 2009;106:9244–49. doi: 10.1073/pnas.0900908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnes AB, Paëpe GD, Wel PCAvd, Hu KN, Joo CG, Bajaj VS, Mak-Jurkauskas ML, Sirigiri JR, Herzfeld J, Temkin RJ, Griffin RG. Applied Magnetic Resonance. 2008;34:237–263. doi: 10.1007/s00723-008-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiller M, Krabben L, Vinothkumar KR, Castellani F, van Rossum BJ, Kühlbrandt W, Oschkinat H. Chembiochem. 2005;6:1679–84. doi: 10.1002/cbic.200500132. [DOI] [PubMed] [Google Scholar]
- 31.Walz T, Smith BL, Zeidel ML, Engel A, Agre P. J Biol Chem. 1994;269:1583–6. [PubMed] [Google Scholar]
- 32.Hite RK, Gonen T, Harrison SC, Walz T. Pflugers Archiv-European Journal of Physiology. 2008;456:651–661. doi: 10.1007/s00424-007-0353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seelig A, Seelig J. Biochemistry. 1974;13:4839–4845. doi: 10.1021/bi00720a024. [DOI] [PubMed] [Google Scholar]
- 34.Seelig J. QUARTERLY REVIEWS OF BIOPHYSICS. 1977;10:353–418. doi: 10.1017/s0033583500002948. [DOI] [PubMed] [Google Scholar]
- 35.Davis JH. Biophysical Journal. 1979;27:339–358. doi: 10.1016/S0006-3495(79)85222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shan X, Davis JH, Chu JWK, Sharom FJ. Biochimica Et Biophysica Acta-Biomembranes. 1994;1193:127–137. doi: 10.1016/0005-2736(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 37.Hite RK, Li ZL, Walz T. Embo Journal. 2010;29:1652–1658. doi: 10.1038/emboj.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis JH, Jeffrey KR, Bloom M, Valic MI, Higgs TP. Chemical Physics Letters. 1976;42:390–394. [Google Scholar]
- 39.WITTEBORT R, OLEJNICZAK E, GRIFFIN R. J Chem Phys. 1987;86:5411–5420. [Google Scholar]
- 40.Rosenke K, Sillescu H, Spiess H. Polymer. 1981;21:757–763. [Google Scholar]
- 41.Spiess H. Characterization of Polymers in the Solid State I: Part A: NMR and Other Spectroscopic Methods Part B: Mechanical Methods. 1985:23–58. [Google Scholar]
- 42.Blume A, Griffin RG. Biochemistry. 1982;21:6230–6242. doi: 10.1021/bi00267a031. [DOI] [PubMed] [Google Scholar]
- 43.Blume A, Rice DM, Wittebort RJ, Griffin RG. Biochemistry. 1982;21:6220–6230. doi: 10.1021/bi00267a030. [DOI] [PubMed] [Google Scholar]
- 44.Blume A, Wittebort RJ, Dasgupta SK, Griffin RG. Biochemistry. 1982;21:6243–6253. doi: 10.1021/bi00267a032. [DOI] [PubMed] [Google Scholar]
- 45.Speyer JB, Weber RT, Dasgupta SK, Griffin RG. Biochemistry. 1989;28:9569–9574. doi: 10.1021/bi00451a003. [DOI] [PubMed] [Google Scholar]
- 46.Huang TH, Lee CWB, Dasgupta SK, Blume A, Griffin RG. Biochemistry. 1993;32:13277–13287. doi: 10.1021/bi00211a041. [DOI] [PubMed] [Google Scholar]
- 47.de Planque MR, Greathouse DV, Koeppe RE, Schäfer H, Marsh D, Killian JA. Biochemistry. 1998;37:9333–45. doi: 10.1021/bi980233r. [DOI] [PubMed] [Google Scholar]
- 48.Daily AE, Greathouse DV, Wel PCAvd, REK Biophysical Journal. 2008;94:480–491. doi: 10.1529/biophysj.106.097543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Groot HJ, Smith SO, Courtin J, van den Berg E, Winkel C, Lugtenburg J, Griffin RG, Herzfeld J. Biochemistry. 1990;29:6873–83. doi: 10.1021/bi00481a017. [DOI] [PubMed] [Google Scholar]
- 50.Schneider R, Ader C, Lange A, Giller K, Hornig S, Pongs O, Becker S, Baldus M. J Am Chem Soc. 2008;130:7427–35. doi: 10.1021/ja800190c. [DOI] [PubMed] [Google Scholar]
- 51.Seidel K, Andronesi OC, Krebs J, Griesinger C, Young HS, Becker S, Baldus M. Biochemistry. 2008;47:4369–76. doi: 10.1021/bi7024194. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Berthold DA, Frericks HL, Gennis RB, Rienstra CM. Chembiochem. 2007;8:434–42. doi: 10.1002/cbic.200600484. [DOI] [PubMed] [Google Scholar]
- 53.Colombini M. Ann N Y Acad Sci. 1980;341:552–63. doi: 10.1111/j.1749-6632.1980.tb47198.x. [DOI] [PubMed] [Google Scholar]
- 54.Colombini M. Mol Cell Biochem. 2004;256–257:107–15. doi: 10.1023/b:mcbi.0000009862.17396.8d. [DOI] [PubMed] [Google Scholar]
- 55.Benz R. Biochimica Et Biophysica Acta-Reviews on Biomembranes. 1994;1197:167–196. doi: 10.1016/0304-4157(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 56.Colombini M, Blachly-Dyson E, Forte M. In: Ion Channels. Narashi T, editor. Vol. 4. Plenum; New York: 1996. pp. 169–202. [DOI] [PubMed] [Google Scholar]
- 57.Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ujwal R, Cascio D, Colletier JP, Faham S, Zhang J, Toro L, Ping PP, Abramson J. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17742–17747. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bayrhuber M, Meins T, Habeck M, Becker S, Giller K, Villinger S, Vonrhein C, Griesinger C, Zweckstetter M, Zeth K. Proc Natl Acad Sci USA. 2008;105:15370–5. doi: 10.1073/pnas.0808115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Colombini M. Journal of Membrane Biology. 1989;111:103–111. doi: 10.1007/BF01871775. [DOI] [PubMed] [Google Scholar]
- 61.Rostovtseva TK, Bezrukov SM. J Bioenerg Biomembr. 2008;40:163–70. doi: 10.1007/s10863-008-9145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rostovtseva T, Colombini M. Biophysical Journal. 2005;72:1954–1962. doi: 10.1016/S0006-3495(97)78841-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dolder M, Zeth K, Tittmann P, Gross H, Welte W, Wallimann T. Journal of Structural Biology. 1999;127:64–71. doi: 10.1006/jsbi.1999.4141. [DOI] [PubMed] [Google Scholar]
- 64.Mannella CA. Journal of Cell Biology. 1982;94:680–687. doi: 10.1083/jcb.94.3.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mannella CA. Science. 1984;224:165–166. doi: 10.1126/science.6322311. [DOI] [PubMed] [Google Scholar]
- 66.Mannella CA. J Bioenerg Biomembr. 1997;29:525–31. doi: 10.1023/a:1022489832594. [DOI] [PubMed] [Google Scholar]
- 67.Rostovtseva TK, Kazemi N, Weinrich M, Bezrukov SM. Journal of Biological Chemistry. 2006;281:37496–37506. doi: 10.1074/jbc.M602548200. [DOI] [PubMed] [Google Scholar]
- 68.Mlayeh L, Chatkaew S, Léonetti M, Homblé F. Biophysical Journal. 2010;99:2097–2106. doi: 10.1016/j.bpj.2010.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poston CN, Duong E, Cao Y, Bazemore-Walker CR. BIOCHEMICAL AND BIOPHYSICAL RESEARCH COMMUNICATIONS. 2011:1–6. doi: 10.1016/j.bbrc.2011.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thuduppathy GR, Craig JW, Kholodenko V, Schon A, Hill RB. J Mol Biol. 2006;359:1045–58. doi: 10.1016/j.jmb.2006.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schneider R, Etzkorn M, Giller K, Daebel V, Eisfeld J, Zweckstetter M, Griesinger C, Becker S, Lange A. Angew Chem Int Ed Engl. 2010;49:1882–5. doi: 10.1002/anie.200906241. [DOI] [PubMed] [Google Scholar]
- 72.Malia TJ, Wagner G. Biochemistry. 2007;46:514–525. doi: 10.1021/bi061577h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hiller S, Wagner G. Current Opinion in Structural Biology. 2009;19:396–401. doi: 10.1016/j.sbi.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sternin E, Bloom M, Mackay AL. Journal of Magnetic Resonance. 1983;55:274–282. [Google Scholar]
- 75.Bennett AE, Ok JH, Griffin RG, Vega S. J Chem Phys. 1992;96:8624–8627. [Google Scholar]
- 76.Bennett AE, Rienstra CM, Griffiths JM, Zhen WG, Lansbury PT, Griffin RG. J Chem Phys. 1998;108:9463–9479. [Google Scholar]
- 77.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. J Chem Phys. 1995:1–8. [Google Scholar]
- 78.Thurber KR, Tycko R. Journal of Magnetic Resonance. 2008;195:179–186. doi: 10.1016/j.jmr.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zizi M, Byrd C, Boxus R, Colombini M. Biophysical Journal. 1998;75:704–713. doi: 10.1016/S0006-3495(98)77560-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shanmugavadivu B, Apell HJ, Meins T, Zeth K, Kleinschmidt JH. J Mol Biol. 2007;368:66–78. doi: 10.1016/j.jmb.2007.01.066. [DOI] [PubMed] [Google Scholar]
- 81.Schein SJ, Colombini M, Finkelstein A. J Membr Biol. 1976;30:99–120. doi: 10.1007/BF01869662. [DOI] [PubMed] [Google Scholar]
- 82.Henzler-Wildman KA, Martinez GV, Brown MF, Ramamoorthy A. Biochemistry. 2004;43:8459–8469. doi: 10.1021/bi036284s. [DOI] [PubMed] [Google Scholar]
- 83.Vist MR, Davis JH. Biochemistry. 1990;29:451–64. doi: 10.1021/bi00454a021. [DOI] [PubMed] [Google Scholar]
- 84.Thilo L, Träuble H, Overath P. Biochemistry. 1977;16:1283–90. doi: 10.1021/bi00626a007. [DOI] [PubMed] [Google Scholar]
- 85.Grisham CM, Barnett RE. Biochemistry. 1973;12:2635–7. doi: 10.1021/bi00738a013. [DOI] [PubMed] [Google Scholar]
- 86.Kleemann W, McConnell HM. Biochim Biophys Acta. 1976;419:206–22. doi: 10.1016/0005-2736(76)90347-3. [DOI] [PubMed] [Google Scholar]
- 87.Jaworsky M, Mendelsohn R. Biochemistry. 1985;24:3422–8. doi: 10.1021/bi00335a005. [DOI] [PubMed] [Google Scholar]
- 88.East JM, Lee AG. Biochemistry. 1982;21:4144–51. doi: 10.1021/bi00260a035. [DOI] [PubMed] [Google Scholar]
- 89.Pfaller R, Freitag H, Harmey MA, Benz R, Neupert W. J Biol Chem. 1985;260:8188–93. [PubMed] [Google Scholar]
- 90.Colombini M. Journal of Membrane Biology. 1983;74:115–121. doi: 10.1007/BF01870500. [DOI] [PubMed] [Google Scholar]
- 91.Colombini M. Trends in Biochemical Sciences. 2009;34:382–9. doi: 10.1016/j.tibs.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 92.Forte M, BlachlyDyson E, Colombini M. Organellar Ion Channels and Transporters. 1996;51:145–154. 275. [Google Scholar]
- 93.Smack DP, Colombini M. Plant Physiol. 1985;79:1094–7. doi: 10.1104/pp.79.4.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shao L, Kinnally KW, Mannella CA. Biophysical Journal. 1996;71:778–86. doi: 10.1016/S0006-3495(96)79277-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nestorovich EM, Rostovtseva TK, Bezrukov SM. Biophysical Journal. 2003;85:3718–3729. doi: 10.1016/S0006-3495(03)74788-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vescovi A, Knoll A, Koert U. Org Biomol Chem. 2003;1:2983. doi: 10.1039/b303249n. [DOI] [PubMed] [Google Scholar]
- 97.Schmidt D, Cross SR, Mackinnon R. Journal of Molecular Biology. 2009;390:902–912. doi: 10.1016/j.jmb.2009.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rostovtseva TK, Liu TT, Colombini M, Parsegian VA, Bezrukov SM. Proc Natl Acad Sci USA. 2000;97:7819–22. doi: 10.1073/pnas.140115397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nagle JF, Tristram-Nagle S. Biochim Biophys Acta. 2000;1469:159–95. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marcelja S. Biochimica Et Biophysica Acta. 1976;455:1–7. doi: 10.1016/0005-2736(76)90149-8. [DOI] [PubMed] [Google Scholar]
- 101.Kang SY, Gutowsky HS, Hsung JC, Jacobs R, King TE, Rice D, Oldfield E. Biochemistry. 1979;18:3257–3267. doi: 10.1021/bi00582a010. [DOI] [PubMed] [Google Scholar]
- 102.Jacobs RE, Oldfield E. Progress in Nuclear Magnetic Resonance Spectroscopy. 1981;14:113–136. [Google Scholar]
- 103.Mouritsen OG, Bloom M. Biophysical Journal. 1984;46:141–153. doi: 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roux M, Auzely-Velty R, Djedaini-Pilard F, Perly B. Biophysical Journal. 2002;82:813–822. doi: 10.1016/S0006-3495(02)75443-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rinia HA, Kik RA, Demel RA, Snel MM, Killian JA, van Der Eerden JP, de Kruijff B. Biochemistry. 2000;39:5852–8. doi: 10.1021/bi000010c. [DOI] [PubMed] [Google Scholar]
- 106.Oldfield E, Meadows M, Rice D, Jacobs R. Biochemistry. 1978;17:2727–2740. doi: 10.1021/bi00607a006. [DOI] [PubMed] [Google Scholar]
- 107.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 108.LONG J, SUN B, BOWEN A, Griffin R. J Am Chem Soc. 1994;116:11950–11956. [Google Scholar]
- 109.Maus D, Copie V, SUN B, Griffiths J, Griffin R, Luo S, Schrock R, Liu A, Seidel S, Davis W, Grohmann A. J Am Chem Soc. 1996;118:5665–5671. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.