Abstract
The exchange of genetic information between donor and acceptor DNA molecules by homologous recombination (HR) depends on the cleavage of phosphodiester bonds. Although double-stranded and single-stranded DNA breaks (SSBs) have both been invoked as triggers of HR, until very recently the focus has been primarily on the former type of DNA lesions mainly due to the paucity of SSB-based recombination models. Here, to investigate the role of nicked DNA molecules as HR-initiating substrates in human somatic cells, we devised a homology-directed gene targeting system based on exogenous donor and chromosomal target DNA containing recognition sequences for the adeno-associated virus sequence- and strand-specific endonucleases Rep78 and Rep68. We found that HR is greatly fostered if a SSB is not only introduced in the chromosomal acceptor but also in the donor DNA template. Our data are consistent with HR models postulating the occurrence of SSBs or single-stranded gaps in both donor and acceptor molecules during the genetic exchange process. These findings can guide the development of improved HR-based genome editing strategies in which sequence- and strand-specific endonucleolytic cleavage of the chromosomal target site is combined with that of the targeting vector.
INTRODUCTION
Depending to some degree on the cell cycle stage, the restoration of broken DNA chains can be accomplished by two of the major DNA repair pathways of the cell, i.e. error-prone non-homologous end-joining (NHEJ) or error-free homologous recombination (HR) (1). In eukaryotes, HR promotes genetic exchange and aids in proper chromosomal segregation in gametes during the first meiotic division, whereas in mitotically active somatic cells it ensures faithful homologous template-assisted DNA repair. Although nicks or single-stranded DNA breaks (SSBs) were originally postulated as the initiators of HR (2–4), subsequent models invoking double-stranded DNA breaks (DSBs) instead have acquired in due course preeminence [see e.g. (5,6]). Possible reasons for this are 2-fold: (i) the difficulty in ruling out the possibility that nicks are converted into DSBs prior to HR initiation and (ii) the compelling evidence for DSBs as HR stimuli gathered from studies of, amongst others, meiotic recombination, mating-type switch in Saccharomyces cerevisiae and group I intron homing catalyzed by DSB-inducing sequence-specific meganucleases (e.g. I-SceI). The finding that sequence-specific DSBs elicit homology-directed gene repair provided a rationale for engineering meganucleases (7,8), zinc-finger nucleases (9,10) and, more recently, transcription activator-like effector nucleases (11,12). Depending on the specific pathway that is utilized to repair the resulting DSBs (i.e. HR, NHEJ or single-strand annealing) different targeted genomic modifications can be introduced. Indeed, sequence-specific DSB-based genome editing technologies have greatly increased the efficiency of homology-directed gene targeting in cells of higher eukaryotes and, as a result, are opening new perspectives for fundamental and applied research (e.g. plant biotechnology and gene therapy).
Experiments based on an intrachromosomal recombination system and the site-specific nicking endonuclease gIIp from a filamentous bacteriophage provided clues about the potential role of nicks as recombination-initiating lesions in dividing Saccharomyces cerevisae cells (13). However, the first clear indications that SSBs as such, i.e. without evolving into DSBs, can serve as triggers for HR were only more recently obtained through a study on V(D)J recombination in mammalian cells (14). Critical to this study was the use of a reporter gene rescue assay and panels of RAG1 and RAG2 mutants that nick instead of cleave their cognate target sites (14). Subsequent research in different laboratories including ours provided further evidence that SSBs at predefined loci elicit HR in mammalian cells and may thus be of use in the context of homology-directed gene editing approaches (15–18). Using enzymes that nick instead of break double-stranded DNA in order to induce HR may have the advantage of being less harmful to target cells. This conjecture is supported by a recent study demonstrating that a nicking mutant of the homing endonuclease I-AniI was significantly less cytotoxic to 293 cells harboring a target reporter gene than the parental DSB-inducing protein (17). This is presumably related to the lower number of DSBs caused by the I-AniI ‘nickase’ than by its DSB-inducing counterpart as inferred from the 143-fold lower frequency of NHEJ-mediated disruption of the engineered I-AniI recognition site in cells expressing the SSB-generating I-AniI variant (17). The aforementioned studies on nick-mediated homology-directed DNA editing in mammalian cells deployed natural or engineered sequence- and strand-specific endonucleases recognizing either endogenous or recombinant loci. Despite these differences in experimental setup, in all cases nicks were exclusively introduced in only one of the two recombination substrates (14–18).
In this study, we asked whether introduction of SSBs in both donor and acceptor DNA molecules would facilitate HR in human cells as posited by nick-based HR models (2–4). To this end, we built on the previous finding that nicking of the major integration site of adeno-associated virus (AAV) in the human genome (i.e. the AAVS1 locus embedded in the PPP1R12C gene at 19q13.42-qter) by the AAV sequence- and strand-specific endonucleases Rep78 and Rep68 stimulates HR-mediated gene targeting (16). While in the previous study an AAVS1-targeting construct without Rep recognition sequences was used, here we employed a panel of donor targeting constructs that, in addition to a reporter gene, contain wild-type or mutant Rep78/68 target DNA elements. Using this experimental system, we show that sequence-specific nicking of donor and acceptor DNA molecules triggers HR in human cells to higher levels than those resulting from the introduction of a SSB exclusively in the chromosomal target locus. Our results lend support to the notion that nicks in both recombination partners play a role in the initiation of mitotic HR in mammalian cells and may aid in the design of improved HR-based methodologies to edit, in a predictable manner, the genomes of higher eukaryotes.
MATERIALS AND METHODS
Cells
HeLa cells (American Type Culture Collection [ATCC]) and adenovirus type 5 early region 1 (E1)-expressing human retinoblasts 911 cells (19) were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 5% fetal bovine serum (Invitrogen). All cells were cultured at 37°C in an atmosphere of 10% CO2 in humidified air.
DNA transfections
DNA transfections of 8 × 104 HeLa cells seeded in wells of 24-well plates (Greiner Bio-One) were performed by using ExGen 500 (Fermentas) as detailed before (16).
Cell sorting and clonal expansion
After transfection with pA1.p5.GFP.A2 and pGAPDH.Rep78/68 (Supplementary Data), HeLa cells were sub-cultured for 44 days to remove the input episomal DNA. Next, the cells were subjected to green fluorescent protein (GFP)-based fluorescence-activated single-cell sorting and clonally expanded as detailed elsewhere (16).
Molecular characterization of DNA junctions between exogenous and endogenous DNA
The techniques and primers to PCR amplify and clone ‘telomeric’ and ‘centromeric’ junctions between exogenous and endogenous DNA have been previously described (16). Nucleotide sequence analysis was carried out with the aid of a 3730xl DNA Analyzer and BigDye Terminator v3.1 Cycle Sequencing Kit (both from Applied Biosystems).
Flow cytometry and light microscopy
The frequency of GFP-positive cells and the fluorescent signal intensity in the GFP-positive cells were determined by using a BD LSR II flow cytometer (BD Biosciences). Data were analyzed with the aid of BD FACSDiva 5.0.3 software (BD Biosciences). Untransfected HeLa cells were used to set the background level of fluorescence. Ten to one hundred thousand viable single cells were analyzed per sample/time point. For the light microscopic analysis of cell cultures, an IX51 inverse fluorescence microscope equipped with an XC30 Peltier-cooled digital color camera (both from Olympus) was used. Images were processed using CellF 3.4 imaging software (Olympus).
In vivo nicking assay
911 cells (19) were seeded in six-well plates (Greiner Bio-One) at a density of 106 cells per well. After an overnight incubation period, the cells in each well were transfected in regular culture medium with a total amount of 8 µg of DNA dissolved in 150 mM NaCl by using 21 µl of ExGen500 (Fermentas). The DNA mixtures consisted of 6 µg of AAVS1-targeting vector pA1.p5.GFP.A2, pA1.RBE/trs.GFP.A2 or pA1.RBE/Δtrs.GFP.A2 (Supplementary Data) mixed with 2 µg of pGAPDH.Rep68 or with 2 µg of pGAPDH.Rep68(Y156F) (16). After an overnight incubation period, the AAV helper functions provided by the adenovirus E2A, E4ORF6, VAI and VAII gene products were introduced into the transfected 911 cells by exposing them for 3 h to the E1-deleted adenovirus vector Ad.floxedΨ.F50 (25 infectious units per cell) (20). Four days post-transfection extrachromosomal DNA was isolated as detailed elsewhere (21). Subsequently, half (i.e. 20 µl) of the extrachromosomal DNA solution was incubated with NcoI and DpnI. NcoI was used to generate diagnostic 4.96-kb DNA fragments containing the humanized Renilla reniformis GFP sequence, whereas DpnI was employed to selectively digest the prokaryotic input DNA and thus discriminate it from DpnI-resistant templates synthesized de novo in the transfected 911 cells. The resulting DNA fragments were separated in a 1.0% agarose gel in 1× Tris–acetate–EDTA buffer. Next, the DNA was transferred by capillary action onto an Amersham Hybond-XL membrane (GE Healthcare) using a standard Southern blot technique. For the detection of newly replicated episomal DNA, a 739-bp DNA probe encompassing the complete GFP open reading frame (ORF) was used. This probe was labeled with EasyTide [α-32P] dCTP (3000 Ci/mmol, 10 mCi/ml; Perkin Elmer) using the DecaLabel DNA Labeling Kit (Fermentas). A Storm 820 PhosphorImager (Amersham Biosciences) was deployed for the detection of the radiolabeled DNA. Images were acquired using the Storm Scanner Control 5.03 software and processed using ImageQuant Tools 3.0 software (both from Amersham Biosciences).
Statistical analysis
Statistical parameters were computed using Graph Pad Prism 4.03. Student's t-test was applied to compare data sets with P < 0.05 considered significant.
RESULTS
The incorporation of a nicking-competent cis-acting element in donor DNA stimulates HR at an endogenous human locus with an SSB
The prototypic AAV serotype 2 (hereinafter referred to as AAV) rep gene encodes, in addition to the Rep52 and Rep40 proteins, the sequence- and strand-specific endonucleases Rep78 and Rep68. Biochemically, the two large Rep proteins are virtually indistinguishable. During AAV DNA replication, they bind to replicative intermediates at so-called Rep-binding elements (RBEs) in the AAV inverted terminal repeats (ITRs) (22) and catalyze the nicking at an adjacent terminal resolution site (trs). This gives rise to a free 3′-hydroxyl group necessary for the re-initiation of DNA synthesis (23). An RBE and trs at a proper distance of each other to allow Rep78/68-mediated nicking are also present in the AAV promoter at map position 5 (p5) (24) and in the human AAVS1 locus at 19q13.42-qter. In cells of humans and other primates, these cis-acting elements together with the Rep78/68 proteins mediate insertion of AAV genomic DNA at 19q13.42-qter via a poorly defined, HR-independent mechanism (25,26).
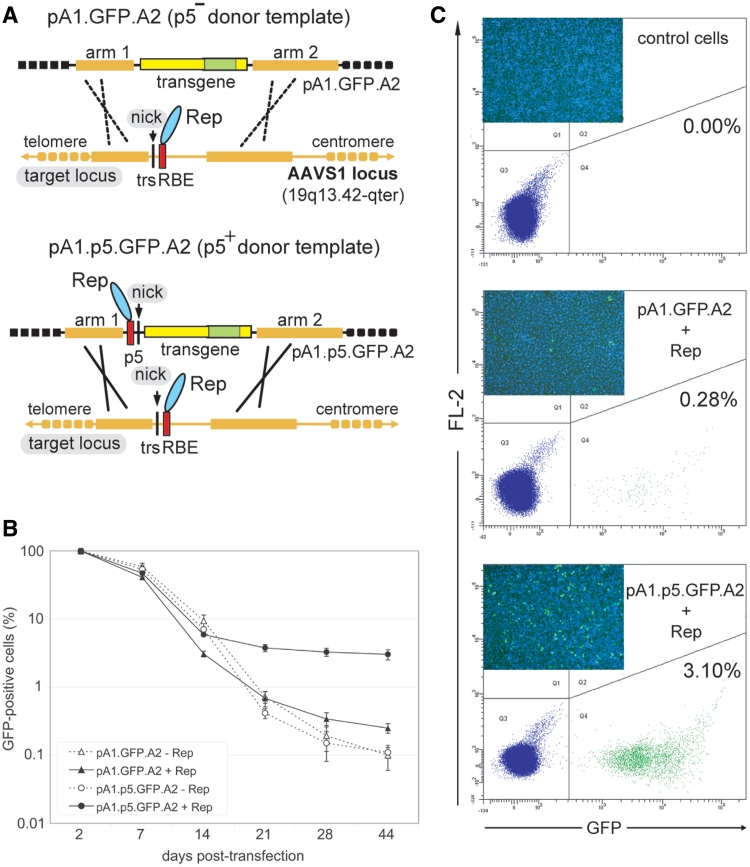
The ability of AAV Rep78/68 to nick the trs of AAVS1 was exploited in a previous study from our laboratory, to show that homology-directed gene targeting could ensue following the introduction of a SSB at a predefined human endogenous locus (16) (Figure 1A, upper panel). Herein, we build on this in vivo model system to investigate the impact of concomitant nicking of donor and acceptor DNA molecules on homology-directed gene targeting in human somatic cells (Figure 1A, lower panel). On the basis of the p5-less donor plasmid pA1.GFP.A2 (16), a new targeting construct named pA1.p5.GFP.A2 was generated. The latter plasmid contains p5 sequences (i.e. AAV genome positions 153 through 300) and a 4.1-kb humanized Renilla reniformis GFP transcription unit framed by human chromosome 19 sequences homologous to those bracketing the RBE and trs in AAVS1 (Figure 1A). The GFP gene permitted us to trace, accurately quantify and sort genetically modified cells regardless of the mechanism by which they arose. This is noteworthy since it avoids the exclusive detection of HR-dependent gene targeting events in contrast to assays based on the rescue of reporter gene expression.
Figure 1.
Stable genetic modification of human cells with p5-negative or -positive targeting vectors containing DNA sequences homologous to the genomic region framing the RBE and trs at AAVS1. (A) Experimental setup deployed to investigate the role of sequence- and strand-specific cleavage of donor and acceptor DNA molecules on mitotic HR at an endogenous human locus. The donor template pA1.GFP.A2 differs from pA1.p5.GFP.A2 by lacking p5. Both targeting constructs contain a 4.1-kb transcription unit consisting of the EF1α promoter (large yellow box), the GFP ORF (green box) and the SV40 pA signal (small yellow box). Immediately upstream of the EF1α promoter in pA1.p5.GFP.A2 lies the nicking-competent p5 element, whose RBE and trs are indicated by a red bar and vertical thin black line, respectively. The GFP gene in both donor plasmids is bracketed by DNA segments homologous to those framing the trs (vertical thin black line) and RBE (red box) at the chromosomal target site (i.e. the AAVS1 locus embedded in the PPP1R12C gene at 19q13.42-qter). The arbitrarily designated homology ‘arms’ 1 and 2 (thick yellow lines) are 2063 and 4381 bps in length, respectively. The AAV endonucleases Rep78 and Rep68 are represented by a cyan oval. (B) Flow cytometric quantification of the frequency of GFP-positive HeLa cells at different times after co-transfection with pA1.GFP.A2 or pA1.p5.GFP.A2 and either the AAV rep78/68 expression plasmid pGAPDH.Rep78/68 (+Rep) or an ‘empty’ control vector (−Rep). The frequencies of GFP-positive cells at the different time points in each of the experimental groups are plotted relative to those measured at 2 days post-transfection. Bars represent means ± SD of three independent experiments. (C) Representative flow cytometry dot plots corresponding to untransfected HeLa cells (control cells) and to HeLa cells initially co-transfected with pA1.GFP.A2 and pGAPDH.Rep78/68 (pA1.GFP.A2 + Rep) or with pA1.p5.GFP.A2 and pGAPDH.Rep78/68 (pA1.p5.GFP.A2 + Rep) at 44 days post-transfection. The frequency of stably transfected cells in each of the cell populations is indicated. The insets show direct fluorescence micrographs of each of the three types of HeLa cell populations. The GFP-specific signals (green) are overlaid with those of the DNA-binding dye Hoechst 33342 (blue).
HeLa cells were co-transfected with pA1.GFP.A2 or with pA1.p5.GFP.A2 and either the AAV rep78/68 expression plasmid pGAPDH.Rep78/68 (27) or an ‘empty’ control vector. Upon extensive sub-culturing, to dilute the exogenous episomal DNA, stably transfected cells in each of these four cell populations were identified by virtue of their green fluorescence. Flow cytometric measurements revealed that in the cultures initially exposed to pA1.p5.GFP.A2 and pGAPDH.Rep78/68 the frequency of GFP-positive cells stabilized at a 12.1-fold higher value than in those incubated with pA1.GFP.A2 and pGAPDH.Rep78/68 (i.e. 3.02 versus 0.25%) (P = 0.001; n = 3) (Figure 1B and Figure 1C). The cell populations that were transfected with pA1.GFP.A2 or with pA1.p5.GFP.A2 but did not receive the AAV rep78/68 expression plasmid contained the lowest frequencies of stably transfected cells. In addition, these percentages did not significantly differ from each other (P = 0.587; n = 3) (Figure 1B).
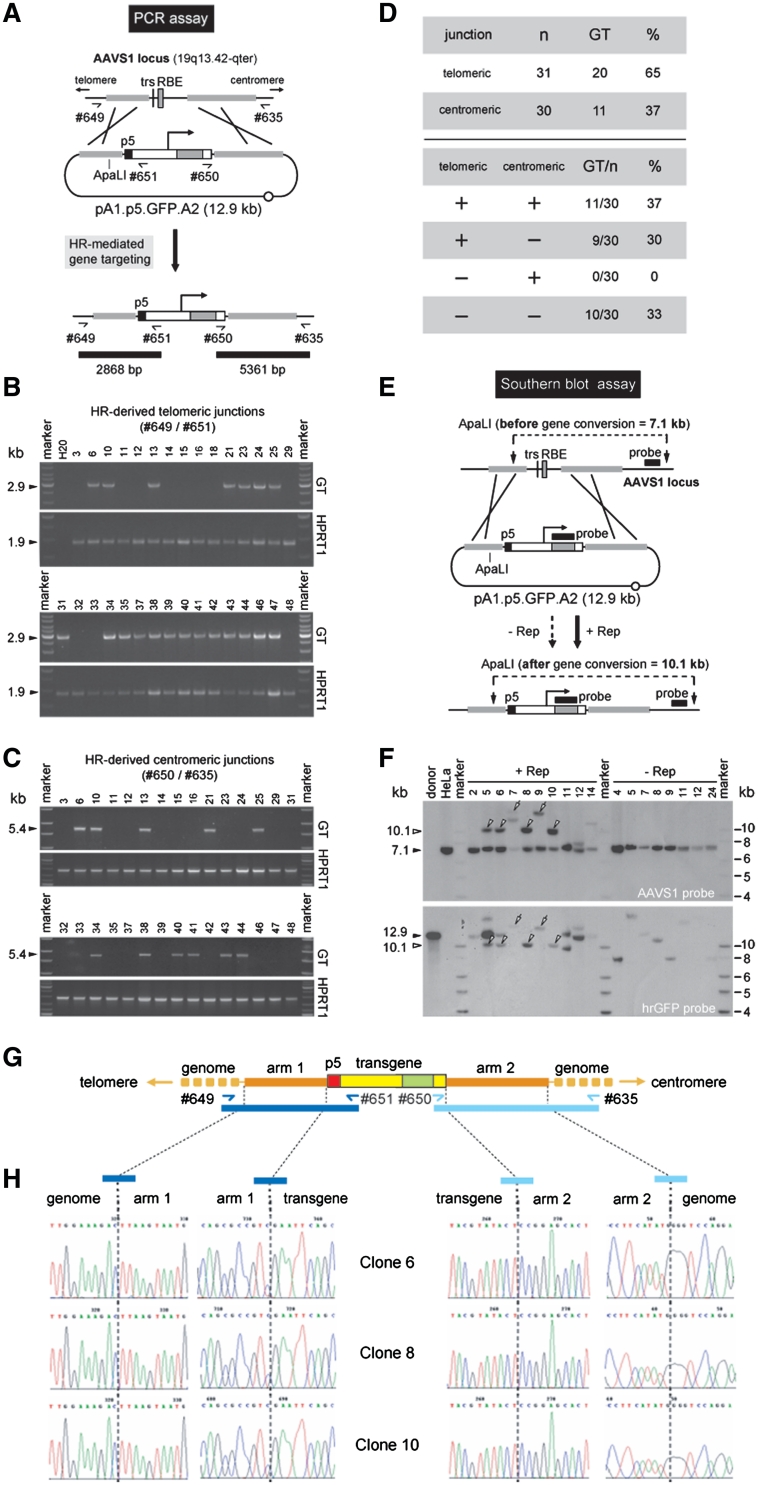
We previously established the occurrence of HR-mediated gene targeting events at AAVS1 in HeLa cells co-transfected with the p5-negative donor plasmid pA1.GFP.A2 and with AAV rep78/68 or rep68 expression vectors (16). To find out whether the same phenomenon occurs with the p5-positive targeting construct pA1.p5.GFP.A2, GFP-positive HeLa cells derived from the pGAPDH.Rep78/68- and pA1.p5.GFP.A2-transfected cultures were sorted and individually expanded. Subsequently, genomic DNA extracted from 31 randomly selected single-cell clones was subjected to the PCR assay depicted in Figure 2A using primers specific for AAVS1 and the human eukaryotic translation elongation factor 1 alpha (EF1α) promoter (#649 and #651, respectively). HR-mediated GFP gene addition should give rise to a 2868-bp amplicon representing junctions between the ‘telomeric’ part of AAVS1 and exogenous DNA sequences (Figure 2A). PCR products consistent with this process could readily be identified in 65% (i.e. 20 out of 31) of the samples (Figure 2B, GT panels). Conversely, in agreement with the previous results obtained with pA1.GFP.A2 (16) and with the well-established very low incidence of HR in HeLa cells (28,29), chromosomal DNA extracted from 28 clones derived from pA1.p5.GFP.A2-transfected HeLa cell cultures that had received the ‘empty’ plasmid, did not yield HR-specific PCR products (not shown). Next, to complement these data, the chromosomal DNA from the various clones was screened by using primers recognizing the simian virus 40 polyadenylation (SV40 pA) signal and AAVS1 sequences (#650 and #635, respectively). This primer set was designed to amplify junctions between the exogenous DNA and the ‘centromeric’ part of AAVS1 and should yield a 5361-bp PCR product diagnostic for HR-mediated GFP insertion (Figure 2A). DNA species consisted with this process could be identified in 37% (i.e. 11 out of 30) of the samples analyzed (Figure 2C, GT panels). The collective PCR data summarized in Figure 2D highlight the fact that exchange of genetic information involved primarily the ‘telomeric’ region of sequence identity. Clones containing exclusively ‘centromeric’ DNA junctions were not detected in the panel of 30 randomly selected samples that were analyzed. Indeed, in those clones where HR-dependent gene targeting led to the generation of ‘centromeric’ DNA junctions, these were invariably accompanied by the presence of their respective ‘telomeric’ counterparts (Figure 2D). Taken together these data suggest that exchange of genetic information takes place predominantly in DNA regions proximal to the site at which the SSB lesion occurs.
Figure 2.
Detection of homology-directed gene targeting events. (A) Diagram of the PCR assay deployed to identify cells genetically modified through HR-mediated GFP gene addition. The primer pairs #649/#651 and #650/#635 allow the detection of HR events at the AAVS1 of human cells transfected with the targeting construct pA1.p5.GFP.A2 by yielding diagnostic 2868-bp and 5361-bp PCR amplicons, respectively (horizontal black bars). Half arrows, primers #649, #651, #650 and #635 drawn in relation to their respective target sequences; thin black line, AAVS1 chromosomal region; horizontal thick grey lines, sequences shared by target and donor DNA; grey bar and vertical black line, RBE and trs, respectively; black box, nicking-prone p5 element; open box with broken arrow, EF1α promoter; large grey box, GFP ORF; open box, SV40 pA signal; open circle, prokaryotic origin of DNA replication. (B) PCR screening of clones derived from stably transfected HeLa cell populations initially co-transfected with pA1.p5.GFP.A2 and pGAPDH.Rep78/68. The panels labeled GT display the results of amplification reactions carried out with primers #649 and #651. PCR amplification of a 1.9-kb segment of the hypoxanthine phosphoribosyltransferase 1 gene (HPRT1) was performed in parallel to ascertain the integrity of the various genomic DNA templates (panels marked HPRT1). Marker, Gene Ruler DNA Ladder Mix (Fermentas); H20, PCR performed with nuclease-free water instead of chromosomal DNA. The positions (arrowheads) and sizes (in kb) of the PCR products are indicated at the left. (C) PCR screening of clones derived from stably transfected HeLa cell populations originally co-transfected with pA1.p5.GFP.A2 and pGAPDH.Rep78/68. The panels marked GT correspond to the PCR assay performed with primers #650 and #635 whereas those labeled HPRT1 are for the purpose specified in the legend of Figure 2B. (D) Summary of the data presented in Figure 2B and 2C, which resulted from the PCR assays depicted in Figure 2A. (E) Schematic representation of the Southern blot assay with ApaLI-digested genomic DNA from randomly selected clones of pA1.p5.GFP.A2-transfected HeLa cells. Unmodified target loci should yield a 7.1-kb AAVS1-specific restriction fragment while ‘two-sided’ HR should give rise to a DNA species of 10.1 kb hybridizing to the AAVS1- as well as the GFP-specific probe (black horizontal bars). Both probes are drawn in relation to their respective target DNA sequences. For an explanation of the other elements and symbols see the legend of Figure 2A. (F) Southern blots of ApaLI-treated genomic DNA of untransfected HeLa cells (HeLa) and of HeLa cell clones derived from cultures co-transfected with pA1.p5.GFP.A2 and pGAPDH.Rep78/68 (+Rep) or with pA1.p5.GFP.A2 and ‘empty’ plasmid (−Rep). The 12.9-kb ApaLI-linearized pA1.p5.GFP.A2 DNA (donor) served as an internal reference. Marker, Gene Ruler DNA Ladder Mix (Fermentas). (G) Diagram of the GFP expression unit (yellow and green boxes) inserted at 19q13.42-qter (horizontal yellow lines) upon homology-directed gene targeting deploying pA1.p5.GFP.A2 as donor template. Primers used to amplify the left- and right-hand junctions (dark and light blue half arrows, respectively) are drawn in relation to their recognition sequences. The 2.9- and 5.4-kb PCR amplicons specific for ‘telomeric’ and ‘centromeric’ DNA junctions are indicated by dark and light blue bars, respectively. (H) Nucleotide sequence analysis of ‘telomeric’ and ‘centromeric’ junctions between endogenous and exogenous DNA resulting from Rep78/68-induced HR events. The nucleotide sequences of the transition regions between pA1.p5.GFP.A2 sequences and flanking genomic or transgene DNA for three different clones that underwent HR are shown.
To further substantiate these results, we used Southern blot analyses of ApaLI-digested chromosomal DNA. Non-targeted AAVS1 alleles should give rise to 7.1-kb DNA species whereas homology-directed gene targeting at AAVS1 should yield 10.1-kb DNA fragments (Figure 2E). The chromosome 19-specific probe revealed the 10.1-kb fragments predicted to result from HR events at AAVS1 involving both ‘arms’ of homology in 4 out of the 10 analyzed samples of pA1.p5.GFP.A2- and pGAPDH.Rep78/68-transfected cells (Figure 2F, upper panel, open arrowheads). Importantly, genomic DNA of GFP-positive clones isolated from cell cultures that had not experienced AAV Rep78/68 activity did not yield this HR-specific DNA fragment. In fact, in these samples, the AAVS1-specific probe recognized exclusively the 7.1-kb fragments corresponding to unmodified AAVS1 alleles (Figure 2F, upper panel, solid arrowhead). Next, the membrane was stripped and incubated with a GFP-specific probe. The hybridization of the 10.1-kb species to this new probe confirmed that they resulted from HR-mediated gene conversion at AAVS1 (Figure 2F, lower panel, open arrowheads). It is noteworthy mentioning the binding of >10.1-kb DNA fragments to both probes in samples corresponding to GFP-positive clones expanded from pA1.p5.GFP.A2- and pGAPDH.Rep78/68-transfected cell cultures (Figure 2F, open arrows). This may be the result of ‘classical’ AAV Rep78/68-induced and HR-independent chromosomal insertion of pA1.p5.GFP.A2 DNA sequences (30). Although the details of this process remain poorly defined, it is well-established that p5 sequences constitute efficient substrates for Rep78/68-mediated DNA integration into AAVS1 [see e.g. (30)]. A hallmark of this non-HR pathway is the generation of integrants that give rise to DNA fragments with disparate sizes in Southern blot-based assays. Southern blot analyses of ApaLI-digested genomic DNA of pA1.p5.GFP.A2-transfected cells not exposed to pGAPDH.Rep78/68 led to the detection of a variety of GFP probe-binding DNA fragments with sizes different from 10.1 kb reflecting random chromosomal insertion events (Figure 2F).
To check whether the AAV nicking endonuclease-mediated gene targeting at AAVS1 had been accurate, as expected from legitimate ‘two-sided’ HR, we performed PCR amplifications on chromosomal DNA from HeLa cell clones 6, 8 and 10 (Figure 2). By using primer pairs #649/#651 and #635/#650, both left (i.e. ‘telomeric’) and right (i.e. ‘centromeric’) junctions between endogenous and exogenous DNA were targeted (Figure 2G). Nucleotide sequence analysis of the amplification products demonstrated that the DNA sequences surrounding the ‘left’ and ‘right’ terminus of the ‘telomeric’ and ‘centromeric’ arm of homology, respectively, were co-linear with those of the endogenous target locus (Figure 2H). Moreover, the DNA sequences at the junctions between the reporter gene and the ‘arms’ of homology were identical to those in the donor plasmid (Figure 2H). These data confirm at the nucleotide level that genetic modification of HeLa cell clones 6, 8 and 10 occurred through bona fide HR events at AAVS1 involving pA1.p5.GFP.A2 donor sequences. Collectively, the results described above suggest that the efficiency of SSB-induced HR at an endogenous human locus may be increased by donor DNA template nicking.
SSB formation in both HR partners yields higher gene targeting levels than those resulting from the exclusive nicking of the chromosomal acceptor DNA
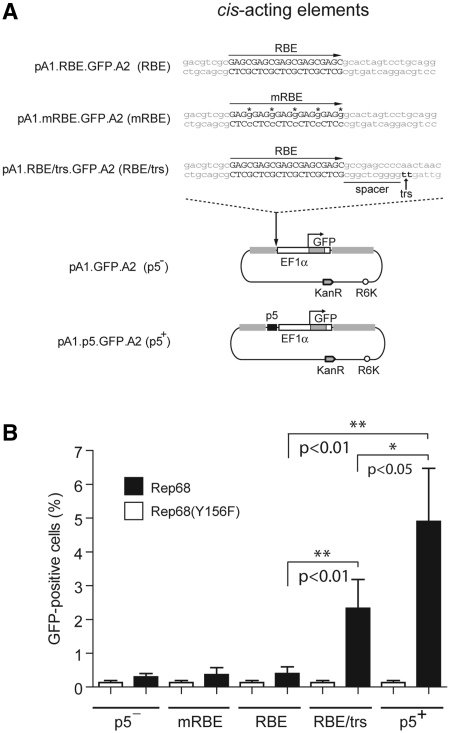
The 148-bp p5 element in pA1.p5.GFP.A2 not only contains an RBE and a trs but also a TATA box and binding sites for the transcription factors YY1 and USF1/2 (31). At this point, we can thus not rule out the possibility that cellular factors are the cause of or contribute to p5-dependent HR enhancement. Accordingly, to investigate the effect of donor DNA nicking per se on HR at AAVS1, we substituted the p5 element in pA1.p5.GFP.A2 by the minimal DNA sequence susceptible to endonucleolytic cleavage by the two large Rep proteins. This gave rise to targeting construct pA1.RBE/trs.GFP.A2 (Figure 3A). We also generated donor plasmids pA1.RBE.GFP.A2 and pA1.mRBE.GFP.A2 by replacing the p5 sequences with a wild-type RBE or a mutant RBE in which the cytosines in each of the four Rep-binding GAGC repeats were exchanged for guanines in order to abolish Rep binding (32) (Figure 3A).
Figure 3.
Stable genetic modification of human cells with AAVS1-targeting vectors containing minimal AAV Rep endonuclease recognition sequences. (A) Structures of the p5-negative pA1.GFP.A2 and the p5-positive pA1.p5.GFP.A2 donor constructs and of the targeting plasmids pA1.RBE.GFP.A2, pA1.mRBE.GFP.A2 and pA1.RBE/trs.GFP.A2 harboring the minimal AAV Rep endonuclease recognition sites RBE, mutant RBE (mRBE) and RBE/trs, respectively. The nucleotide sequence corresponding to the wild-type RBE is shown in black uppercase letters whereas the trs (i.e. the position at which Rep78/68-mediated nicking takes place) is indicated by black lowercase letters and a vertical arrow. The DNA sequence between the RBE and the trs is dubbed the spacer. The nucleotide sequence of the mRBE contains guanines (shown in lowercase and marked with asterisks) in place of cytosines. R6K, prokaryotic origin of DNA replication; KanR, transposon Tn5 neomycin phosphotransferase II gene conferring resistance to kanamycin. For an explanation of the other symbols and elements see the legend of Figure 2A. (B) Stable transfection levels in cultures of Hela cells initially co-transfected with the targeting vector pA1.GFP.A2 (p5−), pA1.mRBE.GFP.A2 (mRBE), pA1.RBE.GFP.A2 (RBE), pA1.RBE/trs.GFP.A2 (RBE/trs) or pA1.p5.GFP.A2 (p5+) and either pGAPDH.Rep68(Y156F) (white bars) or pGAPDH.Rep68 (black bars). Flow cytometric analysis of 104 viable cells per sample was performed at 37 days post-transfection. Results shown correspond to means ± SD from three independent experiments.
Next, HeLa cells were co-transfected with pA1.GFP.A2, pA1.p5.GFP.A2, pA1.RBE/trs.GFP.A2, pA1.RBE.GFP.A2 or pA1.mRBE.GFP.A2 and either pGAPDH.Rep68 or pGAPDH.Rep68(Y156F) (16). The latter two expression plasmids encode nicking-proficient and nicking-defective AAV Rep68 proteins, respectively. Again, after extensive sub-culturing, the frequency of stably transfected cells was determined by flow cytometric quantification of the percentage of GFP-positive cells. Results depicted in Figure 3B show that, regardless of the targeting construct employed, cell cultures initially transfected with pGAPDH.Rep68(Y156F) contained approximately the same very low frequency of stably transfected cells (i.e. 0.1% on average). Consistent with previous results (16), transfection of the p5-less donor plasmid pA1.GFP.A2 together with pGAPDH.Rep68 led to a measurable increase in the frequency of GFP-positive cells to 0.3 ± 0.1% (n = 3). In the presence of functional Rep68 molecules, the targeting constructs endowed with wild-type or mutant RBEs not flanked by a trs yielded percentages of GFP-positive cells that were not significantly different from those corresponding to cell populations co-transfected with pA1.GFP.A2 and pGAPDH.Rep68 (Figure 3B). Cell cultures initially exposed to pGAPDH.Rep68 and either of the two targeting constructs with cis-acting elements susceptible to Rep78/68 nicking (i.e. pA1.p5.GFP.A2 and pA1.RBE/trs.GFP.A2), displayed the highest frequencies of GFP-positive cells (Figure 3B). Of note, HeLa cells that had been co-transfected with pA1.p5.GFP.A2 and pGAPDH.Rep68 contained an ∼2-fold higher percentage of GFP-positive cells than those originally exposed to pA1.RBE/trs.GFP.A2 and pGAPDH.Rep68. Together, these data suggest that concerted nicking of donor and chromosomal acceptor DNA leads to higher levels of homology-directed transgene insertion than when SSB induction is restricted to the target chromosomal DNA.
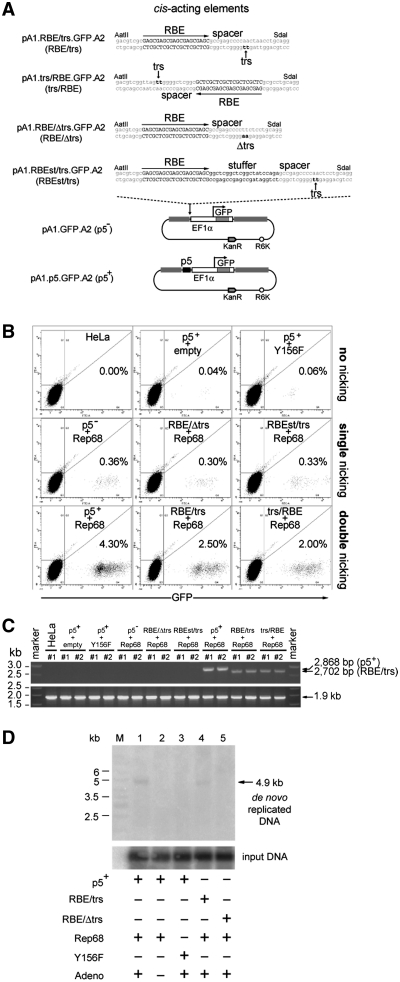
The unhindered nicking of DNA molecules by the AAV Rep78 and Rep68 proteins is dependent on a proper spacing between RBE and trs (33). Thus, to further verify the stimulatory effect caused by AAV Rep68 nicking of donor DNA templates on HR, targeting plasmids pA1.RBEst/trs.GFP.A2, pA1.RBE/Δtrs.GFP.A2 and pA1.trs/RBE.GFPA2 were constructed (Figure 4A). In pA1.RBEst/trs.GFP.A2 the RBE and trs are separated from each other by the regular 11-bp spacer plus an additional ‘stuffer’ DNA segment of 21 bp (st), whereas pA1.RBE/Δtrs.GFP.A2 contains the wild-type spacer but has the dinucleotide at which Rep78/68-mediated nicking takes place mutated from TT to AA (Figure 4A). The increased distance between RBE and trs in pA1.RBEst/trs.GFP.A2 as well as the mutation of the trs in pA1.RBE/Δtrs.GFP.A2, should minimize the susceptibility of these donor plasmids to AAV Rep78/68 endonucleolytic cleavage (33). Targeting construct pA1.trs/RBE.GFP.A2, on the other hand, should undergo unhindered nicking by AAV Rep68 since it contains the same structure and arrangement of cis-acting elements as that of pA1.RBE/trs.GFP.A2, albeit in the opposite orientation (Figure 4A).
Figure 4.
Effect of DNA sequences susceptible or unsusceptible to AAV Rep-mediated nicking on the stable transfection levels with AAVS1-targeting vectors. (A) Structures of the nicking-competent targeting plasmids pA1.p5.GFP.A2, pA1.RBE/trs.GFP.A2 and pA1.trs/RBE.GFP.A2 and that of the nicking-resistant donor constructs pA1.GFP.A2, pA1.RBE/Δtrs.GFP.A2 and pA1.RBEst/trs.GFP.A2. The nucleotide sequence corresponding to the wild-type RBE is shown in black uppercase letters whereas the trs (i.e. the position at which Rep78/68-mediated nicking takes place) is indicated by black lowercase letters and a vertical arrow. The DNA sequence between the RBE and the trs is called the spacer. Targeting plasmid pA1.RBE/Δtrs.GFP.A2 has the dinucleotide at which Rep endonuclease-mediated nicking occurs mutated from TT to AA while in pA1.RBE.st/trs.GFP.A2 a 21-bp stuffer positions the trs at a bigger distance from RBE-bound AAV Rep molecules. For an explanation of the other symbols and elements see the legend of Figure 3A. (B) Representative dot plots of flow cytometric analysis of the frequency of GFP-modified HeLa cells at 37 days post-transfection in cultures initially exposed to pA1.p5.GFP.A2 and ‘empty’ plasmid (p5++empty), pA1.p5.GFP.A2 and pGAPDH.Rep68(Y156F) (p5++Y156F), pA1.GFP.A2 and pGAPDH.Rep68 (p5−+Rep68), pA1.RBE/Δtrs.GFP.A2 and pGAPDH.Rep68 (RBE/Δtrs+Rep68), pA1.RBEst/trs.GFP.A2 and pGAPDH.Rep68 (RBEst/trs+Rep68), pA1.p5.GFP.A2 and pGAPDH.Rep68 (p5++Rep68), pA1.RBE/trs.GFP.A2 and pGAPDH.Rep68 (RBE/trs+Rep68) or to pA1.trs/RBE.GFP.A2 and pGAPDH.Rep68 (trs/RBE+Rep68). Untransfected HeLa cells were used to set the background of the assay at 0.00% GFP-positive cells (HeLa). For each sample, 105 viable single cells were analyzed. (C) PCR analysis using primer set #649/#651 on chromosomal DNA extracted from untransfected HeLa cells (HeLa) and from HeLa cells co-transfected with pGAPDH.Rep68 (Rep68) and targeting constructs pA1.GFP.A2 (p5−), pA1.RBE/Δtrs.GFP.A2 (RBE/Δtrs), pA1.RBEst/trs.GFP.A2 (RBEst/trs), pA1.p5.GFP.A2 (p5+), pA1.RBE/trs.GFP.A2 (RBE/trs) or pA1.trs/RBE.GFP.A2 (trs/RBE). HeLa cells were also co-transfected with pA1.p5.GFP.A2 (p5+) and pGAPDH.Rep68(Y156F) or with an ‘empty’ control plasmid (empty). The genomic DNA was isolated at 43 days post-transfection. HPRT1-specific PCRs served as control for the integrity of the input DNA. (D) In vivo nicking assay based on Southern blot analysis of DpnI-resistant extrachromosomal DNA. Episomal DNA was isolated at 4 days post-transfection from 911 cells co-transfected with pGAPDH.Rep68 and pA1.p5.GFP.A2 (lanes 1 and 2), pGAPDH.Rep68 and pA1.RBE/trs.GFP.A2 (lane 4) or pGAPDH.Rep68 and pA1.RBE/Δtrs.GFP.A2 (lane 5). Episomal DNA isolated from 911 cells co-transfected with pGAPDH.Rep68(Y156F) and pA1.p5.GFP.A2 (lane 3) served as a negative control. All cell cultures except for the one represented by lane 2 were exposed to Ad.floxedΨ.F50 at a multiplicity of infection of 25 infectious units per cell. Lane M, Gene Ruler DNA Ladder Mix. Prior to Southern blot analysis, the DNA was digested with DpnI (to fragment the input prokaryotic DNA) and with NcoI. Southern blots were exposed to a radiolabeled GFP-specific probe. The position of the 4.9-kb GFP-containing NcoI fragments derived from de novo synthesized DNA is indicated by an arrow at the right of the autoradiogram.
HeLa cells were co-transfected in duplicate with pGAPDH.Rep68 and either pA1.GFP.A2, pA1.RBEst/trs.GFP.A2, pA1.RBE/Δtrs.GFP.A2, pA1.trs/RBE.GFP.A2, pA1.RBE/trs.GFP.A2 or pA1.p5.GFP.A2. Negative controls consisted of untransfected HeLa cells and of HeLa cells exposed to pA1.p5.GFP.A2 and pGAPDH.Rep68(Y156F) or to pA1.p5.GFP.A2 and an ‘empty’ control plasmid. The percentages of stably transfected cells were determined by flow cytometry at 37 days post-transfection after extensive sub-culturing (Figure 4B). In the presence of Rep68, pA1.RBEst/trs.GFP.A2 and pA1.RBE/Δtrs.GFP.A2 yielded similar frequencies of stably transfected cells as the RBE/trs-less construct pA1.GFP.A2 (i.e. on average 0.33 and 0.30 versus 0.36%), whereas the DNA templates containing nicking-competent elements (i.e. pA1.p5.GFP.A2, pA1.RBE/trs.GFP.A2 and pA1.trs/RBE.GFP.A2) gave rise to the highest percentages of GFP-modified cells (i.e. on average 4.3, 2.5 and 2.0%, respectively) (Figure 4B). PCR analysis on genomic DNA extracted from the duplicates of the different HeLa cell cultures readily revealed the presence of HR-specific products in the samples corresponding to cells co-transfected with pGAPDH.Rep68 and either of the nicking-prone constructs pA1.p5.GFP.A2, pA1.RBE/trs.GFP.A2 or pA1.trs/RBE.GFP.A2 (Figure 4C). These results directly correlate with those presented in Figures 3B and 4B and, together, confirm that concomitant nicking of donor and chromosomal acceptor DNA leads to higher homology-directed gene targeting levels than the exclusive nicking of the target locus. Finally, to confirm the susceptibility of AAVS1-targeting vectors harboring the cis-acting elements p5 or RBE/trs to AAV Rep78/68-mediated cleavage, we set up an in vivo nicking assay. This functional assay is based on the fact that AAV-dependent DNA synthesis relies on the generation of free 3′-hydroxyl groups through nicking by Rep78/68. Indeed, in the presence of these endonucleases and the adenovirus helper functions necessary for AAV-dependent DNA synthesis (i.e. the E1, E2A, E4ORF6, VAI and VAII gene products) (23), productive SSB formation can be assessed by Southern blot analysis of de novo generated AAV replicative intermediates in the extrachromosomal DNA fraction of host cells. Hence, E1-expressing 911 cells (19) were co-transfected with pGAPDH.Rep68 and either pA1.p5.GFP.A2, pA1.RBE/trs.GFP.A2 or pA1.RBE/Δtrs.GFP.A2 (Figure 4D). The missing AAV helper functions were provided by exposing the transfected cells to the E1-deleted adenovirus vector Ad.floxedΨ.F50 (20) (Figure 4D, lanes 1, 3, 4 and 5). Mock-infected 911 cells co-transfected with pA1.p5.GFP.A2 and pGAPDH.Rep68 and Ad.floxedΨ.F50-infected 911 cells co-transfected with pA1.p5.GFP.A2 and pGAPDH.Rep68(Y156F) served as negative controls (Figure 4D, lanes 2 and 3, respectively). The autoradiogram presented in Figure 4D shows that de novo replicated plasmid DNA was only detected in the samples of 911 cells transfected with pA1.p5.GFP.A2 or with pA1.RBE/trs.GFP.A2 and exposed to nicking-proficient Rep68 plus helper adenovirus vector transduction (Figure 4D, lanes 1 and 4, respectively). Thus, the AAVS1-targeting constructs that in the presence of the functional large AAV Rep proteins give rise to the highest stable transfection levels and best serve as HR substrates (i.e. pA1.p5.GFP.A2, pA1.RBE/trs.GFP.A2 and pA1.trs/RBE.GFP.A2) are indeed susceptible to Rep endonuclease-dependent nicking. We conclude that homology-directed gene targeting is stimulated in human somatic cells by the generation of SSBs in both recombination partners.
DISCUSSION
Although the role of DSBs in meiotic and mitotic HR in lower and higher eukaryotes is firmly established, only recently have experimental results been obtained indicating that SSBs at chromosomal acceptor loci can also serve as HR-initiating lesions in somatic mammalian cells (14–17). Interestingly, initial HR models have invoked SSBs or single-stranded DNA gaps, not only in acceptor, but also in donor DNA molecules, at some stage during the genetic exchange process (2–4). Surprisingly, no experimental evidence supporting this postulate in higher eukaryotes is hitherto available. In this study, we build on a recently developed HR assay system based on AAVS1-targeting vectors and on the bona fide sequence- and strand-specific AAV Rep endonucleases (16) to investigate in human cells whether nicks in donor and chromosomal acceptor DNA constitute HR stimuli. In the presence of functional AAV Rep78/68 molecules, of all donor DNA templates tested, those endowed with the whole p5 element or with minimal nicking-prone RBE/trs or trs/RBE sequences gave rise to the highest frequencies of stably transfected cells with homology-directed gene targeting events being readily detected by Southern blot- and PCR-based assays. These data indicate that coordinated nicking of both recombination partners turns them into efficient HR substrates lending support to the notion that homologous sequence-bearing double-stranded DNA molecules with SSBs constitute preferred templates for mitotic HR in human cells.
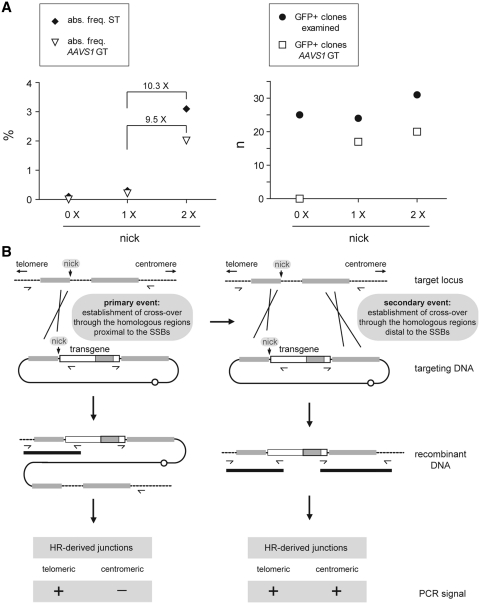
In a previous study (16), our research group demonstrated by using pA1.GFP.A2 as donor template that AAV Rep78/68-mediated nicking at AAVS1 could lead to a modest increase in the absolute frequency of stably transfected cells (these data are again presented in Figure 5A, left graph; compare the solid diamond at the leftward column [0× nick] with that in the middle column [1× nick]). Most significantly, however, PCR analysis of these cells revealed that most of them (i.e. 71%) had been genetically modified through homology-directed gene targeting at AAVS1 (Figure 5A, right graph, solid circle and open square in the middle column), representing an absolute AAVS1-targeting frequency of 0.2% (Figure 5A, left graph, open triangle in the middle column). On the contrary, GFP-modified cells whose progenitors had not been exposed to AAV Rep endonucleases gave rise to undetectable levels of HR-dependent gene insertion (Figure 5A, right graph, solid circle and open square in the leftward column). Crucially, from the results obtained in the current study with the nicking-prone donor template pA1.p5.GFP.A2 using an equivalent experimental system and the same readout assay as before, we conclude that AAV Rep endonuclease-mediated nicking of donor and acceptor chromosomal DNA increases the absolute frequencies of stably transfected cells and of AAVS1-targeted cells (Figure 5A, left graph, solid diamond and open triangle in the rightward column, respectively). These values are in fact one order of magnitude higher than those measured when the SSB is restricted to the target chromosomal locus. This finding supports the notion that SSBs in homology-bearing donor and target DNA promote mitotic HR in human cells as postulated in ‘classic’ nick-initiated HR models [for a brief overview see (34)].
Figure 5.
(A) Summary of the data resulting from the HR-specific PCR assay presented in a previous study (16; values plotted in the leftward and middle columns [i.e. 0× nick and 1×nick, respectively]) and in the current work (values plotted in the rightward columns [i.e. 2× nick]). The results are shown in terms of the absolute frequencies of stably transfected (ST) cells (left graph, solid diamonds) and of AAVS1-targeted cells (left graph, open triangles) as well as in terms of the total numbers of reporter-positive clones analyzed (right graph, solid circles) and of those that were targeted at AAVS1 through HR (right graph, open squares). (B) Working model for nick-initiated mitotic HR in human cells (see text for details).
PCR screening of chromosomal DNA extracted from GFP-positive clones whose progenitors had been exposed to both pA1.p5.GFP.A2 and AAV Rep endonucleases showed that the HR-dependent exchange of genetic information involved primarily the ‘telomeric’ AAVS1 sequences. In fact, all clones with ‘centromeric’ exogenous DNA/AAVS1 junctions also possessed their ‘telomeric’ counterparts (Figure 2D). These data gathered from independent HR-mediated DNA insertion events suggest that sequence exchange between both recombination partners occurs preponderantly at or near the nicking sites. Thus, on the basis of these experimental results a working model for the nick-initiated mitotic HR process in human cells can be submitted (Figure 5B). In this model, juxtaposed or quasi-juxtaposed nicks in two parental DNA duplexes sharing sequence identity can be locally processed giving rise to substrates that are competent for displacement DNA synthesis and invasion/annealing of complementary acceptor DNA sequences. Ligation of the resulting DNA heteroduplexes leads to the establishment of a Holliday junction that, upon resolution, yields recombinant DNA molecules (Figure 5B, left panel). In other instances, a secondary Holliday junction emerges at a distal region of shared sequence identity due to displacement DNA synthesis followed by invasion/annealing ‘back to the donor’ template or by branch migration. In this case, upon resolution of the ‘distal’ Holliday junction, another type of recombinant DNA molecule arises (Figure 5B, right panel). Clearly, although our experimental results are consist with the emergence of both types of HR products (Figure 5B, lower part of both panels), further experiments will be necessary to elucidate in detail the series of molecular events underlying their generation.
Interestingly, ∼2-fold higher stable transfection levels were obtained with pA1.p5.GFP.A2 than with pA1.RBE/trs.GFPA2 or with pA1.trs/RBE.GFP.A2. The most parsimonious explanation for this difference lies in the fact that the RBE, spacer and trs in pA1.p5.GFP.A2 are derived from the p5 element, whereas those in pA1.RBE/trs.GFP.A2 and pA1.trs/RBE.GFP.A2 are from the AAVS1 ortholog of the African green monkey (35). Although the terminal resolution sites in the AAV ITR, chromosomal target loci and p5 are all susceptible to nicking, their surrounding RBE and spacer sequences are, to some extent, degenerate. Of note, previous results indicate that AAV Rep endonucleases display different binding and/or nicking activities depending on the origin of their cis-acting elements. For instance, Amiss and colleagues reported that although the sequence spanning the RBE, spacer and trs of the human AAVS1 locus displays 98% nucleotide sequence identity with that of its monkey ortholog, Rep endonucleases bind with higher affinity to the simian DNA (35). Interestingly, an in vivo functional assay based on co-transfection of COS-7 and HeLa cell cultures with a GFP-encoding AAV shuttle vector and an AAV rep68 expression plasmid revealed that stable transfection levels were ∼2-fold higher in the monkey COS-7 cells (i.e. 4.6% versus 2.1%) (35). It is also possible that, in comparison with the nicking-competent cis-acting elements in the minimal RBE/trs, the p5 element with its extra sequences and transcription factor-binding sites, such as those for the YY1 and USF1/2 proteins, might lead to a stronger stabilization of Rep78/68–DNA complexes and/or DNA bending. Under certain experimental conditions, both YY1 and USF1/2 have the capacity to bend DNA (36,37). It is thus enticing to hypothesize that bending of p5 DNA sequences in vivo enhances the formation of the hairpin structure that positions the trs at the single-stranded apical loop necessary for DNA strand-specific nicking (38,39).
HR-mediated gene targeting is a very rare event in most mammalian cell types. For instance, in HeLa and HT-1080 cells it has an incidence of ∼10−7 to 10−8 (28,29) and 10−6 to 10−7 (29,40,41), respectively, while in human fibroblasts it occurs at a frequency of ∼10−7 (42). Moreover, following the introduction of foreign DNA into human somatic cells, the frequency of random chromosomal integration is at least 100-fold higher than that corresponding to accurate homology-directed gene targeting. Accordingly, a great deal of effort is currently being put in developing artificial endonucleases that, by site-specifically introducing DSBs at predefined chromosomal sequences of choice, can increase the frequency of HR-mediated genome editing by several orders of magnitude (often up to between 0.1 and 10%) (7–12). An unresolved issue with DSB-based gene targeting procedures, however, concerns the fact that these lesions can be channeled for repair through the NHEJ pathway instead of that of HR. NHEJ is an error-prone process that can lead to deleterious chromosomal translocations and various other types of unintended genomic mutations (1). An emerging, potentially less genotoxic approach (17) to induce HR-mediated genome editing consists in exploring endonucleases that generate nicks instead of DSBs (15–17). However, the data gathered hitherto comparing SSBs versus DSBs for targeted genome modifications indicate that the former DNA insult is less effective at initiating HR in mammalian cells (15,17). Thus, our finding that SSB formation in donor and chromosomal DNA leads to substantially higher HR levels than when nicking is restricted to the latter molecules might guide the development of improved, potentially safer, gene targeting strategies. These new genome editing approaches may be based either on natural or de novo engineered nicking endonucleases (15–18,43).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary methods, Supplementary references [16,27,35,44,45]
FUNDING
Funding for open access charge: The research leading to these results has received funding from the European Community's 7th Framework Programme for Research and Technological Development under grant agreement number 222878 (PERSIST).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Martijn Everts for PCR amplification and cloning of the p5 element.
REFERENCES
- 1.Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holliday R. A mechanism for gene conversion in fungi. Genet. Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 3.Meselson MS, Radding CM. A general model for genetic recombination. Proc. Natl Acad. Sci. USA. 1975;72:358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radding CM. Homologous pairing and strand exchange in genetic recombination. Annu. Rev. Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- 5.Resnick MA. The repair of double-strand breaks in DNA; a model involving recombination. J. Theoret. Biol. 1976;59:97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- 6.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 7.Redondo P, Prieto J, Muñoz IG, Alibés A, Stricher F, Serrano L, Cabaniols JP, Daboussi F, Arnould S, Perez C, et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008;456:107–111. doi: 10.1038/nature07343. [DOI] [PubMed] [Google Scholar]
- 8.Silva G, Poirot L, Galetto R, Smith J, Montoya G, Duchateau P, Pâques F. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr. Gene Ther. 2011;11:11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y-G, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl Acad. Sci. USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Händel EM, Cathomen T. Zinc-finger nuclease based genome surgery: it's all about specificity. Curr. Gene Ther. 2011;11:28–37. doi: 10.2174/156652311794520120. [DOI] [PubMed] [Google Scholar]
- 11.Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2010;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahfouz MM, Li L, Shamimuzzaman M, Wibowo A, Fang X, Zhu JK. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc. Natl Acad. Sci. USA. 2011;108:2623–2628. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galli A, Schiestl RH. Effects of DNA double-strand and single-strand breaks on intrachromosomal recombination events in cell-cycle-arrested yeast cells. Genetics. 1998;49:1235–1250. doi: 10.1093/genetics/149.3.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 15.McConnell-Smith A, Takeuchi R, Pellenz S, Davis L, Maizels N, Monnat RJ, Jr, Stoddard BL. Generation of a nicking enzyme that stimulates site-specific gene conversion from the I-AniI LAGLIDADG homing endonuclease. Proc. Natl Acad. Sci. USA. 2009;106:5099–5104. doi: 10.1073/pnas.0810588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Nierop GP, de Vries AAF, Holkers M, Vrijsen KR, Gonçalves MAFV. Stimulation of homology-directed gene targeting at an endogenous human locus by a nicking endonuclease. Nucleic Acids Res. 2009;37:5725–5736. doi: 10.1093/nar/gkp643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metzger MJ, McConnell-Smith A, Stoddard BL, Miller AD. Single-strand nicks induce homologous recombination with less toxicity than double-strand breaks using an AAV vector template. Nucleic Acids Res. 2011;39:926–935. doi: 10.1093/nar/gkq826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan SH, Stoddard BL, Xu SY. Natural and engineered nicking endonucleases-from cleavage mechanism to engineering of strand-specificity. Nucleic Acids Res. 2011;39:1–18. doi: 10.1093/nar/gkq742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fallaux FJ, Kranenburg O, Cramer SJ, Houweling A, Van Ormondt H, Hoeben RC, van der Eb AJ. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum Gene Ther. 1996;7:215–222. doi: 10.1089/hum.1996.7.2-215. [DOI] [PubMed] [Google Scholar]
- 20.Gonçalves MAFV, Holkers M, Cudré-Mauroux C, van Nierop GP, Knaän-Shanzer S, van der Velde I, Valerio D, de Vries AAF. Transduction of myogenic cells by retargeted dual high-capacity hybrid viral vectors: robust dystrophin synthesis in duchenne muscular dystrophy muscle cells. Mol. Ther. 2006;13:976–986. doi: 10.1016/j.ymthe.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 21.Gonçalves MAFV, Pau MG, de Vries AAF, Valerio D. Generation of a high-capacity hybrid vector: packaging of recombinant adenoassociated virus replicative intermediates in adenovirus capsids overcomes the limited cloning capacity of adenoassociated virus vectors. Virology. 2001;288:236–246. doi: 10.1006/viro.2001.1073. [DOI] [PubMed] [Google Scholar]
- 22.McCarty DM, Pereira DJ, Zolotukhin I, Zhou X, Ryan JH, Muzyczka N. Identification of linear DNA sequences that specifically bind the adeno-associated virus Rep protein. J. Virol. 1994;68:4988–4997. doi: 10.1128/jvi.68.8.4988-4997.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonçalves MAFV. Adeno-associated virus: from defective virus to effective vector. Virol. J. 2005;2:e43. doi: 10.1186/1743-422X-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang XS, Srivastava A. A novel terminal resolution-like site in the adeno-associated virus type 2 genome. J. Virol. 1997;71:1140–1146. doi: 10.1128/jvi.71.2.1140-1146.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young SM, Jr, Samulski RJ. Adeno-associated virus (AAV) site-specific recombination does not require a Rep-dependent origin of replication within the AAV terminal repeat. Proc. Natl Acad. Sci. USA. 2001;98:13525–13530. doi: 10.1073/pnas.241508998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henckaerts E, Dutheil N, Zeltner N, Kattman S, Kohlbrenner E, Ward P, Clément N, Rebollo P, Kennedy M, Keller GM, et al. Site-specific integration of adeno-associated virus involves partial duplication of the target locus. Proc. Natl Acad. Sci. USA. 2009;106:7571–7576. doi: 10.1073/pnas.0806821106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonçalves MAFV, van Nierop GP, Tijssen MR, Lefesvre P, Knaän-Shanzer S, van der Velde I, van Bekkum DW, Valerio D, de Vries AAF. Transfer of the full-length dystrophin-coding sequence into muscle cells by a dual high-capacity hybrid viral vector with site-specific integration ability. J. Virol. 2005;79:3146–3162. doi: 10.1128/JVI.79.5.3146-3162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itzhaki JE, Porter AC. Targeted disruption of a human interferon-inducible gene detected by secretion of human growth hormone. Nucleic Acids Res. 1991;19:3835–3842. doi: 10.1093/nar/19.14.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porter AC, Itzhaki JE. Gene targeting in human somatic cells. Complete inactivation of an interferon-inducible gene. Eur. J. Biochem. 1993;218:273–281. doi: 10.1111/j.1432-1033.1993.tb18375.x. [DOI] [PubMed] [Google Scholar]
- 30.Philpott NJ, Gomos J, Berns KI, Falck-Pedersen E. A p5 integration efficiency element mediates Rep-dependent integration into AAVS1 at chromosome 19. Proc. Natl Acad. Sci. USA. 2002;99:12381–12385. doi: 10.1073/pnas.182430299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang L-S, Shi Y, Shenk T. Adeno-associated virus P5 promoter contains an adenovirus E1A-inducible element and a binding site for the major late transcription factor. J. Virol. 1989;63:3479–3488. doi: 10.1128/jvi.63.8.3479-3488.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wonderling RS, Owens RA. Binding sites for adeno-associated virus rep proteins within the human genome. J. Virol. 1997;71:2528–2534. doi: 10.1128/jvi.71.3.2528-2534.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamartina S, Ciliberto G, Toniatti C. Selective cleavage of AAVS1 substrates by the adeno-associated virus type 2 Rep68 protein is dependent on topological and sequence constraints. J. Virol. 2000;74:8831–8842. doi: 10.1128/jvi.74.19.8831-8842.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith GR. How homologous recombination is initiated: unexpected evidence for single-strand nicks from V(D)J site-specific recombination. Cell. 2004;117:146–148. doi: 10.1016/s0092-8674(04)00338-1. [DOI] [PubMed] [Google Scholar]
- 35.Amiss TJ, McCarty DM, Skulimowski A, Samulski RJ. Identification and characterization of an adeno-associated virus integration site in CV-1 cells from the African green monkey. J. Virol. 2003;77:1904–1915. doi: 10.1128/JVI.77.3.1904-1915.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Natesan S, Gilman MZ. DNA bending and orientation-dependent function of YY1 in the c-fos promoter. Genes Dev. 1993;7:2497–2509. doi: 10.1101/gad.7.12b.2497. [DOI] [PubMed] [Google Scholar]
- 37.Fisher DE, Parent LA, Sharp PA. Myc/Max and other helix-loop-helix/leucine zipper proteins bend DNA toward the minor groove. Proc. Natl Acad. Sci. USA. 1992;89:11779–11783. doi: 10.1073/pnas.89.24.11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brister JR, Muzyczka N. Rep-mediated nicking of the adeno-associated virus origin requires two biochemical activities, DNA helicase activity and transesterification. J. Virol. 1999;73:9325–9336. doi: 10.1128/jvi.73.11.9325-9336.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brister JR, Muzyczka N. Mechanism of Rep-mediated adeno-associated virus origin nicking. J. Virol. 2000;74:7762–7771. doi: 10.1128/jvi.74.17.7762-7771.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ganguly A, Smelt S, Mewar R, Fertala A, Sieron AL, Overhauser J, Prockop DJ. Targeted insertions of two exogenous collagen genes into both alleles of their endogenous loci in cultured human cells containing the promoters and the 5’ ends of the genes. Proc. Natl Acad. Sci. USA. 1994;91:7365–7369. doi: 10.1073/pnas.91.15.7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thyagarajan B, Johnson BL, Campbell C. The effect of target site transcription on gene targeting in human cells in vitro. Nucleic Acids Res. 1995;23:2784–2790. doi: 10.1093/nar/23.14.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown JP, Wei W, Sedivy JM. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 43.Sanders KL, Catto LE, Bellamy SR, Halford SE. Targeting individual subunits of the FokI restriction endonuclease to specific DNA strands. Nucleic Acids Res. 2009;37:2105–2115. doi: 10.1093/nar/gkp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laird PW, Zijderveld A, Linders K, Rudnicki MA, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Russell DW. 3rd edn. Plainview, NY: Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.