Abstract
In order to enhance DNA triple helix stability synthetic oligonucleotides have been developed that bear amino groups on the sugar or base. One of the most effective of these is bis-amino-U (B), which possesses 5-propargylamino and 2′-aminoethoxy modifications. Inclusion of this modified nucleotide not only greatly enhances triplex stability, but also increases the affinity for related sequences. We have used a restriction enzyme protection, selection and amplification assay (REPSA) to isolate sequences that are bound by the heavily modified 9-mer triplex-forming oligonucleotide B6CBT. The isolated sequences contain An tracts (n = 6), suggesting that the 5′-end of this TFO was responsible for successful triplex formation. DNase I footprinting with these sequences confirmed triple helix formation at these secondary targets and demonstrated no interaction with similar oligonucleotides containing T or 5-propargylamino-dU.
INTRODUCTION
Triple-stranded nucleic acids are generated by the sequence-specific binding of oligonucleotides within the duplex major groove (1–3). The third strand bases form specific hydrogen bond contacts with the Hoogsteen face of the duplex purines and these structures are therefore usually restricted to oligopurine.oligopyrimidine tracts. The third strand can bind either parallel or antiparallel to the duplex purine strand, depending on its sequence. CT-containing oligonucleotides bind in a parallel arrangement, generating C+.GC and T.AT triplets (4,5), while GT- or GA-containing third strands bind antiparallel forming A.AT, G.GC and T.AT triplets (6). Although the parallel motif requires conditions of low pH, necessary for protonation of the third strand cytosine, this has been more widely studied than the antiparallel form as purine-rich oligonucleotides can form other competing structures.
Although triplex formation occurs with high specificity, these complexes are usually less stable than their duplex counterparts, largely due to electrostatic repulsion between the three negatively charged strands. A number of strategies have been employed to enhance the binding, including attachment of other DNA binding groups, such as intercalators (7–10), addition of exogenous triplex-binding ligands (11–15) or inclusion of modified nucleotides that bear charged groups on either the sugar (16,17) or the base (18–22). The most successful of these nucleotide analogues is bis-amino-U (B), Figure 1A, which contains a propargylamino group attached to the 5-position of the base and a 2′-aminoethoxy group attached to the sugar (23–27). This analogue combines the two modifications, which had previously been independently shown to enhance triplex formation. The propargylamino group is thought to interact with the adjacent 5′-phosphate on the third strand, while the aminoethoxy group interacts with a phosphate in the duplex purine strand (19). Inclusion of a single B residue within an 18-mer triplex-forming oligonucleotide (TFO) increased the melting temperature by 8°C at pH 6.0 (25) and facilitated triplex formation at higher pH values (28). Studies on the specificity of TFOs containing one B residue showed that the B.GC triplet is weaker than B.AT (11°C lower melting temperature), but the interaction with TA and CG was no stronger than the mismatched triplets formed with T (25). B therefore has enhanced discrimination against pyrimidines, but has the same selectivity between A and G as the natural base T. These previous studies on specificity used TFOs that only contain one B residue; we were concerned to examine the specificity of TFOs that contain multiple B-residues, since these may possess a net positive charge and might therefore show reduced selectivity, interacting with secondary sites.
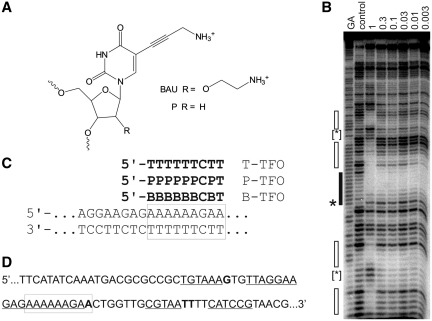
Figure 1.
(A) Chemical structures of bis-amino-U and propargylamino-dU. (B) DNase I footprint showing the interaction of B-TFO with the tyrT(43-59) fragment. The experiment was performed in 50 mM sodium acetate pH 5.0; TFO concentrations (µM) are shown at the top of each gel lane. The DNA was labelled at the 3′-end; the gel therefore runs 5′–3′ from top to bottom. The lane labelled control shows digestion of the DNA in the absence of oligonucleotide; GA is a marker specific for purines. The exact target site is shown by the filled box and the accompanying enhancement is indicated by an asterisks. Secondary binding sites are shown by the unfilled boxes and the enhancements are indicated by bracketed asterisks. (C) Sequence of the oligopurine tract in tyrT(43–59) and its interaction with the modified TFOs. The exact target site is boxed. (D) Sequence of a part of the tyrT(43–59) fragment showing the position of the secondary binding sites, which are underlined and the enhancements, which are shown in bold. The exact target site is boxed.
One method for identifying the preferred sites of DNA-binding ligands is Restriction Endonuclease Protection, Selection and Amplification (REPSA), which was first described by Hardenbol and van Dyke (29–33). This technique has previously been used to study the interaction of proteins, small molecules and oligonucleotides with DNA. Since it does not require the physical separation of the bound and unbound templates, it is ideal for studying non-covalent interactions. Like SELEX, REPSA uses a population of synthetic DNA templates, which contains a central region of randomized bases that are challenged by the ligand. A recognition site for a type IIS restriction enzyme, such as Fok I (GGATG), is included in the template adjacent to this random region. Since type IIS restriction enzymes cut at a fixed distance from their binding site they can be used to cleave a region with an unknown sequence. This makes REPSA superior to other similar methods as no prior knowledge of the target sequence is required. When these random duplex targets are incubated with the ligand, a small number will contain the correct binding sites and will be protected from enzyme cleavage. The majority of the templates will not be bound by the ligand and so will be cleaved. The reaction mixture is then subjected to polymerase chain reaction (PCR), in which only the bound (uncleaved) templates will undergo exponential amplification. Each round of selection therefore enriches the oligonucleotide pool for templates that contain the ligand-binding site and after several rounds of selection the remaining templates are cloned and sequenced.
This study examines the binding of a short 9-mer triplex forming oligonucleotide, which contains 7-bis-amino-U residues. We first examine its interaction with a fragment that contains its predicted binding site, and then use REPSA to isolate other sequences to which it binds. The interaction with these secondary binding sites is then confirmed by DNase I footprinting and compared with oligonucleotides that contain T or propargylamino-dU.
MATERIALS AND METHODS
Oligonucleotides
All oligonucleotides were synthesized on an Applied Biosystems ABI 394 automated DNA/RNA synthesizer on the 0.2- or 1-µmol scale using the standard cycles of acid-catalysed detritylation, coupling, capping and iodine oxidation procedures. Phosphoramidite monomers and reagents were purchased from Applied Biosystems or Link Technologies. The phosphoramidites for B and propargylamino-U (P) were prepared as previously described.(19,27) The 9-mer triplex-forming oligonucleotide used in the REPSA experiments had the sequence 5′-BBBBBBCBT, while 5′-TTTTTTCTT and 5′-PPPPPPCPT were also used in the footprinting experiments. The naphthylquinoline triplex-binding ligand was provided by Dr L. Strekowski, Dept. Chemistry, Georgia State University, Atlanta.
REPSA
REPSA was performed as previously described (29). The selection template used for these experiments had the sequence 5′-GTAGGATCCTGACTGGATGAAN15TCTTCCTGACAAGCTTCAG-3′ (the underlined bases show the FokI recognition sequence). Double-stranded DNA was generated from this by annealing the primer 5′-CTGAAGCTTGTCAGGAAGA-3′, which had been labelled with 32P at the 5′-end, and extending with Taq polymerase. The double-stranded template containing the 15-bp random sequence was then purified on a 6% non-denaturing polyacrylamide gel and resuspended in 10 µl Fok I buffer (50 mM potassium acetate, 20 mM Tris-acetate pH 7.9, 10 mM magnesium acetate, 1 mM dithiothreitol). Ten microlitres of 10 -µM oligonucleotide 5′-BBBBBBCBT, dissolved in 50 mM sodium acetate pH 5.0, was then added and the mixture was equilibrated for 1 h at 20°C (the final pH was about 5.6). The complex was then cleaved with 8 U FokI for 5 min at 37°C and the products of the reaction were subjected to PCR. Unbound sequences were cleaved by the enzyme and were not amplified by PCR while those protected by the triplex-forming oligonucleotide were selectively amplified. By including the FokI recognition sequence (GGATG) in the primer sequence, we avoided the selection of any nuclease-resistant products that arise from mutations in the restriction enzyme recognition site. PCR amplification was performed by mixing 10 µl of the cleavage reaction with 15 µl of the labelled forward primer 5′-CTGAAGCTTGTCAGGAAGA, 2 µl of unlabelled 5′-GTAGGATCCTGACTGGATG (80 µM), 1 µl 25 mM dNTPs, 5 µl 10xTaq buffer and 5 U of Taq polymerase in a total volume of 50 µl. The mixture was subject to 15 cycles of 93°C (30 s) and 50°C (3 min). A further two cycles of PCR were then performed after adding 1 µl dNTPs, 2.5 µl unlabelled forward primer and 5 U of Taq polymerase. This pulse-chase procedure, using a low concentration of the radiolabelled primer for the initial cycles, ensures efficient incorporation of the radiolabel, while preventing the formation of any mismatched duplexes, which would have arisen from the annealing of PCR products. The amplified DNA was run on an 8% non-denaturing polyacrylamide gel from which the full-length products were purified. The purified template, enriched in binding sites for the TFO, was redissolved in 10 µl of FokI buffer and subjected to a further round of REPSA selection. The procedure was repeated 10 times. After the tenth round of selection, the products were cleaved with BamHI and HindIII and ligated into pUC19 that had also been cleaved with BamHI and HindIII. The ligated plasmid was transformed into Escherichia coli TG2 and plated on agar plates containing 0.02% X-gal, 1 mM IPTG and 100 µg/ml carbenicillin. White colonies were picked from these plates and sequenced. Colonies were first sequenced manually using a T7 sequencing kit (USB), although the sequences of all the fragments used for the footprinting experiments were subsequently confirmed by Eurofins MWG Operon, Ebersberg, Germany.
DNA fragments for footprinting
Plasmids containing the REPSA templates were cleaved with EcoRI and labelled at the 3′-end with α-32P[dATP] using reverse transcriptase. The fragment of interest was then released by cutting with HindIII, purified on a 6% polyacrylamide gel and redissolved in 10 mM Tris–HCl pH 7.5, containing 0.1 mM EDTA at a concentration of at least 10 c.p.s per microlitre (as determined using a hand-held Geiger counter). The tyrT(43-59) restriction fragment, which contains an 18-bp oligopurine tract that includes the target site for these 9-mer TFOs, was prepared as previously described (34).
DNase I footprinting
DNase I footprinting was performed as previously described (35). Briefly, 1.5 µl of radiolabelled DNA was mixed with 3 µl of oligonucleotide dissolved in 50 mM sodium acetate pH 5.0 to give a final concentration of between 0.03 and 1 µM and incubated overnight at 20°C. The mixture was cleaved by adding 2 µl DNase I (typically 0.01 U/ml) dissolved in 20 mM NaCl, 2 mM MgCl2 and 2 mM MnCl2, stopping the reaction after 2 min with 4.5 µl formamide containing 10 mM EDTA and bromphenol blue. The products of the reaction were separated on a 9% denaturing polyacrylamide gel containing 8M urea. After electrophoresis the gels were fixed, dried and subjected to autoradiography using a phosphorimager screen (Kodak), which was scanned with a Storm phosphorimager.
RESULTS
The 18-bp oligopurine tract in tyrT(43–59) has often been used to examine the binding of modified triplex-forming oligonucleotides (19,24,25,36,37). Single base mutations within this target have been used to examine the binding selectivity of oligonucleotides that contain one or more bis-amino-U (B) residues (25). These studies have shown that B has a very high affinity for AT relative to all other Watson–Crick base pairs in DNA and that each B.AT triplet increases the melting temperature by about 8°C relative to T.AT. Binding to GC is much weaker and B-modified oligonucleotides show enhanced discrimination against YR compared with T. However, we were concerned that oligonucleotides that contain multiple B substitutions, generating TFOs with a net positive charge, might show reduced selectivity and interact with other secondary sites. We therefore examined the interaction of the 9-mer TFO 5′-BBBBBBCBT, which is expected to have a formal charge of +5 at physiological pH, with its target site in tyrT(43-59) (Figure 1C) by DNase I footprinting and the results are shown in Figure 1B. As expected, a clear footprint is produced at the intended target (indicated by the filled box), which persists to nanomolar concentrations. As previously noted, this footprint is accompanied by enhanced DNase I cleavage at the 3′-(lower) end of the target site (19,24,36,38). In addition to this footprint, other regions of protection are evident at the highest TFO concentration (1 µM) as indicated by the open boxes, and the primary footprint is much larger, extending throughout the entire oligopurine tract. Enhanced cleavage is also observed at the 3′-ends of each of these secondary sites. The location of these secondary sites is shown in Figure 1D. No secondary binding was observed with the unmodified TFO 5′-TTTTTTCTT or the propargylamino-U-modified 5′-PPPPPPCPT (data not shown). Visual inspection of these secondary sites does not reveal any obvious common sequence characteristics. REPSA experiments were therefore carried out to select the preferred binding sites for this oligonucleotide from a pool of random templates.
Since the conditions of the REPSA experiments are limited by the pH requirement of the selecting type IIS restriction enzyme (FokI) we first examined the pH dependence of these secondary sites (data not shown). Although the primary footprint was still evident at pH 7.0, the secondary sites had largely disappeared at pH 6.0 and above (Supplementary Figure S1). These secondary sites also disappeared within 1 min when the pH was rapidly raised from pH from 5.0 to 6.0 or 7.0. A low pH was therefore employed in these experiments in order to observe the secondary sites, although we observed that FokI cleavage was at least 100-fold slower at pH 5.0 and 6.5 than at pH 7.9.
REPSA
REPSA selection was performed with FokI using the random template in the presence of 5 µM B-substituted TFO. This TFO concentration is higher than might normally be employed in gene inhibition studies, but was used in order to exaggerate the proportion of secondary binding sites. Out of 77 colonies picked for sequencing, 45 contained an insert that could not be fully sequenced (possibly because they adopt unusual structures). The other 32 colonies were sequenced and the sequences of the N15 random regions are shown in Supplementary Table S1. As the aim of this work was to compare the secondary binding sites with the exact target sequence, we only examined templates that showed some similarity to the exact target. Fourteen of these sequences, each of which contained an oligo(dA) tract, were therefore chosen for footprinting (Table 1). Sequence 6 is included as it was originally thought to have a run of six As but was re-sequenced when the B-TFO failed to produce a footprint and was found to contain a pyrimidine interruption. We considered that the other sequences were less likely to bind the TFO and are likely to be random sequences, which were not screened out by REPSA or which were refractory to FokI cleavage for other reasons. Such sequences might have been eliminated with further rounds of REPSA, but we were concerned to avoid introducing mutations in the template sequences. Indeed, it can be seen that some of the sequences are not the same length as the original random template; this is an acknowledged limitation of the REPSA technique. The relatively high level of unselected sequences may also arise because FokI is not producing efficient double-strand cleavage under these conditions, allowing the persistence of some unbound templates.
Table 1.
Sequences of the central regions of clones obtained from REPSA selection, corresponding to the N15 region in the random template
| Number | Sequence |
|---|---|
| 1 | CAATCAAAAAAACAA |
| 2 | AAAAAAAAGTTGCTCT |
| 3 | CCCCGAACAAAAAAA |
| 4 | CAAAAAAAGAGAGAC |
| 5 | TATTGCTAAAAAAGG |
| 6 | AAATAAACGGCCATTG |
| 7 | CCAGAGCACAAAAAACAA |
| 8 | GGTAAAAAACCGGCA |
| 9 | CAACTCAAAAAATCA |
| 10 | CCCTAAAAATAACAA |
| 11 | GCAGTAAAAAAACTA |
| 12 | AAGAAAAGCAATGGGATCT |
| 13 | CACACGCAAAAAAG |
| 14 | AAGAAAAAACAGCTCCTAC |
For sequences 2, 4, 12 and 14 the first two As correspond to the bases in the flanking sequence.
The strand shown is the one that is labelled on the footprinting gels and is the complementary strand to the original template strand. The oligo(dA) tracts are given in bold.
An interesting pattern is evident in these oligo(dA)-containing sequences; the selected templates were all in the same orientation, with the purine-rich strand opposite to the GGATG of the FokI site (i.e. on the same strand as CATCC). As a consequence, all the footprinting experiments described below display the purine-rich strand of the target. None of the isolated fragments were in the opposite orientation with oligo(T) on the GGATG-containing strand. This is presumably a property of the selecting enzyme FokI and will be considered further in the ‘Discussion’ section.
DNase I footprinting
Radiolabelled DNA fragments containing 14 of the REPSA-selected sequences were prepared and subjected to DNase I footprinting in the presence of varying concentrations of 5′-BBBBBBCBT (B-TFO), 5′-PPPPPPCPT (P-TFO) or 5′-TTTTTTCTT (T-TFO). In each case, the radiolabelled DNA fragment and the TFO were incubated overnight at 20°C in 50 mM sodium acetate pH 5.0. The results for the B-substituted TFO are shown in Figures 2 and 3. Since no footprints were observed with the P-substituted TFO at concentrations up to 30 µM, these experiments were repeated in the presence of 10 µM naphthylquinoline triplex-binding ligand and these results are presented in Figure 4. No footprints were evident with the unmodified T-TFO on any of these sequences at concentrations up to 30 µM, even in the presence of the triplex-binding ligand (data not shown). The results for the various targets will be described in turn, while the positions of the various footprints are summarized in Table 2. The addition of 5 mM MgCl2 generally made no difference to the position of the footprints, merely enhancing the strength of interaction with the P-TFO. In a few instances, this produced slightly different cleavage patterns with P-TFO in the presence of the triplex ligand and these are presented in Supplementary Figure S2.
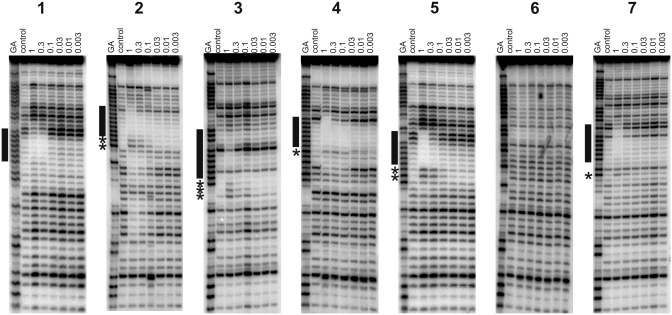
Figure 2.
DNase I footprints showing the interaction of B-TFO with REPSA-selected sequences 1–7. The experiments were performed in 50 mM sodium acetate pH 5.0; TFO concentrations (µM) are shown at the top of each gel lane. The DNA was labelled at the 3′-end; the gel therefore runs 5′–3′ from top to bottom. The lanes labelled ‘control’ show digestion of the DNA in the absence of oligonucleotide; tracks labelled ‘GA’ are markers specific for purines. The locations of the footprints are indicated by the filled boxes and any accompanying enhancements are indicated by asterisks.
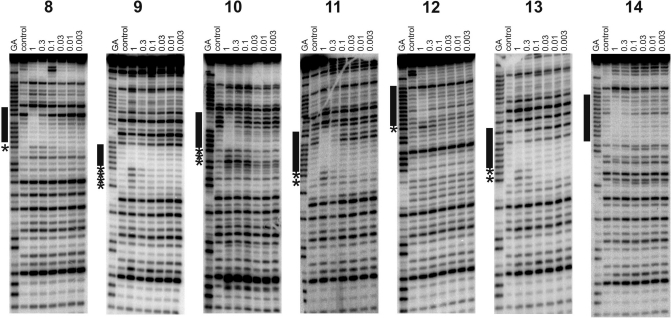
Figure 3.
DNase I footprints showing the interaction of B-TFO with REPSA-selected sequences 8–14. The experiments were performed in 50 mM sodium acetate pH 5.0; TFO concentrations (µM) are shown at the top of each gel lane. The DNA was labelled at the 3′-end; the gel therefore runs 5′–3′ from top to bottom. The lanes labelled ‘control’ show digestion of the DNA in the absence of oligonucleotide; tracks labelled ‘GA’ are markers specific for purines. The locations of the footprints are indicated by the filled boxes and any accompanying enhancements are indicated by asterisks.
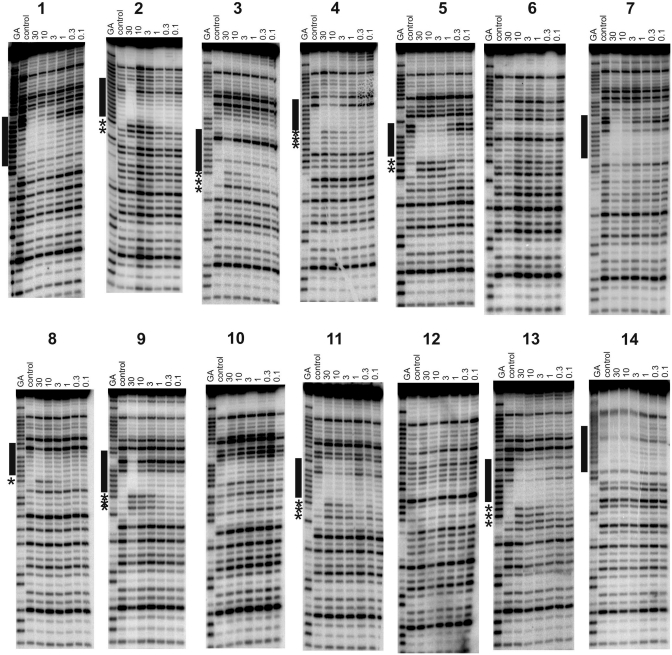
Figure 4.
DNase I footprints showing the interaction of P-TFO with REPSA-selected sequences 1–14 in the presence of 10 µM naphthylquinoline triplex-binding ligand. The experiments were performed in 50 mM sodium acetate pH 5.0; P-TFO concentrations (µM) are shown at the top of each gel lane. The lanes labelled ‘control’ show digestion of the DNA in the absence of oligonucleotide; tracks labelled ‘GA’ are markers specific for purines. The locations of the footprints are indicated by the filled boxes and any accompanying enhancements are indicated by asterisks.
Table 2.
Locations of the footprints and enhancements observed with the REPSA footprinting fragments
| Number | Sequence |
|---|---|
| 1 | AGGAAGACAATCAAAAAAACAATTCATCC |
| 2 | AGGAAGAAAAAAAAGTTGCTCTTTCATCC |
| 3 | AGGAAGACCCCGAACAAAAAAATTCATCC |
| 4 | AGGAAGACAAAAAAAGAGAGACTTCATCC |
| 5 | AGGAAGATATTGCTAAAAAAGGTTCATCC |
| 7 | AGGAAGACCAGAGCACAAAAAACAACCCTCATCC |
| 8 | AGGAAGAGGTAAAAAACCGGCATTCATCC |
| 9 | AGGAAGACAACTCAAAAAATCATTCATCC |
| 10 | AGGAAGACCCTAAAAATAACAATTCATCC |
| 11 | AGGAAGAGCAGTAAAAAAACTATTCATCC |
| 12 | AGGAAGAAAAGCAATGGGATCTTTCATCC |
| 13 | AGGAAGACACACGCAAAAAAGTTCATCC |
| 14 | AGGAAGAAAAAACAGCTCCTACTTCATCC |
The selected sequences are shown in large type, while the adjacent flanking regions in the template are shown in smaller type and italics.
Bases shown in bold correspond to positions of enhanced cleavage, while the footprints are underlined.
Sequence 1
This sequence contains the closest match to the perfect target site with a single G-to-C mutation, and contains a run of seven contiguous As followed by CAA. DNase I cleavage patterns show that the B-TFO produces a footprint down to a concentration of around 0.3 µM, even though this fragment does not contain its correct binding site, confirming that REPSA had successfully selected a TFO-binding site. Unusually, compared to many of the sequences described below, no enhanced cleavage is evident at the 3′-(lower) triplex–duplex junction. This fragment contains a run of seven consecutive As and the lower end of this footprint is coincident with the 3′-end of this tract. The upper end of the footprint extends beyond the A-tract by 2–3 bases, as shown in Table 2. Triple helix protection from DNase I cleavage does not arise from direct steric blockage, as the enzyme and third strand are located in different grooves. A typical triplex footprint extends for 3–4 bases above (5′-) the target site, while the lower (3′) end of the footprint matches the end of the triplex, and is often accompanied by enhanced cleavage of the terminal base at the triplex–duplex junction (19,24,36,38). Based on these observations, it seems reasonable to suggest that the B-TFO is bound to the A7 tract and this will be considered further in the ‘Discussion’ section. In contrast, no footprints were observed with the T- or P-TFOs at concentrations up to 30 µM, although addition of 10 µM of the naphthylquinoline triplex-binding ligand induces a clear footprint in the same region with the P-TFO, which persists to a concentration of about 3 µM (Figure 4). This interaction is further enhanced by the addition of magnesium chloride (5 mM); in these conditions, the footprint persists to about 0.3 µM and is accompanied by enhanced cleavage at the terminal A in the sequence CAA (Supplementary Figure S2).
Sequence 2
This sequence contains nine contiguous As followed by a G. The B-substituted TFO produces a footprint at the highest concentration (1 µM), which is accompanied by two enhancements at the 3′-end of the oligopurine tract. A similar footprint can be seen with the P-TFO in the presence of the triplex ligand (at a TFO concentration of 30 µM), although the two enhancements are shifted one base further down compared with the B-TFO. The presence of two enhanced bands may indicate that the TFO can be bound in more than one location, each of which produces enhanced cleavage at its 3′-end.
Sequence 3
This sequence contains seven contiguous As followed by three pyrimidines (TTC). The B-TFO produces a footprint at the highest concentration (1 µM), although one band is still evident within this region (Figure 2). This protection is accompanied by three enhanced bands at the lower (3′) end of this region, which may again suggest multiple binding locations on this target. The P-TFO does not produce a footprint with this sequence, although addition of the triplex ligand produces a footprint in a similar location at the highest concentration (30 µM). Three enhanced cleavage products are again evident at the 3′-end of this footprint, although their relative intensities are different to those seen with the B-TFO alone.
Sequence 4
This sequence contains seven contiguous As followed by three purines (GAG). The B-TFO produces a clear footprint on this template at a concentration of 1 µM (Figure 2), which is accompanied by a single enhancement at the guanine at the 3′-end of the A-tract. An unusual feature is that three bands below this enhancement also show attenuated cleavage. The B- and T-TFOs did not produce footprints on this sequence, although the addition of the triplex ligand generated a footprint with 10 µM P-TFO. This ligand-induced footprint is accompanied by several enhanced bands and is one base shorter at the 3′-end than with the B-TFO.
Sequence 5
This sequence contains six contiguous As followed by GGT. The B-TFO produces a clear footprint on this template at a concentration of 1 µM (Figure 2), which is accompanied by two enhanced cleavage products at the 3′-(lower) end of the footprint corresponding to the terminal A and the following G. The P-TFO does not produce a footprint on this template, even at a concentration of 30 µM, although the addition of 10 µM triplex ligand produces a clear footprint, which persists to about 3 µM, and is very similar to that produced by the B-TFO alone. Further addition of 5 mM MgCl2 causes the footprint to extend one base further in the 3′-(lower) direction, with enhancement in the two guanines (Supplementary Figure S2).
Sequence 6
This sequence contains an interrupted A-tract (AAATAAAC). None of the TFOs altered the DNase I cleavage pattern of this template under any conditions (Figures 2 and 4).
Sequence 7
This sequence is similar to sequence 1, but the A-tract is one base shorter (A6CAA). Once again the B-TFO produces a footprint within the REPSA-selected region, which is evident at the highest TFO concentration (1 µM). This is accompanied by a faint enhancement at the terminal A. No footprints were produced with the T- and P-TFOs alone, although the addition of 10 µM triplex ligand produced a clear footprint with 10 µM P-TFO, with no evident enhanced cleavage (as noted with sequence 1).
Sequence 8
This sequence contains six contiguous As followed by CCG. The B-TFO produces a DNase I footprint at the highest concentration (1 µM) (Figure 3) which is accompanied by enhanced cleavage at the terminal A of the A6-tract. The T- and P-TFO did not produce footprints, although in the presence of 10 µM triplex ligand 30 µM P-TFO generated a similar footprint to that produced by B-TFO alone.
Sequence 9
This sequence contains six contiguous As followed by TCA. The B-TFO produces attenuated DNase I cleavage within this A-tract, which is accompanied by four enhancements at the 3′-end (Figure 3). The T- and P-TFOs do not affect the cleavage pattern at concentrations as high as 30 µM, but a clear footprint is produced with 30 µM P-TFO in the presence of 10 µM triplex ligand, which is also accompanied by several enhancements.
Sequence 10
This sequence contains only five contiguous As followed by TAA. The B-TFO again produces a footprint with this target at a concentration of 1 µM, which is accompanied by three bands of enhanced cleavage at the 3′-end. In this case, no footprints are produced by the T- or P-TFOs even in the presence of 10 µM triplex ligand, although further addition of 5 mM MgCl2 to the P-TFO plus triplex ligand produces a footprint that is similar to that seen with B-TFO alone (Supplementary Figure S2).
Sequence 11
This sequence contains seven contiguous As followed by CTA. The B-substituted TFO produces a footprint at a concentration of 0.3 µM with two enhanced bands at the 3′-end of the A-tract. No footprints were produced with T- and P-TFO alone. The addition of 10 µM triplex ligand produces a footprint with P-TFO at a concentration of 3 µM in the same position as seen with B-TFO alone. This is accompanied by enhanced cleavage though the precise pattern of these enhancements is not the same as that with B-TFO.
Sequence 12
In this sequence the oligo-A tract is interrupted by a single guanine and so the longest A-tract contains only four contiguous As. The B-TFO attenuates DNase I cleavage within this purine tract at a concentration of 1 µM and produces a single enhanced cleavage product at a G at its 3′-end. The P- and T-TFOs did not affect the DNase I cleavage pattern, even in the presence of the triplex ligand.
Sequence 13
This sequence contains six contiguous As followed by GTT. The B-TFO produces a footprint on this template at a concentration of 1 µM, which is accompanied by two bands of enhanced cleavage that are coincident with the terminal AG. In the presence of 10 µM triplex ligand the P-TFO produces a similar footprint that persists to a concentration of 3 µM. The same two enhancements as seen with the B-TFO are also evident.
Sequence 14
This sequence contains six contiguous As followed by CAG. The B-TFO produces a clear footprint within this oligopurine tract, although no enhanced cleavage products are evident. A similar pattern is evident with P-TFO in the presence of 10 µM triplex ligand.
DISCUSSION
The results presented in this paper clearly show that this heavily modified B-substituted TFO can bind to secondary sites. The interaction is not non-selective, but high affinity binding is observed at DNA sites that contain triplex mismatches and several of the REPSA-selected targets bind the B-TFO at sub-micromolar concentrations, even though they do not contain the exact target site. This is particularly striking in view of the short length of the TFO, which is only a 9-mer. This interaction appears to be restricted to the bis-amino-U modification since T- or P-containing TFOs showed no interaction with these target sites at concentrations up to 30 µM (although some sequences showed interaction with P-TFO in the presence of the triplex-binding ligand).
The exact location of the TFO cannot be accurately deduced from these footprints as DNase I cleavage is generally poor within these oligopurine tracts. However, most, though not all, of the selected sequences contain an A-tract that is 6 bp long. We assume that this corresponds to interaction with the six consecutive B-residues in the TFO. Sequence 6, for which the longest A-tract is only 3 bp, shows no evidence of triplex formation. The two A3 tracts in this sequence, which are separated by a single T, are clearly not sufficient to generate a secondary binding site. Sequence 12 only contains an A4 tract, although this is preceded by AAG and one of the bis-amino-U residues in the B6-tract must be interacting with this GC base pair. This seems reasonable since our previous work has shown that B can bind to an isolated GC interruption (about 15-fold weaker than to AT, but much stronger than to TA or CG) (25). The position of the enhancement with this sequence suggests that the 3′-G is the terminal base of this triplex, forming a standard C+.GC triplet. Sequence 10 also contains a shorter A-tract (A5), although this is followed by TAA. The enhancements seen with this footprint suggest that the TFO can bind in more than one location, although the strongest enhancement is at the first A following the TA interruption. In contrast, sequence 9 also possesses an A6 tract, yet produced one of the weakest footprints.
These results suggest that the binding of this TFO is dominated by the run of six consecutive bis-amino-U residues. The last three bases at the 3′-end of the TFO (CBT) seem to be less important for sequence recognition. This is emphasized by the observation that several of the selected A-tracts are flanked by a 3′-G, at which there is enhanced DNase I cleavage (sequences 2, 4, 5, 12 and 13), suggesting that even the C is not involved in the secondary triplex binding site. The lower stringency at the 3′-end of the TFO contrasts with earlier work that employed REPSA to examine the selectivity of a 19-mer GT-oligonucleotide, for which the 13 bases at the 3′-end of the TFO were shown to be sufficient for binding (30). However, this previous study concerned antiparallel triplexes, and so also involved the 5′-end of the target site. It also employed natural nucleotides with a lower affinity, whereas in the present study the 5′-end of the TFO is more heavily modified than the 3′-end to achieve high-affinity interactions.
Previous work with bis-amino-U studied its interaction with single base mismatches and showed that, as well as forming strong complexes with AT, it also formed a weaker triplet with GC, but showed enhanced discrimination against CG and TA, compared with the unmodified nucleotide T (25). It is therefore not surprising that these secondary sites are all based on A-tracts; other mismatched triplet combinations do not appear to benefit from addition of the 2′-aminoethoxy or 5-propargylamino-dU substitutions. The specific benefits of these additions are also evident from the observation that B-containing TFOs do not stabilize the triplexes formed with parallel or antiparallel GT-containing oligonucleotides. It should be noted that non-specific binding of triplex-forming oligonucleotides can also occur by looping out regions of the TFO (39). This is unlikely with this short 9-mer oligonucleotide, but we cannot exclude the possibility that one or more of the third strand nucleotides is looped out from the duplex.
This study successfully isolated a number of clones, which clearly represent secondary binding sites for this TFO. However, none of these sequences represents the exact target site (A6GAA), although sequence 1 is only one base different (changing a G to C). At first sight this is surprising, although it is likely to be the result of the TFO concentration (5 µM) used in these experiments, which is much higher than the affinity for the exact target site. These results further demonstrate the very high affinity of B for A-rich duplex DNA. Low concentrations of B-containing TFOs will therefore need to be used in potential in vivo applications, reducing the interaction with any secondary sites.
Enhanced DNase I cleavage is often observed at the triplex–duplex boundary (19,24,38). This is only seen on the duplex purine strand (never on the pyrimidine strand), but is not observed with all sequences. For example, sequences 1 and 14 show no evidence of enhanced DNase I cleavage. Although both these sequences contain an adjacent ApC, which is not present in any of the other sequences, this alone cannot be sufficient to explain the lack of enhancement as the original target site in tyr(43-59) is also flanked by ApC. Although we are not sure of the origin of this difference, which may result from the A–B transition in the underlying duplex at the triplex–duplex junction, it has proved very useful in these experiments for delineating the 3′-end of the target site. The presence of multiple enhancements has not been observed in previous triplex footprints and we interpret this as indicating that the TFO can bind in different positions within these targets.
It is also interesting to note that all the cloned sequences are in the same orientation, in which the purine strand of the duplex is evident on the footprinting gels (the T-containing strand is on the same strand as the FokI site GGATG). There is no reason in principle why the strands could not be reversed, producing fragments with a Tn tract on the labelled strand. This must be a function of the way in which FokI cleaves the target triplex. This enzyme is known to interact with two copies of its target site before it can cleave the DNA; DNA cleavage only occurs after two DNA-bound enzymes interact with each other (40). Since our fragments only contain one FokI binding site, this interaction must occur between two separate DNA fragments, either or both of which might be bound by the TFO. The effect cannot be explained by suggesting that the enzyme only cleaves one strand of the underlying duplex, as the remaining strand would still be amplified by the PCR. It therefore appears that under these conditions FokI may still be able to cleave both duplex strands of the triplex formed at GGATG … An, while a similar triplex generated at GGATG … Tn is refractory to the enzyme. It is also possible that this strand selectivity might be caused by the effect of low pH on the FokI enzyme, if the protonation state of the active site of a FokI monomer on one strand is different to the protonation state on other monomer.
It should be noted that in all these experiments the triplex was formed at pH 5.0, before digesting with FokI at a pH of about 5.6. These non-physiological conditions were chosen in order to enhance the affinity of this short (9-mer) oligonucleotide for its target. Stronger binding could have been achieved by using longer B-containing oligonucleotides, which would normally be employed in biological applications, but this would have increased the number of potential secondary binding sites. The affinity of this TFO for its exact target site is pH dependent, as a result of the single third strand cytosine, with dissociation constants of <100 nM at pH 5.0, about 0.3 µM at pH 6.0 and 1 µM at pH 7.0. The footprinting experiments show that the affinities for these secondary sites are typically 0.3–1 µM at pH 5.0. Binding to these sites decreases at higher pH values, although we assume that the relative discrimination between target and non-target sites is not pH sensitive. For in vivo applications of modified triplex-forming oligonucleotides, the concentrations employed may well be lower than the affinity for these secondary sites, and so may not be a problem for such short sequences. However, longer TFOs may show significant interaction with secondary sites at these concentrations. Other applications of triplexes, for instance, in detection/diagnostics or nanostructure formation (41), may well involve conditions of low pH, as they are not limited by physiological constraints. These applications with heavily modified TFOs might be affected by annealing to secondary binding sites.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1 and 2.
FUNDING
ASC was supported by a research studentship from the BBSRC. Funding for Open Access charge: The University of Southampton.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Felsenfeld G, Davies DR, Rich A. Formation of a three-stranded polynucleotide molecule. J. Am. Chem. Soc. 1957;79:2023–2024. [Google Scholar]
- 2.Fox KR. Targeting DNA with triplexes. Curr. Med. Chem. 2000;7:17–37. doi: 10.2174/0929867003375506. [DOI] [PubMed] [Google Scholar]
- 3.Thuong NT, Hélène C. Sequence-specific recognition and modification of double-helical DNA by oligonucleotides. Angew. Chem. Intl. Ed. Engl. 1993;32:666–690. [Google Scholar]
- 4.Doan TL, Perrouault L, Praseuth D, Habhoub N, Decout JL, Thuong NT, Lhomme J, Hélène C. Sequence-specific recognition, photo-cross-linking and cleavage of the DNA double helix by an oligo-[alpha]-thymidylate covalently linked to an azidoproflavine derivative. Nucleic Acids Res. 1987;15:7749–7760. doi: 10.1093/nar/15.19.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moser HE, Dervan PB. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 6.Beal PA, Dervan PB. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991;251:1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- 7.Stonehouse TJ, Fox KR. DNase-I footprinting of triple-helix formation at polypurine tracts by acridine-linked oligopyrimidines – stringency, structural-changes and interaction with minor-groove binding ligands. Biochim. Biophys. Acta Gene Struct. Expr. 1994;1218:322–330. doi: 10.1016/0167-4781(94)90184-8. [DOI] [PubMed] [Google Scholar]
- 8.Li H, Broughton-Head VJ, Peng GM, Powers VEC, Ovens MJ, Fox KR, Brown T. Triplex staples: DNA double-strand cross-linking at internal and terminal sites using psoralen-containing triplex-forming oligonucleotides. Bioconjug. Chem. 2006;17:1561–1567. doi: 10.1021/bc0601875. [DOI] [PubMed] [Google Scholar]
- 9.Sun J-S, François J-C, Montenay-Garestier T, Saison-Behmoaras T, Roig V, Thuong NT, Hélène C. Sequence-specific intercalating agents: intercalation at specific sequences on duplex DNA via major groove recognition by oligonucleotide-intercalator conjugates. Proc. Natl Acad. Sci. USA. 1989;86:9198–9202. doi: 10.1073/pnas.86.23.9198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takasugi M, Guendouz A, Chassignol M, Decout JL, Lhomme J, Thuong NT, Hélène C. Sequence-specific photo-induced cross-linking of the two strands of double-helical DNA by a psoralen covalently linked to a triple helix-forming oligonucleotide. Proc. Natl Acad. Sci. USA. 1991;88:5602–5606. doi: 10.1073/pnas.88.13.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Escudé C, Nguyen CH, Kukreti S, Janin Y, Sun JS, Bisagni E, Garestier T, Hélène C. Rational design of a triple helix-specific intercalating ligand. Proc. Natl. Acad. Sci. USA. 1998;95:3591–3596. doi: 10.1073/pnas.95.7.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandler SP, Strekowski L, Wilson WD, Fox KR. Footprinting studies on ligands which stabilize DNA triplexes – effects on stringency within a parallel triple-helix. Biochemistry. 1995;34:7234–7242. doi: 10.1021/bi00021a039. [DOI] [PubMed] [Google Scholar]
- 13.Wilson WD, Tanious FA, Mizan S, Yao SJ, Kiselyov AS, Zon G, Strekowski L. DNA triple-helix specific intercalators as antigene enhancers – unfused aromatic cations. Biochemistry. 1993;32:10614–10621. doi: 10.1021/bi00091a011. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen CH, Marchand C, Delage S, Sun JS, Garestier T, Hélène C, Bisagni E. Synthesis of 13H-benzo[6,7]- and 13H-benzo[4,5]indolo[3,2- c]quinolines: a new series of potent specific ligands for triplex DNA. J. Am. Chem. Soc. 1998;120:2501–2507. [Google Scholar]
- 15.Mergny JL, Duval-Valentin G, Nguyen CH, Perrouault L, Faucon B, Rougée M, Montenay-Garestier T, Bisagni E, Hélène C. Triple helix specific ligands. Science. 1992;256:1681–1684. doi: 10.1126/science.256.5064.1681. [DOI] [PubMed] [Google Scholar]
- 16.Blommers MJJ, Natt F, Jahnke W, Cuenoud B. Dual recognition of double-stranded DNA by 2′-aminoethoxy-modified oligonucleotides: the solution structure of an intramolecular triplex obtained by NMR spectroscopy. Biochemistry. 1998;37:17714–17725. doi: 10.1021/bi9816352. [DOI] [PubMed] [Google Scholar]
- 17.Cuenoud B, Casset F, Hüsken D, Natt F, Wolf RM, Altmann KH, Martin P, Moser HE. Dual recognition of double-stranded DNA by 2′-aminoethoxy-modified oligonucleotides. Angew. Chem. Intl. Ed. Eng. 1998;37:1288–1291. doi: 10.1002/(SICI)1521-3773(19980518)37:9<1288::AID-ANIE1288>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 18.Gowers DM, Bijapur J, Brown T, Fox KR. DNA triple helix formation at target sites containing several pyrimidine interruptions: stabilization by protonated cytosine or 5-(1-propargylamino)dU. Biochemistry. 1999;38:13747–13758. doi: 10.1021/bi9911637. [DOI] [PubMed] [Google Scholar]
- 19.Bijapur J, Keppler MD, Bergqvist S, Brown T, Fox KR. 5-(1-propargylamino)-2′-deoxyuridine (U-P): a novel thymidine analogue for generating DNA triplexes with increased stability. Nucleic Acids Res. 1999;27:1802–1809. doi: 10.1093/nar/27.8.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganesh KN, Rajeev KG, Pallan PS, Rana VS, Barawkar DA, Kumar VA. Modulation of DNA triplex stability through nucleobase modifications. Nucleosides Nucleotides. 1997;16:1271–1278. [Google Scholar]
- 21.Rusling DA, Peng G, Srinivasan N, Fox KR, Brown T. DNA triplex formation with 5-dimethylaminopropargyl deoxyuridine. Nucleic Acids Res. 2009;37:1288–1296. doi: 10.1093/nar/gkn1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lou CG, Xiao QA, Brennan L, Light ME, Vergara-Irigaray N, Atkinson EM, Holden-Dye LM, Fox KR, Brown T. Synthesis and properties of triplex-forming oligonucleotides containing 2′-O-(2-methoxyethyl)-5-(3-aminoprop-1-ynyl)-uridine. Bioorg. Med. Chem. 2010;18:6389–6397. doi: 10.1016/j.bmc.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Rusling DA, Powers VEC, Ranasinghe RT, Wang Y, Osborne SD, Brown T, Fox KR. Four base recognition by triplex-forming oligonucleotides at physiological pH. Nucleic Acids Res. 2005;33:3025–3032. doi: 10.1093/nar/gki625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sollogoub M, Darby RAJ, Cuenoud B, Brown T, Fox KR. Stable DNA triple helix formation using oligonucleotides containing 2′-aminoethoxy,5-propargylamino-U. Biochemistry. 2002;41:7224–7231. doi: 10.1021/bi020164n. [DOI] [PubMed] [Google Scholar]
- 25.Osborne SD, Powers VEC, Rusling DA, Lack O, Fox KR, Brown T. Selectivity and affinity of triplex-forming oligonucleotides containing 2′-aminoethoxy-5-(3-aminoprop-1-ynyl)uridine for recognizing AT base pairs in duplex DNA. Nucleic Acids Res. 2004;32:4439–4447. doi: 10.1093/nar/gkh776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rusling DA, Broughton-Head VJ, Tuck A, Khairallah H, Osborne SD, Brown T, Fox KR. Kinetic studies on the formation of DNA triplexes containing the nucleoside analogue 2′-O-(2-aminoethyl)-5-(3-amino-1-propynyl)uridine. Org. Biomol. Chem. 2008;6:122–129. doi: 10.1039/b713088k. [DOI] [PubMed] [Google Scholar]
- 27.Sollogoub M, Dominguez B, Fox KR, Brown T. Synthesis of a novel bis-amino-modified thymidine monomer for use in DNA triplex stabilisation. Chem. Commun. 2000:2315–2316. [Google Scholar]
- 28.Rusling DA, Le Strat L, Powers VEC, Broughton-Head VJ, Booth J, Lack O, Brown T, Fox KR. Combining nucleoside analogues to achieve recognition of oligopurine tracts by triplex-forming oligonucleotides at physiological pH. FEBS Lett. 2005;579:6616–6620. doi: 10.1016/j.febslet.2005.10.056. [DOI] [PubMed] [Google Scholar]
- 29.Van Dyke MW, Van Dyke N, Sunavala-Dossabhoy G. REPSA: general combinatorial approach for identifying preferred ligand-DNA binding sequences. Methods. 2007;42:118–127. doi: 10.1016/j.ymeth.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Hardenbol P, VanDyke MW. Sequence specificity of triplex DNA formation: analysis by a combinatorial approach restriction endonuclease protection selection and amplification. Proc. Natl Acad. Sci. USA. 1996;93:2811–2816. doi: 10.1073/pnas.93.7.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardenbol P, Wang JC, VanDyke MW. Identification of preferred distamycin–DNA binding sites by the combinatorial method REPSA. Bioconjug. Chem. 1997;8:617–620. doi: 10.1021/bc970066s. [DOI] [PubMed] [Google Scholar]
- 32.Hardenbol P, Wang JC, VanDyke MW. Identification of preferred hTBP DNA binding sites by the combinatorial method REPSA. Nucleic Acids Res. 1997;25:3339–3344. doi: 10.1093/nar/25.16.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen J, Wang JC, Van Dyke MW. Identification of preferred actinomycin-DNA binding sites by the combinatorial method REPSA. Bioorg. Med. Chem. 2001;9:2285–2293. doi: 10.1016/s0968-0896(01)00146-8. [DOI] [PubMed] [Google Scholar]
- 34.Brown PM, Fox KR. DNA triple-helix formation on nucleosome core particles – effect of length of the oligopurine tract. Eur. J. Biochem. 1999;261:301–310. doi: 10.1046/j.1432-1327.1999.00279.x. [DOI] [PubMed] [Google Scholar]
- 35.Hampshire AJ, Rusling DA, Broughton-Head VJ, Fox KR. Footprinting: a method for determining the sequence selectivity, affinity and kinetics of DNA-binding ligands. Methods. 2007;42:128–140. doi: 10.1016/j.ymeth.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Keppler MD, Fox KR. Relative stability of triplexes containing different numbers of T.AT and C+.GC triplets. Nucleic Acids Res. 1997;25:4644–4649. doi: 10.1093/nar/25.22.4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Rusling DA, Powers VEC, Lack O, Osborne SD, Fox KR, Brown T. Stable recognition of TA interruptions by triplex forming oligonucleotides containing a novel nucleoside. Biochemistry. 2005;44:5884–5892. doi: 10.1021/bi050013v. [DOI] [PubMed] [Google Scholar]
- 38.Cassidy SA, Strekowski L, Wilson WD, Fox KR. Effect of a triplex-binding ligand on parallel and antiparallel DNA triple helices using short unmodified and acridine-linked oligonucleotides. Biochemistry. 1994;33:15338–15347. doi: 10.1021/bi00255a015. [DOI] [PubMed] [Google Scholar]
- 39.Fox KR, Flashman E, Gowers D. Secondary binding sites for triplex-forming oligonucleotides containing bulges, loops, and mismatches in the third strand. Biochemistry. 2000;39:6714–6725. doi: 10.1021/bi992773+. [DOI] [PubMed] [Google Scholar]
- 40.Bath AJ, Milsom SE, Gormley NA, Halford SE. Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem. 2002;277:4024–4033. doi: 10.1074/jbc.M108441200. [DOI] [PubMed] [Google Scholar]
- 41.Tumpane J, Kumar R, Lundberg EP, Sandin P, Gale N, Nandhakumar IS, Albinsson B, Lincoln P, Wilhelmsson LM, Brown T, et al. Triplex addressability as a basis for functional DNA nanostructures. Nano Lett. 2007;7:3832–3839. doi: 10.1021/nl072512i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.