Abstract
Small nucleolar RNAs (snoRNAs) function mainly as guides for the post-transcriptional modification of ribosomal RNAs (rRNAs). In recent years, several studies have identified a wealth of small fragments (<35 nt) derived from snoRNAs (termed sdRNAs) that stably accumulate in the cell, some of which may regulate splicing or translation. A comparison of human small RNA deep sequencing data sets reveals that box C/D sdRNA accumulation patterns are conserved across multiple cell types although the ratio of the abundance of different sdRNAs from a given snoRNA varies. sdRNA profiles of many snoRNAs are specific and resemble the cleavage profiles of miRNAs. Many do not show characteristics of general RNA degradation, as seen for the accumulation of small fragments derived from snRNA or rRNA. While 53% of the sdRNAs contain an snoRNA box C motif and boxes D and D′ are also common in sdRNAs (54%), relatively few (12%) contain a full snoRNA guide region. One box C/D snoRNA, HBII-180C, was analysed in greater detail, revealing the presence of C′ box-containing sdRNAs complementary to several pre-messenger RNAs (pre-mRNAs) including FGFR3. Functional analyses demonstrated that this region of HBII-180C can influence the alternative splicing of FGFR3 pre-mRNA, supporting a role for some snoRNAs in the regulation of splicing.
INTRODUCTION
Small nucleolar RNAs (snoRNAs) are a class of conserved RNAs identified as guides for site-specific post-transcriptional modifications in ribosomal RNA (rRNA) (1–4). snoRNAs are ubiquitous throughout eukaryotes and have also been detected in a subset of archaea (5–6). Two main classes of snoRNAs have been characterized: the box C/D snoRNAs, most of which guide 2′-O-ribose methylation of their RNA targets and the box H/ACA snoRNAs that guide pseudouridine modifications.
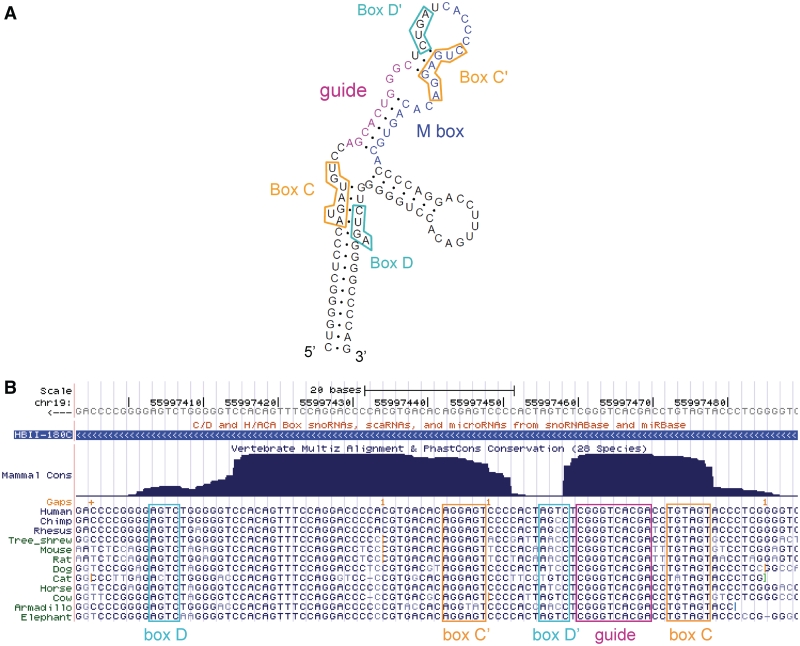
Human box C/D snoRNA molecules are typically 70–120 nt in length and mainly encoded in the introns of protein-coding genes. They can be excised from introns through at least two distinct pathways, then are further processed and bound by conserved proteins including the 2′-O-methyl transferase fibrillarin (4,7). Box C/D snoRNAs are characterized by the presence of two, short conserved motifs, the C box (UGAUGA) and the D box (CUGA), found near the 5′ and 3′-ends of the molecule, respectively (Figure 1A). Both boxes are required for snoRNA processing and localization (4). In the folded box C/D snoRNA molecule, boxes C and D come into close proximity and serve as a binding site for interacting proteins. A second pair of boxes, referred to as C′ and D′, can often be found closer to the middle of box C/D snoRNAs, but display lower conservation than the boxes C and D (2,8) (Figure 1B). The guide region that is complementary to the RNA target is located immediately 5′ to the box D or D′ regions; also called an antisense box, the guide sequence base pairs with the target forming an RNA–RNA duplex. The nucleotide targeted for methylation is usually base paired with the fifth residue upstream from the box D or D′ [reviewed in refs (2,8)]. Although targets have been found for many human box C/D snoRNAs, mainly in rRNA, numerous orphan snoRNAs have also been described, for which no target has been identified (4,9). Furthermore, recent reports suggest additional roles for snoRNAs and indicate that a subset of snoRNAs may be processed into smaller fragments.
Figure 1.
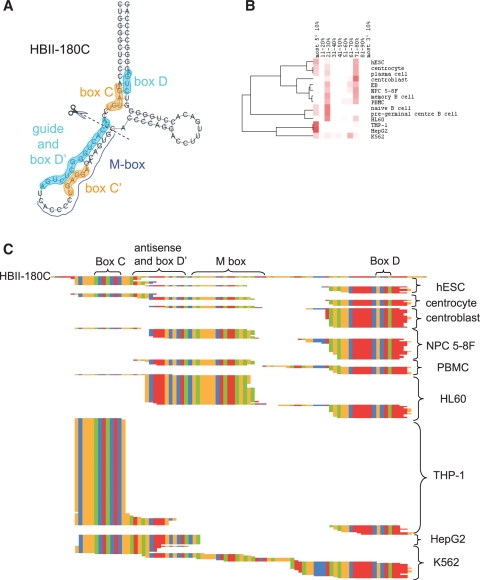
Predicted secondary structure and conservation of the human HBII-180 box C/D snoRNAs. (A) The characteristic features of the box C/D snoRNA HBII-180C are illustrated including conserved C and D motifs indicated by orange and cyan boxes, respectively. The guide region (complementary to 28 S rRNA) is displayed in pink and the position of the M-box region is shown in blue. (B) Screenshot of the UCSC Genome Browser (10) displaying the mammalian conservation and alignment of HBII-180C, encoded in chromosome 19 in an intron of gene C19orf48.
The largest family of orphan box C/D snoRNAs described to date in human is the HBII-52 family that consists of 47 members and is expressed from the SNURF–SNRPN locus (11,12). Most members of the HBII-52 family display complementarity to the serotonin receptor 2C transcript and cause changes in its splicing patterns in transfection experiments (13). More recently, the MBII-52 family, the mouse homologue of the HBII-52 family, was shown to be processed into smaller fragments, some of which regulate the alternative splicing of five distinct endogenous transcripts (11). In addition to the HBII-52 family involved in splicing regulation, several human box C/D snoRNAs were shown to play a role in translation regulation, another non-canonical function for an snoRNA. Originally described for a subset of human box H/ACA snoRNAs (14), several studies have now established a relationship between specific snoRNAs and microRNAs (miRNAs), in both human and Giardia lamblia. These include snoRNAs and miRNAs found to be colocalized in the genome, as well as snoRNAs with miRNA capabilities and miRNA precursors with snoRNA-like features (15–18).
miRNAs are short RNAs of ∼22 nt in length, processed out of longer hairpins, and typically involved in translation repression of mRNAs, generally mediated in animals by base pairing to the 3′-UTR of their targets (19). Consistent with reports of a relationship between miRNAs and snoRNAs, processed fragments derived from snoRNAs (sno-derived RNAs referred to as sdRNAs), with a similar size to mature miRNAs, have been detected in numerous small RNA sequencing data sets [for example refs (20–22)]. Analysis of human small RNAs detected from the acute monocytic leukemia cell line THP-1 and from frozen prefrontal cortex tissue showed stronger accumulation of small fragments from the 5′-end of box C/D snoRNAs than the 3′-end (22,23). A smaller scale study of processed box C/D snoRNAs showed accumulation of functionally tested miRNA-like fragments derived from both the 5′- and 3′-ends of the snoRNA (15). Consistent with a precursor–product relationship between snoRNAs and certain miRNAs, several reports have identified miRNAs accumulating in the nucleus and even specifically in the nucleolus (18,24–26).
Here, we explore the diversity and conservation of processing and accumulation of small RNAs <35 nt (referred to as sdRNAs) derived from box C/D snoRNAs by analysis of multiple deep-sequencing small RNA data sets. We also describe a box C/D snoRNA, HBII-180C, processed into sdRNAs and potentially involved in the regulation of splicing.
MATERIALS AND METHODS
Data sets
The 14 small RNA data sets considered are described in Table 1. For the NPC 5-8F data set, the nuclear and cytoplasmic sequence counts were combined and considered simultaneously. Small RNA sequences from these 14 data sets were mapped to known human box C/D snoRNA sequences downloaded from snoRNAbase (27). When mapping, we required perfect matching for the entire fragment but did not require the fragments to map uniquely to the human genome, because many box C/D snoRNA families include either identical, or near-identical copies.
Table 1.
Small RNA data sets considered
| Data set name | GEO series and platforms or data set link | Cell type details | References |
|---|---|---|---|
| hESC | Link from Genome Research website, see publication | H9 hESCs | (21) |
| EB | H9 human embryoid bodies | ||
| Centroblast | Derived from human tonsil | (28) | |
| Centrocyte | |||
| Naïve B cell | |||
| Plasma cell | |||
| Pre-germinal centre B cell | |||
| Memory B cell | |||
| THP-1 | Human monocytic leukaemia cell line | (22,29) | |
| NPC 5-8F | Human NPC 5-8F cells | (25) | |
| PBMC | Human normal PBMCs | (30) | |
| K562 | Human chronic myelogenous leukaemia cell line | ||
| HL60 | Human acute promyelocytic leukaemia cell line | ||
| HepG2 | Human liver carcinoma cell line | (31) |
Secondary structure, conservation, annotation and alignment of box C/D snoRNAs
RNA secondary structures were predicted by RNAstructure 4.6 (32) and annotated using RnaViz 2.0 (33). The mammalian conservation of HBII-180C was calculated and visualized using the Vertebrate Multiz Alignment and PhastCons Conservation utilities (34–36). The sequence and position of characteristic features of box C/D snoRNAs were obtained from snoRNABase (27) and sno/scaRNAbase (37). Alignment of sdRNAs and their corresponding snoRNA were visualized using Jalview (38).
Counts of fragments containing characteristic snoRNA features
To determine the proportion of sdRNAs-containing characteristic box C/D snoRNA features, the sequences of the boxes C, D and D′, as well as the guide regions, were manually obtained from the sno/scaRNAbase (37), when available. snoRNAs were only considered if they were represented by at least 10 sdRNA counts in at least 10 of the deep-sequencing data sets (Table 1). In total, 87 box C/D snoRNAs were analysed. The proportion of counts containing each of the characteristic features were calculated for each snoRNA and averaged over all snoRNAs considered.
Processing patterns of box C/D snoRNAs
To systematically investigate processing patterns of box C/D snoRNAs, all full-length snoRNAs were divided 5′–3′ into 10% sequence blocks as previously done (22), thus normalizing for varying length. For a given cell type, all sdRNAs detected were mapped to parental snoRNAs and counted in the 10% block from which the 5′-end of the sdRNA originates. The counts for each block were then normalized by the total number of sdRNA counts detected in the cell type examined, yielding a relative abundance value for each block. When an sdRNA mapped to more than one snoRNA, it was only considered once to avoid count duplication. sdRNAs typically map to more than one snoRNA from the same family, which display very similar lengths, thus a random assignment of an sdRNA to one of several parental snoRNAs of the same family is possible for this analysis. However, for the average processing analysis (Figure 2B), because every snoRNA profile is considered, sdRNAs were assigned to all snoRNAs to which they map. When comparing absolute counts of sdRNAs mapped to a specific full-length snoRNA (Figure 4), counts were normalized by counts per million reads mapped to the human genome for each data set. For a given data set, the number of reads mapped to the human genome (NCBI build 37) was determined using Bowtie (39) with the option ‘–n 0’.
Figure 2.
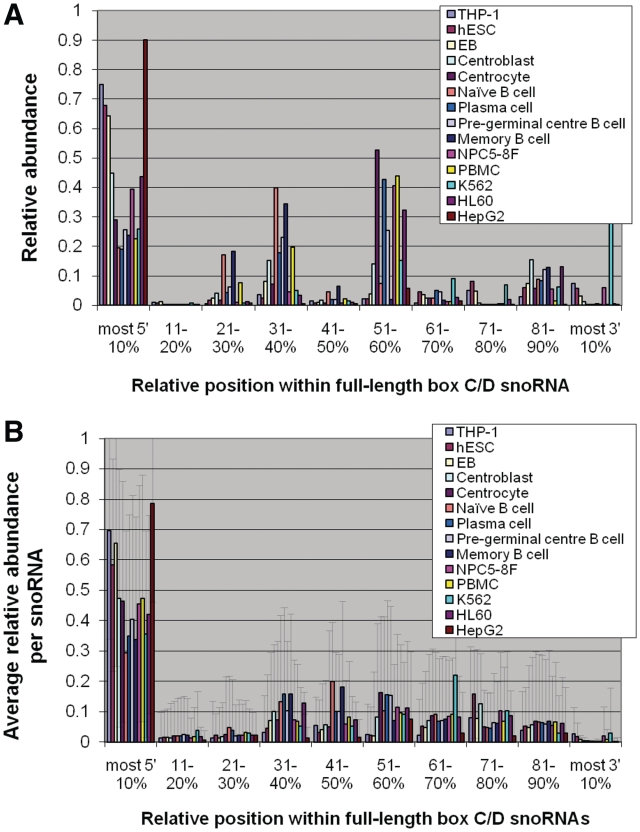
Provenance of sdRNAs from within full-length box C/D snoRNAs in 14 diverse human cell types. (A) Relative abundance of the 5′ position of sdRNAs within full-length box C/D snoRNAs. To normalize for non-uniform distribution of snoRNA length, the 5′-ends of all box C/D sdRNAs detected in a given cell type were mapped and counted in 10% blocks of their respective full-length snoRNAs. For each cell type investigated, the abundance of sdRNAs mapped to a specific block was normalized by the total number of counts of all sdRNAs detected in that cell type and this relative abundance was plotted as a function of relative position within the full-length snoRNAs, as described in the ‘Materials and Methods’ section. (B) To investigate average sdRNA abundance as a function of snoRNA position for all snoRNAs individually, the relative abundance per block was calculated as above for each snoRNA and averaged over all snoRNAs. The error bars represent standard deviation. Only snoRNAs with sdRNA counts of at least 10 were considered for this analysis.
Figure 4.
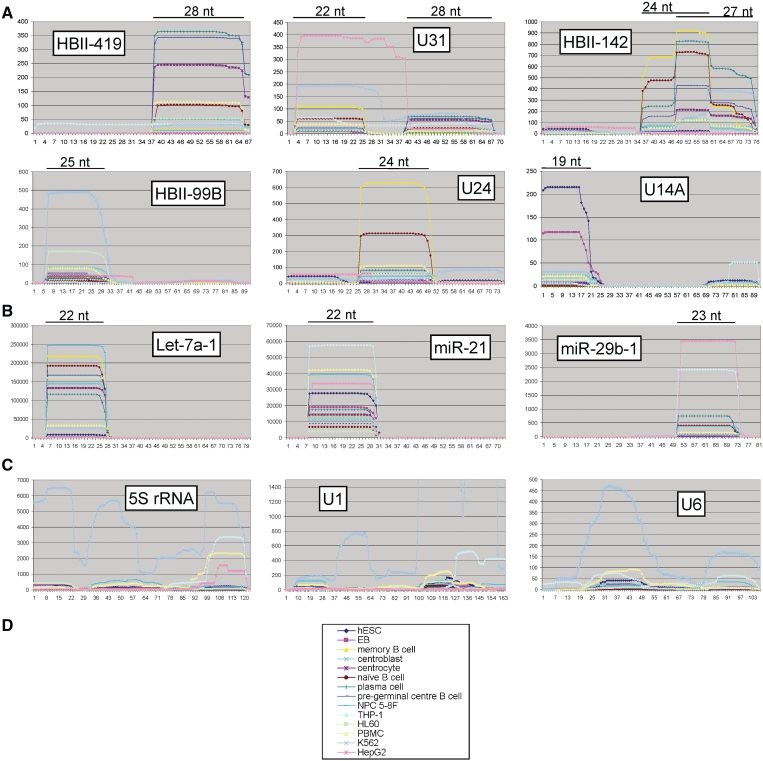
Accumulation profiles of diverse small RNAs. The accumulation profiles of subsets of (A) snoRNAs, (B) miRNAs and (C) rRNA as well as snRNAs were examined across a range of cell types (D). The x-axis on all graphs represents residue positions in the full-length (precursor) molecule. The y-axis represents the number of smaller fragments detected, that contain a specific residue in the full-length molecule, normalized by counts per million reads mapped. The miRNAs chosen (B) are those with the highest number of validated targets according to miRecords (40) that were detected in at least 10 of the 14 cell types examined. snRNAs were also chosen to ensure representation in at least 10 cell types. The lengths of the predominant fragments are shown in panels A and B above the graphs.
Cell culture and transfection
HeLa, WI-38 and HepG2 cells were maintained in Dulbecco's-modified Eagle's medium supplemented with 10% fetal bovine serum (FBS). THP-1, K562 and HL60 cells were maintained in RPMI 1640 with l-glutamine and 10% FBS. All plasmid transfections were performed using effectin (Invitrogen) as described by the supplier.
RNase protection assays
RNase protection assays were performed using the mirVana™ miRNA Detection Kit (Ambion). Full-length HBII-99B, U31, HBII-419, HBII-142, U14A and U24 snoRNAs were 32P labelled according to the manufacturer's protocol. Labelled probes were mixed with HepG2, THP-1, K562 or HL60 cell total RNAs, respectively, and RNase treatment was performed according to the manufacturer's protocol.
Detection of FGFR3 isoform
RNA was isolated by the TRIzol method with DNase I treatment, according to the manufacturer's instructions (Invitrogen). RT–PCR was performed to detect target RNAs. Reverse transcription and PCR were performed with the following gene-specific primers:
FGFR3: 5′-TGGACGTGCTGGAGCGCTCCCCGC-3′ and 5′-CCCAGGGTCAGCCGGGCCCGAGACAG-3′,
FGFR3Δ8–10: 5′-TGGACGTGCTGGAGCGCTCCCCGC-3′ and 5′-CCCAGGGTCAGCCGGGCCCGAGACAG-3′,
GAPDH: 5′-CGCATCTTCTTTTGCGTCGCCAG-3′ and 5′-GGTCAATGAAGGGGTCATTGATGGC-3′,
HBII-180C: 5′-CTCCCATGATGTCCAGCACT-3′ and 5′-CTCAGACCCCCAGGTGTCAA-3′,
U3: 5′-AGAGGTAGCGTTTTCTCCTGAGCG-3′ and 5′-ACCACTCAGACCGCGTTCTC-3′.
To decide the linearity of cycles, we performed real-time PCR using the Superscript III Platinum SYBR Green one-step qRT–PCR kit (Invitrogen) and Rotor-Gene RG-3000 system (Corbett Research). The same amount of RNA was used as templates for RT–PCR reactions. Each experiment was repeated three times independently. We also performed real time PCR using the QuantiFast SYBR Green RT–PCR kit (QIAGEN) and LightCycler 480 II (Roche). FGFR3 Δ8–10 signals were normalized by U3 signals as a loading control. Each experiment was repeated three times independently.
RESULTS
Relative position of stably accumulating sdRNAs
To characterize the processing of box C/D snoRNAs and investigate whether the distribution of stably accumulating snoRNA-derived fragments is cell type specific, we analysed the contents of 14 diverse publically available human small RNA deep-sequencing data sets (described in the ‘Materials and Methods’ section and in Table 1). To facilitate comparison of the different data sets, we considered only small RNAs mapping perfectly to box C/D snoRNAs and determined for each data set the relative abundance of such fragments as a function of their position in the full-length snoRNA. SnoRNAs were only considered if they had at least 10 counts in at least 10 of the different data sets and thus 87 of the 269 human box C/D snoRNAs were investigated for this analysis.
As shown in Figure 2A, while several data sets display a predominant accumulation of sdRNAs from the 5′-end of the full-length snoRNAs [as reported previously for the THP-1 cell line (22)], others show a stronger accumulation of sdRNAs from the middle of the full-length molecules. This analysis does not preclude that fragments derived from a small number of snoRNAs represent the bulk of the counts and the distribution shown might not be representative of the majority of snoRNAs. To investigate this possibility and gain a better understanding of the distribution of box C/D snoRNA processing, we considered the relative abundance and position of sdRNAs for each snoRNA and averaged over all box C/D snoRNAs.
As shown in Figure 2B, across all data sets examined, the processing patterns and accumulation of sdRNAs display significant differences between snoRNAs, as demonstrated by the large standard deviations. For 10 of the data sets considered, averaged over all snoRNAs, <50% of the small fragments are derived from the 5′-end of the snoRNA, while at least as many sdRNAs originate from the middle and 3′-end of the molecule. The variability of origin of the sdRNAs mapping to the 3′ side of the main hairpin likely finds its source in the diversity of structure of snoRNAs.
Processing patterns of box C/D snoRNAs in different cell types
We next sought to investigate whether snoRNA processing is conserved in different cell types, on a per snoRNA basis. To do so, the abundance versus position profiles of sdRNAs from the 14 data sets investigated above were compared for individual snoRNAs. For a given snoRNA, data sets were included only if counts of at least 10 sdRNAs were detected.
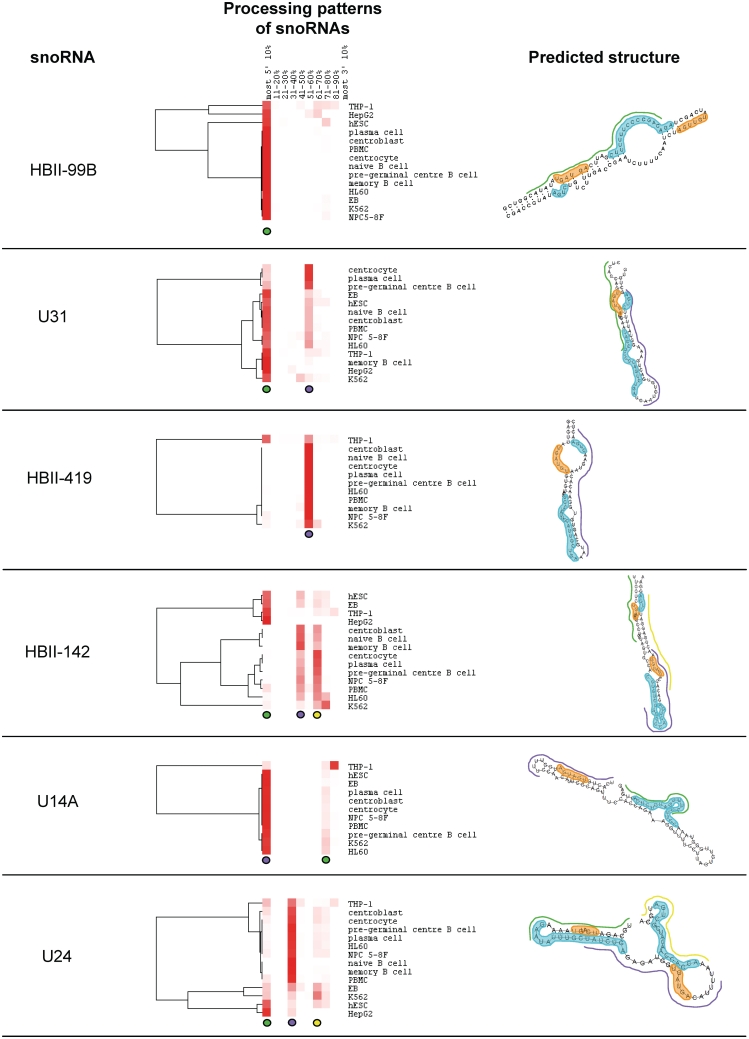
For most box C/D snoRNAs, sdRNAs originate from a small number of positions that are conserved between the different data sets, suggesting the processing pathways in use are common for the cell types considered (Figure 3). However, the relative abundance of the sdRNAs varies between the data sets and typically, while sdRNAs from a specific region on the 5′ side of the main hairpin show the highest abundance in some cell types, sdRNAs mapping to a specific region on the 3′ side of the main hairpin accumulate more strongly in other cell types. For example, for the box C/D snoRNA U31, H9 embroid bodies (EB), H9 human embryonic stem cells (hESC), naïve B cells, centroblasts, peripheral blood mononuclear cells (PBMC), nasopharyngeal carcinoma cells (NPC 5-8F), HL60, THP-1, memory B cells, HepG2 and K562 cells display a strong accumulation of sdRNAs from the 5′- end of the molecule (shown with the green line in the predicted structure). In contrast, centrocytes, plasma cells and pre-germinal B cells display a stronger accumulation from the 3′ side of the hairpin (beginning at positions 51–60% of the full-length molecule, the approximate position of this sdRNA within the full-length snoRNA is shown with a purple line in the predicted structure). These patterns indicate that while the snoRNAs undergo cleavage and processing in a conserved way in different cell types, the ratio of accumulation of specific fragments is cell type specific.
Figure 3.
Processing patterns of representative box C/D snoRNAs. The relative abundance of the 5′-ends of sdRNAs as a function of position in the full-length snoRNA was calculated as described for Figure 2 and in the ‘Materials and Methods’ section for each cell line and cell culture considered and then plotted as a heatmap. Red indicates a high proportion of sdRNAs whose 5′-end maps to a specific 10% block while white indicates an absence of sdRNAs mapping to a specific block. Predominant sdRNAs are typically represented by a red vertical trace indicating a high proportion of sdRNA 5′-ends map to this block in most cell types. Predominant sdRNAs are colour-coded using green, purple and yellow circles below the processing patterns section, which correspond to a line of matching colour in the predicted structure, indicating the approximate position of the sdRNA in the structure. Characteristic snoRNA features derived from refs (27,37) are annotated on the predicted structures including boxes C and C′ highlighted in orange and boxes D′, D and guide regions highlighted in cyan.
The predominant sdRNAs often contain a box C or C′ (for example the sdRNAs shown with green lines in HBII-99B, HBII-142, U24 and U31 and the sdRNAs shown with purple lines in HBII-142, U14A and U24 in Figure 3). Averaged over all snoRNAs that have counts of at least 10 in at least 10 of the data sets considered, 53% of sdRNAs contain the box C and 54% contain a box D′ or D. In contrast, regions of complementarity to rRNA are either absent, or only present at low frequency in the most abundant sdRNAs (seen for all snoRNAs examined in Figure 3 except HBII-99B) and on average, only 12% of sdRNAs contain complete-guide regions.
Conserved processing versus degradation
Next, we computationally addressed whether the sdRNAs are likely to result from general RNA degradation, rather than specific processing resulting in functional smaller molecules. To do this, sdRNA accumulation profiles of individual snoRNAs were analysed on a per residue basis and, as a control, compared with profiles of other abundant, structured small nuclear RNAs. sdRNA accumulation profiles are often highly conserved across the cell types examined and display well-defined sdRNAs with conserved start and end positions, both within and across most cell types (Figure 4A and Supplementary Figure S1). Several of the snoRNA profiles, in particular of HBII-419, U24 and HBII-99B (Figure 4A), resemble processing profiles of well-validated miRNAs (Figure 4B), consistent with directed cleavage. In contrast, processing profiles displayed by abundant nuclear RNAs transcribed by either RNA polymerase II or III, and not known to serve as precursors for smaller functional RNA molecules (rRNA and snRNA; Figure 4C) are poorly conserved between cell types and highly variable, with no strong accumulation of either identical, or highly overlapping, small RNA molecules. They instead have profiles more consistent with general cleavage and exonuclease digestion (for example the highest peak in the U6 plot, Figure 4C). In contrast, over all box C/D snoRNAs considered, approximately half display only one predominant and well-defined sdRNA type conserved in at least 10 data sets, as seen for miRNAs in Figure 4B.
We also examined whether the sdRNA fragments correlate specifically with high-GC content hairpin regions that may be generally more resistant to degradation. The average GC content of full-length box C/D snoRNAs with at least 10 counts in 10 of the data sets considered is 0.43 ± 0.06. The average GC content of their sdRNAs is also 0.43 ± 0.08. Therefore sdRNAs do not arise specifically from high-GC content regions. In summary, the data are not consistent with all sdRNAs arising though general RNA degradation.
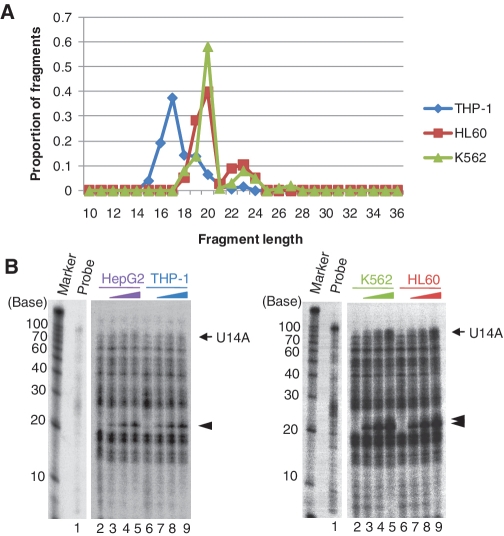
As the 14 sequencing data sets considered were generated in several different laboratories, we also sought to confirm our analyses of the processing of a subset of snoRNAs using an independent method. We thus carried out RNase A/T1 protection assays for 6 box C/D snoRNAs (HBII-419, U31, HBII-142, HBII-99B, U24 and U14A) for four of the commercially available cell lines considered in our deep sequencing analysis (HepG2, THP-1, K562 and HL60). The results show that the length of the sdRNA fragments identified by deep-sequencing (shown in Figure 5A and Supplementary Figures S2–S6) typically match closely with the sizes of the fragments protected in the RNase protection assays (shown by arrow heads of Figure 5B and Supplementary Figures S2–S6). In some cases, the results are more difficult to interpret because of the expression level of sdRNAs and also the presence of non-specific bands of the same size as the expected fragments. Overall, however, the RNase protection data support the results obtained above from analysis of RNA deep-sequencing data.
Figure 5.
Detection of endogenous U14A sdRNA fragments. The fragments processed from the box C/D snoRNA U14A as identified by deep sequencing were compared to endogenous U14A sdRNAs detected by RNase A/T1 protection assays. (A) Distribution of fragment lengths obtained by deep sequencing for U14A in the THP-1, HL60 and K562 cell lines. (B) Detection of endogenous sdRNA fragments by RNase A/T1 protection assay in the HepG2, THP-1, HL60 and K562 cell lines. As a control for the RNase protection assays, the diluted antisense probe against U14A was loaded without RNase digestion (probe lane). For each cell line, the probes were incubated with increasing amounts of total RNA (0, 1, 5, 10 µg, respectively, for lanes 2–5, and for lanes 6–9). Both the mature snoRNAs (arrow) and shorter fragments (arrow heads) were protected from RNase A/T1 digestion.
HBII-180C processing
The HBII-180s are a family of closely related human box C/D snoRNAs, which contain a region of complementarity to 28S rRNA immediately upstream from the box D′ that is common to all members of this family. Three human HBII-180 members (A–C) are encoded in separate introns of the same gene, C19orf48 (41). Though the exons of this gene display low conservation throughout mammals, HBII-180 snoRNAs are well conserved as illustrated in Figure 1B. In addition to the characteristic conserved boxes, HBII-180 members also contain a region of near perfect complementarity to endogenous pre-messenger RNA (pre-mRNA) sequences, termed the M-box (41). The M-box region is not highly conserved among HBII-180 members.
While snoRNAs HBII-180A and HBII-180B are expressed at low levels in all cell types examined, HBII-180C displays higher expression and accumulation of sdRNAs. Analysis of small RNA data sets demonstrates that three main sdRNA forms accumulate from HBII-180C, (Figure 6B and C). While some cell types display almost uniquely the 5′ sdRNA form (see THP-1 and HepG2 in Figure 6B and C), others show a strong accumulation of either the middle, or 3′ fragments. Similar to numerous other box C/D snoRNAs, including those shown in Figure 3, HBII-180C sdRNAs are derived from regions containing not only the boxes D′ and D but also from regions containing the boxes C and C′ (Figure 6C). A subset of fragments detected from HBII-180C contain either a full, or partial, M-box as reported previously (26), and this accumulation is cell type specific. We recently reported a detailed analysis of the M-box fragment, i.e. production of sdRNA from HBII-180C, including an RNase protection assay, expression of the HBII-180C M box fragment for both endogenous and overexpression of HBII-180C and analysis of the localization pattern of both the M box fragment and full-length HBII-180C (26). Careful examination of the sequences of the sdRNAs (Figure 6C) suggests a potential cleavage position for the processing of the full-length HBII-180C (drawn in Figure 6A).
Figure 6.
Processing pattern of HBII-180C. (A) The predicted structure of HBII-180C is shown with boxes C and C′ highlighted in orange and boxes D′, D and guide regions highlighted in cyan. (B) Processing patterns of HBII-180C, derived as described in Figure 3. (C) Sequence alignment of HBII-180C and all its sdRNAs detected in cell types considered. Nucleotides are colour-coded: A (green), C (orange), G (red) and T (blue).
HBII-180C targets and alternative splicing of FGFR3
Although potential functional relationships between snoRNA M-box sequences and the endogenous cellular RNAs to which they are complementary remain to be established, we have shown that it is possible to reduce expression of both the mRNA and protein levels of a targeted gene of interest by altering the snoRNA M-box region to make it complementary to the chosen gene (41).
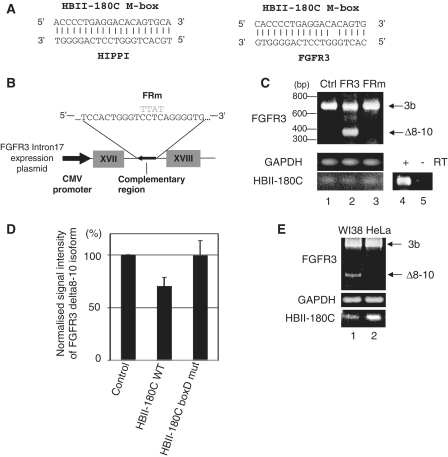
A scan of the genomic sequences of all human protein coding genes using the full-length HBII-180C snoRNA reveals that the top two reverse complementary hits correspond to intronic sequences in the genes HIPPI (refseq nucleotide accession number NM_018010) and FGFR3 (NM_000142) (41). These regions display either perfect, or near-perfect, complementarity to the M-box of HBII-180C, as shown in Figure 7A. As the expression of the gene HIPPI is generally low in most cell types, making experimental investigation difficult, we therefore concentrated on testing the possibility of a functional relationship between HBII-180C snoRNA and FGFR3 pre-mRNA.
Figure 7.
HBII-180C targets and the regulation of splicing of FGFR3. (A) The M-box region of HBII-180C is complementary to intronic regions in the HIPPI and FGFR3 genes. (B) Sequence and diagram of the antisense plasmid (wild-type and mutant), designed to suppress expression/activity of endogenous HBII-180C snoRNA. For the mutant construct the complementary region of FGFR3 was changed from CCTC to TTAT (grey, FRm). (C) The endogenous FGFR3 alternatively spliced mRNA isoform pattern expressed in HeLa cells was changed by overexpression of the FGFR3 mini gene described in B. The gel image shows the results of RT–PCR with transfected empty plasmid pcDNA3.1 (Ctrl, lane 1), wild-type antisense RNA (FR3, lane 2) mutant antisense RNA (FRm, lane 3), and reverse transcription control for HBII-180C (RT ± , lanes 4 and 5). FGFR3 RNA isoforms are indicated by arrows. Loading controls correspond to GAPDH mRNA and HBII-180C snoRNA, respectively. (D) Decrease of the expression level of endogenous FGFR3 Δ8–10 isoform by overexpression of HBII-180C in HeLa cells. The graph shows the signal intensity ratio of FGFR3 Δ8–10 isoform by qRT–PCR after overexpression of either wild-type (WT) HBII-180C or mutant HBII-180C (HBII-180C box D mut). (E) Comparison of the alternative splicing pattern of endogenous FGFR3 in WI38 and HeLa cells. WI38 cells express endogenous HBII-180C at significantly lower levels than HeLa cells and display accumulation of the Δ8–10 isoform.
First, we investigated whether altering expression of HBII-180C snoRNA can influence the expression of alternatively spliced FGFR3 isoforms using an antisense approach. Plasmid vectors encoding either a wild-type (FR3), or mutant (FRm), sequence that spans the region within FGFR3 intron 17 targeted by HBII-180C (described in Figure 7B), were transiently expressed in HeLa cells under the control of the cytomegalovirus (CMV) promoter. PCR analysis of FGFR3 mRNA expression showed that transient overexpression of the intron 17 fragment that is complementary to HBII-180C resulted in an increase in the levels of a spliced FGFR3 isoform called Δ8–10 (42) that is normally expressed at low levels in HeLa cells (Figure 7C, compare lanes 1 and 2). This change in the expression pattern of FGFR3 isoforms is not observed upon expression of either the empty vector alone, or of the vector expressing the same intron 17 sequence with a 4 nt change in the middle of the region complementary to HBII-180C (Figure 7C, lanes 1 and 3). In addition, the overexpression of wild-type HBII-180C reduced the expression level of the FGFR3 Δ8–10 isoform but the overexpression of the HBII-180C box D mutant did not (Figure 7D). We next investigated the expression levels of FGFR3 isoforms in WI38 (human lung primary cells) and HeLa cells to see if there is any correlation between FGFR3 splicing pattern and HBII-180C expression without transient transfection. As shown in Figure 7E, the presence of the smaller isoform of FGFR3 inversely correlates with the abundance of HBII-180C.
In summary, we conclude that the presence of HBII-180C snoRNA can affect the alternative splicing of FGFR3 pre-mRNA by decreasing the accumulation of the Δ8–10 isoform.
DISCUSSION
Full-length snoRNAs have been extensively investigated and their role in the site-specific, post-transcriptional modification of rRNA and other nuclear RNAs described. In the past 3 years, independent studies have identified the stable accumulation of short RNA fragments derived from snoRNAs. These studies range from experimental characterizations of individual snoRNAs [for example refs (11,14)] to large scale analyses of small RNA data sets [for example (20,22,23)], and the description of snoRNA-derived fragments resembling other known small RNAs, such as miRNAs (15,17,18,24,25). While originally regarded only as likely degradation products, the diversity of organisms in which snoRNA-derived fragments have been detected and the abundance of the fragments raise the possibility that they may play a functional role, at least for some snoRNA molecules. Indeed, there is experimental evidence that a small number of these sdRNAs may have a functional role in the regulation of either splicing, or translation (11,14,15,17). Here, we investigated the characteristics of sdRNAs in diverse human cell types and the potential effects of specific sdRNAs on expression of separate spliced isoforms.
Through detailed analysis of small RNA data sets derived from various human cell types we detected conservation of box C/D snoRNA-processing patterns (Figures 3, 4 and 6). This agrees with the results of recent experiments from other studies, suggesting that some highly conserved components from the RNA silencing processing machinery might be involved in the generation of sdRNAs (14,15,22). Interestingly, however, different species of sdRNAs from a given snoRNA display variable accumulation in a cell-specific manner (Figures 3, 4 and 6). While RNA data sets for each cell line and cell culture examined were represented by only one replicate, the snoRNA-processing and sdRNA-accumulation patterns were conserved between groups of related cell types, suggesting that the trends observed are representative. The binding partners of the sdRNAs (both target nucleic acid sequences and binding proteins), which are present in varying and cell-specific amounts, will likely influence the stability and the strength of the sdRNA accumulation. As more binding partners and targets are identified, it will become possible to investigate this hypothesis in a cell-specific manner.
Origin of sdRNA molecules
When small RNAs derived from snoRNAs were initially identified, they were widely dismissed as RNA degradation products. If this is correct, it predicts that sdRNA profiles will be similar to the degradation profiles of other abundant RNAs and different to miRNA profiles. We tested this and our analyses support the view that at least some sdRNAs arise via directed processing rather than degradation and also shows their accumulations are conserved across different cell types for a large number of snoRNAs. Simple RNA degradation profiles would be expected to display a higher degree of randomness, lack conservation and show a stronger accumulation of stable duplex-forming regions. All these characteristics are visible in the rRNA and snRNA-processing profiles examined. In contrast, many snoRNA-processing profiles resemble instead miRNA-processing profiles, showing a strong accumulation of one well-defined region of the full-length molecule (generally a portion of one side of the main hairpin), which is conserved across either most, or all, cell types examined. These snoRNA-processing profiles suggest sdRNAs arise from specific cleavage and protection from further processing, as is seen for miRNAs, rather than degradation. The conservation of processing patterns has recently been independently reported for a subset of box C/D snoRNAs (members of the HBII-52 and HBII-85 families) (11,43). Consistent with these analyses, a small number of sdRNAs derived from diverse snoRNAs have been recently shown to display functionality and affect gene expression (11,14,15,17,18,26).
A comparison of the read counts of sdRNAs to those of longer forms of snoRNAs (>50 nt) as available from ref. (44) also provides clues about the prominence and relative abundance of sdRNAs with respect to other snoRNA forms. Among the snoRNAs with high-sdRNA read counts (at least 10 read counts in at least 10 data sets), approximately half (52%) express moderate-to-high levels of longer forms including full-length snoRNA molecules. The remaining half (48%) express low or non-detectable levels of longer forms and are of particular interest as they represent genomic loci displaying low accumulation of full-length snoRNAs. These genomic loci thus appear to serve either predominantly, or uniquely, for the production of sdRNAs. Indeed, many sdRNAs encoded in these loci display accumulation profiles resembling those of miRNAs with stable and conserved accumulation of specific regions of the snoRNA, as seen in Figure 4.
Conversely, another group of snoRNAs display low levels of sdRNAs. Among these, approximately half originate from genomic loci that express strongly longer forms of snoRNAs (>50 nt), and might represent genomic regions serving mainly in the production of full-length snoRNAs, or long forms of processed snoRNAs such as those resulting from the HBII-52 locus as described in ref. (11). The remaining snoRNAs originate from genomic loci expressing low levels of all products (both long and short forms). Thus as described previously (22), the levels of full-length snoRNAs often do not correlate with the levels of their corresponding sdRNAs, likely reflecting variability in expression and processing regulation. While some genomic loci appear to serve principally in the production of full-length snoRNAs, others might mainly produce sdRNAs.
Similarly, the analysis of snoRNA processing provides clues as to the potential functional role of the different snoRNA fragments. While the functional specificity of classical full-length snoRNAs is conferred by the guide (antisense) region, a large majority of sdRNAs (88% averaged over all snoRNAs considered) do not contain the full-guide region from their parental snoRNA (Figures 3 and 6). In contrast, other characteristic features, in particular the box C, are highly represented in sdRNAs. This is in agreement with recent studies that identify box C in many sdRNAs (23), and in particular in several sdRNAs capable of gene silencing (15). Regions containing box D and in particular those not harboring a known guide sequence immediately upstream, are also represented in sdRNAs. Thus in general, snoRNA regions that carry out classical snoRNA guide functions can differ from those generating stably accumulating sdRNAs.
snoRNAs have been described as mobile genetic elements capable of copying themselves to other genomic locations (45,46), thus providing large numbers of potential sdRNA precursors. As a consequence, many families of snoRNAs include several identical and near identical copies. It is possible that this redundancy ensures a sufficient amount of full-length guide molecules for the targeted, site-specific post-transcriptional modification of rRNA and other such substrates, while also providing starting material for the generation of sdRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6.
FUNDING
Wellcome Trust (Programme Grant 073980/Z/03/Z to AIL and infrastructure grant WT083481); MRC Milstein Award (G0801738 to A.I.L.); Scottish Funding Council (Scottish Bioinformatics Research Network to G.J.B.). Funding for open access charge: Wellcome Trust (Programme Grant 073980/Z/03/Z to AIL and infrastructure grant WT083481).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our colleagues for helpful discussions and suggestions. A.I. Lamond is a Wellcome Trust Principal Research Fellow. M.S.S. is supported by a postdoctoral fellowship from the Caledonian Research Foundation. A.E. is supported by a fellowship from the Japanese Society for the Promotion of Science (JSPS).
REFERENCES
- 1.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 3.Weinstein LB, Steitz JA. Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 4.Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 5.Gaspin C, Cavaille J, Erauso G, Bachellerie JP. Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J. Mol. Biol. 2000;297:895–906. doi: 10.1006/jmbi.2000.3593. [DOI] [PubMed] [Google Scholar]
- 6.Omer AD, Lowe TM, Russell AG, Ebhardt H, Eddy SR, Dennis PP. Homologs of small nucleolar RNAs in Archaea. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- 7.Hirose T, Shu MD, Steitz JA. Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol. Cell. 2003;12:113–123. doi: 10.1016/s1097-2765(03)00267-3. [DOI] [PubMed] [Google Scholar]
- 8.Henras AK, Dez C, Henry Y. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr. Opin. Struct. Biol. 2004;14:335–343. doi: 10.1016/j.sbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Huttenhofer A, Kiefmann M, Meier-Ewert S, O'Brien J, Lehrach H, Bachellerie JP, Brosius J. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, Beach C, Nicholls RD, Zavolan M, Stamm S. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum. Mol. Genet. 2010;19:1153–1164. doi: 10.1093/hmg/ddp585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum. Mol. Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 13.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 14.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A Human snoRNA with MicroRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 15.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–686. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ono M, Scott MS, Yamada K, Avolio F, Barton GJ, Lamond AI. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res. 2011;39:3879–3891. doi: 10.1093/nar/gkq1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput. Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 20.Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langenberger D, Bermudez-Santana CI, Stadler PF, Hoffmann S. Identification and classification of small rnas in transcriptome sequence data. Pac. Symp. Biocomput. 2010:80–87. doi: 10.1142/9789814295291_0010. [DOI] [PubMed] [Google Scholar]
- 24.Politz JC, Hogan EM, Pederson T. MicroRNAs with a nucleolar location. RNA. 2009;15:1705–1715. doi: 10.1261/rna.1470409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, Chen YQ, Qu LH. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3' trailers. PLoS One. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ono M, Scott MS, Yamada K, Avolio F, Barton GJ, Lamond AI. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res. 2011;39:3879–3891. doi: 10.1093/nar/gkq1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 30.Vaz C, Ahmad HM, Sharma P, Gupta R, Kumar L, Kulshreshtha R, Bhattacharya A. Analysis of microRNA transcriptome by deep sequencing of small RNA libraries of peripheral blood. BMC Genomics. 2010;11:288. doi: 10.1186/1471-2164-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Affymetrix_ENCODE_Transcriptome_Project. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Rijk P, Wuyts J, De Wachter R. RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics. 2003;19:299–300. doi: 10.1093/bioinformatics/19.2.299. [DOI] [PubMed] [Google Scholar]
- 34.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl Acad. Sci. USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie J, Zhang M, Zhou T, Hua X, Tang L, Wu W. Sno/scaRNAbase: a curated database for small nucleolar RNAs and cajal body-specific RNAs. Nucleic Acids Res. 2007;35:D183–D187. doi: 10.1093/nar/gkl873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ono M, Yamada K, Avolio F, Scott MS, van Koningsbruggen S, Barton GJ, Lamond AI. Analysis of human small nucleolar RNAs (snoRNA) and the development of snoRNA modulator of gene expression vectors. Mol. Biol. Cell. 2010;21:1569–1584. doi: 10.1091/mbc.E10-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomlinson DC, L'Hote CG, Kennedy W, Pitt E, Knowles MA. Alternative splicing of fibroblast growth factor receptor 3 produces a secreted isoform that inhibits fibroblast growth factor-induced proliferation and is repressed in urothelial carcinoma cell lines. Cancer Res. 2005;65:10441–10449. doi: 10.1158/0008-5472.CAN-05-1718. [DOI] [PubMed] [Google Scholar]
- 43.Shen M, Eyras E, Wu J, Khanna A, Josiah S, Rederstorff M, Zhang MQ, Stamm S. Direct cloning of double-stranded RNAs from RNase protection analysis reveals processing patterns of C/D box snoRNAs and provides evidence for widespread antisense transcript expression. Nucleic Acids Res. 2011;39:9720–9730. doi: 10.1093/nar/gkr684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castle JC, Armour CD, Lower M, Haynor D, Biery M, Bouzek H, Chen R, Jackson S, Johnson JM, Rohl CA, et al. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS One. 2010;5:e11779. doi: 10.1371/journal.pone.0011779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weber MJ. Mammalian small nucleolar RNAs are mobile genetic elements. PLoS Genet. 2006;2:e205. doi: 10.1371/journal.pgen.0020205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo Y, Li S. Genome-wide analyses of retrogenes derived from the human box H/ACA snoRNAs. Nucleic Acids Res. 2007;35:559–571. doi: 10.1093/nar/gkl1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.