Abstract
Synthetic biology offers great promise to a variety of applications through the forward engineering of biological function. Most efforts in this field have focused on employing living cells, yet cell-free approaches offer simpler and more flexible contexts. Here, we evaluate cell-free regulatory systems based on T7 promoter-driven expression by characterizing variants of TetR and LacI repressible T7 promoters in a cell-free context and examining sequence elements that determine expression efficiency. Using the resulting constructs, we then explore different approaches for composing regulatory systems, leading to the implementation of inducible negative feedback in Escherichia coli extracts and in the minimal PURE system, which consists of purified proteins necessary for transcription and translation. Despite the fact that negative feedback motifs are common and essential to many natural and engineered systems, this simple building block has not previously been implemented in a cell-free context. As a final step, we then demonstrate that the feedback systems developed using our cell-free approach can be implemented in live E. coli as well, illustrating the potential for using cell-free expression to fast track the development of live cell systems in synthetic biology. Our quantitative cell-free component characterizations and demonstration of negative feedback embody important steps on the path to harnessing biological function in a bottom-up fashion.
INTRODUCTION
The field of synthetic biology, which aims to forward engineer biological functionality, has made rapid progress (1–4). A variety of synthetic systems have been implemented in living organisms, including oscillators (5,6), bistable switches (7), digital logic (8), counters (9) and even simple ecological systems (10,11). The ability to engineer biological function holds great promise for applications such as bioenergy production (12), drug synthesis (13), bioremediation (14) and biosensor development (15). However, for the most part, synthetic biology is still tied to the living cell. While live cells offer the remarkable capability of self-replication, a key disadvantage of using natural cells is the complexity of the cellular context. Interactions between synthetic components and the host cell's endogenous pathways pose a major challenge to engineering complex systems (1), and many of the cell's pathways are often extraneous to the desired application. For example, whole-cell biosensors are widely useful for environmental monitoring, but quite often only one pathway of the cell is actually needed to detect the chemical of interest (16–18).
In contrast, cell-free systems offer the potential of simpler and better defined contexts for engineering (19–21). In addition to simplicity, another key advantage is flexibility. A wider range of components, including novel amino acids, synthetic nucleotides and proteins toxic to living cells, can be incorporated and utilized in a cell-free context. As a result, cell-free systems facilitate bottom-up approaches to understanding and engineering biological networks and pathways. Initial demonstrations of synthetic cell-free systems include switches (22) and a predator–prey system (23) implemented using DNA and RNA interactions coupled with transcription. Systems that additionally incorporate translation have been made by cascading different polymerases and repressors (24,25), and a system for emulating embryonic pattern formation has been developed by interfacing different zinc-finger repressible T7 and SP6 promoters (26). Still, efforts in the area of developing well-characterized components for cell-free contexts and assembling these components to form regulatory networks lag behind similar efforts in live cells.
In this article, we focus on developing a toolkit for implementing cell-free regulatory systems as outlined in Figure 1. We first build and characterize genetic components. These components are assembled into desired systems, which are then characterized and optimized in an iterative fashion. While most synthetic biology efforts have focused on implementing systems in live cells, the process of engineering cell-free regulatory systems can not only advance applications of cell-free gene expression (27) but also serve as an aid for fast-tracking the deployment of live cell systems. Although live cell systems will require additional testing and optimization in the cell, an initial stage of cell-free system development offers many benefits. These include fast screening due to omission of a transformation step, the ease of direct quantification of devices by addition of purified regulatory proteins and an overall simplified context.
Figure 1.
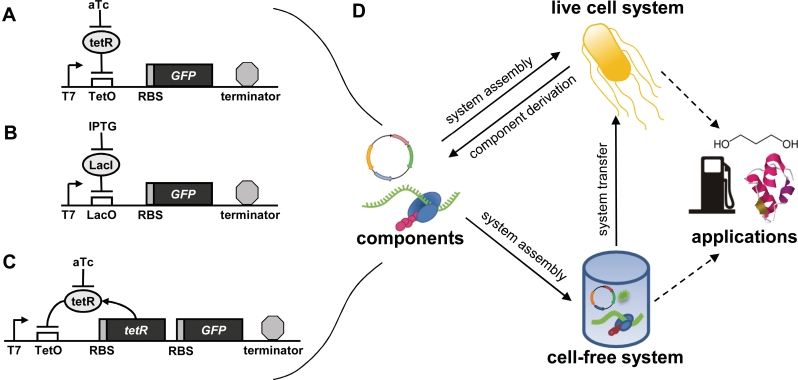
Genetic components characterized and role of cell-free systems in synthetic biology. (A) T7 Tet system. The repressor TetR binds the operator TetO and represses GFP expression. The inducer aTc may be added to relieve repression. (B) T7 Lac system. The repressor LacI binds the operator LacO and represses GFP expression. The inducer IPTG may be added to relieve repression. (C) Tet negative feedback system. A TetR repressible T7 promoter expresses TetR and GFP. A similar system can be assembled using a LacI repressible T7 promoter. (D) Most efforts in synthetic biology have focused on assembling biological components to form systems that are introduced into living cells for applications such as chemical synthesis, drug production, biosensors and energy production. This procedure of engineering biological systems can also be carried out in a cell-free context. Cell-free systems may be directly used in applications. Alternatively, initial deployment of synthetic systems in a cell-free context can help to fast track the development of live cell synthetic systems.
The task of engineering complex synthetic gene networks, either in vivo or in vitro, is facilitated by the availability of a library of well-characterized genetic parts suited to the desired context of operation (28,29). Inducible promoters are a useful starting point, as the ability to add different inducer concentrations provides a simple means of altering system parameters through non-genetic means (30–32). Consequently, we first build and characterize different T7 promoters for cell-free function. For cell-free applications, T7 promoters are commonly chosen due to their high processivity, their high specificity (33,34), and the simplicity of T7 polymerase (a monomer of approximately 100 kDa) (35). Previously, TetR repressible T7 promoters (T7tet) have been employed in a few systems for inducible expression in live protozoans (36–41) but have not been employed in bacterial- or cell-free systems. On the other hand, several studies have examined LacI repressible T7 promoters (42–45); however, like T7tet promoters, T7lac promoters have not been well characterized in cell-free expression contexts. For the Tet system (Figure 1A), we insert the TetR-binding domain, TetO, in different locations downstream of the T7 promoter, allowing repression of target genes such as GFP, which we use here to quantify expression. Similarly, for the Lac system (Figure 1B), we insert the LacI-binding domain LacO downstream of the T7 promoter allowing repression of GFP by LacI.
In addition to the availability of well-characterized genetic components, the assembly of functional gene networks requires a working knowledge of how to tune system parameters to achieve the desired behavior. This includes altering transcription and translation rates, degradation rates and the concentrations of activators/repressors required for response. Accordingly, using the expression systems described in Table 1, we explore the genetic determinants of cell-free expression efficiency, including the ribosome-binding sites, the plasmid backbone and transcriptional terminator sequences. We then examine different approaches for assembling regulatory networks and compare multi-cistronic systems to multi-plasmid systems.
Table 1.
Plasmids used for characterizing repression and for optimizing expression
| Plasmid | Promoter | RBS | Reporter | Backbone | Terminator |
|---|---|---|---|---|---|
| pT7 | T7 | ZE21 | GFPmut3.1(ASV) | pZE21-MSC-2 | T1 |
| pT7tet | T7tet13 | ZE21 | GFPmut3.1(ASV) | pZE21-MSC-2 | T1 |
| pT7tet2 | T7tet19 | ZE21 | GFPmut3.1(ASV) | pZE21-MSC-2 | T1 |
| pT7tet-RBSII | T7tet13 | RBSII | GFPmut3.1(ASV) | pZE21-MSC-2 | T1 |
| pT7tet-RBSg10 | T7tet13 | g10 | GFPmut3.1(ASV) | pZE21-MSC-2 | T1 |
| pT7tet-RBS35 | T7tet13 | BBa_B0035 | GFPmut3.1(ASV) | pZE21-MSC-2 | T1 |
| pT7tet-RBSA | T7tet13 | A | GFPmut3.1(ASV) | pZE21-MSC-2 | T1 |
| pUC-T7tet | T7tet13 | g10 | EGFP | pUC-19 | vsv |
| pUC-T7tet-T7term | T7tet13 | g10 | EGFP | pUC-19 | T7 |
| pT7tetKS | T7tet13 | g10 | EGFP | pBluescript-KS | T7 |
| pT7tetKS-SF | T7tet13 | g10 | EGFP | pBluescript-KS | T7 |
| pKSGFP | T7 | g10 | EGFP | pBluescript-KS | T7 |
| pLacOIDGFP | T7lacO1 | g10 | EGFP | pBluescript-KS | T7 |
| pLacOGFP | T7lacOID | g10 | EGFP | pBluescript-KS | T7 |
| pDEST17-EGFP | T7 | g10 | EGFP | pDEST-17 | T7 |
Finally, to demonstrate our procedure for the development of cell-free systems, we build aTc inducible negative feedback systems, whereby TetR represses its own production as shown in Figure 1C. Negative feedback is a common motif in natural genetic control systems, can speed response times (46), can minimize gene expression noise (47–49) and is critical to the function of many engineered systems such as genetic oscillators (5,6). These important aspects of negative feedback, coupled with the fact that even simple feedback systems have not been demonstrated in cell-free protein expression systems, make it a logical starting point towards the future construction of more complex cell-free networks. We demonstrate our negative feedback systems in cell extracts, in the PURE system and in live cells and compare functionality in each context.
MATERIALS AND METHODS
Plasmids and strains
All plasmids used in this study were constructed using standard methods. These plasmids are described in Supplementary Tables S1–S6. DNA used in cell-free experiments was prepared using Qiagen Plasmid Midi or Maxi prep kits. Escherichia coli strain BL21-AI (Invitrogen Inc., WI, USA) was used for protein purification and for live cell expression experiments. LB media with 100 µg/ml ampicillin was used to culture cells for protein purification, and cells were initially grown at 37°C shaking at 250 rpm. For live cell expression experiments, M9 media prepared as follows was used: M9 salts with Casamino acids (Amresco), 2 mM MgSO4, 0.5% glycerol, 300 µM thiamine and 100 µg/ml ampicillin. As later described, cells for these experiments were initially grown at 37°C shaking at 250 rpm.
Purification of TetR
Escherichia coli (Invitrogen Inc.) expressing TetR-His6 from pET-TetRHis were grown in 250 ml LB media with 100 µg/ml ampicillin at 37°C with shaking at 250 rpm. After reaching 0.4 OD600, the cells were induced using 0.2% l-arabinose (Sigma) and cultured for 2.5 h at 30°C. The induced cells were then pelleted and frozen. The cells were resuspended in 8 ml binding buffer (50 mM sodium phosphate buffer pH 8.0, 300 mM NaCl, 10 mM imidzole) and lysed using sonication. The lysate was centrifuged at 10 000 rpm in a Sorvall SS-34 rotor (12 000 g) for 15 min and supernatant was applied to a Ni-NTA column (Sigma) that had been equilibrated with the binding buffer. The column was subsequently washed with buffer (50 mM sodium phosphate buffer pH 8.0, 300 mM NaCl, 50 mM imidazole). TetR-His6 was then eluted with elution buffer (50 mM sodium phosphate buffer pH 8.0, 300 mM NaCl, 500 mM imidazole). The protein was concentrated and buffer exchanged into 20 mM sodium phosphate pH 7.2 and 50 mM NaCl by ultrafiltration (Vivaspin 500, GE healthcare). The purified protein was analyzed using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and protein was subsequently stored in −80°C till further use.
Cell-free expression experiments
The Promega S30 T7 High-Yield Expression System kit (Promega TM306) was used for the experiments depicted in Figures 2–4 and 5B and C, and the PURExpress In Vitro Protein Synthesis kit (New England Biolabs) was used for the experiment in Figure 5D. Reactions were set up following manufacturer's instructions (Supplementary Figure S3), except that the final reaction volume was 15 μL, and 15 μL mineral oil was added to prevent drying. For induction experiments, aTc (Acros Organics) was added in the denoted concentrations, and we verified that the fluorescence of aTc was negligible compared to GFP fluorescence from our constructs (Supplementary Figure S8). Reactions were set up in Corning CLS3820 plates. Samples were incubated at 30°C (Supplementary Figure S1) with shaking and measured every 6 min in a Biotek Synergy 2 plate reader. For the measurements, excitation was 485/20 nm, emission was 528/20 nm, the optics position was set at ‘Top 510,’ and the sensitivity was set at 40. For each experiment, samples consisting of reaction mix with no DNA were assayed to quantify background fluorescence of the reaction mix, and samples with pDEST17-EGFP (50) were assayed for the purpose of normalizing fluorescence values. ‘Normalized fluorescence’ (NFU) for a given sample was calculated by subtracting background fluorescence of the reaction mix from that sample's fluorescence value and then dividing by the background corrected fluorescence of the benchmark construct pDEST17-EGFP. Results depicting final yield (Figures 2–4 and 5B) are the normalized fluorescence values after 10 h of expression. For the dynamics shown in Figure 5C, fluorescence values at all time points were normalized using the fluorescence of pDEST17-EGFP after 10 h of expression. Error bars in all figures represent standard deviation of at least three replicates.
Figure 2.
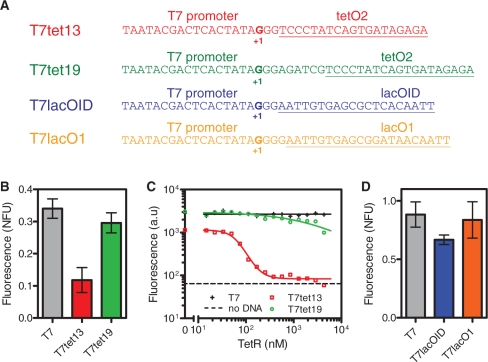
Promoter sequences and dosage responses. (A) Sequences of TetR repressible promoters T7tet13 and T7tet19 and LacI repressible promoters T7lacO1 and T7lacOID, (B) Fluorescence after 10 h of expression from the T7, T7tet13 and T7tet19 promoters using constructs pT7, pT7tet and pT7tet2. (C) Dosage responses of T7, T7tet13 and T7tet19 constructs to purified TetR. (D) Fluorescence after 10 h of expression from the T7, T7lacO1 and T7lacOID promoters using constructs pKSGFP, pT7lacO1 and pT7lacOID. Error bars in (B) and (D) depict standard deviation of triplicate measurements. Note that the T7 controls in (B) and (D) correspond to plasmids (pT7 and pKSGFP, respectively) with different backbones, ribosome binding sites, GFP variants and terminators as described in Table 1.
Figure 3.
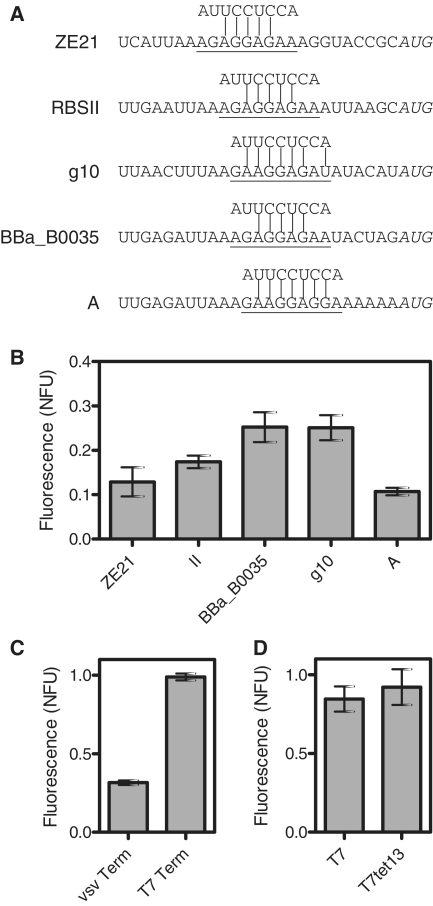
Optimization results. (A) Upstream sequences. Underlined base pairs are ribosome binding sites, and start codons are shown in italics. The 3′ end of the 16S rRNA sequence is shown above each ribosome binding site. (B) Effect of these different RBS sequences on EGFP production as measured by fluorescence after 10 h of expression. (C) Effect of transcriptional terminator on EGFP yield using plasmids pUCT7tet and pUCT7tet-T7term. pUCT7tet has the vsv terminator, while pUCT7tet-T7term has a T7 terminator. These plasmids are otherwise identical. (D) Comparison of T7 and T7tet13 promoter expression after incorporation of g10RBS and the T7 terminator into a pBluescript backbone. Error bars depict standard deviation of triplicate measurements.
Figure 4.
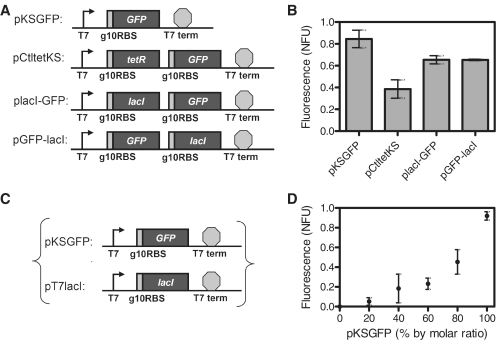
Plasmids and results for exploring different system composition approaches. (A) Constitutive T7 construct pKSGFP, and bicistronic constructs pCtltetKS, placI-GFP and pGFP-lacI. (B) Fluorescence after 10 h of EGFP expression from these constructs. (C) To investigate multi-plasmid systems, separate plasmids containing either GFP (pKSGFP) or lacI (pT7lacI) were constructed. (D) Results for co-expression of pKSGFP and pT7lacI for different percentages of pKSGFP by molar concentration. Error bars depict standard deviation of triplicate measurements.
Figure 5.
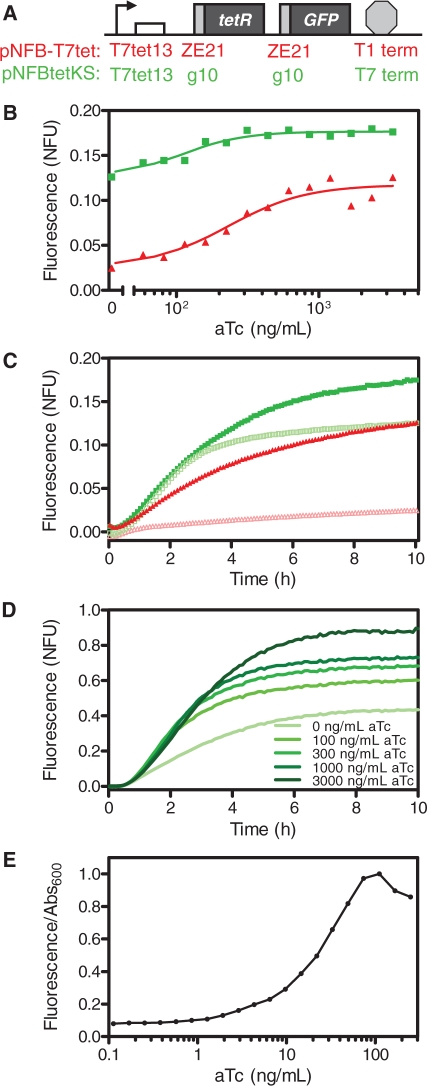
Negative feedback system and results. (A) Negative feedback constructs pNFB-T7tet and pNFBtetKS. pNFB-T7tet has the ZE21 RBS for tetR and GFP along with the T1 terminator, while pNFBtetKS has the g10 RBS for tetR and GFP along with the T7 terminator. (B) Responses of pNFB-T7tet (red triangles) and pNFBtetKS (green squares) to different aTc concentrations. (C) Response dynamics of pNFB-T7tet with no aTc (open red triangles), pNFB-T7tet with 3300 ng/ml aTc (closed red triangles), pNFBtetKS with no aTc (open green squares), pNFBtetKS with 3300 ng/ml aTc (closed green squares). (D) Response dynamics of pNFBtetKS to different aTc concentrations in the PURE system. (E) Responses of E. coli BL21-AI harboring pNFBtetKS to inducer aTc.
Live E. coli experiments
The negative feedback plasmid pNFBtetKS was transformed into chemically competent E. coli. A single colony from the transformation plate was inoculated into 2 ml LB with 100 µg/ml ampicillin and grown at 37°C with shaking at 250 rpm. This starter culture was then diluted 1:100 into 2 ml M9 media prepared as follows: M9 salts with Casamino acids (Amresco), 2 mM MgSO4, 0.5% glycerol, 300 µM thiamine and 100 µg/ml ampicillin. This culture was incubated at 37°C with shaking at 250 rpm until log phase was reached. Then, the culture was diluted to an OD of ∼0.01, and 10 mM l-arabinose was added to induce the expression of T7 polymerase. Aliquots (200 µl) were dispensed into the wells of a 96-well plate (Corning 3370). The inducer aTc (Agros Scientific) was added as indicated in Figure 5E. About 50 µl of mineral oil was added to prevent evaporation. Samples were incubated at 37°C with shaking in a Biotek Synergy 2 plate reader, and both fluorescence and absorbance at a 600 nm wavelength were measured. For the fluorescence measurements, excitation was 485/20 nm, emission was 528/20 nm, the optics position was set at ‘Top 510’ and the sensitivity was set at 60. The measurements reported in Figure 5E were taken after 4 h of growth. Fluorescence values were corrected for background fluorescence, and absorbance readings at 600 nm were used to normalize for cell density.
RESULTS
Control elements
A key issue associated with developing tightly repressible T7 promoters for operation in cell-free contexts is operator placement, and we characterized variants of different Tet and Lac promoters. First, we characterized two different TetR repressible T7 promoters shown in Figure 2A. The T7tet13 promoter has the TetO operator centered at +13, and the T7tet19 promoter has TetO centered at +19. We first compared the expression of these two promoters using the plasmids pT7tet for the T7tet13 promoter and pT7tet2 for the T7tet19 promoter. For comparison, we also quantified expression from the constitutive T7 promoter using plasmid pT7. These three plasmids differ only in the promoter regions (Table 1). As shown in Figure 2B, fluorescence resulting from expression of the T7tet13 construct (pT7tet) after 10 h of incubation at 30°C was ∼3-fold lower than that from the T7tet19 construct (pT7tet2). This is likely due to a reduced transcriptional efficiency of the T7tet13 promoter, as later discussed. Fluorescence from the T7tet19 construct was similar to that of the constitutive T7 promoter construct (pT7).
Figure 2C shows the dosage responses of the T7tet13 and T7tet19 constructs to purified TetR. Though its maximal response is weaker, the T7tet13 promoter exhibits tight repression, as exhibited by the fact that the fluorescence at high TetR concentrations is indistinguishable from the background fluorescence of the cell extract. In addition, expression is cooperative, since the average Hill coefficient determined from fits of three TetR dosage experiments is 2.2 with a standard deviation of 0.68. The concentration of TetR required for half maximal repression is 73 nM, with a standard deviation of 24 nM. In contrast, the T7tet19 promoter does not exhibit tight repression for the range of TetR concentrations explored, and higher concentrations of TetR (3000 nM) are required for half-maximal repression.
Although the effects of operator placement for lac repressible T7 promoters have been characterized in vitro (42–44), we examined two different lac repressible promoters, T7lacOID and T7lacO1, for completeness. The operator of the T7lacOID promoter is centered 13.5 bp downstream of the transcription start site, and the operator sequence begins at +4 bp, just as with the T7tet13 promoter. The operator of the T7lacO1 promoter is centered 15 bp downstream of the transcription start site. We compared the expression of the T7lacOID, T7lacO1 and T7 promoters using the plasmids pLacOIDGFP, pLacOGFP and pKSGFP, respectively. These three plasmids differ only in the promoter regions (Table 1). As observed with the T7tet promoters, closer placement of the operator to the transcription start site resulted in a decreased yield (Figure 2D). Note that the yield from the T7 expression construct in Figure 2B (pT7) differs considerably from the yield of the T7 expression construct in Figure 2D (pKSGFP). Given these differences, we next examined the determinants of expression efficiency.
Optimization of cell-free protein expression
To develop a deeper understanding of how to tune expression efficiency, we quantified the effect of different ribosome binding sites (RBSs), plasmid backbones and terminators on cell-free protein expression. We characterized five purportedly efficient RBSs: the ZE21 RBS (51), RBSII (52), the Biobrick BBa_B0035, g10 RBS from T7 phage (53) and the E. coli consensus RBS A (7). Upstream sequences containing these RBS's are shown in Figure 3A, and the RBS regions, as determined by minimum energy alignment to the last 9 bp of E. coli 16S rRNA (54), are underlined. As shown in Figure 3B, the strongest RBS's are BBa_B0035 and g10 RBS from T7 phage.
Next, we examined the influence of the plasmid backbone on gene expression. The backbone could potentially affect transcription by altering the degree of supercoiling, the amount of non-coding DNA and the efficiency of transcriptional termination. We constructed a minimal backbone based on a reduced pUC-19 vector with the vsv terminator inserted (55). We then inserted the T7tet13 promoter, the strong g10 RBS and EGFP (Supplementary Figure S2). Expression was weaker than expected, as shown in Figure 3C. We hypothesized that this was due to inefficient transcriptional termination. Indeed, exchanging the vsv terminator in pUC-T7tet for a T7 terminator (pUCT7tet-T7term in Figure 3C) improved yield to match that of the benchmark construct.
For comparison purposes, we then inserted each of the sequences T7-g10 RBS-EGFP-T7terminator and T7tet13-g10 RBS-EGFP-T7terminator into a pBluescript backbone to create the plasmids pKSGFP and pT7tetKS (Table 1). These two constructs exhibited similar yields, which were only slightly lower than those realized with the minimal backbone.
System composition
As a step towards creating a negative feedback system and ultimately larger and more complex synthetic systems, we explored the effect of different system composition approaches on expression efficiency. One system composition approach, which is commonly employed in living cells, involves the use of multi-cistronic sequences for co-regulating subsets of genes in the system. As a simple investigation into expression efficiency in multi-cistronic sequences, we constructed three bicistronic sequences. The first, pCtltetKS, consists of tetR inserted into the control construct pKSGFP upstream of GFP (Figure 4A). The second, placI-GFP, consists of lacI inserted upstream of GFP, and the third, pGFP-lacI, consists of lacI inserted downstream of GFP. As expected, fluorescence measurements for all of the bicistronic sequences were significantly lower than for the pKSGFP control. Fluorescence of the tetR-GFP construct pCtltetKS was slightly less than half that of the pKSGFP control (Figure 4B), while fluorescence of the lacI bicistronic constructs was ∼25% lower than pKSGFP. No significant difference was observed between the insertions of lacI upstream versus downstream of GFP.
An alternative to multi-cistronic sequences is to use a separate plasmid for each gene in the system. As later discussed, this approach is particularly amenable to cell-free systems. However, it is important to understand the effect of altering the concentrations of the different plasmids on expression. To this end, we combined a plasmid expressing only GFP (pKSGFP) and a plasmid expressing only LacI (pT7lacI) in different molar ratios, while keeping the sum of the molar concentrations fixed at 8 nM. The resulting GFP fluorescence exhibited a non-linear increase as a function of the percentage of the GFP plasmid. Specifically, the effect of increasing the percentage of pKSGFP grew more pronounced at higher pKSGFP percentages.
To compare GFP expression for the two different system composition approaches, we expressed the bicistronic constructs pLacI-GFP and pGFP-LacI at the concentrations shown in Supplementary Figure S4, and we also co-expressed pKSGFP and pLacI such that the molar concentrations of each plasmid were also as shown in the figure. Thus each concentration on the x-axis corresponds to the same number of copies of the EGFP and lacI genes for each approach. Lower EGFP expression, as measured by fluorescence after 10 h of expression, was realized with the two-plasmid approach.
Negative feedback
We compared two different T7-driven TetR negative feedback systems. In these systems, shown in Figure 1C, a TetR repressible promoter expresses TetR and GFP. The inducer aTc can be added to bind and inactivate TetR repression. In the feedback construct pNFB-T7tet (Figure 5A), the T7tet13 promoter drives expression of tetR and GFP, each of which has the ZE21 RBS. The T1 terminator was used in this construct. The other feedback construct pNFBtetKS also drives expression from the T7tet13 promoter but utilizes the strong g10 RBS for TetR and EGFP along with the T7 terminator.
With the pNFB-T7tet feedback construct, an over 5-fold difference between uninduced and induced expression was observed (Figure 5B), although the maximally induced normalized fluorescence was ∼0.1. Incorporating the g10 RBS and T7 terminator to form pNFBtetKS resulted in a significantly higher fully induced yield, showing the benefit of using the optimal RBS and terminator. However, fluorescence in the uninduced state increased as well. Dynamics are shown in Figure 5C for the uninduced and the maximally induced cases. The pNFB-T7tet construct exhibits slower kinetics in the induced case than pNFBtetKS does with or without induction. For the pNFBtetKS construct, fluorescence initially rises rapidly regardless of the inducer concentration. However, without the aTc inducer, the rate of fluorescence increase eventually decreases more rapidly than with the inducer after ∼2 h.
We also characterized pNFBtetKS in the minimal PURE system (Figure 5D). Fluorescence values here were normalized to pDEST17-EGFP expressed in the PURE system. An over 2-fold difference was observed between uninduced and induced conditions. In addition, for comparison purposes (Figure 5E), we also demonstrated the in vivo function of pNFBtetKS in E. coli BL21-AI cells. As expected, a much wider range of expression was realized.
DISCUSSION
While the field of synthetic biology has enjoyed rapid growth in recent years, the complexity of regulatory networks implemented has plateaued (3). Efforts to engineer complex systems would greatly benefit from both simpler contexts and new bottom-up approaches. Towards this end, our results serve as groundwork for the process of cell-free system development shown in Figure 1D.
The first step in this process consists of characterizing basic genetic components such as the different repressible T7 promoters, RBSs and terminators characterized here in cell extract. Next, these components are assembled into regulatory systems, and multi-cistronic and multi-plasmid approaches are compared in Figure 4 and Supplementary Figure S4. Finally, we demonstrate inducible, negative feedback in cell extracts, in the PURE system and in live E. coli. More complex systems will likely require iterative rounds of system characterization and optimization. However, inducible feedback systems offer an important step. Even simple feedback constructs have not been previously demonstrated in cell-free protein expression systems, despite their functional importance and widespread utilization in live cell gene networks. Also, whereas addition of T7 lysozyme has been used to tune T7 transcription in the past (56), our system is autoregulatory and will be useful in applications such as cell-free biosensors (57).
Beyond offering a simplified context, several features of cell-free synthetic biology are appealing for direct applications and also for the initial prototyping of both genetic devices and assembled systems in live cells. Our characterization of the repressible T7 promoter variants in Figure 2 highlights one of the ultimate benefits of forward engineering biological functionality in cell-free contexts. Specifically, direct quantitative characterization of transcriptional regulation is greatly facilitated. In contrast, in live cells, such dosage responses must be inferred indirectly through careful analysis of single cell responses with consideration of the significant amounts of noise in gene expression (58). Another advantage of cell-free systems is that they enable fast screening of construct libraries, as transformation is not required. Also, as shown in Figure 4, multi-plasmid systems may be implemented without regard to backbone compatibility, and DNA concentration is easily tunable, unlike in cells. Finally, testing regulatory systems in a context free of mutation and recombination can simplify initial system development and can aid troubleshooting efforts in live cells systems.
We first constructed and characterized different repressible T7 promoters with the aim of achieving tight regulation. Figure 2 reveals that a key issue in achieving tight repression is operator placement. With E. coli promoters, repressors placed close to the promoter region can interfere with RNA polymerase binding and transcription initiation, and operators placed hundreds of base pairs downstream of the transcription start site have been shown to terminate transcription (59,60). However, T7 promoters exhibit high processivity compared to E. coli promoters and, as a result, often require different strategies for achieving efficient repression. For example, several studies have examined LacI repression of T7 promoters by insertion of LacI operators in different locations with respect to the promoter (42–44). These studies have demonstrated the requirement for much closer placement of the operator to the transcriptional start site of the T7 promoter than is necessary when using E. coli promoters (42–44).
With the T7tet13 promoter, strong repression is achieved at the cost of a reduced rate of transcription in the absence of a repressor (Figure 2B and C). In particular, we note that the sequences of natural T7 promoters are highly conserved between base pairs −17 and +6 relative to the transcriptional start site (61), and the T7tet13 promoter does not preserve base pairs +4 through +6, while the T7tet19 promoter does (Figure 2A). Interestingly, previous studies characterizing different TetR repressible T7 promoters in protozoans observed either leaky repression or a reduction of fully induced expression (38–40). These findings, as well as the results in Figure 2, portray a tradeoff between expression efficiency and controllability.
In addition to tight repression with the T7tet13 promoter, high sensitivity of repression is also observed in cell extract, as indicated by the Hill coefficient of approximately two. Sensitivity is important for the engineering of a wide variety of systems, including oscillators (5,6), bistable switches (7,62) and digital logic cascades (63,64). Ultimately, development of repressible T7 promoters by optimized placement of binding domains downstream of the T7 transcription initiation site could be extended to other DNA-binding proteins to construct a diverse library of repressible T7 promoters.
Assembling functional complex gene networks using components such as these inducible T7 promoters requires an understanding of how to tune network parameters. An appealing characteristic of using T7 transcription is that promoter mutants with a broad range of transcriptional efficiencies have been characterized, and polymerase mutants with different processivities have also been characterized (65,66). To optimize transcriptional efficiency, we examined the potential importance of the plasmid backbone and transcriptional termination. Initially, we created a minimal backbone and explored the use of the vsv terminator in this backbone (55). Previous results in E. coli extract showed a slightly decreased protein yield when the vsv terminator was used instead of the T7 terminator (55). Our results in Figure 3C show a more significant (3-fold) decrease in yield with the vsv terminator. This decrease in yield is partially explained by a decrease in mRNA stability associated with the vsv terminator (Supplementary Figure S6c), and also by gene-dependent termination efficiency (55). These results highlight the importance of efficient transcriptional termination. Further research into the specific requirements for achieving efficient termination of T7 transcription with the vsv or other terminators would benefit efforts to engineer complex, multi-gene and multi-promoter systems (55).
Besides the ability to tune transcription rates, it is also important to control translation rates. Thus, we aimed to find the most efficient RBS, as measured by GFP yield. Out of the five candidates that we characterized in cell extract (Figure 3A), the g10 RBS and BBa_B0035 were the strongest (Figure 3B). These RBS's combined with mutants that knock down efficiency (7,64) can be used to achieve a wide range of translation rates. It should be noted that for some genes, problematic RNA structures can form that obscure the RBS and inhibit translation. The fact that two different sequences, g10 RBS and BBa_B0035, result in efficient translation can be helpful for achieving strong expression when one of these normally strong RBS's does not perform well (67,68).
When an optimal RBS and the T7 terminator are used in conjunction with the T7tet13 promoter, little or no decrease in yield is observed in comparison to constructs with the constitutive wild-type T7 promoters. Specifically, pUC-T7tet-T7term (Figure 3C) and pT7tetKS (Figure 3D) have normalized fluorescence yields of close to 1. This indicates that their yields are essentially the same as the benchmark construct. These high yields despite potentially lowered transcription rates are likely explained by saturation of translational machinery. Such saturation of translational machinery has been previously observed in T7 expression systems both in vivo (33) and in cell-free expression systems (25).
Having characterized repressible T7 promoters as well as investigated the genetic determinants of cell-free expression efficiency, we sought to examine different approaches for assembling regulatory networks in cell-free systems. When implementing large gene networks in live E. coli, one option is to integrate the genetic components into the chromosome. However, with this approach, it is cumbersome to explore a large number of combinations of different network variants. Thus, most live E. coli systems in synthetic biology have relied on the use of plasmids, but at most two or three different plasmid types can be used. On the other hand, with cell-free systems, the same backbone can be used for different system components. This enables an approach for constructing large synthetic gene networks, whereby each component is encoded on a separate plasmid, and different plasmids are combined in cell-free extract to form the final system. The DNA copy number of each network component can be easily and precisely tuned, whereas in live cells, copy number can only be coarsely tuned for each plasmid by using different origins of replication. Thus, with this multi-plasmid approach, a large number of network variants can be quantified without the need for chromosomal integrations or transformations.
As expected, when either tetR or lacI was inserted in a bicistronic sequence with GFP, fluorescence decreased due to sharing of expression capacity between the repressor and GFP (Figure 4B). Insertion of tetR reduced expression by approximately half, while insertion of lacI reduced expression by approximately a quarter. This difference in the effects of inserting tetR vs. lacI implies that inserting a gene in a bicistronic sequence impacts relative expression in a manner that is dependent on the particular gene inserted. Trading the order of GFP and lacI had no impact on fluorescence.
By comparison to the multi-cistronic approach, when each gene is expressed on a separate plasmid, normalized fluorescence is reduced by half even when the EGFP expressing plasmid comprises 80% of the total plasmid concentration (Figure 4D). In general, one tradeoff with this approach is that the flexibility of easily tuning relative gene copy numbers can potentially come at the cost of weaker expression, as shown in Supplementary Figure S4. For example, due to the higher ratio of promoters to genes, the effects of inefficiency in transcriptional termination may be more pronounced. Nonetheless, it has previously been shown that, with the multiple plasmid approach, properly tuning the ratio of plasmids, along with the use of common downstream box sequences, can help to achieve efficient expression of all genes in the system (69). Interestingly, we observed a non-linear relationship between relative plasmid ratio and expression (Figure 4D). This is potentially due to a competition between the constructs for translational resources (69). Future experiments to quantify the yield of both proteins will help to elucidate further the cause of this non-linearity.
To demonstrate simple cell-free gene networks, we implemented different TetR-negative feedback systems. In these systems (Figure 5A), TetR repressible promoters express TetR and GFP, and the inducer aTc can be added to relieve repression. Our initial T7-driven feedback system, pNFB-T7tet, exhibited an over 5-fold response to aTc, but the normalized fluorescence was ∼0.1 when maximally induced. When the strong g10 RBS was used for EGFP and TetR in conjunction with a T7 terminator in pNFBtetKS, significantly higher fluorescent levels were observed under full induction. However, yield was also much higher in the uninduced case. This is due to overshoot, whereby a burst of transcripts is initially produced before enough functional TetR dimers have been formed to repress the T7tet13 promoter. These initially produced transcripts decay slowly in the cell extract (Supplementary Figures S5 and S6c), and since EGFP also has a long half-life, fluorescence even after 10 h appears high when no initial inducer is added. The wider range of expression realized with pNFBtetKS in live cells in Figure 5E supports this hypothesis. The turnover rates of mRNA and proteins are higher in live cells due to active degradation of mRNA and dilution via growth. In addition, from the start of the experiment, the cells have TetR, unlike the cell-free experiments. This prevents the previously described initial burst of transcripts that occurred in the cell-free reaction.
All cell-free reactions were performed in batch mode, which prevents the influx of nutrients and the efflux of waste products and consequently limits the reaction dynamics (70–75). Extension to a continuous flow system would help to harness the advantages of repressible T7 promoter variants (21,76). As exemplified by the dynamics shown in Figure 5C, the T7 promoters exhibit strong expression in the first few hours, followed by a decrease in the rate of expression. The flow of fresh nutrients and removal of waste products would help to preserve the initially strong rate of expression. The additional incorporation of mechanisms to actively degrade mRNA and EGFP (77) would mitigate the previously described overshoot problem with pNFBtetKS and would reduce the high yield in the absence of inducer.
Accordingly, when implemented in a growing population of live E. coli cells, this optimized negative feedback system does in fact exhibit a greater range of fluorescence in non-induced vs. fully induced conditions (Figure 5E). In general, the tighter repression of the improved repressible T7 promoters is likely also useful in live cell applications which require stringent control of target gene expression to prevent toxic effects or inclusion bodies. In addition, our development of a regulatory system using cell-free expression followed by successful demonstration in live cells exemplifies the future applicability of using cell-free characterization to fast track the deployment of live cell systems.
Our development and use of repressible T7 constructs facilitated characterization in the minimal PURE system (78). We demonstrated feedback with pNFBtetKS in the PURE system in Figure 5D. Performance in the PURE system clearly differs from cell extracts, although the system works in both contexts. The response to aTc in the PURE system was over 2-fold, yielding a greater dynamic range than in cell extracts. A lower yield of EGFP was realized for the control construct pDEST17-EGFP in the PURE system than in the cell extracts (not shown due to normalization). However, in the PURE system, unlike in cell extracts, fluorescence of the fully induced feedback construct was close to that of the control.
Alternative approaches to engineering regulation in cell-free contexts avoid the use of translational machinery, thus further simplifying the engineering of fast, complex systems (79,80). At the same time, protein expression is clearly useful for a number of applications such as production of protein-based therapeutics and chemical sensors. Ultimately, the expression components and simple feedback systems that we present can be interfaced to more complex regulatory networks based on a simplified set of mechanisms (81).
Our results contribute to bottom-up approaches to engineering biological function (82). The simplified context and the facilitation of direct, quantitative component characterization offered by cell-free systems will aid efforts to transcend the complexity of systems currently engineered in living cells (3). Our characterization of the tightly and cooperatively repressible T7tet13 promoter in Figure 2C exemplifies the ability to quantify directly the performance of genetic components in cell-free systems. The library of RBS's along with additional mutants can be used to finely tune translational efficiency, which is important in interfacing components of larger regulatory systems (64,83). For assembling regulatory systems in cell-free contexts, both the traditional multi-cistronic approach and an approach whereby a separate plasmid is used for each gene appear to be viable. Furthermore, the successful demonstration of inducible negative feedback embodies an initial step towards more complex regulatory systems. In the future, coupling the ability to forward engineer cell-free genetic regulation with efforts to compartmentalize reaction components in small liposomes (84–86) or nanofabricated wells (50,87,88) will help to close the gap between harnessing the unique capabilities of living cells and capitalizing on the comparative ease of engineering in simpler contexts.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–6, Supplementary Figures 1–8 and Supplementary References [89–93].
FUNDING
The Center for Nanophase Materials Sciences that is sponsored by the Scientific User Facilities Division, Office of Science, U.S. Department of Energy (to D.K.K., M.L.S., M.J.D.) and National Institutes of Health (EB000657 to S.I. and M.J.D.); Funding for open access charge: National Institutes of Health (grant number EB000657).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Dale A. Pelletier and Dr Jennifer Morrell-Falvey for helpful comments. This research was performed at Oak Ridge National Laboratory (ORNL). ORNL is managed by UT-Battelle, LLC, for the U.S. Department of Energy under contract DE-AC05-00OR22725.
REFERENCES
- 1.Andrianantoandro E, Basu S, Karig DK, Weiss R. Synthetic biology: new engineering rules for an emerging discipline. Mol. Syst. Biol. 2006;2:2006 0028. doi: 10.1038/msb4100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 3.Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nat. Rev. Mol. Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 4.Sprinzak D, Elowitz MB. Reconstruction of genetic circuits. Nature. 2005;438:443–448. doi: 10.1038/nature04335. [DOI] [PubMed] [Google Scholar]
- 5.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 6.Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 8.Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotech. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 9.Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balagadde FK, Song H, Ozaki J, Collins CH, Barnet M, Arnold FH, Quake SR, You L. A synthetic Escherichia coli predator-prey ecosystem. Mol. Syst. Biol. 2008;4:187. doi: 10.1038/msb.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brenner K, Karig DK, Weiss R, Arnold FH. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc. Natl Acad. Sci. USA. 2007;104:17300–17304. doi: 10.1073/pnas.0704256104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 2008;19:556–563. doi: 10.1016/j.copbio.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 14.Bowen TA, Zdunek JK, Medford JI. Cultivating plant synthetic biology from systems biology. New Phytol. 2008;179:583–587. doi: 10.1111/j.1469-8137.2008.02433.x. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ. Programmable cells: interfacing natural and engineered gene networks. Proc. Natl Acad. Sci. USA. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simpson ML, Sayler GS, Applegate BM, Ripp S, Nivens DE, Paulus MJ, Jellison GE. Bioluminescent-bioreporter integrated circuits form novel whole-cell biosensors. Trends Biotechnol. 1998;16:332–338. [Google Scholar]
- 17.Simpson ML, Sayler GS, Patterson G, Nivens DE, Bolton EK, Rochelle JM, Arnott JC, Applegate BM, Ripp S, Guillorn MA. An integrated CMOS microluminometer for low-level luminescence sensing in the bioluminescent bioreporter integrated circuit. Sens. Actuat. B Chem. 2001;72:134–140. doi: 10.1016/s0925-4005(00)00641-9. [DOI] [PubMed] [Google Scholar]
- 18.Yagi K. Applications of whole-cell bacterial sensors in biotechnology and environmental science. Appl. Microbiol. Biotechnol. 2007;73:1251–1258. doi: 10.1007/s00253-006-0718-6. [DOI] [PubMed] [Google Scholar]
- 19.Doktycz MJ, Simpson ML. Nano-enabled synthetic biology. Mol. Syst. Biol. 2007;3:125. doi: 10.1038/msb4100165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jewett MC, Calhoun KA, Voloshin A, Wuu JJ, Swartz JR. An integrated cell-free metabolic platform for protein production and synthetic biology. Mol. Syst. Biol. 2008;4:220. doi: 10.1038/msb.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katzen F, Chang G, Kudlicki W. The past, present and future of cell-free protein synthesis. Trends Biotechnol. 2005;23:150–156. doi: 10.1016/j.tibtech.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Kim J, White KS, Winfree E. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol. Syst. Biol. 2006;2:68. doi: 10.1038/msb4100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wlotzka B, McCaskill JS. A molecular predator and its prey: coupled isothermal amplification of nucleic acids. Chem. Biol. 1997;4:25–33. doi: 10.1016/s1074-5521(97)90234-9. [DOI] [PubMed] [Google Scholar]
- 24.Ishikawa K, Sato K, Shima Y, Urabe I, Yomo T. Expression of a cascading genetic network within liposomes. FEBS Lett. 2004;576:387–390. doi: 10.1016/j.febslet.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 25.Noireaux V, Bar-Ziv R, Libchaber A. Principles of cell-free genetic circuit assembly. Proc. Natl Acad. Sci. USA. 2003;100:12672–12677. doi: 10.1073/pnas.2135496100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isalan M, Lemerle C, Serrano L. Engineering gene networks to emulate Drosophila embryonic pattern formation. PLoS Biol. 2005;3:e64. doi: 10.1371/journal.pbio.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hodgman CE, Jewett MC. Cell-free synthetic biology: thinking outside the cell. Metab. Eng. 2012;14:261–269. doi: 10.1016/j.ymben.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat. Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- 29.Shetty RP, Endy D, Knight TF., Jr Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SK, Chou HH, Pfleger BF, Newman JD, Yoshikuni Y, Keasling JD. Directed evolution of AraC for improved compatibility of arabinose- and lactose-inducible promoters. Appl. Environ. Microbiol. 2007;73:5711–5715. doi: 10.1128/AEM.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SK, Keasling JD. A propionate-inducible expression system for enteric bacteria. Appl. Environ. Microbiol. 2005;71:6856–6862. doi: 10.1128/AEM.71.11.6856-6862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 34.Tabor S, Richardson CC. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl Acad. Sci. USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chamberlin M, Ring J. Characterization of T7-specific Ribonucleic Acid Polymerase. J. Biol. Chem. 1973;248:2235–2244. [PubMed] [Google Scholar]
- 36.DaRocha WD, Otsu K, Teixeira SM, Donelson JE. Tests of cytoplasmic RNA interference (RNAi) and construction of a tetracycline-inducible T7 promoter system in Trypanosoma cruzi. Mol. Biochem. Parasitol. 2004;133:175–186. doi: 10.1016/j.molbiopara.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Kushnir S, Gase K, Breitling R, Alexandrov K. Development of an inducible protein expression system based on the protozoan host Leishmania tarentolae. Protein Expr. Purif. 2005;42:37–46. doi: 10.1016/j.pep.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 38.Taylor M, Kelly J. pTcINDEX: a stable tetracycline-regulated expression vector for Trypanosoma cruzi. BMC Biotechnol. 2006;6:32. doi: 10.1186/1472-6750-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wen LM, Xu P, Benegal G, Carvaho MR, Butler DR, Buck GA. Trypanosoma cruzi: exogenously regulated gene expression. Exp. Parasitol. 2001;97:196–204. doi: 10.1006/expr.2001.4612. [DOI] [PubMed] [Google Scholar]
- 40.Wirtz E, Hoek M, Cross GA. Regulated processive transcription of chromatin by T7 RNA polymerase in Trypanosoma brucei. Nucleic Acids Res. 1998;26:4626–4634. doi: 10.1093/nar/26.20.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao C, Luo J, Hsiao CH, Donelson JE, Wilson ME. Leishmania chagasi: a tetracycline-inducible cell line driven by T7 RNA polymerase. Exp. Parasitol. 2007;116:205–213. doi: 10.1016/j.exppara.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dubendorff JW, Studier FW. Controlling basal expression in an inducible T7 expression system by blocking the target T7 promoter with lac repressor. J. Mol. Biol. 1991;219:45–59. doi: 10.1016/0022-2836(91)90856-2. [DOI] [PubMed] [Google Scholar]
- 43.Giordano TJ, Deuschle U, Bujard H, McAllister WT. Regulation of coliphage T3 and T7 RNA polymerases by the lac repressor-operator system. Gene. 1989;84:209–219. doi: 10.1016/0378-1119(89)90494-0. [DOI] [PubMed] [Google Scholar]
- 44.Lopez PJ, Guillerez J, Sousa R, Dreyfus M. On the mechanism of inhibition of phage T7 RNA polymerase by lac repressor. J. Mol. Biol. 1998;276:861–875. doi: 10.1006/jmbi.1997.1576. [DOI] [PubMed] [Google Scholar]
- 45.Peranen J, Rikkonen M, Hyvonen M, Kaariainen L. T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- 46.Rosenfeld N, Elowitz MB, Alon U. Negative autoregulation speeds the response times of transcription networks. J. Mol. Biol. 2002;323:785–793. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- 47.Thattai M, van Oudenaarden A. Stochastic gene expression in fluctuating environments. Genetics. 2004;167:523–530. doi: 10.1534/genetics.167.1.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simpson ML, Cox CD, Sayler GS. Frequency domain analysis of noise in autoregulated gene circuits. Proc. Natl Acad. Sci. USA. 2003;100:4551–4556. doi: 10.1073/pnas.0736140100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Austin DW, Allen MS, McCollum JM, Dar RD, Wilgus JR, Sayler GS, Samatova NF, Cox CD, Simpson ML. Gene network shaping of inherent noise spectra. Nature. 2006;439:608–611. doi: 10.1038/nature04194. [DOI] [PubMed] [Google Scholar]
- 50.Siuti P, Retterer ST, Doktycz MJ. Continuous protein production in nanoporous, picolitre volume containers. Lab Chip. 2011;11:3523–3529. doi: 10.1039/c1lc20462a. [DOI] [PubMed] [Google Scholar]
- 51.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O, and AraC/I1-I2 regulatory elements. Nucl Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bujard H, Gentz R, Lanzer M, Stueber D, Mueller M, Ibrahimi I, Haeuptle MT, Dobberstein B. A T5 promoter-based transcription-translation system for the analysis of proteins in vitro and in vivo. Methods Enzymol. 1987;155:416–433. doi: 10.1016/0076-6879(87)55028-5. [DOI] [PubMed] [Google Scholar]
- 53.Olins PO, Devine CS, Rangwala SH, Kavka KS. The T7 phage gene 10 leader RNA, a ribosome-binding site that dramatically enhances the expression of foreign genes in Escherichia coli. Gene. 1988;73:227–235. doi: 10.1016/0378-1119(88)90329-0. [DOI] [PubMed] [Google Scholar]
- 54.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotech. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Du L, Gao R, Forster AC. Engineering multigene expression in vitro and in vivo with small terminators for T7 RNA polymerase. Biotechnol. Bioeng. 2009;104:1189–1196. doi: 10.1002/bit.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moffatt BA, Studier FW. T7 lysozyme inhibits transcription by T7 RNA polymerase. Cell. 1987;49:221–227. doi: 10.1016/0092-8674(87)90563-0. [DOI] [PubMed] [Google Scholar]
- 57.Pellinen T, Huovinen T, Karp M. A cell-free biosensor for the detection of transcriptional inducers using firefly luciferase as a reporter. Anal. Biochem. 2004;330:52–57. doi: 10.1016/j.ab.2004.03.064. [DOI] [PubMed] [Google Scholar]
- 58.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Accurate prediction of gene feedback circuit behavior from component properties. Mol. Syst. Biol. 2007;3 doi: 10.1038/msb4100185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deuschle U, Gentz R, Bujard H. lac Repressor blocks transcribing RNA polymerase and terminates transcription. Proc. Natl Acad. Sci. USA. 1986;83:4134–4137. doi: 10.1073/pnas.83.12.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horowitz H, Platt T. Regulation of transcription from tandem and convergent promoters. Nucleic Acids Res. 1982;10:5447–5465. doi: 10.1093/nar/10.18.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 62.Cherry JL, Adler FR. How to make a biological switch. J. Theor. Biol. 2000;203:117–133. doi: 10.1006/jtbi.2000.1068. [DOI] [PubMed] [Google Scholar]
- 63.Hooshangi S, Thiberge S, Weiss R. Ultrasensitivity and noise propagation in a synthetic transcriptional cascade. Proc. Natl Acad. Sci. USA. 2005;102:3581–3586. doi: 10.1073/pnas.0408507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weiss R, Basu S. NSC-1: The First Workshop of Non Silicon Computing. Boston, MA; 2002. [Google Scholar]
- 65.Bonner G, Lafer EM, Sousa R. Characterization of a set of T7 RNA polymerase active site mutants. J. Biol. Chem. 1994;269:25120–25128. [PubMed] [Google Scholar]
- 66.Bonner G, Patra D, Lafer EM, Sousa R. Mutations in T7 RNA polymerase that support the proposal for a common polymerase active site structure. EMBO J. 1992;11:3767–3775. doi: 10.1002/j.1460-2075.1992.tb05462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramesh V, De A, Nagaraja V. Engineering hyperexpression of bacteriophage Mu C protein by removal of secondary structure at the translation initiation region. Protein Eng. 1994;7:1053–1057. doi: 10.1093/protein/7.8.1053. [DOI] [PubMed] [Google Scholar]
- 68.Schumann W, Ferreira LCS. Production of recombinant proteins in Escherichia coli. Genet. Mol. Biol. 2004;27:442–453. [Google Scholar]
- 69.Keum JW, Ahn JH, Choi CY, Lee KH, Kwon YC, Kim DM. The presence of a common downstream box enables the simultaneous expression of multiple proteins in an E. coli extract. Biochem. Biophys. Res. Commun. 2006;350:562–567. doi: 10.1016/j.bbrc.2006.09.072. [DOI] [PubMed] [Google Scholar]
- 70.Calhoun KA, Swartz JR. Energizing cell-free protein synthesis with glucose metabolism. Biotechnol. Bioeng. 2005;90:606–613. doi: 10.1002/bit.20449. [DOI] [PubMed] [Google Scholar]
- 71.Jewett MC, Swartz JR. Substrate replenishment extends protein synthesis with an in vitro translation system designed to mimic the cytoplasm. Biotechnol. Bioeng. 2004;87:465–472. doi: 10.1002/bit.20139. [DOI] [PubMed] [Google Scholar]
- 72.Jewett MC, Swartz JR. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol. Bioeng. 2004;86:19–26. doi: 10.1002/bit.20026. [DOI] [PubMed] [Google Scholar]
- 73.Kim DM, Swartz JR. Prolonging cell-free protein synthesis by selective reagent additions. Biotechnol. Prog. 2000;16:385–390. doi: 10.1021/bp000031y. [DOI] [PubMed] [Google Scholar]
- 74.Michel-Reydellet N, Calhoun K, Swartz J. Amino acid stabilization for cell-free protein synthesis by modification of the Escherichia coli genome. Metab. Eng. 2004;6:197–203. doi: 10.1016/j.ymben.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 75.Nakano H, Shinbata T, Okumura R, Sekiguchi S, Fujishiro M, Yamane T. Efficient coupled transcription/translation from PCR template by a hollow-fiber membrane bioreactor. Biotechnol. Bioeng. 1999;64:194–199. doi: 10.1002/(sici)1097-0290(19990720)64:2<194::aid-bit8>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 76.Spirin AS. High-throughput cell-free systems for synthesis of functionally active proteins. Trends Biotechnol. 2004;22:538–545. doi: 10.1016/j.tibtech.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 77.Shin J, Noireaux V. Study of messenger RNA inactivation and protein degradation in an Escherichia coli cell-free expression system. J. Biol. Eng. 2010;4:9. doi: 10.1186/1754-1611-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 79.Kim J, White KS, Winfree E. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim J, Winfree E. Synthetic in vitro transcriptional oscillators. Mol. Syst. Biol. 2011;7 doi: 10.1038/msb.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karig DK, Simpson ML. Tying new knots in synthetic biology. HFSP J. 2008;2:124–128. doi: 10.2976/1.2907240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guido NJ, Wang X, Adalsteinsson D, McMillen D, Hasty J, Cantor CR, Elston TC, Collins JJ. A bottom-up approach to gene regulation. Nature. 2006;439:856–860. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- 83.Basu S, Mehreja R, Thiberge S, Chen MT, Weiss R. Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl Acad. Sci. USA. 2004;101:6355–6360. doi: 10.1073/pnas.0307571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Monnard PA. Liposome-entrapped Polymerases as Models for Microscale/Nanoscale Bioreactors. J. Membr. Biol. 2003;191:87–97. doi: 10.1007/s00232-002-1046-0. [DOI] [PubMed] [Google Scholar]
- 85.Noireaux V, Libchaber A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl Acad. Sci. USA. 2004;101:17669–17674. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pereira de Souza T, Stano P, Luisi PL. The minimal size of liposome-based model cells brings about a remarkably enhanced entrapment and protein synthesis. Chembiochem. 2009;10:1056–1063. doi: 10.1002/cbic.200800810. [DOI] [PubMed] [Google Scholar]
- 87.Fletcher BL, Hullander ED, Melechko AV, McKnight TE, Klein KL, Hensley DK, Morrell JL, Simpson ML, Doktycz MJ. Microarrays of Biomimetic Cells Formed by the Controlled Synthesis of Carbon Nanofiber Membranes. Nano Lett. 2004;4:1809–1814. [Google Scholar]
- 88.Karig DK, Siuti P, Dar RD, Retterer ST, Doktycz MJ, Simpson ML. Model for biological communication in a nanofabricated cell-mimic driven by stochastic resonance. Nano Commun. Netw. 2011;2:39–49. doi: 10.1016/j.nancom.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 90.Karig D. Ph.D. Thesis. Princeton University: 2007. Engineering multi-signal synthetic biological systems. [Google Scholar]
- 91.Fukuda H, Arai M, Kuwajima K. Folding of green fluorescent protein and the cycle3 mutant. Biochemistry. 2000;39:12025–12032. doi: 10.1021/bi000543l. [DOI] [PubMed] [Google Scholar]
- 92.Sacchetti A, Cappetti V, Marra P, Dell'Arciprete R, El Sewedy T, Crescenzi C, Alberti S. Green Fluorescent Protein variants fold differentially in prokaryotic and eukaryotic cells. J. Cell. Biochem. 2001;81:117–128. doi: 10.1002/jcb.1091. [DOI] [PubMed] [Google Scholar]
- 93.Selinger DW, Saxena RM, Cheung KJ, Church GM, Rosenow C. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 2003;13:216–223. doi: 10.1101/gr.912603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.