Abstract
The moss Physcomitrella patens is unique among plant models for the high frequency with which targeted transgene insertion occurs via homologous recombination. Transgene integration is believed to utilize existing machinery for the detection and repair of DNA double-strand breaks (DSBs). We undertook targeted knockout of the Physcomitrella genes encoding components of the principal sensor of DNA DSBs, the MRN complex. Loss of function of PpMRE11 or PpRAD50 strongly and specifically inhibited gene targeting, whilst rates of untargeted transgene integration were relatively unaffected. In contrast, disruption of the PpNBS1 gene retained the wild-type capacity to integrate transforming DNA efficiently at homologous loci. Analysis of the kinetics of DNA-DSB repair in wild-type and mutant plants by single-nucleus agarose gel electrophoresis revealed that bleomycin-induced fragmentation of genomic DNA was repaired at approximately equal rates in each genotype, although both the Ppmre11 and Pprad50 mutants exhibited severely restricted growth and development and enhanced sensitivity to UV-B and bleomycin-induced DNA damage, compared with wild-type and Ppnbs1 plants. This implies that while extensive DNA repair can occur in the absence of a functional MRN complex; this is unsupervised in nature and results in the accumulation of deleterious mutations incompatible with normal growth and development.
INTRODUCTION
DNA double-strand breaks (DSBs) represent one of the most cytotoxic forms of damage an organism can acquire (1). Such events occur with high frequency resulting from cellular metabolism (such as reactive radicals or stalled replication forks during S phase) and through the action of exogenous agents (such as ionizing radiation or chemical mutagens). Failure to repair such damage can lead to the irrecoverable loss of genetic material, with both immediate and long-term consequences: the onset of cancerous transformation in animal cells, or the failure to transmit genetic information in gametes (especially in plants, where there is no early developmental partitioning of germ-line and somatic cell lineages).
Unsurprisingly, all living organisms have evolved efficient mechanisms that can be deployed to sense DNA DSBs, activate DNA repair, cell-cycle arrest and sometimes apoptosis. Such is the importance of these mechanisms, that the genes encoding many of the essential components of the DNA repair machinery are highly conserved in evolution (2). In particular, this is true of the mechanism by which the broken ends of DNA molecules are recognized and recruited into DNA repair complexes. In eukaryotes, the MRN/MRX complex undertakes this task (3,4). This conserved complex is composed of three proteins, Meiotic recombination 11 (MRE11), Radiation sensitive 50 (RAD50), and Nijmegen Breakage Syndrome 1 (NBS1) (X-ray sensitive 2, XRS2 in the yeast, Saccharomyces cerevisiae). Together, the MRE11, RAD50 and NBS1 proteins form a multisubunit complex (M2R2N1) that binds the ends of broken DNA molecules, and can tether the broken ends through dimerization between adjacent MRN complexes mediated by an association between the RAD50 components (3,5). The formation of MRN–DNA complexes also initiates a cell-cycle checkpoint through interaction with the phosphoinositide 3-kinase-related protein kinases (PIKKs) ATM and ATR (for ‘Ataxia Telangiectasia Mutated’ and ‘Ataxia Telangiectasia mutated-like and Rad 3 related’) and the DNA Protein Kinase catalytic subunit (DNA-PKcs) (6). These proteins phosphorylate multiple targets to initiate a cascade of downstream events leading to DNA DSB repair either by non-homologous end joining (NHEJ), a rapid but occasionally inaccurate mechanism, or through homologous recombination (HR), a conservative mechanism that uses an homologous sequence (e.g. a sister chromatid) as a template to restore the original sequence at the DSB site. In this latter pathway, an Mre11-specific nuclease activity is required (with other components) for the resection of DNA ends necessary for strand invasion (7).
Transgene integration into flowering plant genomes occurs through the agency of endogenous mechanisms that have evolved for the repair of DNA DSBs. In flowering plants, the integration of exogenous DNA whether directly delivered via microprojectile bombardment or protoplast transfection, or delivered by Agrobacterium-mediated transformation occurs predominantly at random positions throughout the genome, whereas gene targeting frequencies remain extremely low (8). Random integration of transgenes requires enzymes from the NHEJ pathway, and the inefficiency of GT probably reflects the prevalence of the NHEJ pathway in repairing DNA DSBs in angiosperms (9–11). In contrast with flowering plants, transformation of the moss, Physcomitrella patens, with DNA containing homology with genomic sequences results in preferential incorporation of the transforming DNA at these homologous sequences (12). This facility for ‘gene targeting’ is similar to that seen in Saccharomyces (13) and suggests a preference for the use of the HR-dependent pathway as the primary means of undertaking DSB repair, although molecular analyses of gene targeting events provide clear evidence for modification of the transforming DNA by both NHEJ and HR reactions upon integration (14,15). Physcomitrella thus represents an excellent model in which to analyse DNA-DSB repair pathways in plants, particularly in regard to its outstanding gene targeting efficiency (12). Previous studies have shown that PpRAD51, the protein at the core of the HR reaction, was required to preserve genome integrity and essential to achieve gene targeting (16,17). The mismatch repair PpMSH2 gene was also shown to be essential to preserve genome integrity and to prevent homeologous gene targeting (18).
We have characterized the role of the Physcomitrella MRN complex in DNA DSB-repair and gene targeting. We find that in moss the major loss of function phenotypes of the MRN complex depends on PpRAD50 or PpMRE11, but not PpNBS1. Inactivation of either PpRAD50 or PpMRE11 reduced GT ∼11-fold in both Pprad50 and Ppmre11 mutants, while illegitimate integration rates only slightly affected. Gene expression studies further show that Ppmre11 and Pprad50 strains display constitutively high expression of the DNA damage response, implying the activation of alternative pathways to minimize endogenous DNA damage in the mutant strains. The mutants exhibit a severe developmental phenotype, possibly associated with early senescence processes, and hypersensitivity to UV-B and bleomycin-induced DNA damage.
MATERIALS AND METHODS
Plant material
Physcomitrella patens (Hedw.) B.S.G. ‘Gransden2004’ was vegetatively propagated as previously described (19). Individual plants were cultured as ‘spot inocula’ on BCD agar medium supplemented with 1 mM CaCl2 and 5 mM ammonium tartrate (BCDAT medium), or as lawns of protonemal filaments by subculture of homogenized tissue on BCDAT agar medium overlain with cellophane for the isolation of protoplasts. Transformation experiments were performed as previously described (20) using linear fragments of DNA generated either by digestion of transforming vectors with restriction enzymes (19) or by polymerase chain reaction (PCR) amplification (14). Growth conditions for the generation of deletion strains were as described previously (17).
Gene identification and isolation
Genomic DNA and total RNA were isolated from Physcomitrella as previously described (19). For verification of gene models, RNA was extracted from a polyribosome-enriched fraction: 7-day subcultured protonemal tissue (∼5 g squeeze-dried chloronemal tissue) was homogenized in 30 ml extraction buffer [200 mM sucrose 40 mM Tris–HCl, pH 8.5, 60 mMKCl, 30 mM MgCl2, 1% (v/v) Triton X-100, 2 mM dithiothreitol] and the extract was clarified at 25 000g for 20 min (Sorvall SS34 rotor). The supernatant was layered over a cushion comprising 1 M sucrose, 40 mM Tris–HCl, pH 8.5, 20 mM KCl, 10 mM MgCl2 and centrifuged for 3 h at 141 000g (Beckman SW28 rotor). The pellet was drained and resuspended in 0.5 ml RNA extraction buffer for aqueous phenol extraction (19). RNA used for RT-PCR was first digested with RQ DNase I (Promega) to remove residual DNA. Physcomitrella genomic sequences encoding the MRE11, RAD50 and NBS1 genes were identified by BLAST search (http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html). The available gene models were used for the design of PCR primers to amplify cognate genomic sequences, which were cloned in the plasmid pBluescript KS+. PCR primers used are listed in Supplementary Table S1. In order to obtain a correct gene model for each sequence, full-length complementary DNA (cDNA) sequences were amplified from Physcomitrella polyribosome-derived RNA by RT-PCR. Total RNA (1 μg) was reverse transcribed using an oligo-dT12–15 primer and AMV reverse transcriptase as supplied in the Promega Reverse Transcription system in a 20 -μl reaction. Following cDNA synthesis, the reaction mixture was diluted by the addition of 80 μl water, and 1 μl aliquots were used for PCR amplification using primers predicted to anneal with 5′- and 3′-untranslated region (UTR) sequences (Supplementary Table S1). PCR products were cloned by blunt-end ligation into the EcoRV site of pBluescript KS+ for sequence analysis (ABI3130) in the DNA sequencing facility of the Leeds University Faculty of Biological Sciences. Predicted polypeptide sequences were aligned with the orthologous genes from Arabidopsis thaliana, Homo sapiens and Saccharomyces cerevisiae using CLUSTALW.
Targeted gene knockout
Gene disruption cassettes were constructed by ligating a selection cassette comprising a neomycin phosphotranferase gene driven by the Cauliflower Mosaic Virus 35S promoter and terminated with the CaMV gene 6 termination sequence (35S-nptII-g6ter) derived from the vector pMBL6 (14) into convenient restriction sites within the cloned PpMRE11, PpRAD50 and PpNBS1 genes to replace endogenous coding sequences. For the PpMRE11 gene, the selection cassette was placed between residues 1763501 (in exon 4) and 1764332 (in exon 8) in JGI Phypa1_1/scaffold 18. For the PpRAD50 gene the selection cassette was placed between residues 1431738 (in intron 13) and 1433260 (in intron 16) in JGI Phypa1_1/scaffold 51. For the PpNBS1 gene, the selection cassette was placed between residues 276622 (in intron 4) and 277547 (in intron 7) in JGI Phypa1_1/scaffold 219. For targeted knockout of the moss genes, fragments of DNA containing these cassettes and flanked by ∼1 kb of 5′- and 3′-flanking genomic sequence were PCR amplified. These linear fragments were used to transform Physcomitrella protoplasts, and stable transformants were selected following regeneration in medium containing 50 μg ml−1 G418 for 2 weeks, followed by subculture onto medium lacking antibiotic for 2 weeks, and a final subculture on selective medium. Targeted replacement of the native genes by the disruption cassette was confirmed by PCR reactions using external, gene-specific primers in combination with ‘outward-pointing’ selection cassette-specific primers (Supplementary Table S1). Single-copy allele replacements were identified by PCR using the external primer pairs, and the absence of additional transgene insertion in the genome was confirmed by Southern blot analysis. Conditions for PCR analysis were as previously described (14) and Southern blot analysis of genomic DNA was carried out as previously described (21), using the 35S-nptII-g6ter cassette as a hybridization probe.
For generation of deletion mutants mre11Δ and rad50Δ, the 5′-and 3′-targeting fragments were amplified from P. patens genomic DNA and cloned upstream (5′) and downstream (3′) of the loxP sites flanking the resistance cassette in plasmid pBNRF (17) to create the plasmids pMRE11delta and pRAD50delta, respectively. For the PpMRE11 gene, a 1009-bp 5′-targeting fragment (coordinates 1763137-1764146 in JGI Phypa1_1/scaffold 18) and an 803-bp 3′-targeting fragment (coordinates 1765334–1766184) were PCR-amplified. For PpRad50, an 831-bp 5′ targeting fragment (coordinates 1426648–1427479 in JGI Phypa1_1/scaffold 51) and an 819-bp 3′-targeting fragment (coordinates 1435197–1436016) were PCR-amplified.
Moss protoplasts were transformed with pMRE11delta digested with BstXI and AseI, or with pRAD50delta digested with XbaI and NsiI. Stable disruptants were selected by successive subculture on selective and non-selective medium and PCR analysis as described above. Clean deletions in the PpMRE11 (encompassing exons 7–10) and PpRAD50 genes (exons 4–20) were obtained by transient Cre recombinase expression (18). Deletions in the recombinant loci were confirmed by PCR amplification using gene-specific external primers MRE11#1 and MRE11#2, and RAD50#1 and RAD50#2, respectively. Primers APT#14 and APT#19 were used as positive controls (Supplementary Table S1).
For gene targeting studies the vectors PpAPT-KO2 (17) and PpAPT-KO3 have been used. To obtain PpAPT-KO3, an internal 1631-bp SalI/BglII fragment containing the 35S:HygR-LoxP marker was deleted in PpAPT-KO2 and replaced by an XhoI/BglII fragment from pBNRF (17) containing the 35S:NeoR-LoxP marker.
Analysis of gene expression in mutants
Transcript abundance in selected knockout lines was determined by RT-PCR of cDNA. Total RNA was isolated from protonemal tissue (19) and 1 µg was reverse-transcribed using a Promega reverse transcription system. The 20 -µl reaction mixture was diluted 25-fold and 5 µl aliquots were used for PCR. Detection of Mre11, Rad50 and Nbs1 mRNA in mutant lines was by RT-PCR using primers indicated in Supplementary Table 1.
For quantitative determination of the relative abundances of transcripts encoding DNA repair genes in wild-type and mutant strains, quantitative real-time PCR was carried out using a Qiagen Rotor-Gene Q instrument and Qiagen SYBR-Green PCR kit. RNA was isolated from 7-day subcultured protonemal tissue from each of three independent lines (wild-type, Ppmre11KO, Pprad50KO and Ppnbs1KO, respectively), with two replicates for each sample. Transcript abundance was estimated by reference to both internal and external reference sequences. As an external reference, Physcomitrella RNA samples were ‘spiked’ with tenfold serial dilutions (10−1–10−4) of an in vitro transcript from a full-length wheat ‘Em’ cDNA (22) prior to reverse transcription. These were used to test a number of candidate internal reference sequences, corresponding to Physcomitrella gene models Phypa1_1:227826 (SAND family endocytosis protein), Phypa1_1:209451 (Clathrin adapter complex subunit), Phypa1_1:224488 (Acyltransferase) and Phypa1_1:163153 (Ribosomal protein S4) for stability of expression in response to bleomycin treatment. Phypa1_1:227826 was subsequently selected as the internal reference standard for the determination of the abundances of PpRad51-1 (Phypa1_1:206066), PpRad51-2 ((Phypa1_1:207856), PpPARP-1 (Phypa1_1:150949), PpPARP-2 (Phypa1_1:188096) PpKu70 (Phypa1_1:60909), PpKu80 (Phypa1_6:23553) and PpCtIP (Phypa1_6:453490) transcripts. Relative transcript abundance was calculated using the ΔΔCt method and normalized to the wild-type value.
Bleomycin and UV-B sensitivity assays
Physcomitrella explants were inoculated as ‘spot inocula’ onto BCDAT-agar plates supplemented with bleomycin (Bleocin inj., Euro Nippon Kayaku GmbH, Germany) at concentrations indicated in the text, to determine sensitivity to chronic exposure to the drug. Plant growth was assessed by measurement of the surface area of each plant at intervals following inoculation by digital photography of the plates. The image analysis software ‘ImageJ’ (23) was used to convert the digital images to binary format and determine the colony area based on counting the number of pixels corresponding to each colony. Colony area determinations based on different photographs were normalized for each colony using the estimated area of the plate.
For acute toxicity testing, protoplast viability and protonemal growth were analysed. Viability was tested when protoplasts of wild-type and mre11, rad50 and nbs1 mutants in BCD liquid medium supplemented with mannitol were treated with bleomycin at concentrations indicated in the text for 1 h. Protoplasts were washed two times and then resuspended in liquid mannitol medium. After 20 h in the dark, the protoplasts were spread on BCD agar medium supplemented with mannitol (∼105 protoplasts per Petri dish). After 6 days regeneration, the number of survivors was counted. We repeated these experiments three times.
Protoplasts of wild-type, mre11 and rad50 mutants were spread (∼50 000/plate) on protoplast agar medium (PpNH4 + 0.5% glucose + 8.5% mannitol). Plates were immediately irradiated with UV-B light (308 nm) in a Stratagene Stratalinker. The intensity of the irradiation was controlled using the internal probe of the Stratalinker and one plate of each strain was treated simultaneously. The experiment was repeated three times. Plates were immediately transferred to darkness for 24 h after treatment then to standard growth conditions for protoplast regeneration. Survival was determined after 1 week by microscopic observation.
Protonemal growth was tested by incubating 7-day-old protonemal tissue in BCDAT liquid medium containing bleomycin at concentrations indicated in the text for 1 h. The tissue was washed three times with medium lacking bleomycin and homogenized. Explants were inoculated onto BCDAT agar medium and recovery following treatment was determined by measuring the increase in plant surface area over a 3-week period.
Evaluation of spontaneous mutation frequency
Mutations in the PpAPT gene confer resistance to 2-Fluoroadenine (2-FA), a toxic compound for cells. The number of 2-FA resistant colonies that grow following protoplast regeneration reflects the frequency of spontaneous mutations. Protoplasts of wild-type, mre11, rad50, nbs1 and msh2 (18) mutants were regenerated for 6 days on BCD agar medium supplemented with mannitol (∼105 protoplasts per Petri dish) and then transferred on to BCD agar medium supplemented with 5 μM 2-FA (Fluorochem). After 2 weeks, the number of resistant clones was counted. Experiments were repeated three times and statistically analysed using Fisher's exact test.
Isolation of apt mutants after bleomycin treatments
One-day-old protonemata prepared from 10 plates of 7-day-old tissue (around 25.106 dividing cells) of wild-type and the Pprad50KO mutants were exposed to sublethal acute doses of bleomycin: 50 µg/ml for 2 h for wild-type and 0.1 µg/ml for 1 h for the Pprad50KO mutant, before being transferred onto cellophane-overlaid BCDAT agar medium supplemented with 2–3 μM 2-FA. After 3 weeks, resistant foci were clearly visible. Cellophane discs bearing resistant colonies were transferred to plates without 2-FA. This process was repeated three times until stable Ppapt clones were established. The results of selection are summarized in Supplementary Table S2. Genomic DNA was isolated as previously described (19) and the mutant Ppapt genes were amplified by PCR and sequenced using the primers listed in Supplementary Table S1 and indicated in Supplementary Figure S4.
Gene targeting assays
Transformation efficiency and APT targeting frequency were measured as previously described (17). Moss protoplasts (4.8 × 105) were transformed with the nonhomologous pBHRF or pBNRF plasmids (17) digested respectively with HindIII or XmaJI to produce a linear fragment containing the 35S::hygR or 35S::neoR markers, or with PpAPT-KO2 or PpAPT-KO3 plasmids digested respectively with BsaAI/HindIII or PvuI/BsrGI to produce the targeting APT fragment containing the 35S::hygR cassette (from pBHRF) or the35S::neoR cassette (from pBNRF) flanked by genomic PpAPT sequences. Targeted integration of PpAPT-KO2 or PpAPT-KO3 at the APT genomic locus confers resistance to 2-FA. We selected primary transformants (unstable + stable) with 25 mgl−1 hygromycin B (Duchefa) for PpAPT-KO2 or with 50 mgl−1 G418 (Duchefa) for PpAPT-KO3. Integrative transformants were isolated following a second round of selection. Protonemal explants from these transformants were then transferred onto medium containing 5 µM of 2-FA to detect APT gene targeting events. Experiments were repeated three times.
DNA-DSB repair assays
Protonemal lawns of wild-type and mutant strains subcultured for 1 week were used to generate protonemal tissue for DNA repair assays by shearing tissue collected from single 9-cm plates with an IKA T2T Digital Ultra Turrax homogenizer at maximum speed (24 krpm) for 1 min in 5 ml of liquid BCD medium. This was spread on BCD-agar medium overlaid with cellophane and grown for 1, 7 or 14 days prior to harvesting for bleomycin treatment.
Protonemata were gently transferred from cellophane to liquid BCD medium in 4-cm wells of a six-well microtitre plate to avoid drying. DSBs were induced by addition of bleomycin to 10, 20, 30 and 50 μg ml−1 for 1 h. Following treatment, the tissue was thoroughly rinsed in H2O in disposable 22 -µm mesh funnels (Partec GmbH, Germany), blotted on filter paper and either flash-frozen in liquid N2 (t = 0) or left to recover on BCD-agar plates overlaid with cellophane for the indicated repair times, before being frozen in liquid N2. All handling and transfer of protonemata was with tweezers.
DNA-DSBs were detected by a neutral comet assay (24) as described previously (25,26). Approximately 100 mg of frozen tissue was cut with a razor blade in 300 µl phosphate-buffered saline (PBS)+10 mM ethylenediaminetetraacetic acid (EDTA) on ice and the tissue debris removed by filtration through 50 -µm mesh funnels (Partec GmbH, Germany) into Eppendorf tubes on ice. Fifty microlitres of nuclear suspension was dispersed in 200 µl of melted 0.7% LMT agarose (15510-027, GibcoBRL, Gaithersburg, USA) at 40° C and four 80 -µl aliquots were immediately pipetted onto each of two agarose coated microscope slides (two duplicates per slide), covered with a 22 × 22-mm cover slip and then chilled on ice for 1 min to solidify the agarose. After removal of cover slips, slides were dipped in lysis solution (2.5 M NaCl, 10 mM Tris–HCl, 0.1 M EDTA, 1% N-lauroyl sarcosinate, pH 7.6) on ice for at least 1 h to dissolve cellular membranes and remove attached proteins. The whole procedure from chopping tissue to dipping into lysis solution takes ∼3 min. After lysis, slides were twice equilibrated for 5 min in Tris–borate–EDTA (TBE) electrophoresis buffer to remove salts and detergents. Comet slides were then subjected to electrophoresis at 1 V/cm (∼12 mA) for 5 min. After electrophoresis, slides were dipped for 5 min in 70 % EtOH, 5 min in 96% EtOH and air-dried.
DNA ‘comets’ were viewed in epifluorescence with a Nikon Eclipse 800 microscope after staining with SybrGold stain (Molecular-Probes Invitrogen, USA) and evaluated by the Comet module of the LUCIA cytogenetics software suite (LIM, Praha, Czech Republic).
Comet assay data analysis
The fraction of DNA in comet tails (% tail-DNA) was used as a measure of DNA damage. Data for the wild-type strain and the three mutant lines (Pprad50, Ppmre11 and Ppnbs1) analysed in this study were obtained in at least three independent experiments. In each experiment, the % tail-DNA was measured at seven time points: 0, 5, 10, 20, 60, 180 and 360 min after treatment and in control tissue without treatment. Measurements included four independent gel replicas of 25 evaluated comets totalling at least 300 comets analysed per experimental point.
The percentage of damage remaining as plotted on figures after given repair time (tx) is defined as:
 |
Repair kinetics following two-phase decay kinetics defined as:
SpanFast=(Y0-Plateau)*PercentFast*0.01
SpanSlow=(Y0-Plateau)*(100-PercentFast)*0.01
Y=Plateau + SpanFast*exp(-KFast*X) + SpanSlow*exp(-KSlow*X)
was analysed by linear regression of experimental data with the Prism v.5 program (GrafPad Software Inc., USA). Goodness of fit characterized by R-squared was better than 0.99.
RESULTS
Identification of MRN complex genes
Sequence homology searches of the draft Physcomitrella genome identified single putative homologues of the MRE11, RAD50, and NBS1 genes on sequence scaffolds 18, 51 and 219, respectively. Whilst EST sequences were available to provide partial support for predicted gene models for the PpMRE11 and PpNBS1 genes, no corroborative evidence was available for the PpRAD50 gene, and the automated gene prediction software had not generated a gene model. We therefore generated gene models for all three genes based on BLASTX similarity to flowering plant proteins (Arabidopsis, rice and maize) to identify putative full-length protein coding sequences, and used these models to design PCR primers for the amplification of full-length protein coding sequences by reverse transcription-PCR of moss polyribosome-derived RNA. The resulting cDNA sequences and genomic models have been deposited in GenBank (Accession Nos: JF820817 and JF820820 for PpMRE11; JF82018 and JF82021 for PpRAD50; JF82019 and JF82022 for PpNBS1) and the curated and structurally annotated gene models entered in the JGI Physcomitrella genome browser in which they were assigned the Protein ID numbers Phypa1_1:235701 (PpMRE11), Phypa1_1:235526 (PpRAD50) and Phypa1_1:235702 (PpNBS1).
The deduced polypeptide sequences were compared with the corresponding human, yeast and flowering plant sequences (Supplementary Figure S1). Like both the Arabidopsis and human genes, the PpMRE11 gene comprises 22 protein-coding exons. There is extensive similarity among all the MRE11 polypeptides (Supplementary Figure S1a) especially within the N-terminal two-thirds of the protein. The Physcomitrella MRE11 protein contains the characteristic phosphoesterase motifs within the nuclease domain, the capping domain and amino acids (N123 and W225) shown to be essential for the MRE11–NBS1 interaction (4).
The RAD50 sequences (Supplementary Figure S1b) are also well conserved at the amino-acid sequence level, and show good conservation of functionally important domains. The Physcomitrella protein contains the characteristic Walker A and Walker B adenosine triphosphatase (ATPase) motifs at either end of the sequence that associate to form a crucially important ATP-binding cassette (27) and that typify the RAD50 protein. These are separated by a long coiled-coil domain with a central CXXC zinc-hook (CPCC in both Physcomitrella and Arabidopsis) by which pairs of RAD50 proteins interact in the tethering of broken chromosome ends by the MRN/MRX complex (28).
The Physcomitrella NBS1 protein (Supplementary Figure S1c) has an N-terminal fork-head associated domain, a partial BRCT domain, and putative SQ-dipeptide phosphorylation sites and conserved MRE11-interacting motifs in the C-terminal region, as identified in all previously identified NBS1 orthologs (4).
Generation of targeted knockouts of the MRN complex genes
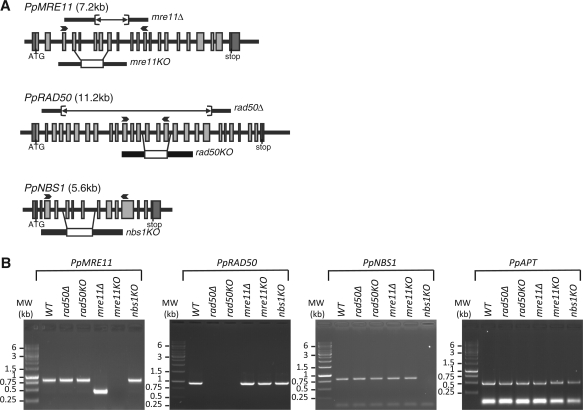
We used gene targeting to generate mutant alleles of the PpMRE11, PpRAD50 and PpNBS1 genes. For the PpMRE11 and PpRAD50 genes, two types of mutant were generated: disruption mutants, designated mre11KO and rad50KO, in which several exons were replaced by an antibiotic selection cassette and deletion mutants, designated mre11Δ and rad50Δ, in which a number of exons were replaced by a selection cassette that was subsequently removed by cre-lox recombination (Figure 1A). For the PpNBS1 gene, we generated a disruption mutant (nbs1KO) and a deletion mutant in which the complete coding sequence was deleted (nbs1Δ). Gene targeting events were identified by PCR and Southern blot analyses to identify lines in which precise modification of the target genes had occurred without additional insertion of the targeting constructs at adventitious loci. For the deletion mutants we confirmed by PCR that a portion of the coding regions was removed using primers that flanked the deletion (Supplementary Figure S2). RT-PCR analysis established that the full-length transcripts were no longer produced in the mutants (Figure 1B). For all further experiments, we used two independent disruption or two independent deletion strains which displayed similar phenotypes.
Figure 1.
Targeted disruption of Physcomitrella MRN genes. (A) Structure of the PpMRE11, PpRAD50 and PpNBS1 genes. Exons are represented by shaded boxes, with 5′- and 3′-UTR sequences in darker grey. The region deleted by cre-lox excision of a selection cassette is shown as a line above each gene. For the replacement constructs (below each gene) the extent of targeting sequence homology is indicated by the line, and the P35S-nptII-g6ter selection cassette is shown as a white box, replacing the genomic region indicated by the lines joining the gene structure diagram and the replacement cassette. Arrows indicate position of primers used for RT-PCR analysis. (B) RT-PCR analysis of MRN transcripts in wild-type and mutant plants. RNA was isolated from protonemal tissue of wild-type and mutant lines for cDNA synthesis and PCR amplification using gene-specific primers (PpMRE11#1 + PpMRE11#2 for MRE11, PpRAD50#1 + PpRAD50#2 for RAD50, PpNBS1#1 + PpNBS1#2 for NBS1). The PpAPT transcript has been used as control (primers: PpAPT#14 + PpAPT#19). Primers are listed in Supplementary Figure S4.
Gene targeting is strongly decreased in mre11 and rad50 mutants
The MRN complex is one of the earliest respondents to DNA-DSBs and plays a central role in controlling repair pathway choice between NHEJ and HR (5). The importance of the MRN complex for DSB repair by HR has been shown in mre11-deficient chicken DT40 cells in which gene targeting efficiency is strongly reduced (29). In contrast, a somatic hyper-recombination phenotype has been described in the Arabidopsis rad50 mutant (30). In order to examine the involvement of the MRN complex in genetic transformation of Physcomitrella we determined transformation and gene targeting rates in wild-type, mre11Δ, rad50Δ and nbs1KO cells after transformation with either an homologous vector designed to inactivate the PpAPT gene (PpAPT-KO2 or PpAPT-KO3) or a vector sharing no homology with the moss genome (pBHRF or pBNRF) to determine the rate of untargeted transgene integration. Relative transformation frequency (RTF) was reduced to approximately one-third of the wild-type level in mre11 and rad50 mutants, but gene targeting (GT) was reduced by at least an order of magnitude in both Ppmre11Δ and Pprad50Δ strains compared to WT, while untargeted integration frequencies were approximately double that observed in WT (Table 1). These data demonstrate that an active MRN complex is required to achieve high GT efficiencies in Physcomitrella, but that a low level of GT is possible in its absence. They also indicate that the untargeted integration of DNA is still supported following the loss of MRN function but that this pathway is not significantly up-regulated as has been observed to occur in Pprad51 mutants (17). Noticeably, RTF, GT and untargeted integration rates were unaffected in the nbs1KO mutant. These observations suggest that both PpRAD50 and PpMRE11, but not PpNBS1, are directly involved in DNA DSB recognition and the targeted integration of transgenes following transformation.
Table 1.
Comparison of transformation and gene targeting efficiencies
| Genotypes | PpAPT-KO |
pBHRF or pBNRF |
||||
|---|---|---|---|---|---|---|
| RTFa | AntibR | 2FAR | GTb | RTFa | AntibR | |
| Wild type | 1 ± 0.1c | 287 (95.7 ± 10.8c) | 212 (70.7 ± 11.2c) | 73.9 ± 3.3c | 0.09 ± 0.04d | 14 (7 ± 2.8c) |
| mre11Δ | 0.37 ± 0.1c | 81 (27 ± 4c) | 6 (2 ± 0.5c) | 7.4 ± 1c | 0.2 ± 0.01d | 34 (17.7 ± 2.8c) |
| rad50Δ | 0.35 ± 0.1c | 76 (25.3 ± 4.5c) | 4 (1,3 ± 0.6c) | 5.3 ± 2.1c | 0.17 ± 0.01d | 31 (15.5 ± 2.5c) |
| nbs1Δ | 0.94 ± 0.1c | 261 (87 ± 5.6c) | 178 (59.3 ± 4.5c) | 68.2 ± 1.4c | 0.17 ± 0.03d | 24 (12 ± 1.4c) |
aRelative transformation frequencies (RTF in 0/00) express the frequency of antibiotic-resistant transgenic strains in the whole regenerated population.
bGT efficiencies (in percentage) express the frequency of 2-FA resistant among the population of antibiotic-resistant transgenic strains.
cAverage and standard deviation was determined from three independent experiments, each of them performed in duplicates.
dAverage and standard deviation was determined from two independent experiments, each of them performed in duplicates.
PpMRE11 and PpRAD50 but not PpNBS1 are essential for normal growth and development
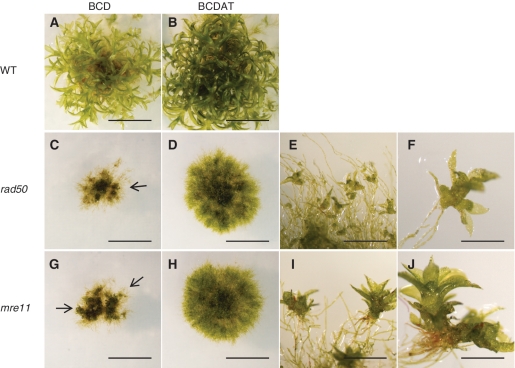
All the plants containing disruptions or deletions in the PpMRE11 and PpRAD50 genes exhibited a severe developmental phenotype (Figure 2). On minimal BCD medium, protonemal growth was strongly reduced and eventually ceased after a month (Figure 2A, C and G). At this stage, colonies comprised both chloronemal and caulonemal cells and carried only a few abortive gametophore initials, whereas WT colonies carried numerous fully differentiated leafy shoots (Figure 2A, C and G). In both mutants, the proportion of chloronemata was enhanced on ammonium tartrate-supplemented medium (BCDAT), which improved protonemal growth (Figure 2B, D and H) and enabled the isolation of numerous protoplasts. The rate of protoplast regeneration was approximately half that of WT (data not shown). Gametophore differentiation was also slightly improved on BCDAT medium, with numerous leafy shoot initials observed in 2-month-old colonies of both mutants (Figure 2E, F, I and J). However, further development into fully expanded leafy shoots was arrested in both strains, although at an earlier stage in rad50 mutants than in mre11 mutants (compare Figure 2F and J) and both mutants were thus unable to differentiate reproductive organs. In contrast, all of the Ppnbs1KO disruptant lines were indistinguishable from wild-type in both growth rate and developmental progression, producing normal gametophores and viable spores, demonstrating that the disruption of PpNBS1 was neither detrimental to development nor to meiosis. These data show that a functional MRN complex is essential for normal completion of processes involved in development. Noticeably, PpNBS1 is not required to complete these processes. The similar phenotype displayed by both rad50 and mre11 mutants argues for the involvement of the whole MRN complex in these processes. Our data indicate that this complex is involved in the coordination between developmental programme and DNA damage repair and/or cell-cycle control, and future experiments will assess the molecular mechanisms underlying these MRN functions.
Figure 2.
Vegetative developmental phenotypes of mre11 and rad50 mutants. WT (A and B), Rad50 7-20 (C–F) and Mre11 1-195 (G–J) 30-day-old colonies grown on BCD (A, C and G) or BCDAT (B, D and H) medium, scale 1 cm. (E, F, I and J) aborted gametophores observed at the edge of 2-month-old colonies grown on BCDAT, scale bar 500 mm in E and I, 200 mm in F and J.
The mre11 and rad50 mutants display increased sensitivity to DNA damage but no significant mutator phenotype
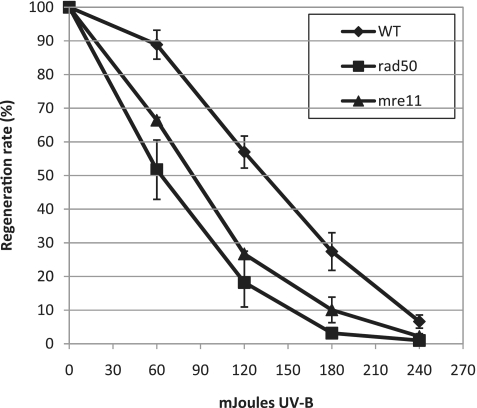
Wild-type and mre11 and rad50 mutant plants were also analysed for their sensitivity to DNA damaging agents. Sensitivity of mutants and WT strains to UV-B (308 nm) was investigated using a protoplast survival assay (17). Both strains displayed increased sensitivity to UV-B compared to the WT (Figure 3). We further investigated sensitivity of the mutants to the DSB inducing agent bleomycin. We first monitored the growth of WT and mutant explants submitted to chronic exposure to different concentrations of bleomycin over a 3-week period. In WT and nbs1KO strains, growth was impaired at low doses (1–40 ng/ml), whilst higher concentrations (200 ng/ml–1 μg/ml) were lethal (Figure 4A and Supplementary Figure S3). In contrast, Ppmre11 and Pprad50 disruption and deletion mutants displayed hypersensitivity to bleomycin. At concentrations below 8 ng/ml, little or no growth took place, although the tissue remained green. At or above this concentration, all mre11 and rad50 mutant lines were killed (Figure 4A, Supplementary Figure S3A and B).
Figure 3.
Hypersensitivity of the rad50 and mre11 mutants to UV-B treatment. Survival curves of wild type and rad50 and mre11 mutant protoplasts regenerating after exposure to UV-B treatment. Wild-type survival is represented with diamonds, rad50 mutant survival is represented with squares and mre11 mutant survival is represented with triangles. Error bars indicate SDs based on at least two independent experiments in all cases.
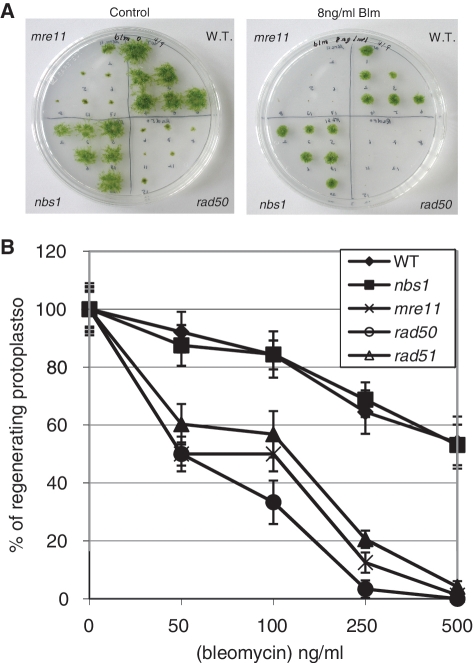
Figure 4.
Hypersensitivity of the rad50 and mre11 mutants to bleomycin. (A) Wild-type and mutant plants were inoculated as six explants into quadrants of plates containing standard growth medium with or without bleomycin at 8 ngml−1. For the mutant strains, each inoculum represents an independent disruption line. The photograph illustrates the extent of growth 10 days following inoculation of the explants. (B) Survival curves of wild type and nbs1, mre11, rad50 and rad51 mutant protoplasts regenerating after exposure to bleomycin treatment. Error bars indicate SD based on at least two independent experiments in all cases.
We tested the acute toxicity of bleomycin in wild type and of the different mutants at the cellular level. Following incubation for 1 h with increasing concentrations of bleomycin, the ability of protoplasts to divide and regenerate into colonies was assessed by subculture on drug-free medium. Survival was calculated as the ratio of protoplasts surviving after 15 days regeneration following treatment to the number of protoplasts undergoing normal regeneration without treatment. The LD50 for the wild type and nbs1 mutant was about 500 ng/ml bleomycin, whereas the mre11 and rad50 cells were more sensitive, with an LD50 of ∼50 ng/ml (Figure 4B). The mre11 and rad50 cells were even more sensitive than the rad51-1-2 double mutant, already described as hypersensitive to bleomycin (16).
Acute treatment of intact protonemal tissue also adversely affected the subsequent growth rate of the recovering protonemata, with the mre11 and rad50 mutants being more sensitive to bleomycin than the wild type (Supplementary Figure S3C).
Mutator phenotypes in the absence of proteins essential for HR have been described in S. cerevisiae or A. nidulans (31,32). We therefore evaluated the mutator phenotype of the moss MRN mutants to assess their ability to repair endogenous DNA damage, using as reporter loss of function of the adenine phosphoribosyl transferase gene (PpAPT) as previously described (18). The frequencies of apt mutations were lower than 3 × 10−7 in wild type, rad50 and nbs1 mutants, was 4 × 10−7 for mre11 but was ∼100-fold higher (3.3 × 10−5) in msh2 mutants (Supplementary Table S3), which is in good accordance with previous results obtained with this mutant (18). These results indicate that loss of proteins of the MRN complex does not lead to a significant mutator phenotype in P. patens. It is most likely that DNA-DSB repair defects in the mre11 and rad50 mutants cause genomic damage so much more severe than the point mutations seen in the msh2 mutant that cell death results.
DNA-DSB repair is not affected in mre11 and rad50 mutants
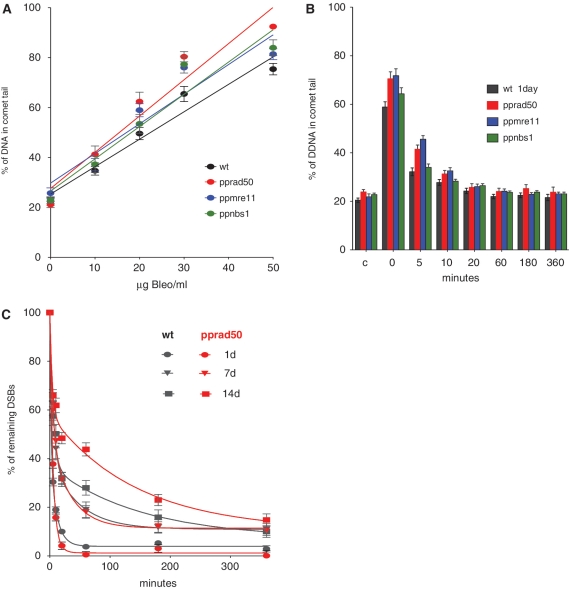
Gene targeting in the mre11 and rad50 mutants was severely impaired, while untargeted integration frequencies were 2-fold higher than those observed in WT. Since the rad50 and mre11 mutants were clearly impaired in growth and hypersensitive to DNA damage, we reasoned that the mutants remained capable of ligating broken ends of DNA molecules, but in an ‘unsupervised’, and therefore inaccurate manner. We tested this by directly estimating the ability of wild-type and mutant strains to repair DNA damage following acute exposure to bleomycin using single nucleus gel electrophoresis (the ‘comet assay’). Treatment with bleomycin for 1 h resulted in a linear, dose-dependent fragmentation of genomic DNA in both wild-type and mutant lines, with the rad50 and mre11 lines exhibiting a greater susceptibility to DNA damage than the wild-type and Ppnbs1 lines, respectively (Figure 5A). The rate of repair of DSBs was determined by measuring the proportion of fragmented DNA at intervals during a recovery period.
Figure 5.
Kinetics of DNA repair in wild-type and mutant plants. (A) Bleomycin dose-response. Protonemal tissue from wild-type and mutant lines was treated with bleomycin for 1 h at the indicated concentrations, prior to nuclear extraction and the analysis of DNA damage by single-cell electrophoresis (the ‘comet assay’). The extent of DNA damage is indicated by the proportion of DNA detected in the fragmented fraction (the ‘comet tail’). The background level of genomic DNA damage in all lines is similar, at between 20 and 30%, indicating that the mutations have no significant effect on natural levels of DNA fragmentation. (B) Repair kinetics in 1-day regenerated protonemata. In both wild-type and mutant lines, the fragmentation of DNA induced by bleomycin is repaired with rapid kinetics (t1/2 between 1 and 4 min). (C) Repair kinetics in relation to protonemal age. As protonemata are regenerated for longer periods (resulting in a concomitant reduction in the proportion of mitotically active apical cells), so the proportion of the rapid phase DNA repair declines. This occurs in both the wild-type and the rad50KO mutant lines.
Both wild-type and mutant lines exhibited similarly high rates of DNA repair with a characteristic biphasic profile: an initial rapid phase (t1/2 1–4 min) accounting for ∼60% of the fragmented DNA, followed by a slower phase (t1/2 7–90 min) accounting for the remainder (Figure 5B, Table 2). The rate of DNA repair was closely correlated with the age of the protonemal tissue following subculture. Tissue that was homogenized and subcultured for only 1 day comprised largely short protonemal fragments, four to seven cells in length. This tissue exhibited the most rapid repair kinetics (Figure 5B, Table 2). Tissue that was subcultured for 1 week comprised longer filaments 15–20 cells in length, whilst after subculture for 2 weeks, the filaments were over 30 cells long. These tissues were progressively slower in their DNA repair kinetics (Figure 5C, Table 2) with an increasing proportion of the DNA-DSBs being repaired with slow-phase kinetics. We ascribe these age-related differences to the relative representation of apical cells within the protonemal population. Physcomitrella protonemata grow by serial division of the apical cells, so that in a 1d-subcultured homogenate, we estimate the proportion of mitotically active apical cells to comprise 30–50% of the total cell population. This proportion will be 10–15% in 7d-subcultured tissue, and ∼3% in 14d-sucbultured protonemata. Thus, the initial rapid phase of DNA repair can be accounted for by processes undertaken in the mitotically competent apical cells, whilst the slow-phase repair kinetics is likely due to processes carried out in mitotically inactive subapical cells.
Table 2.
Kinetics of DNA repair in wild-type and mutant strains
| Genotypes | Tissue age | t1/2fast (min) | % fast | t1/2 slow (min) |
|---|---|---|---|---|
| wild-type | 1d | 1.2 | 61.4 | 7.6 |
| wild-type | 7d | 4.0 | 67.6 | 32.9 |
| wild-type | 14d | 3.8 | 67.7 | 103.4 |
| mre11KO | 1d | 4.1 | 96.5 | 84.1 |
| nbs1KO | 1d | 1.9 | 84.1 | 17.0 |
| rad50KO | 1d | 2.9 | 71.4 | 5.6 |
| rad50KO | 7d | 2.9 | 52.6 | 18.2 |
| rad50KO | 14d | 2.7 | 46.8 | 100.0 |
Although differences can be seen in the rates of DNA repair between mutant and wild-type strains, these are not dramatic. Clearly, the extensive fragmentation of DNA that occurs during the initial bleomycin treatment is being rapidly reversed, even in mutants in which components of the principal DNA surveillance and repair system for both HR and NHEJ-mediated repair (PpMRE11 and PpRAD50) have been eliminated.
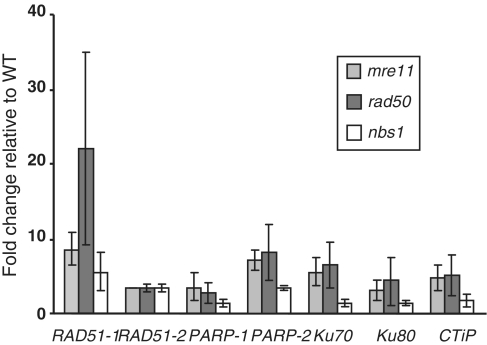
MRN mutants exhibit enhanced repair gene expression
One possibility is that in the absence of a viable MRN complex, DNA-DSBs are repaired, but in an ‘unsupervised’ manner. In the absence of the tethering function to hold broken ends in close proximity, repair may be inaccurate, generating increased numbers of deletions and promoting end joining between inappropriate ends, resulting in increased disruption of essential genes and consequent loss of viability and cell death. Analysis of the expression of a selection of DNA repair genes, implicated in HR, NHEJ and alternative end-joining processes, showed a significantly enhanced accumulation of repair gene transcripts in MRN mutants relative to wild-type, and this was most marked in the Ppmre11KO and Pprad50KO mutants (Figure 6). This suggests that in the absence of a functional MRN complex, the cell initiates an emergency response by the rest of the DNA repair machinery. The levels of induction of the repair genes observed in the mutant lines are comparable to those seen in wild-type protonemal tissue in response to DNA-DSB induction by bleomycin (Whitaker,J., personal communication).
Figure 6.
Gene expression analysis of DNA repair genes in the MRN mutants. Quantitative determination of the relative abundances of transcripts encoding DNA repair genes (PpRad51-1, PpRad51-2, PpPARP-1, PpPARP-2, PpKu70, PpKu80 and PpCtIP) in 7-day-old wild-type, Ppmre11KO, PpRad50KO and Ppnbs1KO strains was done by quantitative real-time PCR. Relative transcript abundance was calculated using the ΔΔCt method and normalized to the wild-type value. Error bars indicate SD based on at least three independent experiments with two replicates for each sample in all cases.
We then directly tested the nature of DNA repair in an MRN mutant line, by screening a series of apt mutants generated by bleomycin treatment of both wild-type and the Pprad50KO mutant. Sublethal doses of bleomycin, determined by growth tests of bleomycin-treated protonemal tissue (Supplementary Figure S3C), were used to generate mutants selected on the basis of resistance to 2-FA. The mutability of the PpAPT gene in the Pprad50KO line was observed to be at least an order of magnitude greater than that in the wild type (Supplementary Table S2). The nature of the induced mutations was examined by PCR-amplification and sequencing of the APT gene from a number of lines. For wild-type, each mutant analysed contained only point mutations within the APT coding sequence and introns (Table 3). In contrast, three of the seven mutants analysed in the Pprad50KO background contained deletions, varying in length between 10 and 747 bp (Table 3). This supports our working hypothesis that MRN-unsupervised repair generates more severe forms of genomic damage.
Table 3.
Mutations identified in the APT genomic sequence in wild-type and Pprad50-KO 2-FA resistant clones
| Clone# | Genotype | Mutations in CDS |
Mutations in introns |
||
|---|---|---|---|---|---|
| Point mutations | Deletionsa | Point mutations | Deletionsa | ||
| 1 | rad50/apt | + T (2095)a | + T (1683)a | – | |
| Δ T (2517)a | |||||
| 2 | rad50/apt | Δ T (1706)a | 1450–1455 | + T (1683)a | – |
| 3 | rad50/apt | A to T (1461)a | 1466–1521 | + T (1683)a | – |
| G to C (1534)a | |||||
| 4 | rad50/apt | T to C (1291)a | Δ G (1524)a | – | |
| C to G (1752)a | + T (1683)a | ||||
| Δ T (1730)a | + T (2517)a | ||||
| 5 | rad50/apt | Δ G (1524)a | – | + T (1683)a | – |
| 7 | rad50/apt | – | 1050–1797 | – | – |
| 11 | RAD50/apt | A to G (1491)a | – | Δ A (1052)a | – |
| A to C (1498)a | T to C (2330)a | ||||
| G to C (2327)a | |||||
| + GT (2328)a | |||||
| 12 | RAD50/apt | – | – | Δ A (1052)a | – |
| T to G (1376)a | |||||
| C to A (1569)a | |||||
| + G (2328)a | |||||
| 13 | RAD50/apt | A to C (2475)a | – | A to T (1591)a | – |
| Δ T (2499)a | |||||
aPosition 1 corresponds to the first nucleotide in the genomic PpAPT sequence DQ117987.
DISCUSSION
GT Efficiency is reduced in the Physcomitrella rad50 and mre11 mutants
In Physcomitrella the protein at the heart of the HR pathway, PpRAD51, is required simultaneously to enable targeted integration by HR, and to repress untargeted insertion by an as yet unidentified molecular mechanism (17). The unique GT efficiency of Physcomitrella suggests that DNA DSBs are predominantly repaired by HR in moss cells. Our analysis of mutants in the principal sensor of DNA DSBs, the MRN complex, further shows that although a fully active MRN complex clearly appears to be necessary for high-efficiency targeted transgene integration, a background level of HR with gene targeting reduced to ∼8.6% of wt is still maintained in the absence of either a functional PpMRE11 or PpRAD50 protein. This contrasts with the complete abolition of gene targeting seen in rad51 null mutants, a component specific to the HR pathway (17). Noticeably the overall frequency of untargeted transgene integration is not reduced in the mre11 and rad50 mutants relative to wild type (Table 1), implying that whatever mechanisms undertake random transgene integration, these are relatively unimpaired in the absence of PpMRE11 or PpRAD50. Together with the observation that a number of DNA repair genes show enhanced expression levels in mre11 and rad50 mutants, our results suggest that while some HR-mediated repair may still operate in mre11 and rad50 mutants, the HR pathway is unlikely to account for the majority of the DNA-DSBs that are rapidly religated in these mutants.
NBS1 is not required for growth and development or for HR in Physcomitrella
Phenotypic analyses of mutants in the MRN complex in moss failed to identify a detectable difference between wild-type and Ppnbs1 knock-outs. In eukaryotes, the MRN-complex proteins act as the ‘gatekeepers’ of the DNA-DSB response, directing the repair of DSBs into either the NHEJ or HR pathways through the activation of the ATM or ATR kinases that (in mammalian cells) are recruited to sites of DNA damage through analogous mechanisms involving conserved interaction motifs (6). The NBS1 protein is involved in the recruitment of ATM to DNA-DSBs and ATRIP is involved in the recruitment of ATR to single-stranded DNA (ssDNA). The recruitment of ATM is mediated by its direct interaction with NBS1 which becomes phosphorylated at residues conserved between the Arabidopsis and Physcomitrella NBS1 sequences (6,33). In A. thaliana, nbs1/atm double mutants appear additive in their negative consequences for growth and fertility relative to the wild-type and single mutants (34). ATM is necessary for the imposition of a cell-cycle checkpoint, and for the induction of DNA-damage-responsive gene expression in Arabidopsis, in which the principal DNA repair pathway is through NHEJ (33,35). DNA repair in Physcomitrella is believed to operate primarily via the HR pathway, which in mammalian cells, at least, depends principally on the activity of the ATR kinase. Thus, impairment of ATM-related signalling in the Ppnbs1KO mutant may have relatively little impact on growth and fertility, if NHEJ is subordinate to HR. This conclusion is also supported by the observation that HR-dependent gene targeting is unaffected in the Ppnbs1KO mutant. In contrast, NBS1 has been shown to be essential to HR in chicken DT40 cells, possibly by processing recombination intermediates (36) and in human cells recruitment of ATR to sites of DNA damage is dependent on ATM (37). This implies that in Physcomitrella NBS1 may not be involved in the production of single-stranded tails that are the substrates for HR and that induction of the HR pathway, potentially by the ATR signalling, is independent of ATM. In this respect Physcomitrella would more resemble budding yeast than mammals, as Tel1, the yeast equivalent of ATM, has only minor effects on end-processing and is not required for focus formation by Mec1, the yeast homolog of ATR (38,39). Alternatively, despite the conservation of the ATM interaction domain in the PpNBS1 protein, ATM activation might be independent of NBS1 in Physcomitrella. In this context, it would be of interest to study the exact roles of ATM and ATR in Physcomitrella.
RAD50 and MRE11 are essential for growth and development in Physcomitrella
Null mutants in any components of the MRN complex are lethal in vertebrates (5) and are severely compromised in both budding (40) and fission yeast (41). This is not the case in plants: in Arabidopsis, AtRad50 and AtMre11 mutants are impaired in growth, fertility and in their ability to recover from genotoxic stress (42,43), whereas Atnbs1 mutant plants grow normally and are fully fertile but are sensitive to the DNA cross-linking agent, mitomycin C (34).
Our analyses show that defects in the MRN complex can adversely affect moss development. While the Ppnbs1KO mutant completed its life cycle normally and displayed wild-type levels of susceptibility to DNA damage, the Ppmre11 and Pprad50 strains displayed a strong and similar developmental phenotype. This included defects in cell viability (reduced protoplast regeneration rates), in cell-cycle progression and cell growth (reduced colony growth) and in the completion of a complex developmental programme (abortive leafy shoot development). Precocious arrest of colony growth was also observed on minimal medium, which most likely reflects early senescence. This pleiotropic phenotype is much stronger than that observed in the HR-deficient Pprad51 mutants (16,17) and implies that genome integrity is more severely impaired by loss of function of the MRN complex than by the inactivation of the HR pathway. The phenotype of Ppmre11 and Pprad50 mutants also differs from that previously reported for Ppmsh2 mutants which do not display a strong juvenile phenotype but accumulate mutations and phenotypic alterations during development (18). Noticeably both Pprad51 and Ppmsh2 mutants also displayed a detectable mutator phenotype that is absent in Ppmre11 and Pprad50, probably because MRN mutants accumulate a more extensive and harmful type of DNA damage that accelerates senescence.
Induced DSBs in Physcomitrella can be repaired via a mechanism independent of the MRN complex
Direct analysis of DNA-DSB repair by single-cell electrophoresis showed little difference in the rate of repair of DNA-DSBs in the mre11 and rad50 mutants compared to wild type. Whilst some HR-mediated transgene integration still occurs in mre11 and rad50 mutants, it is unlikely that single-strand annealing (SSA) or homologous strand exchange (HR), which require end-processing, account for this rapid religation. Therefore, the repair of DSBs in the mre11 and rad50 mutants probably occurs via a pathway related to NHEJ. However, the reduced growth and survival of these mutants indicate that such a pathway reduces the genetic stability characteristic of MRN-supervised DNA repair.
Two different NHEJ pathways have been already described, the highly efficient canonical Ku- and DNA ligase IV-mediated NHEJ pathway (C-NHEJ) in which most ends are successfully rejoined without alteration of DNA sequence information (44) and an evolutionarily conserved alternative end-joining pathway (A-NHEJ) (45) thought to proceed via microhomology-mediated end joining (MMEJ), even if the relationship between A-NHEJ and MMEJ is still unclear (45). A-NHEJ represents a major source of DSB-induced genome rearrangements (translocations, deletions and inversions) (46–48) and appears to utilize binding of DNA ends by PARP-1 (polyADP ribose polymerase) and ligation by DNA Ligase III in a Ku-independent process (49,50) and involve the interaction between the MRN complex and DNA ligase IIIα/XRCC1 (51). The function of DNA ligase III is absent in plants, being substituted by DNA ligase I in base-excision repair (52). It may therefore be significant that the Ppmre11 and Pprad50 mutants show substantially elevated levels of expression of PARP and other DNA-repair associated genes, relative to the wild type, and this elevated gene expression may be responsible for the activity of an A-NHEJ repair pathway in the absence of an active MRN complex. Existence of A-NHEJ in plants has been inferred from observations that although the frequency of transgene insertion was reduced in mutants deficient in NHEJ components such as Atku80 and AtligIV, it was not abolished (53–55), from the observation of illegitimate fusions between chromosome arms in telomerase-deficient Arabidopsis, even in an Atku80/Atmre11 mutant background (56), from the recent demonstration of rapid ligation of bleomycin-induced DNA-DSBs in the NHEJ-deficient Atku80 and Atlig4 mutants (57) and from kinetic measurements of assembly and processing of DSB-specific γ-H2AX complexes in Arabidopsis mutants deficient in core components of the C-NHEJ and A-NHEJ pathways (58). In budding yeast both C-NHEJ and A-NHEJ are MRX-dependent processes, with the exonuclease activity of Mre11 playing an important role (59–62), whilst in vertebrates varying roles for MRN complex components have been reported (63–66). Whatever the role of the MRN complex in C-NHEJ or A-NHEJ in plants, it is likely that an NHEJ-like pathway mediates the rapid DSB repair observed in Physcomitrella mre11 and rad50 mutants. However, because these mutants are clearly hypersensitive to DNA damage yet do not show a mutator phenotype, it would appear that whatever rejoining of DNA ends is occurring, it is ‘unsupervised’ and results in genomic perturbations so severe that cells suffering bleomycin-induced breakage soon die.
The rapid interaction of the MRN complex with DNA-DSBs is essential for their stabilization, through the tethering of the adjacent free ends by the Rad50 coiled-coil/zinc hook domains (5). By retaining broken ends in close proximity, the MRN complex thereby supervises the DNA repair process, ensuring that the correct ends are rejoined, and recruiting additional factors required for either NHEJ or HR-based repair. In the absence of such tethering, unsupervised end-joining by backup pathways might occur between unrelated DNA sequences, with the concomitant accumulation of cytotoxic mutations accounting for the reduced rates of growth and enhanced sensitivity to DNA-damaging agents observed in the Ppmre11 and Pprad50 mutants. Combinations of mutations affecting C- or A-NHEJ (58) with mre11 or rad50 mutations should give us insight into the mechanism behind this DSB DNA repair.
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–4.
FUNDING
Institut National de la Recherche Agronomique, Agence Nationale de la Recherche (Grant number ANR GNP05008G to F.N.); the UK Biotechnology and Biological Sciences Research Council (Grant number BB/I006710/1 to A.C.C. and Y.K.); the Swiss National Science Foundation (Grant number 31003A_127572 to D.S.); and the Ministry of Education, Youth and Sports of the Czech Republic (projects LC06004 and 1M0505; EU 6FP projects TAGIP LSH-2004_1.1.0-1 and COMICS LSHB-CT-2006-037575 to K.J.A., J.K. and M.H.). Funding for open access charge: Agence Nationale de la Recherche (Grant number ANR GNP05008G).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 2.Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair. 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rupnik A, Lowndes NF, Grenon M. MRN and the race to the break. Chromosoma. 2010;119:115–135. doi: 10.1007/s00412-009-0242-4. [DOI] [PubMed] [Google Scholar]
- 5.Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair. 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 6.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 7.Lamarche BJ, Orazio NI, Weitzman MD. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010;584:3682–3695. doi: 10.1016/j.febslet.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanin M, Paszkowski J. Plant genome modification by homologous recombination. Curr. Opin. Plant Biol. 2003;6:157–162. doi: 10.1016/s1369-5266(03)00016-5. [DOI] [PubMed] [Google Scholar]
- 9.Britt AB, May GD. Re-engineering plant gene targeting. Trends Plant Sci. 2003;8:90–95. doi: 10.1016/S1360-1385(03)00002-5. [DOI] [PubMed] [Google Scholar]
- 10.Ray A, Langer M. Homologous recombination: ends as the means. Trends Plant Sci. 2002;7:435–440. doi: 10.1016/s1360-1385(02)02327-0. [DOI] [PubMed] [Google Scholar]
- 11.Tzfira T, Li J, Lacroix B, Citovsky V. Agrobacterium T-DNA integration: molecules and models. Trends Genet. 2004;20:375–383. doi: 10.1016/j.tig.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Schaefer DG, Zryd JP. Efficient gene targeting in the moss Physcomitrella patens. Plant J. 1997;11:1195–1206. doi: 10.1046/j.1365-313x.1997.11061195.x. [DOI] [PubMed] [Google Scholar]
- 13.Schaefer DG. Gene targeting in Physcomitrella patens. Curr. Opin. Plant Biol. 2001;4:143–150. doi: 10.1016/s1369-5266(00)00150-3. [DOI] [PubMed] [Google Scholar]
- 14.Kamisugi Y, Cuming AC, Cove DJ. Parameters determining the efficiency of gene targeting in the moss Physcomitrella patens. Nucleic Acids Res. 2005;33:e173. doi: 10.1093/nar/gni172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamisugi Y, Schlink K, Rensing SA, Schween G, von Stackelberg M, Cuming AC, Reski R, Cove DJ. The mechanism of gene targeting in Physcomitrella patens: homologous recombination, concatenation and multiple integration. Nucleic Acids Res. 2006;34:6205–6214. doi: 10.1093/nar/gkl832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markmann-Mulisch U, Wendeler E, Zobell O, Schween G, Steinbiss HH, Reiss B. Differential requirements for RAD51 in Physcomitrella patens and Arabidopsis thaliana development and DNA damage repair. Plant Cell. 2007;19:3080–3089. doi: 10.1105/tpc.107.054049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaefer DG, Delacote F, Charlot F, Vrielynck N, Guyon-Debast A, Le Guin S, Neuhaus JM, Doutriaux MP, Nogue F. RAD51 loss of function abolishes gene targeting and de-represses illegitimate integration in the moss Physcomitrella patens. DNA Repair. 2010;9:526–533. doi: 10.1016/j.dnarep.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Trouiller B, Schaefer DG, Charlot F, Nogue F. MSH2 is essential for the preservation of genome integrity and prevents homeologous recombination in the moss Physcomitrella patens. Nucleic Acids Res. 2006;34:232–242. doi: 10.1093/nar/gkj423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knight CD, Cove DJ, Cuming AC, Quatrano RS. Molecular Plant Biology. Vol. 2. Oxford: Oxford University Press; 2002. pp. 285–299. [Google Scholar]
- 20.Schaefer D, Zryd JP, Knight CD, Cove DJ. Stable transformation of the moss Physcomitrella patens. Mol. Gen. Genet. 1991;226:418–424. doi: 10.1007/BF00260654. [DOI] [PubMed] [Google Scholar]
- 21.Kamisugi Y, Cuming AC. The evolution of the abscisic acid-response in land plants: comparative analysis of group 1 LEA gene expression in moss and cereals. Plant Mol. Biol. 2005;59:723–737. doi: 10.1007/s11103-005-0909-z. [DOI] [PubMed] [Google Scholar]
- 22.Futers TS, Onde S, Turet M, Cuming AC. Sequence analysis of two tandemly linked Em genes from wheat. Plant Mol. Biol. 1993;23:1067–1072. doi: 10.1007/BF00021821. [DOI] [PubMed] [Google Scholar]
- 23.Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 24.Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat. Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 25.Angelis KJ, Dusinska M, Collins AR. Single cell gel electrophoresis: detection of DNA damage at different levels of sensitivity. Electrophoresis. 1999;20:2133–2138. doi: 10.1002/(SICI)1522-2683(19990701)20:10<2133::AID-ELPS2133>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 26.Menke M, Chen I, Angelis KJ, Schubert I. DNA damage and repair in Arabidopsis thaliana as measured by the comet assay after treatment with different classes of genotoxins. Mutat. Res. 2001;493:87–93. doi: 10.1016/s1383-5718(01)00165-6. [DOI] [PubMed] [Google Scholar]
- 27.Moncalian G, Lengsfeld B, Bhaskara V, Hopfner KP, Karcher A, Alden E, Tainer JA, Paull TT. The rad50 signature motif: essential to ATP binding and biological function. J. Mol. Biol. 2004;335:937–951. doi: 10.1016/j.jmb.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 28.Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi-Iwai Y, Sonoda E, Sasaki MS, Morrison C, Haraguchi T, Hiraoka Y, Yamashita YM, Yagi T, Takata M, Price C, et al. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 1999;18:6619–6629. doi: 10.1093/emboj/18.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gherbi H, Gallego ME, Jalut N, Lucht JM, Hohn B, White CI. Homologous recombination in planta is stimulated in the absence of Rad50. EMBO Rep. 2001;2:287–291. doi: 10.1093/embo-reports/kve069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prakash L. The relation between repair of DNA and radiation and chemical mutagenesis in Saccharomyces cerevisiae. Mutat. Res. 1976;41:241–248. doi: 10.1016/0027-5107(76)90097-x. [DOI] [PubMed] [Google Scholar]
- 32.Seong KY, Chae SK, Kang HS. Cloning of an E. coli RecA and yeast RAD51 homolog, radA, an allele of the uvsC in Aspergillus nidulans and its mutator effects. Mol. Cells. 1997;7:284–289. [PubMed] [Google Scholar]
- 33.Garcia V, Bruchet H, Camescasse D, Granier F, Bouchez D, Tissier A. AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell. 2003;15:119–132. doi: 10.1105/tpc.006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waterworth WM, Altun C, Armstrong SJ, Roberts N, Dean PJ, Young K, Weil CF, Bray CM, West CE. NBS1 is involved in DNA repair and plays a synergistic role with ATM in mediating meiotic homologous recombination in plants. Plant J. 2007;52:41–52. doi: 10.1111/j.1365-313X.2007.03220.x. [DOI] [PubMed] [Google Scholar]
- 35.Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 2006;48:947–961. doi: 10.1111/j.1365-313X.2006.02931.x. [DOI] [PubMed] [Google Scholar]
- 36.Tauchi H, Kobayashi J, Morishima K, van Gent DC, Shiraishi T, Verkaik NS, vanHeems D, Ito E, Nakamura A, Sonoda E, et al. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature. 2002;420:93–98. doi: 10.1038/nature01125. [DOI] [PubMed] [Google Scholar]
- 37.Adams KE, Medhurst AL, Dart DA, Lakin ND. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene. 2006;25:3894–3904. doi: 10.1038/sj.onc.1209426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubrana K, van Attikum H, Hediger F, Gasser SM. The processing of double-strand breaks and binding of single-strand-binding proteins RPA and Rad51 modulate the formation of ATR-kinase foci in yeast. J. Cell. Sci. 2007;120:4209–4220. doi: 10.1242/jcs.018366. [DOI] [PubMed] [Google Scholar]
- 39.Mantiero D, Clerici M, Lucchini G, Longhese MP. Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep. 2007;8:380–387. doi: 10.1038/sj.embor.7400911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wasko BM, Holland CL, Resnick MA, Lewis LK. Inhibition of DNA double-strand break repair by the Ku heterodimer in mrx mutants of Saccharomyces cerevisiae. DNA Repair. 2009;8:162–169. doi: 10.1016/j.dnarep.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raji H, Hartsuiker E. Double-strand break repair and homologous recombination in Schizosaccharomyces pombe. Yeast. 2006;23:963–976. doi: 10.1002/yea.1414. [DOI] [PubMed] [Google Scholar]
- 42.Bundock P, Hooykaas P. Severe developmental defects, hypersensitivity to DNA-damaging agents, and lengthened telomeres in Arabidopsis MRE11 mutants. Plant Cell. 2002;14:2451–2462. doi: 10.1105/tpc.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gallego ME, Jeanneau M, Granier F, Bouchez D, Bechtold N, White CI. Disruption of the Arabidopsis RAD50 gene leads to plant sterility and MMS sensitivity. Plant J. 2001;25:31–41. doi: 10.1046/j.1365-313x.2001.00928.x. [DOI] [PubMed] [Google Scholar]
- 44.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 45.McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi J, Frank KM, Chaudhuri J, Horner J, DePinho RA, Alt FW. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 47.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc. Natl Acad. Sci. USA. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat. Cell. Biol. 2007;9:978–981. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 50.Haber JE. Alternative endings. Proc. Natl Acad. Sci. USA. 2008;105:405–406. doi: 10.1073/pnas.0711334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Della-Maria J, Zhou Y, Tsai MS, Kuhnlein J, Carney JP, Paull TT, Tomkinson AE. Human Mre11/human Rad50/Nbs1 and DNA ligase IIIalpha/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. J. Biol. Chem. 2011;286:33845–33853. doi: 10.1074/jbc.M111.274159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waterworth WM, Kozak J, Provost CM, Bray CM, Angelis KJ, West CE. DNA ligase 1 deficient plants display severe growth defects and delayed repair of both DNA single and double strand breaks. BMC Plant Biol. 2009;9:79. doi: 10.1186/1471-2229-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Friesner J, Britt AB. Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J. 2003;34:427–440. doi: 10.1046/j.1365-313x.2003.01738.x. [DOI] [PubMed] [Google Scholar]
- 54.Li J, Vaidya M, White C, Vainstein A, Citovsky V, Tzfira T. Involvement of KU80 in T-DNA integration in plant cells. Proc. Natl Acad. Sci. USA. 2005;102:19231–19236. doi: 10.1073/pnas.0506437103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Attikum H, Bundock P, Hooykaas PJ. Non-homologous end-joining proteins are required for Agrobacterium T-DNA integration. EMBO J. 2001;20:6550–6558. doi: 10.1093/emboj/20.22.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heacock M, Spangler E, Riha K, Puizina J, Shippen DE. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 2004;23:2304–2313. doi: 10.1038/sj.emboj.7600236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kozak J, West CE, White C, da Costa-Nunes JA, Angelis KJ. Rapid repair of DNA double strand breaks in Arabidopsis thaliana is dependent on proteins involved in chromosome structure maintenance. DNA Repair (Amst) 2009;8:413–419. doi: 10.1016/j.dnarep.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 58.Charbonnel C, Allain E, Gallego ME, White CI. Kinetic analysis of DNA double-strand break repair pathways in Arabidopsis. DNA Repair. 2011;10:611–619. doi: 10.1016/j.dnarep.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 59.Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- 60.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang X, Paull TT. The Mre11/Rad50/Xrs2 complex and non-homologous end-joining of incompatible ends in S. cerevisiae. DNA Repair. 2005;4:1281–1294. doi: 10.1016/j.dnarep.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 63.Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol. Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang J, Dynan WS. Reconstitution of the mammalian DNA double-strand break end-joining reaction reveals a requirement for an Mre11/Rad50/NBS1-containing fraction. Nucleic Acids Res. 2002;30:667–674. doi: 10.1093/nar/30.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 66.Taylor EM, Cecillon SM, Bonis A, Chapman JR, Povirk LF, Lindsay HD. The Mre11/Rad50/Nbs1 complex functions in resection-based DNA end joining in Xenopus laevis. Nucleic Acids Res. 2010;38:441–454. doi: 10.1093/nar/gkp905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.