Abstract
The eukaryotic-like primase from the hyperthermophilic archaeon Sulfolobus solfataricus (SsoPriSL) exhibits a range of activities including template-dependent de novo primer synthesis, primer extension and template-independent terminal nucleotidyl transfer using either rNTPs or dNTPs. Remarkably, the enzyme is able to synthesize products far longer than templates in vitro. Here we show that the long products resulted from template-dependent polymerization across discontinuous templates (PADT) by SsoPriSL. PADT was initiated through either primer synthesis or terminal transfer, and occurred efficiently on templates containing contiguous dCs. Template switching took place when the 3′-end of a growing strand synthesized on one template annealed to another template directly or following the terminal addition of nucleotides, and was subsequently extended on the new template. The key to PADT was the ability of SsoPriSL to promote strand annealing. SsoPriSL catalyzed PADT with either dNTPs or rNTPs as the substrates but preferred the latter. The enzyme remained active in PADT but became inefficient in primer synthesis in vitro when temperature was raised from 55°C to 70°C. Our results suggest that SsoPriSL is capable of bridging noncomplementary DNA ends and, therefore, may serve a role in double-strand DNA break repair in Archaea.
INTRODUCTION
All major replicative DNA polymerases lack the capacity of initiating the synthesis of new DNA strands de novo on single-stranded (ss) templates. DNA primases serve a key role in DNA replication by synthesizing primers, normally short ribonucleotides, that are then elongated by replicative DNA polymerases (1). Most DNA primases fall into two classes based on their structure and relationship with other proteins: the monomeric bacterial DnaG-type primases and the heterodimeric eukaryotic primases (PriSL) complexed with DNA polymerase α (pol α) and the B-subunit (1). In the latter class, the small eukaryotic primase subunit (PriS) contains the active site for RNA synthesis. The function of the large subunit is unclear but may be related to the coordination of primase and polymerase action (1). Typically, bacterial and eukaryotic primases synthesize RNA primers of a defined length (2–16 nt) regardless of the sequence of the template (1). Interestingly, however, Escherichia coli DnaG has been shown to incorporate dNTPs, catalyze terminal transfer and synthesize products much longer than the template under certain conditions (2–6).
Analysis of sequenced genomes has identified homologs of both classes of primases in Archaea. Eukaryotic-like primases from Archaea consist of only PriS and PriL. Archaeal PriSL exhibits promiscuous activities in vitro: de novo synthesis of both DNA and RNA products of various sizes depending on templates, primer extension and terminal nucleotidyl transfer using either rNTPs or dNTPs as the substrates (7–11). The physiological function and regulation of the activities of archaeal PriSL remain to be understood. Archaeal DnaG also shows primase activity in vitro (12). However, unlike archaeal PriSL, the archaeal DnaG appears to synthesize only short RNA primers (e.g. 13-nt primer for DnaG from Sulfolobus solfataricus). PriSL from S. solfataricus has been shown to interact specifically with replication factor C (RFC) and GINS, two essential protein complexes involved in DNA replication (13,14). On the other hand, DnaG from S. solfataricus interacts with components of the exosome, which degrades RNA products during transcriptional turnover (15,16). So the in vivo roles of the two primases in Archaea remain to be understood.
Non-primase proteins homologous to the eukaryal/archaeal primase in sequence and structure have been found in Eukarya and Bacteria (17). For example, DNA polymerases of the Pol X family, a group of small (30–70 kDa) DNA polymerases found in most eukaryotes, resemble the small subunit of the eukaryal/archaeal primase both in primary sequence and crystal structure (18–20). More specifically, the highly conserved catalytic aspartate residues of the archaeal primase were structurally superimposable with those in the catalytic core of Pol β, a member of the Pol X family (19,20). This has led to the suggestion that the archaeal primase may share similarities with the Pol X DNA polymerases in catalytic mechanism (18). Notably, Pol μ, a member of the Pol X family from human, is capable of template-dependent synthesis with either dNTPs or rNTPs as the substrates as well as terminal nucleotidyl transfer (21), as found with archaeal PriSL. Another example is the polymerase domain of LigD, a member of the archaeo–eukaryal primase (AEP) superfamily in bacteria and a homolog of PriS (22–24). LigD consists of ligase, polymerase, phosphoesterase domains and possesses corresponding activities (25). Like archaeal primases, the polymerase domain of LigD synthesizes RNA primers and has DNA-dependent DNA and RNA polymerase as well as 3′-terminal nucleotidyl transferase activities (24). Both Pol μ and LigD are known to function in the nonhomologous end joining (NHEJ) pathway in double-strand break (DSB) repair (26,27). It has been speculated that archaeal PriSL may play a role in DNA repair in addition to DNA replication since the majority of Archaea encode no other proteins homologous to members of either Pol X family or AEP superfamily (17).
In this report, we show that PriSL from S. solfataricus (SsoPriSL) is capable of efficient template-dependent polymerization across discontinuous DNA templates with a minimal requirement for pairing between the growing chain and the new template strand. Therefore, the archaeal primases appear adapted for a role in joining the nonhomologous ends of DNA DSBs in Archaea.
MATERIALS AND METHODS
Overproduction and purification of SsoPriSL
The genes encoding both PriS and PriL subunits (SSO1048 and SSO0557, respectively) of the S. solfataricus primase were overexpressed in E. coli, and the recombinant SsoPriSL complex was purified as described previously (14).
Activity assays for SsoPriSL
The standard assay mixture (20 μl) contained specified concentrations of SsoPriSL, an oligonucleotide template (Supplementary Table S1) and rNTP or dNTP including [α-32P]rNTP or [α-32P]dNTP, respectively, in 50 mM NaOH–glycine (pH 9.1), 10 mM MnCl2 and 20 μg/ml BSA. Reaction time and temperature are indicated in the figure legends. Reaction products were extracted with phenol–chloroform–isoamyl alcohol (PCI), mixed with an equal volume of the gel-loading buffer (98% deionized formamide, 10 mM EDTA and 0.1% bromophenol blue), heated for 5 min at 95°C and cooled rapidly in ice water. The samples were subjected to electrophoresis in polyacrylamide containing 7 M urea in 1× Tris–borate–EDTA (TBE). For product quantification, a known amount of a labeled nucleotide was run along with the samples on the gel. The gel was exposed to X-ray film or quantified by ImageQuant Storm PhosphorImager (Amersham Biosciences).
Nuclease digestion assays
The products of SsoPriSL activity assays were precipitated with ethanol, resuspended in ddH2O and incubated with DNase I (2 U, Fermentas), RNase H (10 U, Fermentas) or RNase T1 (100 U, Fermentas) at 37°C for 30 min in a buffer suggested by the manufacturer. The samples were subjected to electrophoresis in 12% polyacrylamide containing 7 M urea in 1× TBE. The gel was exposed to X-ray film.
Product sequencing
The standard reaction mixture (100 μl) contained 4 μM SsoPriSL and 15 μM oligonucleotide (5′-TTTTTTTCTTTTTTTCCC) in 50 mM NaOH–glycine, pH 9.1, 10 mM MnCl2, 20 μg/ml BSA, 1 mM rATP and 1 mM rGTP. The reaction was incubated at 55°C for 2 h. Following PCI extraction and ethanol precipitation, the products were treated with DNase I. The samples were extracted again with PCI, precipitated with ethanol and subjected to electrophoresis in a 15% polyacrylamide gel containing 7 M urea in 1× TBE. Gel slices corresponding to 18 to 42 nt fragments were excised. The RNA fragments were eluted for 15 h in 0.3 M NaCl at 4°C, precipitated with ethanol and resuspended in ddH2O. A 5′-phosphorylated oligonucleotide (3′-adapter: 5′-pCTGTAGGCACCATCAAT, IDT) was ligated to the 3′-end of the fragments with T4 RNA ligase (10 U, Promega). The ligation products were resolved by electrophoresis in a 15% polyacrylamide gel containing 7 M urea in 1× TBE, and gel slices corresponding to 35 to 60 nt fragments were excised. The fragments were eluted for 15 h in 0.3 M NaCl at 4°C, precipitated with ethanol and resuspended in ddH2O. A 5′- adapter (5′-ATCGTAGGCACCUGAAA, ribonucleotides are underlined, IDT) was ligated to the 5′-end of the fragments with T4 RNA ligase (10 U). The ligation products were subjected to reverse transcription using Superscript III (Invitrogen) with an RT primer (5′-ATTGATGGTGCCTACAG), as described by the manufacturer. The resulting cDNAs were PCR-amplified for 15 cycles with the RT primer and a 5′-primer (5′-ATCGTAGGCACCTGAAA), cloned into the pGEM-T vector (Promega). The vector was then transformed into E. coli JM109 cells. Cells were plated onto indicator agar plates. After incubation for overnight at 37°C, white and light blue colonies were picked for plasmid DNA preparation, and the inserts of the plasmids were sequenced using the SP6 and T7 promoter sequencing primers.
RESULTS
SsoPriSL is capable of template-dependent nucleotide polymerization across discontinuous templates
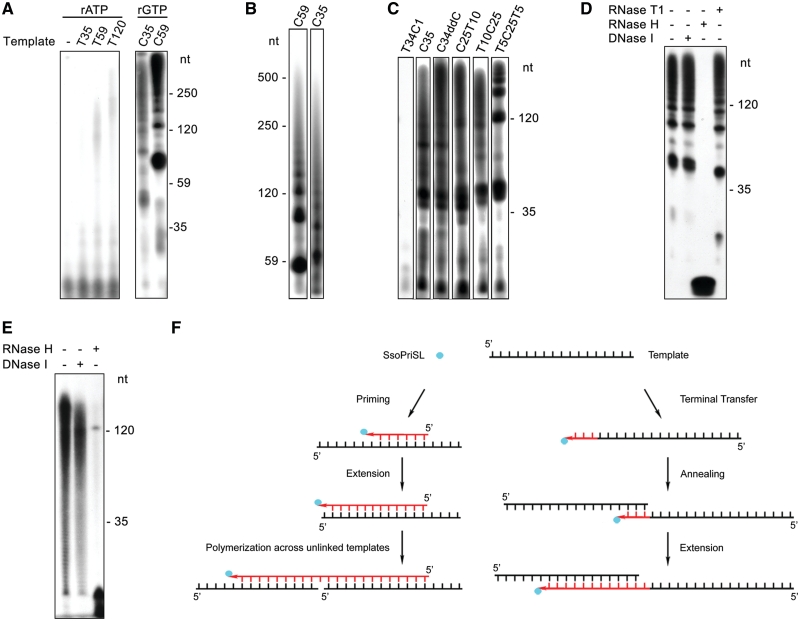
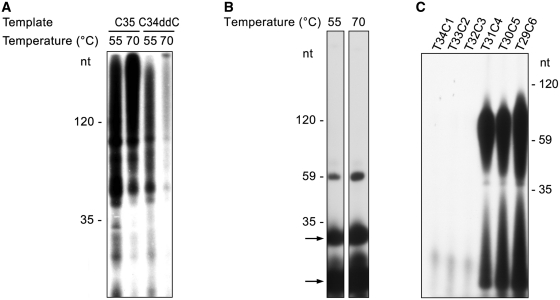
SsoPriSL is known to synthesize products much longer than its oligonucleotide templates (8,9). This ability has been attributed to the terminal nucleotidyl transferase activity of the enzyme (8,9). In addition, SsoPriSL displays significantly higher polymerization activity on an oligo(dC) template than on other homopolymeric DNA templates, especially on an oligo(dA) or oligo(dT) template (9). In the present study, we found that SsoPriSL synthesized substantially more products on oligo(dC) with rGTP as the substrate than on oligo(dT) with rATP as the substrate (Figure 1A). In a typical experiment, incorporation of rGMP on C35 and rAMP on T59 by the enzyme was 83 and 1.6 pmol, respectively, under the standard assay conditions. These values correspond to fractions of template usage (as defined as the molar ratios of the incorporated ribonucleotides to the nucleotides in the total input template molecules) of 47% for C35 and 0.5% for T59. The sizes of the longest products on oligo(dC) (e.g. C59 or C35) were at least ten times longer than those of the templates (Figure 1B). Notably, the enzyme was active in producing a large amount of long products on templates containing a long dC stretch (≥ 25 dCs), whether it existed at the 5′-end, the 3′-end or in the middle of the template (Figure 1C). Even when the template was blocked at the 3′-end by a dideoxy-CMP (C34ddC), synthesis of long products remained. By comparison, synthesis on templates lacking a dC stretch, including one with a single dC at the 3′-end, was drastically reduced. These results indicate that synthesis of the long products may not be attributed entirely to terminal nucleotidyl transfer by SsoPriSL.
Figure 1.
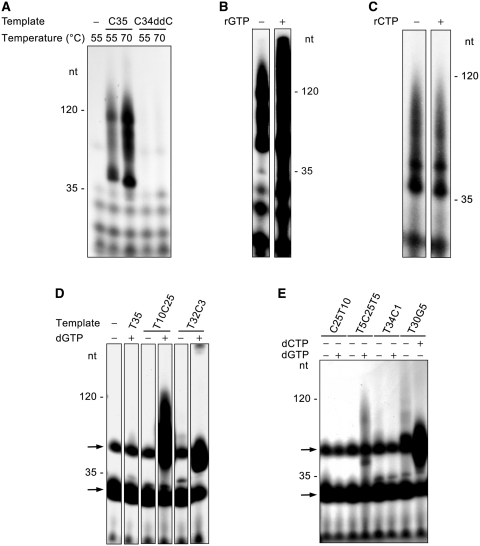
SsoPriSL synthesized products much longer than templates in a template-dependent manner. (A) Comparison of SsoPriSL activity on oligo(dC) and oligo(dT). SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with oligo(dC) (0.25 μM) and [α-32P]rGTP (10 μM, 0.5 Ci/mmol) or oligo(dT) (0.25 μM) and [α-32P]rATP (10 μM, 20 Ci/mmol) in the standard assay mixture. (B) Sizes of the products synthesized by SsoPriSL on oligo(dC) templates. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with templates (0.25 μM) and [α-32P]rGTP (10 μM, 0.5 Ci/mmol) in the standard assay mixture. (C) Dependence of SsoPriSL activity on a dC stretch in template. SsoPriSL (0.5 μM) was incubated for 10 min at 55°C with templates (0.25 μM) and [α-32P]rGTP (10 μM, 5 Ci/mmol) in the standard assay mixture. C34ddC was an oligo(dC) blocked at the 3′-end by a dideoxy-CMP. (D) Nuclease digestion analysis of products synthesized by SsoPriSL on oligo(dC). SsoPriSL (0.5 μM) was incubated for 10 min at 55°C with C35 (0.25 μM) and [α-32P]rGTP (10 μM, 5 Ci/mmol) in the standard assay mixture. Samples were treated with indicated nucleases. (E) Nuclease digestion analysis of products synthesized by SsoPriSL on oligo(dT). SsoPriSL (1.5 μM) was incubated for 60 min at 55°C with T120 (0.25 μM) and [α-32P]rATP (10 μM, 10 Ci/mmol) in the standard assay mixture. All reaction products were subjected to electrophoresis in 12% (A, C, D and E) or 5% (B) polyacrylamide containing 7 M urea. (F) A diagram showing primer synthesis or terminal transfer at the 3′-end of the ssDNA template as the initial step in PADT.
To characterize the products synthesized by SsoPriSL in the above reactions, we treated them with DNase I, RNase H or RNase T1. The products synthesized on oligo(dC) with rGTP as the substrate were completely degraded by RNase H, which cleaves RNA in RNA–DNA hybrids, but were resistant to digestion by DNase I or RNase T1, which degrades ss and double-stranded (ds) DNA or ssRNA at rG residues, respectively (Figure 1D). Therefore, the products appear to be RNA strands hybridized to the DNA template. When products synthesized on oligo(dT) with rATP as the substrate were treated in the same manner, they were largely degraded by RNase H with small amounts of the products of approximately the template size left. The sizes of the products were slightly reduced following treatment with DNase I (Figure 1E). It appears that a portion of these products contained the template DNA sequence linked to RNAs hybridized to the template.
Taken together, our data suggest that SsoPriSL was capable of catalyzing template-dependent polymerization across discontinuous templates (PADT). The observation that the template DNA sequence was found in the polymerization products obtained from some templates (e.g. T120) but not in those from other templates (e.g. C35) suggests that PADT was initiated through either primer synthesis or terminal transfer (Figure 1F). Whether the process was initiated primarily through priming or terminal transfer presumably depended on template sequences and reaction conditions. Template-dependent synthesis of products longer than the template implies that a growing strand from one template was able to anneal to another template, initiating synthesis on the new template.
Efficient PADT by SsoPriSL occurs on templates containing a dC stretch
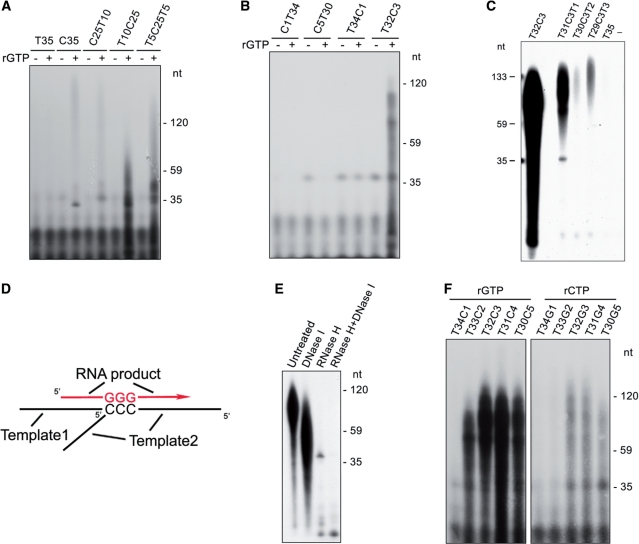
Although SsoPriSL catalyzed PADT with either oilgo(dC) or oligo(dT) as the template, the amounts of synthesis on the two templates differed markedly (Figure 1A). To determine the influence of sequence features of a template on the efficiency of PADT, we performed assays on templates containing dT and dC stretches in various arrangements with [α-32P]rATP and unlabeled rGTP as the substrates.
Very little synthesis was obtained with any of the templates in the absence of rGTP, whereas incorporation of [α-32P]rAMP increased significantly with some of the templates in the presence of unlabeled rGTP. As judged by the amount of synthesis obtained when rGTP was added to the reaction, the templates are classified into three groups (Figure 2A and B). Group 1 includes templates T35, C1T34, C5T30 and T34C1, which supported similarly low levels of product synthesis, suggesting that the dC base(s) present in some of these templates had little effect on the incorporation of [α-32P]rAMP in the presence of rGTP. Group 2, represented by templates C35 and C25T10, allowed slightly more synthesis of the long products than group 1 templates. Synthesis on C35 in the presence of [α-32P]rATP and rGTP indicates that template-independent terminal transfer was involved in the reaction. It is also noticed that the size distribution pattern of the products obtained on C35 in the presence of [α-32P]rATP and rGTP was similar to that in the presence of [α-32P]rGTP (Figure 1C), suggesting that complementary rG products were synthesized on C35 and extended by the addition of [α-32P]rAMP at the 3′-end through terminal transfer. The increase in synthesis on C35 in the presence of rGTP, as compared to that in its absence, points to the possibility that terminal transfer of [α-32P]rAMP to the 3′-end of an rG strand associated with the template strand was more efficient than that to the template dC strand. Indeed, we found that SsoPriSL was significantly more active in adding an rA to a 3′-end rG than to a 3′-end dC by terminal transfer, and such an addition was >10-fold more efficient when the 3′-end was base paired than when it was single-stranded (Supplementary Figure S1). Both the size distribution pattern and amounts of polymerization products on C25T10 were similar to those on C35, indicating that both reactions employed the same mechanism. In other words, the dT stretch in C25T10 was not efficiently used for the synthesis of products by the enzyme. Group 3 includes templates T10C25, T5C25T5 and T32C3. Efficient incorporation of the radiolabel occurred on these templates. For example, the fraction of template usage (i.e. the molar ratio of the incorporated rAs to the dTs in the total input template molecules) reached 73% for T32C3. The size distribution patterns of the products on these templates differed from those when [α-32P]rGTP was used as the sole substrate (Figure 1C). All of these templates contain a run of dTs on the 5′-side of a dC stretch. It is possible that the reaction proceeded in the following manner. SsoPriSL synthesized a short tail of rGs at the 3′-end of the template or the product through terminal transfer and promoted its annealing to the dC stretch in another template molecule. The enzyme then extended the templated rG tail through the dT stretch in the template, incorporating [α-32P]rAMP. Therefore, only dTs on the 5′-side of the dC stretch effectively served as part of the template in the synthesis. An alternative possibility is that the short rG tail folded back upon the template by pairing with the 3′-end dC stretch to initiate synthesis. To test this possibility, we designed templates which were identical to T32C3 except for the addition of 1–3 dTs to its 3′-end. Conceivably, an increase in the distance between the 3′-end newly synthesized rG tail and the dC stretch at the 3′-end of the template would make it easier for the rG tail to fold back upon the template (28,29). However, as shown in Figure 2C, synthesis was drastically reduced when 1–3 dTs were added to the 3′-end of T32C3, ruling out the second possibility. The reduced synthesis was not unexpected since the addition of dTs to the 3′-end of T32C3 would affect the stability of the enzyme-template complex (see below). The proposed scenario for the PADT initiated through terminal transfer, as summarized in Figure 1F and Figure 2D, is consistent with the nuclease digestion analysis of the products from T32C3. Cleavage with DNase I shortened the products. RNase H digested the bulk of the products, leaving behind at the position of the template a band, which disappeared following additional treatment with DNase I (Figure 2E). Therefore, PADT on T32C3 was initiated through terminal transfer.
Figure 2.
Influence of template sequences on PADT by SsoPriSL. (A) and (B) PADT on different templates. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with an indicated template (0.25 μM) and [α-32P]rATP (10 μM, 5 Ci/mmol) in the presence or absence of unlabeled rGTP (10 μM) in the standard assay mixture. (C) Synthesis by SsoPriSL on T32C3 with additional 1–3 dTs at the 3′-end. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with an indicated template (0.25 μM) and [α-32P]rATP (10 μM, 5 Ci/mmol) in the presence of unlabeled rGTP (10 μM) in the standard assay mixture. (D) A diagram showing the proposed role of a dC stretch in a template in PADT. (E) Nuclease digestion analysis of products synthesized by SsoPriSL on T32C3 with [α-32P]rATP and unlabeled rGTP as the substrates. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with T32C3 (0.25 μM), [α-32P]rATP (10 μM, 5 Ci/mmol) and unlabeled rGTP (10 μM) in the standard assay mixture. Samples were treated with indicated nucleases. In the RNase H + DNase I lane, the product was incubated sequentially with RNase H and DNase I. (F) Comparison of the PADT activities of SsoPriSL on templates containing dC and dG stretches of various sizes. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with a template (0.25 μM) and [α-32P]rATP (10 μM, 5 Ci/mmol) in the presence of unlabeled rGTP or rCTP (10 μM, for templates containing dCs or dGs, respectively) in the standard assay mixture. All reaction products were subjected to electrophoresis in 12% polyacrylamide containing 7 M urea.
A dC stretch in a template permits efficient initiation of PADT by SsoPriSL. To determine how the number of dCs in a stretch would affect the efficiency of PADT and if a dG stretch would be able to replace the dC stretch in the reaction, we prepared templates of the same length but with different numbers of dCs in a dC stretch or dGs in a dG stretch. As shown in Figure 2F, little synthesis was found on template T34C1. However, significant amounts of the long products were obtained on template T33C2, and even greater amounts of synthesis were observed on templates T32C3, T31C4 and T30C5. Therefore, a run of at least two dCs was sufficient for efficient PADT by SsoPriSL under the conditions used in the study. On the other hand, a dG stretch played a similar role to that of the dC stretch but was not as efficient as the latter in supporting PADT (Figure 2F). PADT on T33G2 was hardly detectable, and synthesis on T32G3, T31G4 and T30G5 was observed but rather weak, as compared to that in reactions with the same templates except for the replacement of dGs with dCs. It is possible that SsoPriSL is more efficient in PADT when rGTP, instead of rCTP, was used as the substrate (also see below). SsoPriSL was also capable of PADT on dsDNA templates and of strand displacement (Supplementary Figure S2). However, the products were shorter than those obtained on ssDNA templates.
Mechanisms of PADT by SsoPriSL
To understand the mechanism of PADT catalyzed by SsoPriSL, we performed the reaction on an 18-nt oligonucleotide (5′-TTTTTTTCTTTTTTTCCC) in the presence of rATP and rGTP. The reaction products were treated with DNase I, ligated to adaptors at both ends and reverse transcribed. The resulting cDNAs were PCR amplified, and the PCR fragments cloned into pGEM-T. The inserts were sequenced from both ends, and no sequencing errors were detected.
A total of 47 sequences were obtained (Table 1). Among them, 42 sequences had 1–3 dCs at the 5′-end, which were left over from the 3′-end sequence of the template following DNase I digestion. The remaining five sequences all started with contiguous dGs at the 5′-end, as in most of the other sequences downstream from the template dCs. Therefore, synthesis of all 47 products was initiated by terminal transfer.
Table 1.
Sequences of PADT products synthesized by SsoPriSLa
| No. | Sequence | |
|---|---|---|
| 1 | GGGGAAAAAGAAAAAGAAAAAAGAGAAAAAAA | |
| 2 | GGGAAAAAAGAAAAAAAAGGAAAAAAAGAA | |
| 3 | GGAAAAAAAGAAAAAAAAAAAAAAAGAAA | |
| 4 | GGGAAAAAGAAGGGAAAAAAGAAAAA | |
| 5 | GGGAAAAAAAGGAAAAAAGAAAGGGAAAAAA GAAAAAAA | |
| 6 | CGGGAAAAGGGAAAAAGAGAAAAAAAA | |
| 7 | CGGGAAAAAAAGAAAAAAAAGGGAAAAAAGAG | |
| 8 | CGGGGGGAAGGGGGGGGGAAAGAAAAA | |
| 9 | CGGGAAAAAAAGAAAAAAAGGGGGGGGGAAAAA | |
| 10 | CGGGAAAAAAAGGGAAAAAAAGAAAAAAAA | |
| 11 | CGGGAAAAAGAAAAAAAAAAAAAGAAAAGAAA | |
| 12 | CGGGGAAAGGGAAAAAAGAAAAAAAAG | |
| 13 | CGGGAAAAAAGAAAAAAAGGGAAAAGGGAA | |
| 14 | CGGGAAAAAAAGAAAAAAAAAAAGAAGAG | |
| 15 | CGGGAAAAAAAGAAAAAAAAAAAAGAAG | |
| 16 | CGGGAAAAAGAAAAAAAGAAAAAGAA | |
| 17 | CGGGGAAGGGGGGGGGGAAAAAAAGG | |
| 18 | CGGAAAGGGAAAAAAAGAAAAAAAAAAA | |
| 19 | CGGGGAAAAAAGAAAGGGAAAAAAGAAAAAAA | |
| 20 | CGGGGGAAAGGGAAAAAAGAAAGAAAAAG | |
| 21 | CGGGAAAGGGAAAGGGGGAAAAGAAAAAG | |
| 22 | CGGAAAAAAGAAAAGGGAAAAAAGAAAAAAAAA | |
| 23 | CGGAAAAAGGGAAAAAAGAAGAAAAAA | |
| 24 | CAGAAGGGAAAAGGGGAAAAAAGGGA | |
| 25 | CGGAAAAGGGAAAAAAAGAAAAAAAAAA | |
| 26 | CGGAAAGAAGAAAAGGGAAAAAAGAA | |
| 27 | CCGGGAAAAAAAGAAAAAAAGAAAAAAAAA | |
| 28 | CCGGGAAAAAAAGAAAAAAAAAAG | |
| 29 | CCGGGAAAAAAAGAAAAAAAAAAAAGAAAA | |
| 30 | CCGGAAAAAGGGAAAAAGAGAAAAAAAAAAAAA AGAAAA | |
| 31 | CCGGGAAAAAAGAAAAAAAAAGAAAAAAA | |
| 32 | CCGGGAAAAAAAGAAAAAAAGAAAAGAAAA | |
| 33 | CCGGAAAAGGGAAAAAGAAAAAAAAAA | |
| 34 | CCGGGAAAAAAGAAAAAAAGAAAAAGAAA | |
| 35 | CCGGGAAAAAAAGAAAAAAAAAAGGGAAA | |
| 36 | CCAAAAAGAGGGAAAAAAGAAAAAAAA | |
| 37 | CCGGAAAGGAAAAAAGGAAAAAAAGAA | |
| 38 | CCGGGAAGGGAAAAAAGAAAAAAAAAAAA | |
| 39 | CCGGAAAAAGGGAAAAAAAGAAAAAAAAAA | |
| 40 | CCGGGAAAAGGGAAAAAAGAAAAAAAA | |
| 41 | CCGGGAAAAAAAGGGAAAAAAGAAAAAAA | |
| 42 | CCGGAAAAGGGGAAAAAAGAAAAAAAAAAAAAA AAAAGAAAAAAAAGAAAAAAAAAAGAAAAA | |
| 43 | CCGGGAAAAAAAAAGAAAAAAAAAAAAAAG | |
| 44 | CCGGGAAAAAAGAAAAAAAAAAAAAAAG | |
| 45 | CCGGGAAAAAAGAAAAAAAAAAAAAAAAAAGAAAAAA | |
| 46 | CCGGAAAGAAGAAAAAAAAGAAAAAGAAA | |
| 47 | CCCGGAAAAAAGAAAAAAAAAAAAAAGAA | |
aThe 5′-end Cs in some of the sequences are those at the 3′-end of the template, which were left over following digestion of PADT products with DNase I.
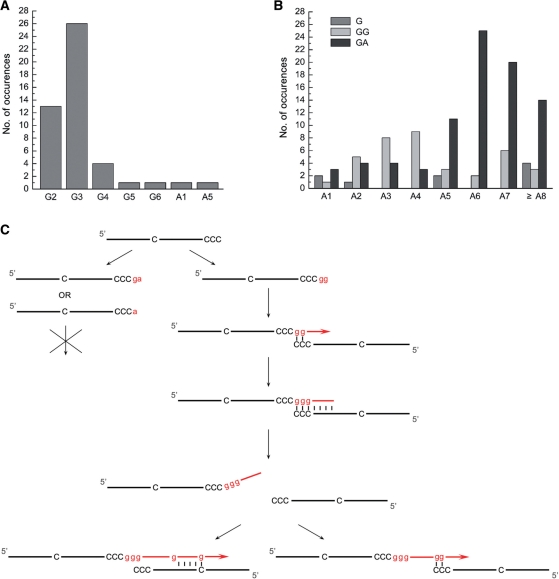
The theoretical sequence of the product of a PADT reaction on the above template was predicted to contain (rG)3(rA)7rG(rA)7 as the basic sequence unit. However, none of the 47 sequences was identical to the theoretical sequence. Careful statistical analysis of these sequences revealed interesting patterns (Table 2). As shown in Figure 3A, the 5′-end sequences, downstream of the template dCs, show a clear pattern of (dG)3 being the most abundant (55%), followed by (dG)2 (28%) and (dG)4 (8.5%). It appeared that, when two or more contiguous rGs were added to the 3′-end of the template DNA, they would anneal to the (dC)3 at the 5′-end of another template, initiating synthesis on the new strand by the enzyme (Figure 3C). Addition of a single rG or 2 nt other than rGs to the 3′-end of the template was apparently insufficient in initiating PADT as very few product sequences (2 out of 47) starting with the corresponding nucleotide(s) were found (Table 2 and Figure 3A). This agrees with the finding that a run of at least two dCs at the 3′-end of templates was sufficient for efficient PADT by SsoPriSL under the conditions used in this study (Figure 2F).
Table 2.
Patterns of the PADT product sequencesa
| Starting sequencesb | Frequency | Middle sequencesc | Frequency |
|||
|---|---|---|---|---|---|---|
| Total | Followed by G | Followed by GG | Followed by GA | |||
| G2 | 13 | A1 | 6 | 2 | 1 | 3 |
| G3 | 26 | A2 | 10 | 1 | 5 | 4 |
| G4 | 4 | A3 | 12 | 0 | 8 | 4 |
| G5 | 1 | A4 | 12 | 0 | 9 | 3 |
| G6 | 1 | A5 | 16 | 2 | 3 | 11 |
| A1 | 1 | A6 | 27 | 0 | 2 | 25 |
| A5 | 1 | A7 | 26 | 0 | 6 | 20 |
| Ending sequencesd | A8 | 5 | 1 | 2 | 2 | |
| A9 | 2 | 0 | 0 | 2 | ||
| A1 | 1 | A10 | 3 | 1 | 1 | 1 |
| A2 | 6 | A11 | 1 | 0 | 0 | 1 |
| A3 | 5 | A12 | 2 | 0 | 0 | 2 |
| A4 | 3 | A13 | 1 | 0 | 0 | 1 |
| A5 | 4 | A14 | 3 | 1 | 0 | 2 |
| A6 | 2 | A15 | 2 | 1 | 0 | 1 |
| A7 | 5 | A18 | 2 | 0 | 0 | 2 |
| A8 | 4 | G1 | 86 | |||
| A9 | 2 | G2 | 4 | |||
| A10 | 3 | G3 | 26 | |||
| A11 | 1 | G4 | 2 | |||
| A12 | 1 | G5 | 1 | |||
| G1 | 9 | G9 | 2 | |||
| G2 | 1 | G10 | 1 | |||
aFrequencies of sequence stretches in the PADT products are calculated. The length of a given nucleotide stretch is indicated by the number following the nucleotide. bRuns of identical nucleotides at the 5′-end of the products downstream of the Cs from the template. cRuns of identical nucleotides between the starting and the ending sequences of the products. Runs of As of a given length are further classified into three groups based on whether they are followed by G (the last nucleotide of the product), GG or GA. dRuns of identical nucleotides at the 3′-end of the products.
Figure 3.
Sequencing analysis of PADT products synthesized by SsoPriSL. (A) Occurrences of indicated sequence stretches at the 5′-end of the PADT products. (B) Occurrences of rA stretches of different lengths and with different downstream nucleotides in the middle portion of the PADT products. (C) An interpretation of the sequencing results. Incorporated ribonucleotides are indicated by letters in lower case.
No predominant ending sequences were found. However, it is worth noting that only a single sequence ended with two dGs in tandem, and no sequences contained a stretch of ≥ 3 dGs at the 3′-end. In other word, rG runs occurred much less frequently at the 3′-end than in the remainder of the PADT products. This again suggests that SsoPriSL promoted strand annealing and subsequent extension efficiently as soon as multiple rGs were sequentially added to the 3′-end of the product strand.
Two types of major sequence variation were observed in the middle portion of the products, as compared to that of the theoretical sequence. First, the number of dAs and dGs corresponding to the sites of (rA)7 and (rG)3 varied from 1 to 18 and 2 to 10, respectively (Table 2 and Figure 3B). However, runs of 6–7 dAs and 3 dGs occurred more frequently at the two sites, respectively, in general agreement with the complementary sequences in the template. Second, a single dG occurred at the site expected for (rG)3 or a run of multiple dGs appeared at the site expected for a single rG in the product sequence. To understand how these sequence variations arose, we examined the frequencies of the contiguous dAs and dGs of different lengths as well as those of nucleotides following a run of dAs of specific length, i.e. contiguous dGs (denoted GG in Table 2) or a single dG (GA) in the middle of the product sequence, or a single dG at the end of the sequence (G). The sequence variations appear to have been generated in the following two pathways: the denature-anneal-extend and the terminal transfer-anneal-extend pathways. As a low-processivity polymerase, SsoPriSL would presumably become dissociated frequently from the template during nucleotide polymerization, and the nascent strand would be partially or completely denatured from the template strand at the reaction temperature (55°C). In the first pathway, the denatured strand annealed to another template strand, initiating a new round of chain extension by SsoPriSL (Figure 3C). If annealing occurred upstream of the single dC site in the template, the subsequent extension would incorporate a single rG, possibly at a site where contiguous rGs were expected, in the product. If annealing occurred downstream of the single dC site, the extension will continue until the strand was dissociated from the template again. As shown in Table 2 and Figure 3B, GA was more frequently preceded by a stretch of ≥ 5 dAs, indicating that annealing between rA and dT stretches required a longer region of paring than that between rG and dC stretches. Conceivably, random termination of strand extension and subsequent strand annealing with a template at a site different from that from which the strand was initially denatured would generate variations in the length of rA runs. In the second pathway, addition of rGs to the 3′-end of the nascent strand occurred. The resulting rG tail annealed to the dC stretch on a new template for subsequent chain extension (Figure 3C). All rG stretches in the middle part of the products were presumably synthesized in this mechanism. Furthermore, rA stretches shorter than expected might be produced in the same manner since a growing rA strand could readily be dissociated from the template. Taken together, our results suggest that SsoPriSL catalyzes PADT by promoting annealing of a denatured product strand with another template, either directly or following terminal transfer of nucleotides at the 3′-end, and subsequent extension of the strand on the new template.
SsoPriSL promotes strand annealing
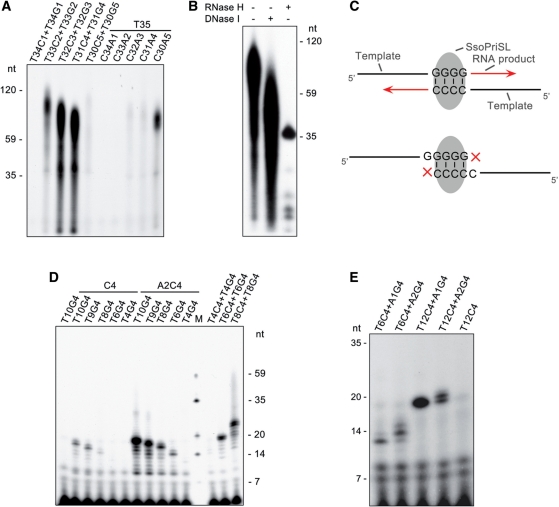
Strand annealing is a key step in PADT by SsoPriSL. A stretch of ≥2 rGs was sufficient to anneal to a run of dCs in the template to initiate strand extension by the primase (Figure 2F). Since sequences with such a short region of complementarity do not anneal spontaneously under our experimental conditions (55°C), we sought to determine the ability of SsoPriSL to promote strand annealing and to extend short template–primers. A series of 35-nt oligonucleotides with runs of dGs or dCs of different lengths at the 3′-end were employed in pair as the template and the primer in SsoPriSL activity assays. As shown in Figure 4A, when two oligonucleotides (T34C1 + T34G1) capable of forming just a single G : C base pair were used, no synthesis was detected. However, when T33C2 and T33G2, which were able to form two G : C pairs, were used as the template and the primer, measurable levels of polymerization were observed. The highest levels of synthesis were obtained with T32C3 and T32G3 or T31C4 and T31G4, which were able to form three or four G : C pairs, respectively. Surprisingly, when T30C5 and T30G5 were used, the amount of synthesis was lower than that obtained with T33C2 and T33G2. Similar low synthesis was observed with T29C6 and T29G6. Analysis of the reaction products by nuclease digestion revealed that the products contained a RNA–DNA hybrid, as expected from an annealing and extension process (Figure 4B). Therefore, we conclude that SsoPriSL is capable of promoting annealing between strands with a very short stretch of paired bases (e.g. 2–4 G : C base pairs). Strands with longer stretches of G : C base pairs may have greater chances of hybridizing imperfectly, thereby reducing the amount of extension (Figure 4C).
Figure 4.
SsoPriSL promoted annealing between strands with short regions of sequence complementarity and subsequent strand extension. (A) Annealing of oligonucleotides with short complementary ends and subsequent strand extension by SsoPriSL. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with a specified pair of oligonucleotides (0.25 μM each) and [α-32P]rATP (10 μM, 5 Ci/mmol) in the standard assay mixture. (B) Nuclease digestion analysis of reaction products from T32C3/T32G3. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with T32C3 and T32G3 (0.25 μM each) in the presence of [α-32P]rATP (10 μM, 5 Ci/mmol) in the standard assay mixture. Samples were treated with indicated nucleases. (C) A diagram depicting the optimal length of matching dC and dG sequences for SsoPriSL-promoted strand annealing. (D) Dependence of SsoPriSL-promoted strand annealing and subsequent extension on the length of the non-paring part of the strands. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with a specified pair of oligonucleotides (0.25 μM each) in the presence of [α-32P]rATP (10 μM, 5 Ci/mmol) in the standard assay mixture. (E) Effect of primer size on the use of a short template by SsoPriSL. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with a primer of different lengths (0.25 μM) and a short template (0.25 μM) in the presence of [α-32P]rUTP (10 μM, 5 Ci/mmol) in the standard assay mixture. All reaction products were subjected to electrophoresis in 12% (A, B) or 15% (D and E) polyacrylamide containing 7 M urea.
When A : T base pairing was tested in SsoPriSL-promoted strand annealing and extension in a similar fashion, we found that synthesis was detectable with oligonucleotides T35 and C32A3 or C31A4, that allowed three or four 3′-end A : T base pairs to form, and increased to a level similar to that obtained with T33C2 and T33G2 when T35 and C30A5 were used (Figure 4A). This is consistent with the observation that a run of >4 rAs was more efficient in strand annealing and hence more frequently followed by rGrA than shorter rA runs (Table 2 and Figure 3B). Furthermore, since SsoPriSL possesses only moderate terminal nucleotidyl transferase activity and is inefficient in adding multiple nucleotides to the 3′-end of ssDNA (Supplementary Figure S1), the requirement of efficient strand annealing for more A : T base pairs than C : G base pairs points to the possibility that runs of dCs or dGs in a template represent hot spots for PADT.
To further examine the ability of SsoPriSL to promote strand annealing, we prepared two sets of oligonucleotides containing a run of dTs of various lengths upstream of either four dGs or four dCs at the 3′-end. Reactions were performed on two oligonucleotides of the same size but with complementary 3′-ends. As shown in Figure 4D, although the region of complementarity (i.e. four G : C pairs) was unchanged, synthesis decreased as the oligonucleotides were shortened. A significant reduction in synthesis was observed when the oligonucleotides were <10 nt. However, when an oligonucleotide of ≥10 nt in size was used as a template, significant synthesis was obtained with a primer <8 nt (Figure 4D). Interestingly, when a primer of ≥10 nt was used, SsoPriSL was able to extend the primer well on a template as short as 5 or 6 nt (Figure 4E). It seems that, once SsoPriSL binds to either a template or a primer of sufficient length, it will be able to utilize a primer or a template that is too short to be used otherwise. These data further support the notion that SsoPriSL is capable of promoting strand annealing.
Effects of temperature and substrate on PADT
SsoPriSL shows little primase activity at physiologically relevant temperatures, e.g. 75°C, in vitro (9). Therefore, the aforementioned experiments were carried out at 55°C, as in previous studies. To understand how temperature would affect PADT, we compared polymerization reactions on C35 or C34ddC with [α-32P]rGTP as the substrate at 70°C with those at 55°C. Both the size and the quantity of the products on C35 increased, while synthesis on C34ddC drastically decreased, when the reaction temperature was raised from 55°C to 70°C (Figure 5A). The remaining products from the 3′-capped template at 70°C probably resulted from synthesis from a very small fraction of the templates that were not capped at the 3′-end. It appears that SsoPriSL failed to synthesize primers at 70°C, but the terminal transferase activity of the enzyme was higher at the higher temperature (Figure 5B).
Figure 5.
Activities of SsoPriSL at 70°C. (A) Synthesis on oligo(dC). SsoPriSL (0.5 μM) was incubated for 30 min at 55 or 70°C with a template (0.25 μM) and [α-32P]rGTP (10 μM, 5 Ci/mmol) in the standard assay mixture. (B) Terminal nucleotidyl transferase activity. SsoPriSL (0.5 μM) was incubated for 30 min at 55 or 70°C with C59 (0.25 μM) and [α-32P]rATP (5 μM, 50 Ci/mmol) in the standard assay mixture. (C) PADT activity. SsoPriSL (0.5 μM) was incubated for 30 min at 70°C with a template (0.25 μM) in the presence of [α-32P]rATP (10 μM, 5 Ci/mmol) and unlabeled rGTP (10 μM) in the standard assay mixture. Reaction products were subjected to electrophoresis in 12% polyacrylamide containing 7 M urea. Bands indicated by an arrow (B) are short products that SsoPriSL was able to synthesize in the absence of a template. These products have been shown to migrate with aberrantly slow mobility. For example, the lower band corresponds to a di-ribonucleotide (9).
We also carried out the assays with oligonucleotides containing dC and dT stretches as the templates and unlabeled rGTP and [α-32P]rATP as the substrates. We found that at least four contiguous dCs at the 3′-end of the template were required for significant synthesis at 70°C (Figure 5C). By comparison, two tandem dCs at the 3′-end of the template were sufficient to initiate strand extension at 55°C (Figure 2F). The observation that PADT remained highly active at 70°C, as compared to that at 55°C, suggests that the more stringent annealing requirement was compensated for by enhanced terminal transferase activity of the enzyme at the higher temperature. Our results indicate that SsoPriSL is capable of PADT at physiologically relevant temperature.
Since SsoPriSL uses both rNTPs and dNTPs as the substrates (8,9), we examined if dNTPs were able to support PADT. As shown in Figure 6A, SsoPriSL synthesized products ranging in size from 35 to 150 nt on C35 with [α-32P]dGTP as the sole substrate at 55°C, and the synthesis increased at 70°C. However, if C34ddC was used as the template, little synthesis was observed at either 55°C or 70°C. It appears that SsoPriSL showed little primase activity but was able to initiate PADT through terminal transfer with dGTP as the sole substrate. In addition, the small average sizes of the reaction products point to a reduced ability of the enzyme to mediate PADT in the presence of dGTP alone. Indeed, when unlabeled rGTP was added to the above reactions, incorporation of [α-32P]dGTP was increased, and the sizes of the products were similar to those obtained on C35 with [α-32P]rGTP as the substrate (Figure 6B and Figure 1A). Therefore, SsoPriSL was more active in catalyzing PADT in the presence of rGTP than in the presence of dGTP but was able to incorporate dGTP efficiently during strand extension. Synthesis on G35 with [α-32P]dCTP as the sole substrate was similar to that in reactions using C35 and [α-32P]dGTP (Figure 6B and C). However, addition of unlabeled rCTP to the reaction produced no drastic changes in synthesis. Therefore, SsoPriSL appeared to use rGTP more efficiently than rCTP in PADT, as shown above (Figure 2F). We also examined PADT on templates containing dC or dG and dT stretches in the presence of [α-32P]dATP and unlabeled dGTP or dCTP (Figure 6D and E). Templates that supported PADT were the same whether rNTP or dNTP was used as substrate. However, the polymerization products were much shorter when dNTP, instead of rNTP, was used as the substrate. These results indicate that SsoPriSL employed the same mechanism, regardless of the substrate, but preferred rNTP over dNTP in PADT.
Figure 6.
Activities of SsoPriSL with dNTPs as the substrates. (A) Synthesis on oligo(dC). SsoPriSL (0.5 μM) was incubated for 30 min at 55 or 70°C with C35 or C34ddC (0.25 μM) and [α-32P]dGTP (10 μM, 5 Ci/mmol) in the standard assay mixture. (B) PADT activity on oligo(dC) with [α-32P]dGTP and unlabeled rGTP as the substrates. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with C35 (0.25 μM) and [α-32P]dGTP (10 μM, 5 Ci/mmol) in the presence or absence of unlabeled rGTP (10 μM) in the standard assay mixture. (C) PADT activity on oligo(dG) with [α-32P]dCTP and unlabeled rCTP as the substrates. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with G35 (0.25 μM) and [α-32P]dCTP (10 μM, 5 Ci/mmol) in the presence or absence of unlabeled rCTP (10 μM) in the standard assay mixture. (D) and (E) PADT activity on various templates with dNTPs as the substrates. SsoPriSL (1.5 μM) was incubated for 30 min at 55°C with a specified template (0.25 μM) and [α-32P]dATP (1 μM, 50 Ci/mmol) in the presence or absence of unlabeled dGTP or dCTP (10 μM, for templates containing dCs or dGs, respectively) in the standard assay mixture. All reaction products were subjected to electrophoresis in 12% polyacrylamide containing 7 M urea. Bands indicated by an arrow (D and E) are short products that SsoPriSL was able to synthesize in the absence of a template.
DISCUSSION
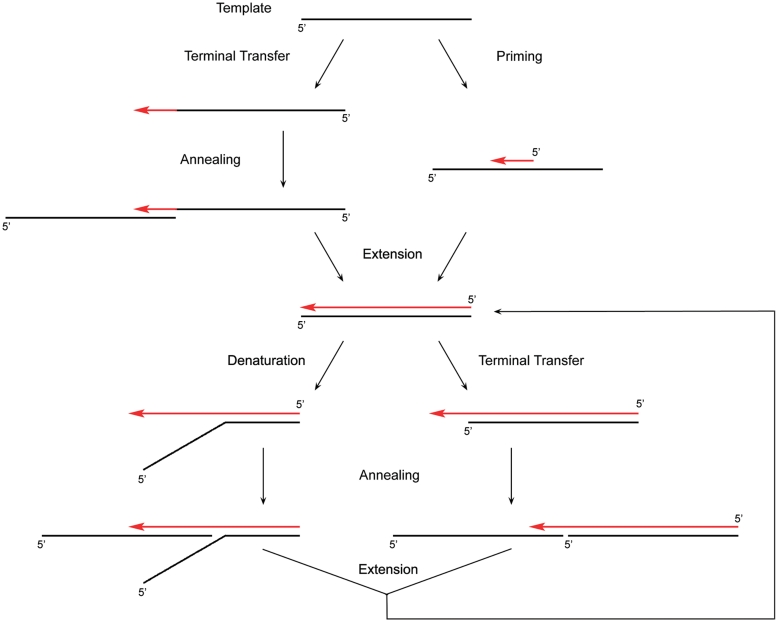
Archaeal primases are known for their versatile nucleotidyl polymerization activities (17). For example, SsoPriSL has been shown to be a primase capable of utilizing both rNTPs and dNTPs as the substrates, a template-dependent polymerase and a template-independent terminal transferase (8,9). This report shows yet another unusual property of SsoPriSL, i.e. the ability to catalyze PADT. PADT is initiated through either terminal transfer or priming. Once initiated, SsoPriSL may polymerize across unlinked templates in either a ‘denature–anneal–extend’ or a ‘terminal transfer–anneal–extend’ mode (Figure 7). The formal mode requires that the nascent chain be readily denatured, at least partially, from the template and the 3′-end portion of the nascent chain has significant sequence matches (e.g. ≥5 A : T base pairs) with the new template. The latter mode becomes efficient when the template contains at least two contiguous dCs. If the contiguous dCs are replaced by contiguous dGs, the efficiency of PADT will decrease since rGTP is the preferred substrate over rCTP for SsoPriSL. dAs/dTs stretches are ineffective in promoting PADT. Among the two modes, PADT through terminal transfer may occur more readily in vivo because of the ready availability of a short stretch of dCs in the genome. Interestingly, Pol μ has also been shown to be capable of PADT in the presence of XRCC4 and DNA ligase IV using a mechanism similar to that of the second mode (30). Terminal transfer and PADT in this case may create microhomology, which is essential to NHEJ in DSB repair (27).
Figure 7.
A model for PADT by SsoPriSL.
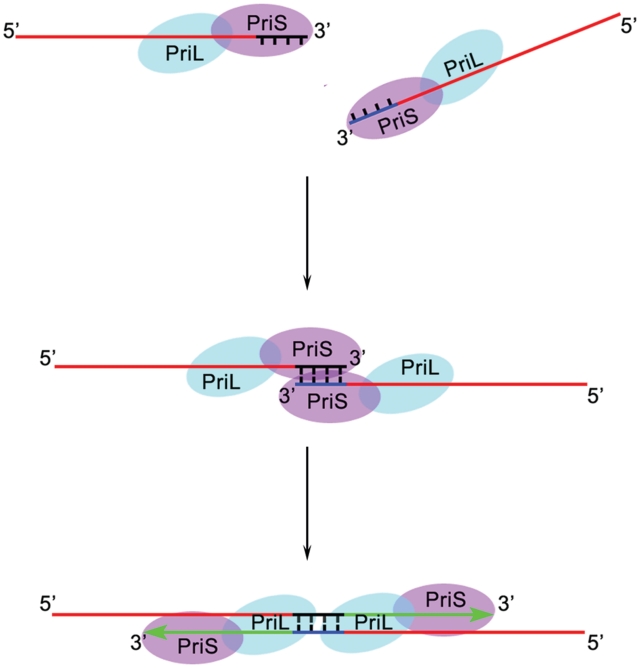
SsoPriSL-promoted strand annealing is a key step in PADT. The enzyme was able to promote annealing between the 3′-end of a strand with a template and extend the annealed 3′-end. Several Sulfolobus enzymes that function in DNA transactions, such as SsoTopo III and SsoDpo I, are known for their ability to enhance the stability of DNA double helix and to promote strand annealing (31,32). This property of the enzymes may represent adaptation of the organism to growth at high temperature. However, annealing promoted by SsoPriSL appears different from that by the other enzymes. Optimal annealing and extension occurred only when the two annealing sequences contained two to four contiguous C : G pairs. An ssDNA that formed five or more contiguous G : C pairs at the 3′-end with the template was not extended as efficiently. In addition, the sizes of the non-pairing regions of the two strands affected the efficiency of strand extension. SsoPriS resembles the polymerase domain of LigD in sequence and structure (23,33). Based on the crystal structure of the Mycobacterium tuberculosis polymerase domain of LigD mediating the synapsis of two non-complementary DNA ends (34), we propose the following mechanistic interpretation for the SsoPriSL-promoted DNA annealing. An SsoPriSL heterodimer binds each of the two ssDNA strands in such an orientation that SsoPriS is bound at the 3′-end, and the two primase–DNA complexes then form a ternary complex (Figure 8). Weak interaction between SsoPriS molecules, as revealed by chemical cross-linking, presumably contributes to the formation of the ternary complex. The complex is optimally stabilized only when the two ssDNA strands form two to four contiguous G : C base pairs at their 3′-end. The annealed 3′-end in the complex is then extended by the enzyme. The stability of the ternary complex will be compromised when a single G : C pair or 1–4 A : T pairs are involved in annealing of the 3′-ends of the two DNA strands. On the other hand, annealing between a longer run of G : C base pairs (e.g. five G : C pairs) would hinder the interaction between two DNA-bound primase heterodimers, reducing the stability of the ternary complex. Presumably, even if there are longer regions of G : C pairing, the two 3′-ends will preferentially form four G : C pairs in the ternary complex, and the resulting mismatched 3′-ends will not be effectively extended. Template binding by SsoPriSL depends primarily on the interaction between the large subunit of the enzyme and the DNA (14). When two ssDNAs to be annealed are too short to be bound tightly by the primase or too short for SsoPriS to be located at their 3′-end even if SsoPriL is bound, the stable ternary complex will not form. However, when one of the two ssDNA strands is sufficiently long (e.g. ≥10 nt), a stable binary complex will form. This complex is capable of catching the shorter ssDNA strand through base pairing at the 3′-end and helping position SsoPriS from the other primase heterodimer at the 3′-end of the shorter DNA through protein-protein interaction. The ternary complex formed in this manner is sufficiently stable to allow primer extension from the 3′-ends of both ssDNA strands. This model also applies to cases where two dsDNAs with 3′-overhangs sharing sequence complementarity anneal.
Figure 8.
A diagram showing the formation of a proposed ternary complex during SsoPriSL-promoted strand annealing.
The various activities of SsoPriSL differ in temperature dependence in vitro. The enzyme was active in primer synthesis and PADT at 55°C. However, it showed extremely low activity in primer synthesis but remained efficient in PADT at 70°C. It may be speculated that the primase-short primer–template complex is too unstable to allow efficient primer synthesis in vitro at 70°C, but the complex is stabilized by other proteins or factors so that primer synthesis can occur in the cell at high temperature. The temperature dependence of SsoPriSL activities reinforced the contention that PADT occurs under conditions optimal for the growth of the cells. SsoPriSL shows preference for rNTP, especially rGTP, over dNTP in PADT. The enzyme could hardly utilize dNTPs in primer synthesis although it was efficiently incorporated in chain extension. Synthesis of primers by SsoPriSL with dNTP as the substrate, as shown in previous studies, probably has been vastly overestimated due to the inclusion of PADT and primer extension in the measured activity. This agrees with the report that Km of the enzyme for dNTP was too high to measure (9). By comparison, DNA polymerases involved in NHEJ pathways, such as mammalian X-family polymerase Pol μ, Pol4 from yeast and Pol domain from bacterial LigD, are all known to be able to use rNTP as the substrate (27). Unlike dNTP, rNTP exists at a high and stable cellular level throughout the cell cycle, allowing the polymerases to repair DSB through NHEJ pathways during different growth phases (35).
Based on its biochemical properties and resemblance to X-family DNA polymerases from Eukarya and the polymerase domain of bacterial LigD, we speculate that SsoPriSL may play a role in the NHEJ pathway of DSB repair. Homologous recombination (HR) and NHEJ are two major DSB repair pathways. The former exists in all forms of life including Archaea, while the latter has been found only in Eukarya and some Bacteria (27). The majority of Archaea lack Ku protein and other enzymes required for NHEJ (26,36,37). Because they live in extremely harsh habitats, most Archaea are probably prone to DNA damage. Therefore, it is conceivable that these organisms have evolved their own NHEJ pathways. SsoPriSL appears to be the only known Sulfolobus polymerase adapted for a role in the NHEJ repair pathway because of its terminal transferase activity, its ability to promote annealing between strands with microhomology and to catalyze PADT as well as its preference for rNTPs as the substrates. Recently, DNA polymerase B1 from Sulfolobus (SsoDpo I) was shown to catalyze terminal transfer in vitro (32). However, this enzyme utilizes only dNTPs as the substrates and possesses a robust 3′–5′ exonuclease activity capable of removing the product of its own terminal transfer activity. Therefore, it appears unlikely that SsoPolB1 plays a central role in NHEJ. Another recent study shows that PriL interacts with Rad50 in Thermococcus kodakarensis (38). Rad50 has been shown to function not only in HR but also in NHEJ pathways, and to serve a regulatory role in DSB repair in yeast (39). In S. solfataricus, the expression level of Rad50 was significantly increased in response to the need for DSB repair following UV irradiation (40). Therefore, we speculate that SsoPriSL is involved not only in DNA replication but also in DNA repair in Sulfolobus. Intriguingly, all Archaea also encode a DnaG-like primase. SsoDnaG exhibits primase activity in vitro, and has been proposed to function in coordination with SsoPriSL in primer synthesis during DNA replication (12). The enzyme has also been shown to participate in RNA degradation (15,16). We hypothesize that LUCA, the last universal common ancestor, employed a dual-primase system consisting of DnaG and PriSL. DnaG and PriSL also served roles in RNA degradation and NHEJ processes, respectively. This system is inherited in Archaea. In Bacteria, DnaG has become the only primase whereas PriSL has given rise to the Pol domain of LigD, which functions in NHEJ. In Eukarya, DnaG is lost, and PriSL has not only become the only primase active in DNA replication but also evolved into Pol X polymerases responsible for the NHEJ pathway.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods, Supplementary Results, Supplementary Table 1, Supplementary Figures 1–2.
FUNDING
National Natural Science Foundation of China (30730003 and 30921065). Funding for open access charge: National Natural Science Foundation of China (30730003).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Fang Yuanyuan for her advice on the preparation of cDNAs from the PADT products.
REFERENCES
- 1.Frick DN, Richardson CC. DNA primases. Annu. Rev. Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 2.Wickner S. DNA or RNA priming of bacteriophage G4 DNA synthesis by Escherichia coli dnaG protein. Proc. Natl Acad. Sci. USA. 1977;74:2815–2819. doi: 10.1073/pnas.74.7.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowen L, Kornberg A. A ribo-deoxyribonucleotide primer synthesized by primase. J. Biol. Chem. 1978;253:770–774. [PubMed] [Google Scholar]
- 4.Swart JR, Griep MA. Primase from Escherichia coli primes single-stranded templates in the absence of single-stranded DNA-binding protein or other auxiliary proteins. Template sequence requirements based on the bacteriophage G4 complementary strand origin and Okazaki fragment initiation sites. J. Biol. Chem. 1993;268:12970–12976. [PubMed] [Google Scholar]
- 5.Swart JR, Griep MA. Primer synthesis kinetics by Escherichia coli primase on single-stranded DNA templates. Biochemistry. 1995;34:16097–16106. doi: 10.1021/bi00049a025. [DOI] [PubMed] [Google Scholar]
- 6.Johnson SK, Bhattacharyya S, Griep MA. DnaB helicase stimulates primer synthesis activity on short oligonucleotide templates. Biochemistry. 2000;39:736–744. doi: 10.1021/bi991554l. [DOI] [PubMed] [Google Scholar]
- 7.Desogus G, Onesti S, Brick P, Rossi M, Pisani FM. Identification and characterization of a DNA primase from the hyperthermophilic archaeon Methanococcus jannaschii. Nucleic Acids Res. 1999;27:4444–4450. doi: 10.1093/nar/27.22.4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Falco M, Fusco A, De Felice M, Rossi M, Pisani FM. The DNA primase of Sulfolobus solfataricus is activated by substrates containing a thymine-rich bubble and has a 3′-terminal nucleotidyl-transferase activity. Nucleic Acids Res. 2004;32:5223–5230. doi: 10.1093/nar/gkh865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lao-Sirieix SH, Bell SD. The heterodimeric primase of the hyperthermophilic archaeon Sulfolobus solfataricus possesses DNA and RNA primase, polymerase and 3′-terminal nucleotidyl transferase activities. J. Mol. Biol. 2004;344:1251–1263. doi: 10.1016/j.jmb.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 10.Liu L, Komori K, Ishino S, Bocquier AA, Cann IK, Kohda D, Ishino Y. The archaeal DNA primase: biochemical characterization of the p41-p46 complex from Pyrococcus furiosus. J. Biol. Chem. 2001;276:45484–45490. doi: 10.1074/jbc.M106391200. [DOI] [PubMed] [Google Scholar]
- 11.Le Breton M, Henneke G, Norais C, Flament D, Myllykallio H, Querellou J, Raffin JP. The heterodimeric primase from the euryarchaeon Pyrococcus abyssi: a multifunctional enzyme for initiation and repair? J. Mol. Biol. 2007;374:1172–1185. doi: 10.1016/j.jmb.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Zuo Z, Rodgers CJ, Mikheikin AL, Trakselis MA. Characterization of a functional DnaG-type primase in archaea: implications for a dual-primase system. J. Mol. Biol. 2010;397:664–676. doi: 10.1016/j.jmb.2010.01.057. [DOI] [PubMed] [Google Scholar]
- 13.Marinsek N, Barry ER, Makarova KS, Dionne I, Koonin EV, Bell SD. GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep. 2006;7:539–545. doi: 10.1038/sj.embor.7400649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu K, Lai X, Guo X, Hu J, Xiang X, Huang L. Interplay between primase and replication factor C in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 2007;63:826–837. doi: 10.1111/j.1365-2958.2006.05535.x. [DOI] [PubMed] [Google Scholar]
- 15.Evguenieva-Hackenberg E, Walter P, Hochleitner E, Lottspeich F, Klug G. An exosome-like complex in Sulfolobus solfataricus. EMBO Rep. 2003;4:889–893. doi: 10.1038/sj.embor.embor929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walter P, Klein F, Lorentzen E, Ilchmann A, Klug G, Evguenieva-Hackenberg E. Characterization of native and reconstituted exosome complexes from the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 2006;62:1076–1089. doi: 10.1111/j.1365-2958.2006.05393.x. [DOI] [PubMed] [Google Scholar]
- 17.Lao-Sirieix SH, Pellegrini L, Bell SD. The promiscuous primase. Trends Genet. 2005;21:568–572. doi: 10.1016/j.tig.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Kirk BW, Kuchta RD. Arg304 of human DNA primase is a key contributor to catalysis and NTP binding: primase and the family X polymerases share significant sequence homology. Biochemistry. 1999;38:7727–7736. doi: 10.1021/bi990247c. [DOI] [PubMed] [Google Scholar]
- 19.Augustin MA, Huber R, Kaiser JT. Crystal structure of a DNA-dependent RNA polymerase (DNA primase) Nat. Struct. Biol. 2001;8:57–61. doi: 10.1038/83060. [DOI] [PubMed] [Google Scholar]
- 20.Sawaya MR, Pelletier H, Kumar A, Wilson SH, Kraut J. Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 21.Moon AF, Garcia-Diaz M, Batra VK, Beard WA, Bebenek K, Kunkel TA, Wilson SH, Pedersen LC. The X family portrait: structural insights into biological functions of X family polymerases. DNA Repair. 2007;6:1709–1725. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koonin EV, Wolf YI, Kondrashov AS, Aravind L. Bacterial homologs of the small subunit of eukaryotic DNA primase. J. Mol. Microbiol. Biotechnol. 2000;2:509–512. [PubMed] [Google Scholar]
- 23.Zhu H, Nandakumar J, Aniukwu J, Wang LK, Glickman MS, Lima CD, Shuman S. Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D. Proc. Natl Acad. Sci. USA. 2006;103:1711–1716. doi: 10.1073/pnas.0509083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pitcher RS, Brissett NC, Picher AJ, Andrade P, Juarez R, Thompson D, Fox GC, Blanco L, Doherty AJ. Structure and function of a mycobacterial NHEJ DNA repair polymerase. J. Mol. Biol. 2007;366:391–405. doi: 10.1016/j.jmb.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 25.Weller GR, Doherty AJ. A family of DNA repair ligases in bacteria? FEBS Lett. 2001;505:340–342. doi: 10.1016/s0014-5793(01)02831-9. [DOI] [PubMed] [Google Scholar]
- 26.Pitcher RS, Brissett NC, Doherty AJ. Nonhomologous end-joining in bacteria: a microbial perspective. Annu. Rev. Microbiol. 2007;61:259–282. doi: 10.1146/annurev.micro.61.080706.093354. [DOI] [PubMed] [Google Scholar]
- 27.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haasnoot CA, den Hartog JH, de Rooij JF, van Boom JH, Altona C. Loopstructures in synthetic oligodeoxynucleotides. Nucleic Acids Res. 1980;8:169–181. doi: 10.1093/nar/8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hilbers CW, Haasnoot CA, de Bruin SH, Joordens JJ, van der Marel GA, van Boom JH. Hairpin formation in synthetic oligonucleotides. Biochimie. 1985;67:685–695. doi: 10.1016/s0300-9084(85)80156-5. [DOI] [PubMed] [Google Scholar]
- 30.Davis BJ, Havener JM, Ramsden DA. End-bridging is required for pol mu to efficiently promote repair of noncomplementary ends by nonhomologous end joining. Nucleic Acids Res. 2008;36:3085–3094. doi: 10.1093/nar/gkn164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L, Huang L. Oligonucleotide cleavage and rejoining by topoisomerase III from the hyperthermophilic archaeon Sulfolobus solfataricus: temperature dependence and strand annealing-promoted DNA religation. Mol. Microbiol. 2006;60:783–794. doi: 10.1111/j.1365-2958.2006.05133.x. [DOI] [PubMed] [Google Scholar]
- 32.Zuo Z, Lin HK, Trakselis MA. Strand annealing and terminal transferase activities of a B-family DNA polymerase. Biochemistry. 2011;50:5379–5390. doi: 10.1021/bi200421g. [DOI] [PubMed] [Google Scholar]
- 33.Lao-Sirieix SH, Nookala RK, Roversi P, Bell SD, Pellegrini L. Structure of the heterodimeric core primase. Nat. Struct. Mol. Biol. 2005;12:1137–1144. doi: 10.1038/nsmb1013. [DOI] [PubMed] [Google Scholar]
- 34.Brissett NC, Pitcher RS, Juarez R, Picher AJ, Green AJ, Dafforn TR, Fox GC, Blanco L, Doherty AJ. Structure of a NHEJ polymerase-mediated DNA synaptic complex. Science. 2007;318:456–459. doi: 10.1126/science.1145112. [DOI] [PubMed] [Google Scholar]
- 35.Gu J, Lieber MR. Mechanistic flexibility as a conserved theme across 3 billion years of nonhomologous DNA end-joining. Genes Dev. 2008;22:411–415. doi: 10.1101/gad.1646608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith P, Nair PA, Das U, Zhu H, Shuman S. Structures and activities of archaeal members of the LigD 3′-phosphoesterase DNA repair enzyme superfamily. Nucleic Acids Res. 2011;39:3310–3320. doi: 10.1093/nar/gkq1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erkel C, Kube M, Reinhardt R, Liesack W. Genome of Rice Cluster I archaea–the key methane producers in the rice rhizosphere. Science. 2006;313:370–372. doi: 10.1126/science.1127062. [DOI] [PubMed] [Google Scholar]
- 38.Li Z, Santangelo TJ, Cubonova L, Reeve JN, Kelman Z. Affinity purification of an archaeal DNA replication protein network. MBio. 2010;1:e00221–10. doi: 10.1128/mBio.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu. Rev. Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 40.Rolfsmeier ML, Laughery MF, Haseltine CA. Repair of DNA double-strand breaks following UV damage in three Sulfolobus solfataricus strains. J. Bacteriol. 2010;192:4954–4962. doi: 10.1128/JB.00667-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.