Abstract
A novel biosensing approach for the label-free detection of nucleic acid sequences of short and large lengths has been implemented, with special emphasis on targeting RNA sequences with secondary structures. The approach is based on selecting 8-aminoadenine-modified parallel-stranded DNA tail-clamps as affinity bioreceptors. These receptors have the ability of creating a stable triplex-stranded helix at neutral pH upon hybridization with the nucleic acid target. A surface plasmon resonance biosensor has been used for the detection. With this strategy, we have detected short DNA sequences (32-mer) and purified RNA (103-mer) at the femtomol level in a few minutes in an easy and level-free way. This approach is particularly suitable for the detection of RNA molecules with predicted secondary structures, reaching a limit of detection of 50 fmol without any label or amplification steps. Our methodology has shown a marked enhancement for the detection (18% for short DNA and 54% for RNA), when compared with the conventional duplex approach, highlighting the large difficulty of the duplex approach to detect nucleic acid sequences, especially those exhibiting stable secondary structures. We believe that our strategy could be of great interest to the RNA field.
INTRODUCTION
RNA detection is an emerging field in molecular biotechnology because RNA plays a fundamental role in cell function. Most RNA applications rely on the analysis and use of protein-coding RNA, but in the recent years the discovery of novel RNA types which does not encode for proteins, the non-coding RNA (ncRNA), have updated the interest for RNA. It is estimated that as much as 97–98% of the transcriptional output of the human genome are ncRNA sequences (1). The amazing roles that such sequences might play in regulating the cell function is now beginning to be elucidated (2,3). With the actual resurgence of RNA studies, there is an increasing demand for novel diagnostic tools that could afford an easy and fast RNA analysis. In addition, the emergence of bacterial contamination on food has triggered the development of novel methods for the rapid detection of potential contamination based on the analysis of bacterial RNA.
One important issue to be taken into account for RNA analysis is that while DNA structures are more likely to be duplexes with full complementarity between two strands, RNA structures are more likely to fold into complex secondary structures, such as hairpins and loops but also into more complex ones such as triplexes (4) and quadruplexes (5). This is in part due to the extra oxygen in the RNA sugar (ribose), which increases the propensity for hydrogen bonding in the nucleic acid backbone. The presence of secondary structures is almost a ‘constant’ in the RNA world. Most RNA sequences, particularly the large ones, usually present these structures. ncRNA is particularly expected to have secondary structures. These structures are mostly considered a ‘mark’ in ncRNA, such us the presence of open reading frames in protein-coding sequences. That is the reason why most RNA predicting bioinformatic tools look for the presence of these structures in a given sequence to carry-out the prediction of the RNA type (6,7). However, the presence of the secondary structures can hinder the hybridization of the RNA target with the probe employed for detection and constitutes one of the main difficulties for RNA analysis. Novel detection methods avoiding this problem should be implemented while providing better sensitivity levels at the same time.
In this line, we have implemented a novel detection method based on the RNA recognition through the formation of a triplex helix and employing an optical biosensor (a home-made surface plasmon resonance [SPR] sensor) for the analysis. Optical biosensing techniques provide the monitoring of biomolecular interactions in real-time without any labelling. Using SPR methods, subnanomolar detection for DNA targets (8–10), including the identification of single mismatches, has been reported (11). RNA biosensing is much less widespread than DNA, not only due to the complexity of detecting a specific RNA sequence in a complex matrix containing hundreds of other RNA sequences but also for the frequent presence of secondary structures. While the detection of short RNA sequences has been demonstrated by SPR (12), detection of longer RNA sequences is not straightforward mainly due to the abundance of different folding structures which RNA can adopt (13,14).
We have chosen the detection of RNA by forming a triplex helix because this approach has been previously demonstrated to be more efficient than duplex strategy in binding RNA with potential secondary structures in solution (15,16). The triplex approach represents a universal approach for targeting nucleic acid sequences by simply binding to a capturing duplex probe; however, the nucleic acid target must have a homopurine–homopyrimidine track within its sequence. This is essential for the Hoogsteen-base pairing, which is responsible for the specific interaction of the third strand with the duplex (17).
Although the need of homopurine–homopyrimidine tracks on the target sequence might be considered at first glance as a restrictive criterion for RNA detection, these motifs can constitute a highly valuable feature. They are largely over-represented on the mammalian genome: 97.8% of known human and 95.2% of known mouse genes have, at least, one potential high-affinity homopurine–homopyrimidine track in the promoter and/or transcribed gene regions. Importantly, 86.5% of known human and 83% of the known mouse genes have at least one motif that is unique to that gene (18). It has been described that triplex structures may play a role in many processes, such as recombination and destabilization of chromosomal DNA (19,20), induction of repair and mutation (19,21–24) or regulation of replication and transcription (19,23,25,26). In addition, it has been recently observed that formation of triplexes can constitute a main mechanism in epigenetic regulation pathways (27) and they might be crucial for ncRNAs to carry-out their regulatory function (28,29). Also, homopurine–homopyrimidine tracks can constitute key motifs for the specific detection of microorganisms (16,30).
All above examples demonstrate that having a strategy for RNA detection based on the formation of triplexes can be highly valuable for the study of many biological processes. In addition, the recognition principle and generation of triplex forming structures are simple and easy to achieve, especially when the triplex structure is additionally stabilized by chemical modifications (i.e. nucleobase, sugar and backbone modifications) and by the introduction of intercalating moieties (31–33). Among these stabilization strategies, we have previously reported that triplexes can be greatly stabilized: (i) by modifying the duplex structure in a hairpin fashion using a parallel stranded tail-clamp and (ii) by introducing one or several 8-aminoadenine nucleotides within its sequence.
A parallel stranded tail-clamp is a parallel stranded clamp bearing an oligonucleotide (a tail sequence complementary to the 5′ flanking sequence of the homopyrimidine track) connected to its Watson–Crick-forming strand. Since the tail region is a sequence matching to an adjacent portion of the homopurine–homopyrimidine track of the target, both the strength and the specificity of the binding becomes increased (15). In a previous work (15,16), we demonstrated that by modifying parallel stranded clamps with this tail the efficiency toward RNA interaction was increased, especially when the RNA is very structured. The tail also appears to help in the triplex formation by interfering with the tendency of the homopyrimidine target to form secondary structures with adjacent sequences. We also observed that the introduction of 8-aminoadenine nucleotides within parallel stranded tail-clamp sequences further contributes to increase the triplex stability allowing RNA detection, not only under acidic but also under neutral and slightly basic pH conditions (15,16,34). Using this approach, robust and stable triple helices can be obtained. We have reported that 45% of Listeria innocua virulence iap mRNA molecules from a total RNA solution were captured under neutral pH conditions (16).
The characteristics of parallel stranded tail-clamps clearly represent an interesting strategy to be implemented in biosensing methods for nucleic acids detection. Therefore, we have combined the triplex detection method and SPR biosensing to target nucleic acid sequences of different lengths, with special emphasis on the detection of RNA samples with predicted secondary structures (Figure 1). The efficiency of parallel tail-clamps was also compared with their homologues duplex-forming oligonucleotides (DFO) to fully demonstrate the feasibility of the triplex approach (Scheme 1).
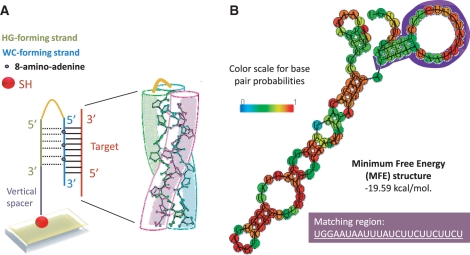
Figure 1.
(A) Design of the amino-modified tail-clamp bioreceptor used in this study. The Watson–Crick and Hoogsteen forming strands are highlighted in blue and green, respectively. The vertical spacer (dark blue) and aminoadenines introduced in some tail-clamps receptors are also indicated. (B) Predicted structure with minimum-free energy folding (MFE) for L. innocua RNA target obtained from The Vienna RNA Website (37). The matching region with the tail-clamp receptor is highlighted in purple color.
Scheme 1.
Schematic representation of the theoretical interaction of iap parallel tail-clamps and duplex forming sequences with their RNA target sequences.
MATERIALS AND METHODS
All experiments have been carried out using a L. innocua virulence iap gene sequence as the target sequence. Two types of targets were designed (Table 1):
RNA-list: a fragment of the iap (mRNA (103-mer) obtained by in vitro transcription. It was selected because it contains a homopyrimidine track of 12 nt within a region of predicted secondary structures (Figure 1B).
DNA-list: a short DNA target of 32-mer containing the same homopyrimidine track but not presenting secondary structures. It was designed to assess the potential of the clamps to detect short targets such as micro-RNAs.
Table 1.
DNA and RNA sequences of this study
| Short name | Long name | Sequence |
|---|---|---|
| Receptors | ||
| DFO-SH | Unmodified Listeria duplex forming oligonucleotide | thiol-5′AGAAGAAGAAGATAAATTATTCCA3′ |
| 3A- DFO-SH | 3A modified Listeria duplex forming oligonucleotide | thiol-5′AGAAGAAGAAGATAAATTATTCCA3′ |
| T15-3A-DFO-SH | T15-3A modified Listeria duplex forming oligonucleotide | thiol-5′TTTTTTTTTTTTTTT-AGAAGAAGAAGATAAATTATTCCA3′ |
| TFC-SH | Unmodified Listeria triplex forming clamp | thiol-3′TCTTCTTCTTCT5′-5′TTTT-AGAAGAAGAAGATAAATTATTCCA3′ |
| 3A-TFC-SH | 3A-modified Listeria triplex forming clamp | thiol-3′TCTTCTTCTTCT5′-5′TTTT-AGAAGAAGAAGATAAATTATTCCA3′ |
| T15-3A-TFC-SH | T15-3A modified Listeria triplex forming clamp | thiol-3′TTTTTTTTTTTTTTT-TCTTCTTCTTCT5′-5′TTTT- AGAAGAAGAAGATAAATTATTCCA3′ |
| Targets | ||
| DNA-List | Listeria DNA target (32-mer) | 5′TGGAATAATTTATCTTCTTCTTCTATTTATGT3′ |
| DNA control | DNA negative control target (33-mer) | 5′ CGTCTTCTTTTTCCACGATGCTCCTCGTGGGTG3′ |
| RNA-List | mRNA from L. Innocua iap | 5′AACGUUAAAAGCGGCGACACAAUUUGGGCAUUAUCCGUGAAGU ACGGUGUUUCUGUUCAAGAUAUUAUGUCAUGGAAUAAUUUAUC UUCUUCUUCUAUUUAUGU3′ |
| RNA control | RNA negative control target | 5′CCUACAUAAAUAGAAGAAGAAGAUAAAUUAUUCCAUGACAUAAU AUCUUGAACAGAAACACCGUACUUCACGGAUAAUGCCCAAAUU GUGUCGCCGCUUUUAACGUU3′ |
A: 8-aminoadenine; underlined: matching sequence containing the homopurine or homopyrimidine motif.
Synthesis of oligonucleotides
The oligonucleotides employed in this study are listed in Table 1 and are described in the following:
- Unmodified oligonucleotides (DNA-List and DNA control) were synthesized on an Applied Biosystems 3400 DNA synthesizer. The sequence was prepared using standard (Bz- or ibu-protected) 3′-phosphoramidites and polystyrene solid supports (LV200) according to the protocols of the manufacturer. Coupling efficiencies were higher than 98%. After the assembly of the sequence, oligonucleotide support was treated with 32% aqueous ammonia at 55°C for 16 h. The solution was concentrated to dryness and the product was desalted on NAP-10 (Sephadex G-25) columns eluted with water. Oligonucleotides were analysed by reversed phase High Performance Liquid Chromatography (HPLC) and mass spectrometry:
- DNA-list: expected for C316H403N98O201P31 9750.4; found Matrix-Assisted Laser Desorption/Ionization (MALDI): 9750.4; purity 86%.
- DNA control: expected for C320H410N106O207P32 10044.5; found (MALDI): 10037.2; purity 84%.
- Thiolated oligonucleotides (DFO-SH, 3A-DFO-SH and T15-3A-DFO-SH) were prepared in a similar way to those above (Scheme 1). The thiol group was added at the 5′-end of the oligonucleotides using 5′-thiol modifier-C6 S-S CE phosphoramidite (Link Technologies). The phosphoramidite of 8-aminoadenine was prepared as described elsewhere (35,36). After the assembly of the sequences, oligonucleotide-supports were treated with aqueous ammonia (32%) containing 0.1 M dithiothreitol (DTT) at 55°C for 16 h. The ammonia solutions were concentrated to dryness and used directly. Before immobilization the products were desalted on NAP-10 (Sephadex G-25) columns and eluted with water to remove the excess of DTT. Oligonucleotides were analysed by reversed-phase HPLC and mass spectrometry:
- DFO-SH: expected for C244H310N104O137P24S 7622.1; found (MALDI): 7618.3; (electrospray): 7622; purity 87%.
- 3A-DFO-SH: expected for C244H310N104O137P24S 7667.2; found (MALDI): 7667; purity 85%.
- T15-3A-DFO-SH: expected for C394H505N134O242P39S 12230.1; found (electrospray): 12260 (M+ Na+); purity 80%.
Listeria parallel-stranded tail-clamps (TFC-SH, 3A-TFC-SH and T15-3A-TFC-SH) designed for the study (Scheme 1 and Figure 1A) carry the purine sequence complementary to the L. innocua iap homopyrimidine target, as well as 12 additional bases at the 3′-end of its Watson and Crick forming strand; connected head-to-head and 5′-ends with the Hoogsteen C,T-sequence. A four thymidines sequence was used for connecting both strands. At the 3′-end a thiol molecule was added for immobilization onto the gold sensor surface and, depending on the sequence type, it is further modified or not with three 8-aminoadenines (3A). They were prepared on an automatic Applied Biosystems 392 DNA synthesizer as described elsewhere (15,16,34,38,39). 5′-5′ Clamps were prepared in three steps. First, the purine part and the four T-linker were assembled using standard phosphoramidites from the natural bases and the 8-aminoadenine phosphoramidites. The phosphoramidite of 8-aminoadenine was prepared as described elsewhere (35,36). Then, the pyrimidine part was prepared using reversed C and T phosphoramidites (Link technologies). Finally, a thiol group was added at the end of the sequence using the 5′-thiol modifier-C6 S-S CE phosphoramidite (Link Technologies).
After the assembly of the sequences, oligonucleotide supports were treated with aqueous ammonia (32%) containing 0.1 M DTT at 55°C for 16 h. The resulting ammonia solutions were concentrated to dryness and used directly. Before immobilization, the products were desalted on NAP-10 (Sephadex G-25) columns and eluted with water to remove the excess of DTT. Oligonucleotides were analysed by reversed-phase HPLC and mass spectrometry:
TFC-SH: expected for C400H511N137O245P40S 12429.2; found (electrospray): 12430; purity 73%.
3A-TFC-SH: expected for C400H514N140O245P40S 12474.3; found (MALDI): 12549 (M+3Na+); purity 70%.
T15-3A-TFC-SH: expected for C550H709N170O350P55S 17028.8; found (electrospray): 17037; purity 67%.
Electrospray ionization mass spectra were recorded on a Fisons VG Platform II spectrometer. Matrix-Assisted Laser Desorption/Ionization-Time-Of-Flight spectra were performed using a Perseptive Voyager DETMRP mass spectrometer, equipped with nitrogen laser at 337 nm using a 3 ns pulse. The matrix used contained 2,4,6-trihydroxyacetophenone (10 mg/ml in ACN/water 1:1) and ammonium citrate (50 mg/ml in water).
Preparation of target RNA transcripts
Two RNA transcripts corresponding to (i) iap-103 (RNA-List target), a fragment located at positions 604–707 of the coding sequence of L. innocua virulence gene iap (Accession Number M80349) and (ii) iap-107(−) (RNA control), the complementary sequence of iap-103 were prepared by in vitro transcription from polymerase chain reaction (PCR) products. Listeria innocua genomic DNA was used for the synthesis of iap-103 and iap-107(−) PCR products with primers T7iap-103f/iap-103 r and iapQf/T7iap-107r, respectively (Table 2).
Table 2.
Primers used for the preparation of RNA transcripts
| Name | Sequence | Use |
|---|---|---|
| T7iap-103f | 5′TAATACGACTCACTATAGGAACGTTAAAAGCGGCGACAC3′ | Synthesis of RNA-List target |
| iap-103r | 5′ACATAAATAGAAGAAGAAGATAAATTATTCCA3′ | |
| T7iap-107r | 5′TAATACGACTCACTATACCTACATAAATAGAAGAAGAAGA3′ | Synthesis of RNA control |
| iapQf | 5′ AACGTTAAAAGCGGCGACAC3′ | Synthesis of RNA control iap qPCR and RT-qPCR (16) |
| iapQr | 5′AATATCTTGAACAGAAACACCGTACTTC3′ | iap qPCR and RT-qPCR (16,56) |
| iapQp | 5′6-FAM-CGGATAATGCCCAAA-MGBNFQ3′ |
Synthesis of RNA targets by in vitro transcription and quality control by qPCR: (real-time PCR) and; RT-qPCR: (reverse transcription real-time PCR); qPCR and RP-qPCR probe was labeled with FAM: (fluorescein) and label; MGBNFQ: (non-fluorescent quencher with minor grove binding activity, which increases the Tm of the probe; MGB) probe with non-fluorescent quencher (NFQ).
RNA targets were in vitro transcribed from 500 ng of DNA template (i.e. PCR product) with the T7 transcription kit (Roche Applied Sciences) for 1.5 h at 37°C. A subsequent DNAse enzymatic treatment was performed using the TURBO DNA-free kit (Ambion). Transcript concentration was determined by spectrophotometry using a NanoDrop device (NanoDrop Tehcnologies).
Sequence-specific real-time qualitative PCR (RT-qPCR) and qPCR assays were carried out to further confirm the identity of RNA sequences and the absence of remaining DNA templates (Table 2). iap-specific reactions were performed with TaqMan® technology as described (16). Reactions were run on an ABI Prism 7300 apparatus (Applied Biosystems) with the following program: 2 min at 50°C, 10 min at 95°C and 40 cycles of 15 s at 95°C and 1 min at 60°C. The reverse transcription was optimized to be included in the same qPCR tube by adding 2.5 U of Moloney murine leukemia virus reverse transcriptase (Applied Biosystems) and substituting the initial 50°C incubation by 30 min at 42°C. qPCR and RT-qPCR assays were evaluated by using sequence detection system software, version 1.7 (Applied Biosystems, Foster City, CA, USA). Negative values or a lack of amplification was set at a cycle threshold (CT) value of 40. All reactions were performed in triplicate.
SPR biosensing
In our experiments, we have used a home-made SPR sensor system. The working principle relies on the detection of the specific biomolecular interaction between a target analyte and its capturing receptor, previously immobilized at the gold sensing surface. The recognition of the target molecule causes a change in the refractive index, which is proportional to its concentration at the sensor surface and is detected through a change of the reflectivity at the SPR sensor interface (41). The changes in refractive index are measured in real time, and the results plotted as response of refractive index units (RIUs) variation versus time. SPR analysis involves rapid and simple procedures that do not require either labelling of targets or amplification steps. The SPR sensor can operate under reversible conditions by regeneration of the receptor layer after the interaction with the target, making possible to analyse a high number of interactions using the same receptor monolayer. The use of self-assembled monolayer technique for receptor immobilization onto the gold surface provides a high reproducibility of the SPR analysis.
The SPR device works at a fixed angle of incidence using monochromatic laser light at 670 nm, p-polarized. The device has two flow cells (300 nl each) for two independent measurements and a sensing area of 3 mm2. The sensor chip is a microscope cover glass slide (10 × 10 × 0.15 mm) coated with 2 nm of chromium and 45 nm of gold. Before the experiments, the sensor chip surface is cleaned using organic solvents (trichloroethylene, acetone and ethanol), rinsed with water and finally treated with piranha solution (70%H2SO4–30% H2O2) during a few seconds, then rinsed again with water and dried under nitrogen flux. The liquid handling system is fully automated and incorporated into the SPR platform. A continuous buffer-flow is delivered onto the sensor surface at a constant rate of 12 µl/min by a peristaltic pump. Injection of the samples is controlled by diaphragm pumps that load precise volumes of 250 µl into each flow cell by its corresponding injection valve. Samples are pumped into the flow cell at a constant rate of 12 µl/min.
To keep conditions of RNAse free, before starting the experiments, the sensor was cleaned by flowing SDS 0.5% for 12 h, 100 mM HCl for 5 h, ethanol for 5 h and finally diethyl pyrocarbonate (DEPC; Sigma-Aldrich) treated H2O for 5 h. All buffer and reagents were autoclaved and DEPC treated.
Immobilization of receptors onto the gold sensor surface was done ex situ by directly dropping on the clean sensor surface 75 µL of a 1-µM solution of the receptor nucleic-type prepared on 50 mM phosphate buffered (PB) solution with 0.5 M NaCl, pH 7. The chip was kept under humid chamber for 3 h. After, it was rinsed in DEPC treated H2O, dried under N2 flux and placed in the sensor system.
Hybridization was done in situ at room temperature by flowing over the sensor surface a sample of the target prepared in 5×-SSC (0.075 M Sodium Saline Citrate/0.75 M NaCl) buffer pH 7. Regeneration after hybridization was possible by injecting a solution of 35% formamide in sterile water.
The experimental detection limit was defined as the refractive index value after hybridization which is at least three times larger than the standard deviation of the DNA control or the RNA control signals.
RESULTS AND DISCUSSION
Detection of short nucleic acids sequences using parallel tail clamps
Detection of short DNA sequences 32-mer long (DNA-list, Table 1) was first analysed using tail clamps receptors previously immobilized at the SPR surface. The length of the target was selected to be representative of short RNA types like miRNA.
Previously reported experiments for detection of this target in bulk, showed that substitution of adenine (A) for aminoadenine (8-AA) produced a large stabilization of the structure allowing triplex formation at neutral pH (16). However, detection on solid supports, as in SPR sensing, imposes steric hindrance conditions that might affect hybridization efficiency. For this reason, the use of tail-clamps as receptors and the effect of aminoadenine on triplex stabilization were explored using unmodified triplex-forming clamps (TFC-SH) and clamps bearing an aminoadenine modification (3A-TFC-SH).
Results for DNA-list target capture on both aminoadenine and unmodified monolayer types are displayed on Figure 2. Results show that triplexes are greatly stabilized by the introduction of the aminoadenine base into the hairpin sequence (Figure 2A). Unmodified triplex gave a detection limit of 1.5 nM (375 fmol), which is 1.8 times lower than that of the modified one (0.85 nM or 212 fmol). At saturation, the different behavior for both types of receptors followed the same trend, as the aminoadenine-modified clamp gave a signal 1.8 larger.
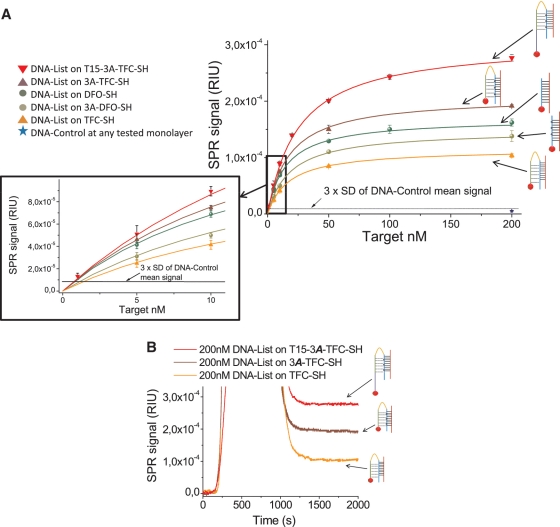
Figure 2.
(A) Calibration curves of DNA List detection using 3A-DFO-SH, DFO-SH, TFC-SH, 3A-TFC-SH and T15-3A-TFC-SH receptors and zoom of the 0–10 nM detection range. DNA control 200 nM on any of the tested monolayers gave a mean signal of 5.4 ± 1.2 × 10−6 RIU, which was equivalent to the baseline noise standard deviation. Our criteria to assess the sensitivity (three times the standard deviation of the negative control) gave a refractive index cut-off value of 9 × 10−6 RIU which is highlighted as a discontinued line in the graph. All target concentrations able to give a signal above this threshold were considered as positive hybridizations. (B) Real-time sensograms of 200 nM DNA-List using the three tail-clamp variants.
Contrary to results in solution, hybridization experiments showed that the non-modified triplex became possible at neutral pH; only a decrease in sensitivity was observed. We suggest that the receptor monolayer itself, with the hairpins in a very close format, might act as a stabilizing agent in the case of non-modified tail-clamps. It has also been reported a dependence of the triplex formation on salt (Na+) concentration for up to 200 mM (42). Hence, the large salt concentration employed during our hybridization experiments (0.5 M) could in addition contribute to the triplex stabilization.
As both tail clamp receptors have demonstrated to be effective for DNA capture at neutral pH, we have compared them with the conventional duplex detection approach to establish if the triplex approach could be superior to the duplex forming one. DNA-list was then captured using unmodified DFO-SH. We also decided to test the aminoadenine-modified duplex forming version (3A-DFO-SH), to analyse if that modification has the same positive effect than observed before. Results are shown on Figure 2A. As can be observed, although it is possible to create the triplex at neutral pH using unmodified tail clamps, in comparison with duplexes, their ability to detect the target was markedly lower. The limit of the detection was 0.9 nM (225 fmol) and 1.24 nM (310 fmol) for unmodified and modified duplexes receptors, respectively; both values are lower than the one from unmodified triplex (1.5 nM). These values clearly demonstrate that at neutral pH, unmodified triplexes are less efficient in target capture than duplexes. Only when the aminoadenine modification is introduced, the triplex forming receptor can effectively compete at neutral pH with their duplex forming oligonucleotide homologue in target capture. Although detection limits for DFO-SH and 3A-TFC-SH were found to be very close (0.9 and 0.85 nM respectively), the largest efficiency of triplexes was evident by the higher SPR values obtained at saturating target concentrations. At 200 nM target concentration, 3A-TFC-SH was 18% more efficient than the DFO-SH and 27% more efficient than the 3A-DFO-SH.
On the other hand, results obtained for DFO provided evidence that duplex formation on the sensor surface was also dependent on the aminoadenine modification at the receptor sequence. However, on the contrary to triplexes, the aminoadenine modification induced a destabilizing effect of the duplex structure leading to lower efficiency in target capture. These findings are supported by previously theoretical calculations (35) which found that the Watson–Crick pairing of A·T is slightly less favorable than of A·T. In addition, there is a large desolvation penalty upon binding of 8-aminoadenine, which together with the disadvantaged pairing binding, justify that duplexes containing A are less stable than those with A.
Finally, the effect of introducing a vertical spacer within the receptor sequence was also explored as a strategy to enhance target accessibility and detection efficiency. The spacing sequence consists on a 15 thymidine track inserted immediately after the thiol modification in the 3A-TFC-SH receptor. Thymidines were chosen because they are the least electrostatically attractive nucleotide among the four nucleic bases with the gold surface (43), helping to reduce the magnitude of physisorption and favoring the accessibility of the target to the matching sequence. This positive effect has been previously observed for standard DNA duplexes (11) but to our knowledge, it has not been analysed on triplexes.
When DNA-list target detection is compared on T15-3A-TFC-SH and 3A-TFC-SH monolayers, a notably increase in sensitivity was found for receptors with the additional vertical spacer sequence (Figure 2). The detection limit of the non-vertically spaced tail-clamp (0.85 nM or 212 fmol), is decreased to 0.8 nM (200 fmol). Although this limit is only slightly lower, a stronger difference is observed at saturation where the T15 modified tail clamp is up to 37.5% more efficient in target capture than the non-modified one.
The specificity of the interactions was analysed for all monolayer types using a 200-nM control DNA sequence, with similar characteristics as the target, such as the presence of a non-complementary 12 nt homopyrimidine track. Measurements were evaluated in triplicate obtaining for all cases a negligible signal equivalent to the baseline noise (Figure 2A).
Detection of large nucleic acids sequences using parallel tail-clamps
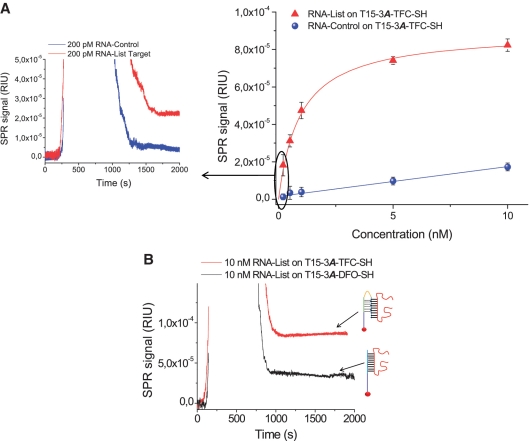
Short targets are usually easier to detect than large nucleic acids targets such as RNA, as they do not normally exhibit secondary structures that could hinder hybridization. As an example, model sequences up to 33 nt but not 50-nt sequences produced triplex structures with conventional clamps (15). However, it has been observed that triplexes with tail-clamps show an enhanced ability to interact with RNA targets displaying stable secondary structures (15,16). For that reason, we have explored the effectiveness of our T15-amino-modified tail-clamps for capturing a 103-nt purified RNA target sequence (RNA-List) with predicted secondary structures. Results are shown on Figure 3A. The sensitivity for RNA detection using the T15-3A-TFC-SH receptor was greatly improved compared to that achieved for short DNA targets. It was possible to detect up to 200 pM (5.5 pg/µL) of RNA concentration, which corresponds to 50 fmol. A RNA control sequence was also tested to check the specificity of the interaction (Figure 3). Minimum detectable SPR signals were obtained specially for concentrations below 5 nM.
Figure 3.
(A) Calibration curve of RNA detection on the T15-3A-TFC-SH monolayer. Inset shows sensograms of the detection of 200 pM RNAList (red) and 200 pM RNA control (blue) on the T15-3A-TFC-SH monolayer. (B) Approximately 10 nM sensograms of RNA-List samples on T15-3A-TFC-SH (red) and T15–DFO-SH (black) monolayers indicating the lower disability of DFO receptors to detect RNA samples.
Finally, as in the solution, the triplex approach works better for RNA than DNA targets detection (15), we have explored the enhanced sensitivity levels found previously using tail-clamps to RNA detection. We have studied if they were related to: (i) the nature of the nucleic acid (RNA versus DNA and whether secondary structures are present or not) or (ii) to the larger molecular weight of the RNA target as compared to the short DNA one, which is a factor that enhances detection in SPR sensing. To confirm it, a 10 nM RNA target was evaluated in triplicate using the T15-3A-TFC-SH and T15-DFO-SH monolayers, respectively (Figure 3B). If the lower sensitivity levels are not related to the nucleic acid nature, but to the molecular weight, the difference in capture efficiency between the duplex and triplex approaches should be kept constant. The difference would be in the range of 18%, which is the same found for short DNA targets.
As depicted in Figure 3B, the T15-DFO-SH was able to give a signal of 3.8 ± 0.7· × 10−5 RIU, while the triplex forming one gave 8.3 ± 1· × 10−5 RIU. Efficiency of the triplex approach was 54.2% higher than that found for the duplex one. This value was similar to the value reported when the same comparison of RNA capture was done in solution (16) and much larger than the expected 18%. This clearly indicates that triplex efficiency is markedly increased for RNA than for the short DNA target. This could be explained by the enhanced mechanism of triplexes to interact with RNA targets rather than DNA ones (44,45). However, we suggest that the differences between both target types are probably more strongly related to the presence of secondary structures, which exist only in RNA. With these results, duplex strategy has a lower ability for detecting targets bearing secondary structures, enforcing the great advantage of tail-clamp-based triplexes for RNA capture.
Examples of label-free SPR-biosensing detection of RNA are scarce in the literature (12–14,46). Strategies to increase sensitivity are mainly based on amplification strategies such as the use of enzymatic reactions and/or nanoparticles (47–50) or by coupling the biosensor to other tools such as 1D and 2D microchannels (51). To our knowledge, SPR has only been used for the label-free analysis of structured RNA targets in the recent work of Mandir et al. (52). This work involved the multiplexed RNA analysis as a function of RNA secondary structures, thereby allowing the identification of RNA accessible sites. Its objective largely differs from our approach, as we aim at the development of a strategy to overcome the difficulties arisen from the detection of structured targets. In addition, the sensitivity levels were considerably worse than our results, at the micromolar range (0.1 µM or 0.2 nmol). Our work constitutes the first example in which SPR has been successfully applied to the label-free detection of RNA targets with predicted secondary structures demonstrating excellent sensitivity levels. Using conventional techniques, parallel tail-clamps could detect up to 25 fmol RNA L. innocua target using radiolabelling and up to 150 fmol using streptavidin-coated magnetic beads and RT-QPCR (16). Comparing those values with our results, which allow detection of up to 50 fmol without any label requirement, the advantage of using a biosensor tool for RNA analysis is clearly demonstrated. In addition, our methodology can be easily adapted to modified oligonucleotides having enhanced hybridization properties such as locked nucleic acids which have demonstrated a superior affinity for the detection of miRNAs (53) than conventional DNA oligomers. In this line, peptide nucleic acids clamps have demonstrated their enhanced potential to detect DNA and RNA from environmental samples (54). It has been described that triple helix formation by parallel clamps carrying 8-aminopurines may be enhanced by substitution of the Hoogsteen strand by 2′-O-methyl-RNA (55) and locked nucleic acids units (56). For all the above reasons, we believe that the results shown in this work may be further improved to enhance duplex and triplex formation. Our methodology constitutes a powerful strategy to analyse RNA targets and it could be of large interest in the field of RNA analysis.
Assay reproducibility
Repeatability and reproducibility of the experiments were analysed by testing the response of 5 nM DNA-List target on T15-3A-TFC-SH monolayers. The values from several measurements on a single SPR chip after several regeneration cycles were compiled and subjected to a statistic analysis (intra-chip analysis). The experimental error estimated as the relative error of the mean absolute deviation did not excess the 15% range, which was an acceptable value, similar to those reported in other SPR applications (57). The same analysis was done with a data collection of consecutive SPR values obtained after target regeneration in two different sensing chips within (i) a given sensor platform and (ii) using two different sensor platforms. Similarly, an interchip analysis within each experimental sensor platform and an intersensor analysis by comparing the values from both sensing platforms were done.
Interchip analysis within a given sensor gave non-statistical difference at the 0.05 level (P = 0.2). The inter-sensor analysis performed by the comparison of the collection of measurements at several sensor chips within each sensor showed a mean SPR value of 5.1 ± 0.6 × ·10−5 RIU and 5 ± 0.7 × 10−5 RIU on each sensor system, respectively. Experimental interchip error within a given sensor was also in the range of 15%, similar to the one obtained on the alternative sensor system and from the intrachip analysis.
The excellent agreement obtained when using different sensors indicates that the reproducibility of the sensor methodology is large and this error is negligible in comparison to the experimental intrachip and interchip errors, which must be the ones taken as reference to evaluate the accuracy of the detection method.
CONCLUSIONS
We have provided a novel approach for the label-free detection of nucleic acids using SPR biosensing. It is possible to create stable triplexes at neutral pH by using parallel-stranded tail-clamp receptors up to a limit of detection of 1.5 nM. The introduction of several 8-aminoadenine modifications in the tail-clamp sequence supplies a strong stability to the triplex structure, achieving lower detection limits for both short and long nucleic acid targets. On the contrary to triplexes, the introduction of aminoadenine modification into the duplex forming receptor sequence has a destabilizing effect.
The 8-aminoadenine modified tail-clamp showed a detection limit of 0.8 nM (200 fmol) for short DNA targets. It was up to 18% more efficient in target detection than the conventional duplex approach. This detection limit was further enhanced by the introduction of a vertical spacer within the tail-clamp sequence. Using this spacer, detection was improved up to 37.5%.
The use of 8-aminoadenine modified and vertically spaced tail-clamps demonstrated excellent capabilities for the detection of RNA sequences with predicted secondary structures. A detection limit of up to 200 pM (50 fmol or 5.5 pg/µl) was achieved. Compared with the duplex homologue receptor, RNA detection was improved by 54% using the parallel tail-clamp, indicating the large disability of the duplex approach to capture this type of target.
RNA targets typically exhibit stable secondary structures that reduce the chance of detection by conventional biological methods. This article demonstrates that our methodology could afford an advanced tool for RNA capture and sensing. In addition, the triplex affinity capture method for RNA detection is simple, fast, label-free and accurate which places it as a promising approach in the RNA detection field.
FUNDING
Project FUNMOL of the EECC (FP7-NMP-213382-2); the Spanish Ministry of Education (BFU2007-63287, CTQ2010-20541, TEC2009-08729 and AGL2010-17181); the project Microplex from CIBER-BBN (CIBER-BBN [The Biomedical Research Networking center in Bioengineering, Biomaterials and Nanomedicine] which is an initiative funded by the VI National R&D&i Plan 2008–2011, Iniciativa Ingenio 2010, Consolider Program, CIBER Actions and financed by the Instituto de Salud Carlos III with assistance from the European Regional Development Fund.); the Generalitat de Catalunya (2009/SGR/208) and M. Botín Foundation. Funding for open access publication charges: Spanish Ministry of Education: CTQ2010-20541, AGL2010-17181, TEC2009-08729.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mattick J. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- 2.Goodrich J, Kugel J. From bacteria to humans, chromatin to elongation, and activation to repression: the expanding roles of noncoding RNAs in regulating transcription. Crit. Rev. Biochem. Mol. Biol. 2009;44:3–15. doi: 10.1080/10409230802593995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mattick J. Deconstructing the dogma: a new view of the evolution and genetic programming of complex organisms. Ann. N Y Acad. Sci. 2009;1178:29–46. doi: 10.1111/j.1749-6632.2009.04991.x. [DOI] [PubMed] [Google Scholar]
- 4.Dinman JD, Richter S, Plant EP, Taylor RC, Hammell AB, Rana TM. The frameshift signal of HIV-1 involves a potential intramolecular triplex RNA structure. Proc. Natl Acad. Sci. USA. 2002;99:5331–5336. doi: 10.1073/pnas.082102199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer I. A practical guide to the art of RNA gene prediction. Brief Bioinform. 2007;8:396–414. doi: 10.1093/bib/bbm011. [DOI] [PubMed] [Google Scholar]
- 7.Raasch P, Schmitz U, Patenge N, Vera J, Kreikemeyer B, Wolkenhauer O. Non-coding RNA detection methods combined to improve usability, reproducibility and precision. BMC Bioinform. 2010;11:491. doi: 10.1186/1471-2105-11-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaisocherová H, Zítová A, Lachmanová M, Tpánek J, Králíková, Liboska R, Rejman D, Rosenberg I, Homola J. Investigating oligonucleotide hybridization at subnanomolar level by surface plasmon resonance biosensor method. Biopolymers. 2005;82:394–398. doi: 10.1002/bip.20433. [DOI] [PubMed] [Google Scholar]
- 9.Song F, Zhou F, Wang J, Tao N, Lin J, Vellanoweth RL, Morquecho Y, Wheeler-Laidman J. Detection of oligonucleotide hybridization at femtomolar level and sequence-specific gene analysis of the Arabidopsis thaliana leaf extract with an ultrasensitive surface plasmon resonance spectrometer. Nucleic Acids Res. 2002;30:e72. doi: 10.1093/nar/gnf072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y-J, Xiang J, Zhou F. Sensitive and Label-Free Detection of DNA by Surface Plasmon Resonance. Plasmonics. 2007;2:79–87. [Google Scholar]
- 11.Carrascosa LG, Calle A, Lechuga LM. Label-free detection of DNA mutations by SPR: application to the early detection of inherited breast cancer. Anal. Bioanal. Chem. 2009;393:1173–1182. doi: 10.1007/s00216-008-2555-1. [DOI] [PubMed] [Google Scholar]
- 12.Sipova H, Zhang S, Dudley AM, Galas D, Wang K, Homola J. Surface Plasmon Resonance Biosensor for Rapid Label-Free Detection of Microribonucleic Acid at Subfemtomole Level. Anal. Chem. 2010;82:10110–10115. doi: 10.1021/ac102131s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson BP, Liles MR, Frederick KB, Corn RM, Goodman RM. Label-free detection of 16S ribosomal RNA hybridization on reusable DNA arrays using surface plasmon resonance imaging. Env. Microbiol. 2002;4:735–743. doi: 10.1046/j.1462-2920.2002.00350.x. [DOI] [PubMed] [Google Scholar]
- 14.Nair TM, Myszka DG, Davis DR. Surface plasmon resonance kinetic studies of the HIV TAR RNA kissing hairpin complex and its stabilization by 2-thiouridine modification. Nucleic Acids Res. 2000;28:1935–1940. doi: 10.1093/nar/28.9.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nadal A, Eritja R, Esteve T, Pla M. Parallel and Antiparallel Tail-Clamps Increase the Efficiency of Triplex Formation with Structured DNA and RNA Targets. Chem. Bio. Chem. 2005;6:1034–1042. doi: 10.1002/cbic.200400358. [DOI] [PubMed] [Google Scholar]
- 16.Nadal A, Coll A, Aviño A, Esteve T, Eritja R, Pla M. Efficient Sequence-Specific Purification of Listeria innocua mRNA Species by Triplex Affinity Capture with Parallel Tail-Clamps. Chem. Bio. Chem. 2006;7:1039–1047. doi: 10.1002/cbic.200500519. [DOI] [PubMed] [Google Scholar]
- 17.Duca M, Vekhoff P, Oussedik K, Halby L, Arimondo PB. The triple helix: 50 years later, the outcome. Nucleic Acids Res. 2008;36:5123–5138. doi: 10.1093/nar/gkn493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Q, Gaddis S, MacLeod M, Walborg E, Thames H, DiGiovanni J, Vasquez K. High-affinity triplex-forming oligonucleotide target sequences in mammalian genomes. Mol. Carcinog. 2007;46:15–23. doi: 10.1002/mc.20261. [DOI] [PubMed] [Google Scholar]
- 19.Bacolla A, Collins J, Gold B, Chuzhanova N, Yi M, Stephens R, Stefanov S, Olsh A, Jakupciak J, Dean M, et al. Long homopurine*homopyrimidine sequences are characteristic of genes expressed in brain and the pseudoautosomal region. Nucleic Acids Res. 2006;34:2663–2675. doi: 10.1093/nar/gkl354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain A, Bacolla A, Chakraborty P, Grosse F, Vasquez K. Human DHX9 helicase unwinds triple-helical DNA structures. Biochemistry. 2010;49:6992–6999. doi: 10.1021/bi100795m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chin J, Schleifman E, Glazer P. Repair and recombination induced by triple helix DNA. Front Biosci. 2007;12:4288–4297. doi: 10.2741/2388. [DOI] [PubMed] [Google Scholar]
- 22.Fox KR. Targeting DNA with triplexes. Curr. Med. Chem. 2000;7:17–37. doi: 10.2174/0929867003375506. [DOI] [PubMed] [Google Scholar]
- 23.Bissler JJ. Triplex DNA and human disease. Front. Biosci. 2007;12:4536–4546. doi: 10.2741/2408. [DOI] [PubMed] [Google Scholar]
- 24.Wang G, Carbajal S, Vijg J, DiGiovanni J, Vasquez K. DNA structure-induced genomic instability in vivo. J. Natl Cancer Inst. 2008;100:1815–1817. doi: 10.1093/jnci/djn385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potaman V, Oussatcheva E, Lyubchenko Y, Shlyakhtenko L, Bidichandani S, Ashizawa T, Sinden R. Length-dependent structure formation in Friedreich ataxia (GAA)n*(TTC)n repeats at neutral pH. Nucleic Acids Res. 2004;32:1224–1231. doi: 10.1093/nar/gkh274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain A, Magistri M, Napoli S, Carbone G, Catapano C. Mechanisms of triplex DNA-mediated inhibition of transcription initiation in cells. Biochimie. 2010;92:317–320. doi: 10.1016/j.biochi.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 27.Schmitz K-M, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chou M, Chang K. An intermolecular RNA triplex provides insight into structural determinants for the pseudoknot stimulator of -1 ribosomal frameshifting. Nucleic Acids Res. 2010;38:1676–1685. doi: 10.1093/nar/gkp1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pieczenik G. A positive-selection function for microRNA: an adaptor hypothesis revisited. Reprod. Biomed. Online. 2006;12:292–297. doi: 10.1016/s1472-6483(10)61000-0. [DOI] [PubMed] [Google Scholar]
- 30.Xu Y, Xu Y, Hou Q, Yang R, Zhang S. Triplex real-time PCR assay for detection and differentiation of Bordetella pertussis and Bordetella parapertussis. APMIS. 2010;118:685–691. doi: 10.1111/j.1600-0463.2010.02644.x. [DOI] [PubMed] [Google Scholar]
- 31.Verhoff P, Oussedik K, Halby L, Arimondo PB. The triple helix: 50 years later, the outcome. Nucleic Acids Res. 2008;36:5123–5138. doi: 10.1093/nar/gkn493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Filichev VV, Pedersen EB. DNA-conjugated organic chromophores on DNA stacking interactions. Wiley Encyclopedia of Chemical Biology. 2008:1–32. doi:10.1002/9780470048672.wecb654. [Google Scholar]
- 33.Robles J, Grandas A, Pedroso E, Luque FJ, Eritja R, Orozco M. Nucleic acid triple helices: stability effects of nucleobase modifications. Curr. Org. Chem. 2002;6:1333–1368. [Google Scholar]
- 34.Aviño A, Frieden M, Morales JC, de la Torre BG, Garcia RG, Azori F, Gelpi JL, Orozco M, Gonzaez C, Eritja R. Properties of triple helices formed by parallel-stranded hairpins containing 8-aminopurines. Nucleic Acids Res. 2002;30:2609–2619. doi: 10.1093/nar/gkf374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Güimil-García R, Ferrer E, Macías MJ, Eritja R, Orozco M. Theoretical calculations, synthesis and base-pairing properties of oligonucleotides containing 8-amino-2′-deoxyadenosine. Nucleic Acids Res. 1999;27:1991–1998. doi: 10.1093/nar/27.9.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frieden M, Aviñó A, Eritja R. Convenient synthesis of 8-amino-2′-deoxyadenosine. Nucleosides Nucleotides Nucleic Acids. 2003;22:193–202. doi: 10.1081/NCN-120019521. [DOI] [PubMed] [Google Scholar]
- 37.Gruber A, Lorenz R, Bernhart S, Neuböck R, Hofacker I. The Vienna RNA Websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aviñó A, Morales JC, Frieden M, de la Torre BG, Güimil-García R, Cubero E, Luque FJ, Orozco M, Azorín F, Eritja R. Parallel-stranded hairpins containing 8-aminopurines. Novel efficient probes for triple-helix formation. Bioorg. Med. Chem. Lett. 2001;11:1761–1763. doi: 10.1016/s0960-894x(01)00295-5. [DOI] [PubMed] [Google Scholar]
- 39.Cubero E, Aviñó A, de la Torre BG, Frieden M, Eritja R, Luque FJ, González C, Orozco M. Hoogsteen-based parallel-stranded duplexes of DNA. The effect of 8-amino derivatives. J. Am. Chem. Soc. 2002;124:3133–3142. doi: 10.1021/ja011928+. [DOI] [PubMed] [Google Scholar]
- 40.Afonina I, Reed M, Lusby E, Shishkina I, Belousov Y. Minor groove binder-conjugated DNA probes for quantitative DNA detection by hybridization-triggered fluorescence. Biotechniques. 2002;32:940–944, 946–949. doi: 10.2144/02324pf01. [DOI] [PubMed] [Google Scholar]
- 41.Homola J. Surface Plasmon Resonance Sensors for Detection of Chemical and Biological Species. Chem. Rev. 2008;108 doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- 42.Plum GE, Park Y-W, Singleton SF, Dervan PB, Breslauer KJ. Thermodynamic characterization of the stability and the melting behavior of a DNA triplex: a spectroscopic and calorimetric study. Proc. Natl. Acad. Sci. USA. 1990;87:9436–9440. doi: 10.1073/pnas.87.23.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimura-Suda H, Petrovykh D, Tarlov M, Whitman L. Base-dependent competitive adsorption of single-stranded DNA on gold. J. Am. Chem. Soc. 2003;125:9014–9015. doi: 10.1021/ja035756n. [DOI] [PubMed] [Google Scholar]
- 44.Lestienne P, Boudsocq F, Bonnet J. Initiation of DNA replication by a third parallel DNA strand bound in a triple helix manner leads to strand invasion. Biochemistry. 2008;47:5689–5698. doi: 10.1021/bi702318g. [DOI] [PubMed] [Google Scholar]
- 45.Bernal-Mendez E, Leumann CJ. Stability and kinetics of nucleic acids triplexes with chimaeric DNA/RNA third strands. Biochemistry. 2002;41:725–735. doi: 10.1021/bi025937m. [DOI] [PubMed] [Google Scholar]
- 46.Nelson B, Grimsrud T, Liles M, Goodman R, Corn R. Surface plasmon resonance imaging measurements of DNA and RNA hybridization adsorption onto DNA microarrays. Anal. Chem. 2001;73:1–7. doi: 10.1021/ac0010431. [DOI] [PubMed] [Google Scholar]
- 47.Zhou W-J, Chen Y, Corn RM. Ultrasensitive Microarray Detection of Short RNA Sequences with Enzymatically Modified Nanoparticles and Surface Plasmon Resonance Imaging Measurements. Anal. Chem. 2011;83:3897–3902. doi: 10.1021/ac200422u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang S, Lee H, Wark A, Corn R. Attomole microarray detection of microRNAs by nanoparticle-amplified SPR imaging measurements of surface polyadenylation reactions. J. Am. Chem. Soc. 2006;128:14044–14046. doi: 10.1021/ja065223p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joung H, Lee N, Lee S, Ahn J, Shin Y, Choi H, Lee C, Kim S, Kim M. High sensitivity detection of 16s rRNA using peptide nucleic acid probes and a surface plasmon resonance biosensor. Anal. Chim. Acta. 2008;630:168–173. doi: 10.1016/j.aca.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 50.Lee H, Wark A, Li Y, Corn R. Fabricating RNA microarrays with RNA-DNA surface ligation chemistry. Anal. Chem. 2005;77:7832–7837. doi: 10.1021/ac0516180. [DOI] [PubMed] [Google Scholar]
- 51.Jin-Lee H, Goodrich T, Corn R. SPR imaging measurements of 1-D and 2-D DNA microarrays created from microfluidic channels on gold thin films. Anal. Chem. 2001;73:5525–5531. doi: 10.1021/ac010762s. [DOI] [PubMed] [Google Scholar]
- 52.Mandir JB, Lockett MR, Phillips MF, Allawi HT, Lyamichev VI, Smith LM. Rapid Determination of RNA Accessible Sites by Surface Plasmon Resonance Detection of Hybridization to DNA arrays. Anal. Chem. 2009;81:8949–8956. doi: 10.1021/ac9015962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kauppinen S, Vester B, Wengel J. Locked nucleic acid: high-affinity targeting of complementary RNA for Rnomics. Hand. Exp. Pharmacol. 2006;173:405–422. doi: 10.1007/3-540-27262-3_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chandler DP, Stults JR, Cebula S, Schuck BL, Weaver DW, Anderson KK, Egholm M, Brockman FJ. Affinity purification of DNA and RNA from environmental samples with peptide nucleic acid clamps. Appl. Environ. Microbiol. 2000;66:3438–3845. doi: 10.1128/aem.66.8.3438-3445.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aviñó A, Grimau MG, Frieden M, Eritja R. Synthesis and triple-helix stabilization properties of branched oligonucleotides carrying 8-aminoadenine moieties. Helv. Chim. Acta. 2004;87:303–316. [Google Scholar]
- 56.Alvira M, Eritja R. Triplex-stabilizing properties of parallel clamps carrying LNA derivatives at the Hoogsteen strand. Chem. Biodivers. 2010;7:376–382. doi: 10.1002/cbdv.200900230. [DOI] [PubMed] [Google Scholar]
- 57.Gaus K, Hall EAH. Surface plasmon resonance sensor for heparin measurements in blood plasma. Biosens. Bioelectron. 1998;13:1307–1315. doi: 10.1016/s0956-5663(98)00094-3. [DOI] [PubMed] [Google Scholar]