Abstract
Most bacteria can only be transformed with circular plasmids, so robust DNA integration methods for these rely upon selection of single-crossover clones followed by counter-selection of double-crossover clones. To overcome the limited availability of heterologous counter-selection markers, here we explore novel DNA integration strategies that do not employ them, and instead exploit (i) activation or inactivation of genes leading to a selectable phenotype, and (ii) asymmetrical regions of homology to control the order of recombination events. We focus here on the industrial biofuel-producing bacterium Clostridium acetobutylicum, which previously lacked robust integration tools, but the approach we have developed is broadly applicable. Large sequences can be delivered in a series of steps, as we demonstrate by inserting the chromosome of phage lambda (minus a region apparently unstable in Escherichia coli in our cloning context) into the chromosome of C. acetobutylicum in three steps. This work should open the way to reliable integration of DNA including large synthetic constructs in diverse microorganisms.
INTRODUCTION
The addition of DNA conferring new or altered properties to microorganisms has underpinned biotechnology for decades. Recently, the potential scope and scale of this approach has grown with the acceleration of genome sequencing, development of commercial de novo DNA synthesis and advent of Synthetic Biology, the new discipline which brings engineering principles to the design and construction of biological systems (1).
DNA can be added to microorganisms using replicative plasmids, but these are inherently unstable, limiting their applied utility. To stabilize exogenous DNA, it must be irreversibly incorporated into a stable DNA molecule inside the cell, usually a chromosome. This can be accomplished in a one-step homologous recombination procedure (often called ‘allele exchange’ or ‘gene replacement’) for those organisms that are efficiently transformed with linear DNA, such as yeast and naturally competent bacteria like Bacillus subtilis (2). A selectable marker gene positioned alongside the DNA sequence of interest within an allele exchange cassette is only retained by the desired recombinant cells, allowing these cells to be specifically selected and easily isolated, typically using an antibiotic. Large or multiple sequences can be inserted at a single locus simply by alternating between two selectable markers in a series of integration steps, as in the ‘domino’ method of Itaya et al. (3).
Most bacteria cannot be transformed with linear DNA, so an integrative plasmid bearing the homologous recombination construct is used instead. As plasmids are circular, a single homologous recombination event can reversibly integrate the entire plasmid into the chromosome, resulting in unstable single-crossover cells with the potential to revert to wild-type. The desired stable double-crossover cells are much rarer, as they result from two homologous recombination events in a single cell or lineage. Double-crossover cells are not easily isolated from single-crossover cells, because both contain the selectable marker in the cassette. This issue can be overcome using a counter-selection marker located on the plasmid ‘backbone’, but identifying a suitable counter-selection marker and appropriate conditions for its use can be one of the most challenging aspects of developing genetic tools for a particular organism (4). We are interested in several bacterial species for which genetic tools are very limited, and the use of counter-selection markers has not been described.
Here we develop generally applicable strategies for the selection of double-crossover clones at certain types of genomic loci without using a plasmid-borne counter-selection marker. Crucially, the expression of a selectable marker gene is coupled to the formation of the desired double-crossover recombinant chromosome, so that double-crossover clones can be isolated using the associated selectable phenotype. We call this approach allele-coupled exchange (ACE). The principles described here bring much of the power and practical simplicity of linear DNA procedures to integration of plasmid-borne DNA, and open the way to the step-by-step insertion of large DNA sequences into diverse microbial chromosomes, as we demonstrate using Clostridium acetobutylicum.
MATERIALS AND METHODS
Bacterial strains
Escherichia coli strains were grown in Luria-Bertani (LB) broth at 37°C with rotary shaking at 200 rpm, or on LB plates. All solid media were prepared by adding 1.5 % w/v agar to the corresponding broth. E. coli strains containing plasmids (Table 1) were cultured in LB broth supplemented with 12.5 μg/ml chloramphenicol, or on LB plates supplemented with 25 μg/ml chloramphenicol. E. coli strain TOP10 (Invitrogen) was used for plasmid cloning and storage, strain CA434 (5) was used as a conjugation donor, and TOP10 containing pAN2 (6) was used for in vivo methylation of plasmid DNA prior to transformation of C. acetobutylicum. Clostridium spp. were grown in static culture at 37°C under an anaerobic atmosphere of N2:H2:CO2 (80:10:10, vol:vol:vol) in an anaerobic workstation (Don Whitley, UK) using media prereduced overnight under the same conditions. Plasmids were transferred into Clostridium spp. by electroporation or conjugation as described previously (7).
Table 1.
List of integration vectors
| Plasmid | Accession number | Organism | Locus | First region of homology | Second region of homology | Element(s) between regions of homology |
|---|---|---|---|---|---|---|
| pMTL-JH1 | HQ875748 | Clostridium acetobutylicum | pyrF | None (to be provided with insert) | 300 bp internal fragment of pyrF | lacZ MCS |
| pMTL-JH2 | HQ875749 | Clostridium acetobutylicum | pyrF | 1200 bp immediately following pyrF | 300 bp internal fragment of pyrF | lacZ MCS |
| pMTL-JH3 | HQ875750 | Clostridium acetobutylicum | pyrF | None (to be provided with insert) | 300 bp internal fragment of pyrF | 3′ part of pyrF, lacZ MCS |
| pMTL-JH4 | HQ875751 | Clostridium acetobutylicum | pyrF | 1200 bp immediately following pyrF | 300 bp internal fragment of pyrF | 3′ part of pyrF, lacZ MCS |
| pMTL-JH11 | HQ875752 | Clostridium acetobutylicum | pyrE | None (to be provided with insert) | 300 bp internal fragment of pyrE | lacZ MCS |
| pMTL-JH12 | HQ875753 | Clostridium acetobutylicum | pyrE | 1200 bp immediately following pyrE | 300 bp internal fragment of pyrE | lacZ MCS |
| pMTL-JH13 | HQ875754 | Clostridium acetobutylicum | pyrE | None (to be provided with insert) | 300 bp internal fragment of pyrE | 3′ part of pyrE, lacZ MCS |
| pMTL-JH14 | HQ875755 | Clostridium acetobutylicum | pyrE | 1200 bp immediately following pyrE | 300 bp internal fragment of pyrE | 3′ part of pyrE, lacZ MCS |
| pMTL-JH15 | HQ875756 | Clostridium acetobutylicum | thl | None (to be provided with insert) | Last 300 bp of thl | ermB lacking promoter, lacZ MCS |
| pMTL-JH16 | HQ875757 | Clostridium acetobutylicum | thl | 1200 bp immediately following thl | Last 300 bp of thl | ermB lacking promoter, lacZ MCS |
| pMTL-JH17 | HQ875758 | Clostridium difficile | pyrE | None (to be provided with insert) | 300 bp internal fragment of pyrE | lacZ MCS |
| pMTL-JH18 | HQ875759 | Clostridium difficile | pyrE | 1200 bp immediately following pyrE | 300 bp internal fragment of pyrE | lacZ MCS |
| pMTL-JH19 | HQ875760 | Clostridium difficile | pyrE | None (to be provided with insert) | 300 bp internal fragment of pyrE | 3′ part of pyrE, lacZ MCS |
| pMTL-JH20 | HQ875761 | Clostridium difficile | pyrE | 1200 bp immediately following pyrE | 300 bp internal fragment of pyrE | 3′ part of pyrE, lacZ MCS |
| pMTL-JH26 | HQ875762 | Clostridium sporogenes | pyrE | None (to be provided with insert) | 300 bp internal fragment of pyrE | lacZ MCS |
| pMTL-JH27 | HQ875763 | Clostridium sporogenes | pyrE | 1200 bp immediately following pyrE | 300 bp internal fragment of pyrE | lacZ MCS |
| pMTL-JH28 | HQ875764 | Clostridium sporogenes | pyrE | None (to be provided with insert) | 300 bp internal fragment of pyrE | 3′ part of pyrE, lacZ MCS |
| pMTL-JH29 | HQ875765 | Clostridium sporogenes | pyrE | 1200 bp immediately following pyrE | 300 bp internal fragment of pyrE | 3′ part of pyrE, lacZ MCS |
| pMTL-JH30 | HQ875766 | Clostridium acetobutylicum | thl | None (to be provided with insert) | Last 300 bp of thl | pyrE lacking promoter, lacZ MCS |
| pMTL-JH31 | HQ875767 | Clostridium acetobutylicum | thl | 1200 bp immediately following pyrE | Last 300 bp of thl | pyrE lacking promoter, lacZ MCS |
The elements of the integration cassette of each plasmid are shown, and are best understood with reference to the text and Figures 1–3. Plasmids pMTL-JH12, 14, 15, 16, 30 and 31 are described in the main text, and pMTL-JH2, 18 and 27 are described in the Supplementary Data. Suitable combinations of plasmids targeting the same locus could be used in a series of steps to integrate several overlapping fragments of a large sequence, as described in the text and shown in Figure 3. MCS denotes a multiple cloning site.
Plasmid construction
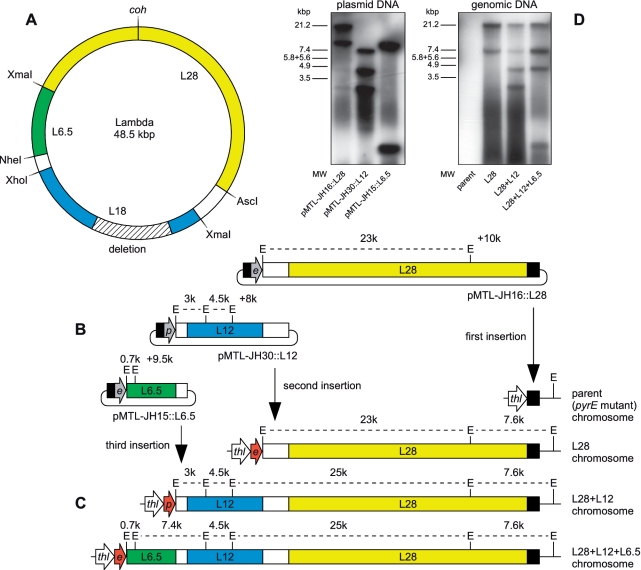
Integration vectors for use in C. acetobutylicum ATCC 824, Clostridium sporogenes NCIMB 10696 and Clostridium difficile 630 are based upon Clostridium–E. coli shuttle plasmids pMTL85141, pMTL85151 and pMTL83151, respectively (7), and differ to those parental plasmids only between the SbfI and AscI sites. The integration vectors and their key functional components are listed in Table 1, and annotated sequences are available from GenBank/EMBL/DDJB. Chromosomal DNA of phage lambda cI857ind 1 Sam 7 was obtained from NEB and fragments were cloned as shown in Figure 3. L12 was derived from L18 by a spontaneous deletion of 5996 bp corresponding to nucleotides 21738–27733 of the lambda cI857ind 1 Sam 7 chromosome.
Figure 3.
Multistep insertion of the chromosome of phage lambda into the chromosome of C. acetobutylicum. (A) Chromosome of phage lambda showing restriction sites used to excise the three overlapping fragments. Yellow, 28 kb XmaI-XmaI fragment L28; blue, 18 kb NheI-AscI fragment L18; green, 6.5 kb XmaI-XhoI fragment L6.5; white, regions of overlap between fragments; cross-shaded deletion, 6 kb region of L18 absent from L12; coh, ligated cohesive ends. (B) Integration plasmids pMTL-JH16::L28, pMTL-JH30::L12 and pMTL-JH15::L6.5. The lambda sequences are colored as in (A). Gray arrows e and p, inactive (non-expressed) ermB and pyrE, respectively; black, homology to thl locus; E, EcoRI sites; dashed lines, EcoRI fragments (except those spanning the plasmid backbone); numeric labels, EcoRI fragment lengths in kilo base pair. (C) The thl locus of C. acetobutylicum before and after 1, 2 and 3 insertions of lambda DNA. Elements are labeled as in (B). The first recombination event at each step, indicated, is directed by a long region of homology. A short region of homology mediates plasmid excision and simultaneously activates ermB or pyrE in alternate steps, shown by red arrows e and p, respectively, by positioning them under the control of the chromosomal thl promoter. (D) Southern blot of EcoRI digests of the plasmids and chromosomes shown in (B) and (C), using lambda DNA as probe. MW, HindIII-digested lambda DNA molecular weight marker.
Growth media for integration procedures in Clostridium acetobutylicum
After electroporation, transformants of C. acetobutylicum were selected on CGM (8) agar plates supplemented with 15 μg/ml thiamphenicol to select for the plasmid-borne resistance marker catP and 20 μg/ml uracil to allow pyrE mutants to grow (except for those experiments which did not involve pyrE mutants, where uracil was not required). CGM agar plates supplemented with 400 μg/ml 5-fluoroorotic acid (FOA) and 1 μg/ml uracil were used to select pyrE-minus clones. These conditions were validated using a previously constructed pyrF mutant (6) which should have the same phenotype as a pyrE mutant. CBM (9) agar plates (which do not contain uracil) were used to select clones able to synthesise their own uracil, which requires a functional pyrE gene. Erythromycin-resistant clones were selected on CGM agar plates supplemented with 40 μg/ml erythromycin.
PCR analysis of chromosomal insertions
Genomic DNA from Clostridium spp. was prepared using the QIAGEN DNeasy Blood and Tissue Kit in accordance with the manufacturer's instructions and recommended pretreatment for Gram-positive bacteria. PCRs were performed using Taq DNA polymerase (NEB) or KOD DNA Polymerase (Merck) or Phusion polymerase (NEB) in accordance with the manufacturer's instructions. Appropriate combinations of oligonucleotide primers are specified in the legends of relevant figures and in the Supplementary Data, and oligonucleotide sequences are listed in Supplementary Table S1.
Analysis of fermentation products
The fermentation products of wild-type C. acetobutylicum ATCC 824 were compared to the adh-expressing recombinant strain in batch culture using CBM broth containing 50 g/l glucose and 5 g/l CaCO3. Starter cultures in the same medium were inoculated using fresh colonies, then when these reached an OD600 of ∼0.5, they were used at a 1% inoculum to start the main cultures. Samples of 1 ml were removed, placed on ice, then centrifuged at 16 000g for 1.5 min. Supernatants were removed and stored at −20°C before analysis by gas chromatography. Ethanol, acetone, butanol, acetic acid and butyric acid were quantified using a Thermo Focus GC equipped with a 30 m TR-FFAP column (0.25 mm internal diameter) and a flame ionization detector. H2 was used as the carrier gas at 0.8 ml/min. The flame was maintained by 35 ml/min H2, 350 ml/min compressed air and 30 ml/min N2. The injector and detector temperatures were 240°C and 270°C, respectively. Peaks were resolved using a column profile of 40°C for 2 min, followed by a 30°C/min ramp to 140°C, then a 45°C/min ramp to 210°C and finally 1 min at 210°C. Samples were extracted before injection by adding an equal volume of ethyl acetate (500 μl) to the supernatant sample, vortexing for 10 s and centrifuging for 5 min at 16 000g. The 300 μl organic phase was removed to a 2 ml sample vial containing a 300 μl deactivated glass insert. Samples of 1 μl were injected.
RESULTS
Specific selection of double-crossovers by switching the state of pyrE
Clostridium acetobutylicum is an industrial organism that naturally produces the excellent biofuel butanol, but the yield, specificity and feedstock utilization of the fermentation process might each be improved by the addition of DNA encoding heterologous enzymes (10). This motivated us to develop a robust DNA integration method for C. acetobutylicum. Our first approach requires a gene which is both positively and negatively selectable, such as pyrE or pyrF, which encode the pyrimidine biosynthesis enzymes orotate phosphoribosyltransferase and orotidine 5-phosphate decarboxylase, respectively. Cells which contain pyrE and pyrF can be selected on growth medium lacking uracil, as they are required for uracil biosynthesis. Conversely, cells that lack either pyrE or pyrF can be selected on growth medium supplemented with FOA, as this compound is highly toxic only to cells which contain this pathway. FOA has been used for counter selection for some time (11) and in various organisms (12–17) including very recently in Clostridium thermocellum (18).
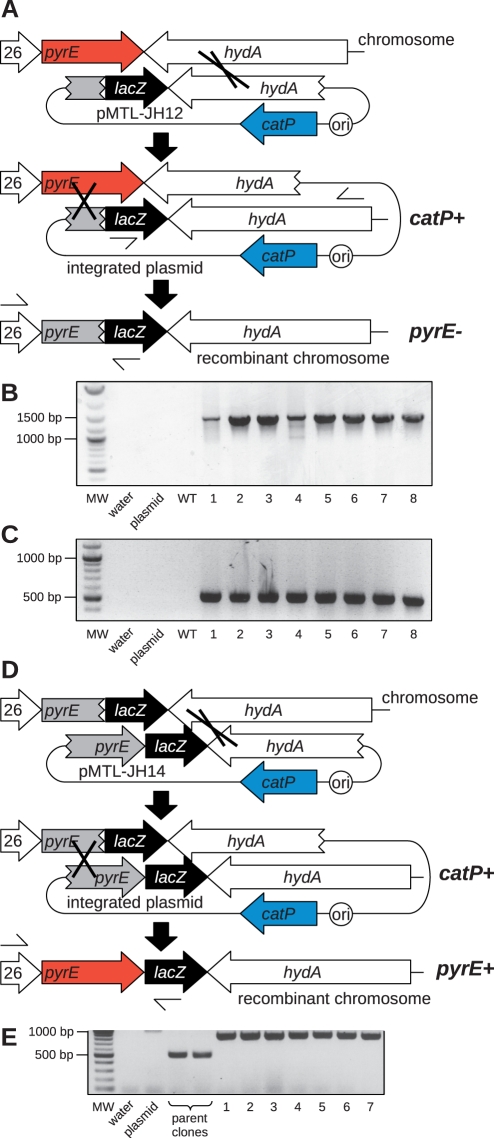
To isolate double-crossover clones by exploiting a change in the state of pyrE, we designed a special allele exchange cassette (Figure 1A). The two regions of homology are of very different lengths, in an attempt to control the order of the recombination events. A long region of homology corresponding to the 1200 bp immediately downstream of pyrE is intended to direct the first recombination event so that a large majority of single-crossover clones are the result of recombination in this region, which would not inactivate pyrE. A subsequent recombination event at a second, much smaller region of homology corresponding to a 300 bp internal portion of pyrE would excise the plasmid, resulting in FOA-resistant (FOAR) double-crossover cells in which the pyrE gene is inactivated, and the DNA insert is stably delivered to the chromosome.
Figure 1.
DNA integration at the pyrE locus of C. acetobutylicum. (A) Selection of stable double-crossover clones using pMTL-JH12. The first recombination event (plasmid integration) is mediated by the long region of homology between pMTL-JH12 and hydA. Single-crossover clones are obtained on medium containing thiamphenicol. The second recombination event (plasmid excision) is mediated by the short region of homology between pMTL-JH12 and an internal portion of pyrE. Double-crossover clones are selected using FOA. (B) PCR screening of eight candidate single-crossover clones using primers lacZa-sF2 and Cac-hydA-sR2, which anneal to the single-crossover chromosome where indicated in (A). MW, 2-Log DNA Ladder (NEB) molecular weight marker; plasmid, pMTL-JH12 plasmid DNA control; WT, wild-type C. acetobutylicum genomic DNA control; 1–8, candidate clones. All eight candidates show the expected 1428 bp band. (C) PCR screening of eight candidate double-crossover clones using primers CAC0026-sF2 and M13F which anneal to the double-crossover chromosome where indicated in (A). MW, 2-Log DNA Ladder (NEB) molecular weight marker; plasmid, pMTL-JH12 plasmid DNA control; WT, wild-type C. acetobutylicum genomic DNA control; 1–8, candidate clones. All eight candidates show the expected 558 bp band. (D) Selection of stable double-crossover clones using pMTL-JH14. The first recombination event (plasmid integration) is mediated by the long region of homology between pMTL-JH14 and hydA/lacZ. Single-crossover clones are obtained on medium containing thiamphenicol. The second recombination event (plasmid excision) is mediated by the short region of homology between pMTL-JH14 and the corresponding portion of pyrE. Double-crossover clones are selected using growth medium lacking uracil. (E) PCR screening of seven candidate double-crossover clones using primers CAC0026-sF2 and M13F which anneal to the double-crossover chromosome where indicated in (C). MW, 2-Log DNA Ladder (NEB) molecular weight marker; plasmid, pMTL-JH14 plasmid DNA control; 1–7, candidate clones. Controls using DNA from two of the pyrE-minus clones obtained in (A) are marked as parent clones. All seven candidates show the expected 861 bp band, and the parent clones show the expected 558 bp band. This increase of 303 bp corresponds to the restoration of the truncated pyrE gene to full length.
We constructed plasmid pMTL-JH12 (Figure 1A), which includes the allele exchange cassette described above, the chloramphenicol/thiamphenicol-resistance marker catP and the origin of replication from Bacillus plasmid pIM13 (19). Like several other Clostridia and many other organisms, C. acetobutylicum is not efficiently transformed by electroporation, so it is not generally practical to use suicide plasmids (that is, typically no colonies appear on selective plates following electroporation of suicide plasmids). Instead, replicative plasmids are used, so transformants are readily obtained. The pIM13 replicon exhibits segregational instability in C. acetobutylicum (7), and single-crossover clones which spontaneously arise within transformant populations can be isolated by subculture under antibiotic selection.
Clostridium acetobutylicum cells were transformed with pMTL-JH12 DNA and plated onto medium supplemented with thiamphenicol and uracil. Typically, 10–100 colonies were visible after 24–48 h incubation, and two clones from each of four independent transformations were subcultured twice on the same medium. The eight resulting clones were screened by PCR, and all were shown to be single-crossover clones in which pMTL-JH12 had integrated into the chromosome via homologous recombination at the long region of homology, as intended (Figure 1B). Next, the eight single-crossover clones were subcultured onto medium supplemented with FOA and uracil to select the desired pyrE-minus double-crossover clones. FOAR colonies were obtained from each of the eight independent subcultures, and one clone from each was purified by subculturing again on the same medium. PCR screening showed the expected double-crossover genotype for all eight clones (Figure 1C), and replica-plating on medium with and without thiamphenicol confirmed the absence of the plasmid-borne catP marker.
The strategy described above effectively replaces a negatively selectable gene with a sequence of interest, which could be achieved using a conventional allele exchange cassette (14). The crucial difference is that the pyrE locus of a recombinant strain constructed using pMTL-JH12 is specially configured to facilitate subsequent genetic modification (Figure 1D). The non-functional pyrE gene in these cells is not completely deleted, it is only truncated at the 3′-end. Homologous recombination between this partial pyrE gene and an appropriate counterpart partial pyrE gene, foreshortened at the 5′-end, would result in a full-length, functional gene. We constructed plasmid pMTL-JH14, which is almost identical to pMTL-JH12, except the 300 bp internal portion of pyrE that comprises the small homology region is followed immediately by the remainder of the pyrE coding sequence (Figure 1D). As before, the long region of homology is intended to direct plasmid integration, without affecting the pyrE phenotype. Subsequent plasmid excision via the short 300-bp homology region will result in the desired double-crossover clones, specifically selectable by their pyrE-positive, uracil prototrophic phenotype. We transformed one of the previously constructed pyrE mutant clones of C. acetobutylicum with pMTL-JH14 DNA, selected transformants on plates supplemented with thiamphenicol and uracil and subcultured them twice on this medium. To select pyrE-positive clones, cells were subcultured onto medium lacking uracil. Seven independent clones were purified and shown to be the intended double-crossovers by PCR (Figure 2D) and thiamphenicol sensitivity.
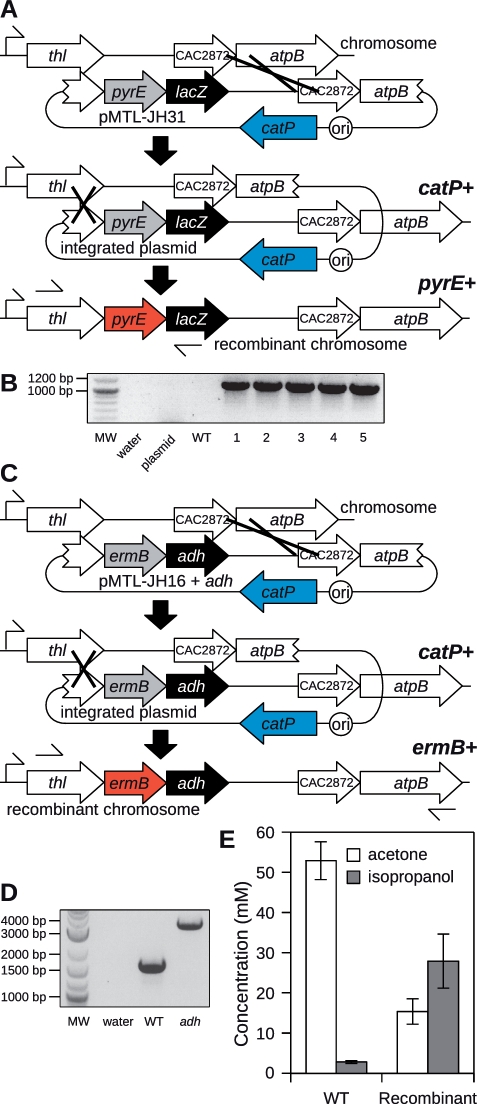
Figure 2.
DNA integration at the thl locus of C. acetobutylicum. (A) Selection of stable double-crossover clones using pMTL-JH31. The first recombination event (plasmid integration) is mediated by the long region of homology between pMTL-JH31 and CAC2872/atpB. Single-crossover clones are obtained on medium containing thiamphenicol. The second recombination event (plasmid excision) is mediated by the short region of homology between pMTL-JH31 and the 3′-end of thl. Double-crossover clones are selected using growth medium lacking uracil. (B) PCR screening of five candidate double-crossover clones using primers Cac-thl-sF1 and M13F which anneal where indicated in (A). MW, 2-Log DNA Ladder (NEB) molecular weight marker; plasmid, pMTL-JH31 plasmid DNA control; WT, wild-type C. acetobutylicum genomic DNA control; 1–5, candidate clones. All five candidates show the expected 1101 bp band. (C) Selection of stable double-crossover clones using pMTL-JH16 containing adh. The first recombination event (plasmid integration) is mediated by the long region of homology between pMTL-JH16 and CAC2872/atpB. Single-crossover clones are obtained on medium containing thiamphenicol. The second recombination event (plasmid excision) is mediated by the short region of homology between pMTL-JH16 and the 3′-end of thl. Double-crossover clones are selected using erythromycin. (D) PCR screening of one candidate adh-expressing double-crossover clone using primers Cac-thl-sF1 and Cac-atpB-sR1 which anneal where indicated in (C). MW, 2-Log DNA Ladder (NEB) molecular weight marker; WT, wild-type C. acetobutylicum genomic DNA control which shows the expected 1660 bp band; adh, candidate clone which shows the expected 3523 bp band. (E) Concentrations of acetone and isopropanol in the supernatant of cultures of wild-type C. acetobutylicum (WT) or the adh-expressing recombinant clone after 72 h growth. The concentrations of both fermentation products differ significantly between the two strains (P < 0.01).
It would be useful to apply the strategy above to Clostridium spp. other than C. acetobutylicum. We constructed plasmids equivalent to pMTL-JH12 and pMTL-JH14 for C. sporogenes NCIMB 10696, which shows potential in novel tumor therapies (20); and strain 630Δerm of the important human pathogen C. difficile (21) (Table 1). Double-crossover clones of both these organisms could be selected using media supplemented with FOA, as described in the Supplementary Data and shown in Supplementary Figure S2. Integration at the pyrF locus of C. acetobutylicum, including insertions of fragments of phage lambda DNA of various sizes, is also described in the Supplementary Data and shown in Supplementary Figure S1.
Coupling expression of heterologous selectable markers to a chromosomal promoter
Using plasmid pMTL-JH14 in the specially configured pyrE mutant constructed using pMTL-JH12, we had effectively fused together two partial pyrE genes to result in one complete, functional pyrE gene via recombination in the middle of the coding sequence. We realized that a recombination event bringing together two partial genes to form one whole gene need not occur in the coding sequence. In an alternative arrangement, the full-length coding sequence of a selectable marker, lacking only a promoter, could be linked by recombination to a promoter situated on the chromosome, completing the gene and leading to its expression. We designed plasmid pMTL-JH31 (Figure 2A) to implement this modified concept by integrating a pyrE gene lacking its own promoter into the chromosome of the previously constructed pyrE mutant of C. acetobutylicum. The insertion was targeted to a site downstream of the thiolase (thl) promoter, which is known to exhibit strong expression throughout growth (22,23). To prevent possible homologous recombination at the pyrE locus, pMTL-JH31 uses the orthologous pyrE gene from C. sporogenes ATCC 15579, which is only 47.6% identical to the C. acetobutylicum gene. As before, a long (1200 bp) region of homology directs plasmid integration without altering the pyrE phenotype, and single-crossover clones are obtained on medium containing thiamphenicol. Subsequently, plasmid excision mediated by a second region of homology places the silent (non-expressed) pyrE gene immediately downstream of the thl gene on the chromosome, leading to its co-expression with thl, and allowing these double-crossover clones to be selected on medium lacking uracil (Figure 2A). To test this scheme, we transformed the pyrE mutant of C. acetobutylicum with pMTL-JH31 DNA, and performed the integration procedure as described above for pMTL-JH14. Five independent clones were screened by PCR (Figure 2B) and thiamphenicol sensitivity, and all were verified, confirming the utility of this modified approach.
The use of recombination to link a chromosomal promoter to a selectable marker within an allele exchange cassette represents a very flexible strategy, not limited to restoration of an auxotrophic mutant to prototrophy. We constructed pMTL-JH16, which is identical to pMTL-JH31, except the ribosome-binding site and coding sequence of pyrE are replaced by those of the macrolide–lincosamide–streptogramin (MLS) antibiotic-resistance marker ermB (24). As before a long (1200 bp) region of homology directs plasmid integration. Next, the recombination event mediating plasmid excision positions ermB immediately downstream of thl, leading to ermB expression and an MLS-resistant phenotype which can be used to specifically select double-crossover clones (Figure 2C). We tested pMTL-JH16 in wild-type C. acetobutylicum using a similar integration procedure as before, culturing transformants initially on thiamphenicol, and were readily able to specifically select double-crossover clones using the macrolide antibiotic erythromycin. Six recombinant clones were screened in the same way as before, and all were confirmed to be the desired recombinants (data not shown).
Integration and expression of an alcohol dehydrogenase
In the previous strategy, the expression of heterologous marker genes was coupled to the formation of a recombinant double-crossover chromosome, providing a means to select these clones. Next we sought to extend this principle to the expression of other heterologous genes of interest, for other purposes. Besides butanol, the other major products of sugar fermentation by C. acetobutylicum are acetone and ethanol, and the process is often called the acetone–butanol–ethanol (ABE) fermentation (25). Butanol is the most valuable product, but ethanol also has value both as a commodity chemical and as a biofuel. Acetone is less valuable and is not suitable as a fuel, so it represents an undesirable by-product or waste product of the industrial ABE process. Clostridium beijerinckii NRRL B593 has a similar physiology to C. acetobutylicum, but reduces the acetone it produces to isopropanol (propan-2-ol) using an NADPH-dependent primary/secondary alcohol dehydrogenase (26). Isopropanol is more valuable than acetone, and could be used as biofuel in a blend with butanol and ethanol, so it would be useful to add the adh gene which encodes this activity to C. acetobutylicum.
We inserted the coding sequence of adh into pMTL-JH16 along with a ribosome-binding site, but did not provide a promoter (Figure 2C). Double-crossover recombinants will therefore contain an artificial thl-ermB-adh operon, with all three genes dependent upon the thl promoter for expression. The integration procedure was performed as before, and double-crossover clones were selected using erythromycin. We picked only one clone of the many obtained to screen for integration of adh, because the experiments described in the previous sections suggested that screening multiple clones was unnecessary. The adh-containing clone was verified as before by PCR (Figure 2D) and thiamphenicol sensitivity. We compared the fermentation products of the adh-containing clone to the wild-type using anaerobic batch cultivation conditions under which C. acetobutylicum exhibits classic bi-phasic growth, switching from organic acid production to acid re-uptake and solvent production as it enters stationary phase. After 72 h growth, supernatants of wild-type cultures contained typical concentrations of fermentation products, including 91.9 ± 8.4 mM butanol, 52.9 ± 4.7 mM acetone and 12.6 ± 1.5 mM ethanol. At the same timepoint, supernatants from cultures of the adh clone contained 15.4 ± 3.1 mM acetone and 27.9 ± 6.7 mM isopropanol (Figure 2E). None of the other fermentation product concentrations differed significantly from the wild-type. This result demonstrates expression of functional adh, and the recombinant strain represents an improvement over the wild-type from an industrial perspective.
Multistep integration of the phage lambda genome
Up to 1.8 kb of heterologous DNA was integrated in the experiments described above, and larger fragments of up to 6.5 kb were integrated in additional experiments described in the Supplementary Data. However, there must be limits to the size of DNA that can be delivered in a single step. For C. acetobutylicum, this limitation may well be imposed by the frequency of transformation, which is low even for small plasmids (7). Sequences too large to be integrated in a single step could be delivered in a series of steps using overlapping subfragments of the desired sequence, as in the domino method for Bacillus subtilis (3). This might be achieved either by alternately switching on and off a positively and negatively selectable gene such as pyrE (Figure 1) or by alternately linking expression of two different heterologous selectable markers to a chromosomal promoter (Figure 2). In either scheme, the short region of homology would remain the same in every step, whereas the long region of homology would target the end of the previous insert in the second and subsequent steps.
To test multistep DNA delivery using ACE, we attempted to insert the entire genome of phage lambda into the chromosome of C. acetobutylicum. We identified three restriction fragments of circular phage lambda DNA (circularized by ligation of the cohesive ends of the linear chromosome) that covered the entire genome and provided regions of overlap suitable to direct the initial recombination events in the second and third ACE steps (Figure 3). We used restriction ligation cloning to insert the 28 kb fragment (L28), 18 kb fragment (L18) and 6.5 kb fragment (L6.5) into ACE vectors pMTL-JH16, pMTL-JH30 and pMTL-JH15, respectively. Vectors pMTL-JH30 and pMTL-JH15 are similar to pMTL-JH31 and pMTL-JH16, but do not include a long region of homology, so these vectors are appropriate when the long region of homology depends upon a previous step, and must therefore be provided with the insert or as part of the insert (Table 1). When we inserted L18 into pMTL-JH30, we obtained clones with deletions in the lambda sequence. It appears that cloning this region (which includes lysis genes) into pMTL-JH30 is toxic to E. coli, so clones with spontaneous deletions are selected. In one of these constructs, 6 kb of DNA is deleted, but the regions required for recombination between fragments are not affected. We used this 12 kb of lambda DNA (L12) instead of L18, and proceeded to the multistep integration.
We integrated each of the three lambda DNA fragments in turn into the chromosome of the pyrE mutant of C. acetobutylicum (Figure 3). Double-crossover clones were selected using erythromycin (ermB+) for the L28 insertion, then medium lacking uracil (pyrE+) for the L12 insertion, then erythromycin again for the L6.5 insertion. At each step, the insertion was initially verified by thiamphenicol sensitivity and PCR. After all three insertions were completed, a Southern blot of EcoRI-digested genomic DNA from the three recombinant strains (containing one, two or three lambda DNA insertions) was performed, using lambda DNA as the probe (Figure 3D). EcoRI-digested genomic DNA from the parental pyrE mutant strain was also included, as was EcoRI-digested plasmid DNA of the three integration plasmids. All samples showed the expected distinguishing pattern of fragments (none in the case of the pyrE strain control) confirming the successful multistep insertion. The insertions were also verified by sequencing.
DISCUSSION
In this work, we addressed the need to reliably and stably deliver DNA to organisms lacking mature genetic tools, which include bacterial species of applied importance. The specialized allele exchange approaches we have demonstrated share two common principles: (i) cells in which a particular homologous recombination event has occurred can be selected if the event interrupts a counter-selectable gene, or appropriately fuses together two parts of a positively-selectable gene; and (ii) regions of homology of very different lengths strongly bias the order of homologous recombination events, and can therefore be exploited to couple acquisition of a selectable phenotype to the second recombination event in a lineage—the formation of a double-crossover clone. The combination of these principles constitutes a novel, generally applicable strategy which we call allele-coupled exchange (ACE). The variant demonstrated using pMTL-JH15, 16, 30 and 31 is particularly interesting and broadly applicable, because providing the entire coding sequence of a heterologous marker on the plasmid means that any genomic locus with a promoter can be a target for integration. Furthermore, using a chromosomal promoter to direct expression of a heterologous gene of interest allows the gene to be cloned without a promoter, precluding toxicity which might otherwise result from over-expression in the cloning host. This variant also allows specific selection of double-crossover clones using only positively selectable markers, like antibiotic-resistance genes, which are more easily applied to diverse organisms than counter-selection markers.
Perhaps the most exciting implication of this work is the potential to iteratively introduce large insertions (beyond the size which could be delivered in a single step) in a similar way to procedures available for organisms which can be transformed with linear DNA (3). We have constructed all the plasmids necessary to undertake such multi-step ACE strategies at each of the loci studied here (Table 1). In our proof-of-principle experiment over 40 kb of lambda DNA was inserted into the chromosome of C. acetobutylicum, which is the largest arbitrary sequence integrated in any Clostridium by an order of magnitude (27–29). After each successful transformation of the lambda constructs, double-crossover clones were obtained just as easily as in the earlier experiments using the empty vectors. However, for both pMTL-JH16::L28 and pMTL-JH30::L12, several electroporation attempts were required before transformant colonies were observed. Apparently, inserts of this size substantially decrease transformation frequency to a level that reduces the practicality of the procedure. This observation suggests that the size of inserts used in multistep strategies, at least in C. acetobutylicum, should strike a balance between the number of steps and the ease of each step. Inserts L28 and L12 appear to be larger than the optimum.
ACCESSION NUMBERS
HQ875748, HQ875749, HQ875750, HQ875751, HQ875752, HQ875753, HQ875754, HQ875755, HQ875766, HQ875757, HQ875758, HQ875759, HQ875760, HQ875761, HQ875762, HQ875763, HQ875764, HQ875765, HQ875766, HQ875767.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary results, Supplementary Table 1, Supplementary Figures 1 and 2, and Supplementary References [30, 31].
FUNDING
Biotechnology and Biological Sciences Research Council (BB/F003390/1, BB/G016224/1, BB/E021271/1); Medical Research Council (G0601176); European Union (HEALTH-F3-2008-223585); TMO Renewables Ltd. Funding for open access charge: The University of Nottingham.
Conflict of interest statement. The University of Nottingham has filed a patent application encompassing some of the work described in this article. The patent application names J.TH. and N.P.M. as inventors.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Alexandra Faulds-Pain and Sarah Kuehne for valuable criticism and suggestions during preparation of the manuscript.
REFERENCES
- 1.Collins JJ, Endy D, Hutchison CA, Roberts RJ. Editorial-synthetic biology. Nucleic Acids Res. 2010;38:2513. doi: 10.1093/nar/gkq221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Itaya M, Tanaka T. Gene-directed mutagenesis on the chromosome of Bacillus subtilis 168. Mol. Gen. Genet. 1990;223:268–272. doi: 10.1007/BF00265063. [DOI] [PubMed] [Google Scholar]
- 3.Itaya M, Fujita K, Kuroki A, Tsuge K. Bottom-up genome assembly using the Bacillus subtilis genome vector. Nat. Methods. 2008;5:41–43. doi: 10.1038/nmeth1143. [DOI] [PubMed] [Google Scholar]
- 4.Reyrat JM, Pelicic V, Gicquel B, Rappuoli R. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect. Immun. 1998;66:4011–4017. doi: 10.1128/iai.66.9.4011-4017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purdy D, O'Keeffe TAT, Elmore M, Herbert M, McLeod A, Bokori-Brown M, Ostrowski A, Minton NP. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 2002;46:439–452. doi: 10.1046/j.1365-2958.2002.03134.x. [DOI] [PubMed] [Google Scholar]
- 6.Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP. The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods. 2007;70:452–464. doi: 10.1016/j.mimet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 7.Heap JT, Pennington OJ, Cartman ST, Minton NP. A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods. 2009;78:79–85. doi: 10.1016/j.mimet.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Hartmanis MGN, Gatenbeck S. Intermediary metabolism in Clostridium acetobutylicum: levels of enzymes involved in the formation of acetate and butyrate. Appl. Environ. Microbiol. 1984;47:1277–1283. doi: 10.1128/aem.47.6.1277-1283.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Brien RW, Morris JG. Oxygen and the growth and metabolism of Clostridium acetobutylicum. J. Gen. Microbiol. 1971;68:307–318. doi: 10.1099/00221287-68-3-307. [DOI] [PubMed] [Google Scholar]
- 10.Papoutsakis ET. Engineering solventogenic clostridia. Curr. Opin. Biotechnol. 2008;19:420–429. doi: 10.1016/j.copbio.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 12.Husson RN, James BE, Young RA. Gene replacement and expression of foreign DNA in mycobacteria. J. Bacteriol. 1990;172:519–524. doi: 10.1128/jb.172.2.519-524.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sander P, Meier A, Böttger EC. rpsL+: a dominant selectable marker for gene replacement in mycobacteria. Mol. Microbiol. 1995;16:991–1000. doi: 10.1111/j.1365-2958.1995.tb02324.x. [DOI] [PubMed] [Google Scholar]
- 14.Knipfer N, Seth A, Shrader TE. Unmarked gene integration into the chromosome of Mycobacterium smegmatis via precise replacement of the pyrF gene. Plasmid. 1997;37:129–140. doi: 10.1006/plas.1997.1286. [DOI] [PubMed] [Google Scholar]
- 15.Sato T, Fukui T, Atomi H, Imanaka T. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 2003;185:210–220. doi: 10.1128/JB.185.1.210-220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bitan-Banin G, Ortenberg R, Mevarech M. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 2003;185:772–778. doi: 10.1128/JB.185.3.772-778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng L, Zhu H, Chen Z, Liang YX, She Q. Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles. 2009;13:735–746. doi: 10.1007/s00792-009-0254-2. [DOI] [PubMed] [Google Scholar]
- 18.Tripathi SA, Olson DG, Argyros DA, Miller BB, Barrett TF, Murphy DM, McCool JD, Warner AK, Rajgarhia VB, Lynd LR, et al. Development of pyrF-based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant. Appl. Environ. Microbiol. 2010;76:6591–6599. doi: 10.1128/AEM.01484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monod M, Denoya C, Dubnau D. Sequence and properties of pIM13, a macrolide-lincosamide-streptogramin B resistance plasmid from Bacillus subtilis. J. Bacteriol. 1986;167:138–147. doi: 10.1128/jb.167.1.138-147.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theys J, Pennington O, Dubois L, Anlezark G, Vaughan T, Mengesha A, Landuyt W, Anné J, Burke PJ, Dürre P, et al. Repeated cycles of Clostridium difficile-directed enzyme prodrug therapy result in sustained antitumour effects in vivo. Br. J. Cancer. 2006;95:1212–1219. doi: 10.1038/sj.bjc.6603367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hussain HA, Roberts AP, Mullany P. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Deltaerm) and demonstration that the conjugative transposon Tn916DeltaE enters the genome of this strain at multiple sites. J. Med. Microbiol. 2005;54:137–141. doi: 10.1099/jmm.0.45790-0. [DOI] [PubMed] [Google Scholar]
- 22.Tummala SB, Welker NE, Papoutsakis ET. Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 1999;65:3793–3799. doi: 10.1128/aem.65.9.3793-3799.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris LM, Welker NE, Papoutsakis ET. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 2002;184:3586–3597. doi: 10.1128/JB.184.13.3586-3597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clewell DB, Yagi Y, Dunny GM, Schultz SK. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J. Bacteriol. 1974;117:283–289. doi: 10.1128/jb.117.1.283-289.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones DT, Woods DR. Acetone-butanol fermentation revisited. Microbiol. Rev. 1986;50:484–524. doi: 10.1128/mr.50.4.484-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ismaiel AA, Zhu CX, Colby GD, Chen JS. Purification and characterization of a primary-secondary alcohol dehydrogenase from two strains of Clostridium beijerinckii. J. Bacteriol. 1993;175:5097–5105. doi: 10.1128/jb.175.16.5097-5105.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, Scott JC, Minton NP. The ClosTron: Mutagenesis in Clostridium refined and streamlined. J. Microbiol. Methods. 2010;80:49–55. doi: 10.1016/j.mimet.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Caruso L, McClane B, Fisher D, Gupta P. Disruption of a toxin gene by introduction of a foreign gene into the chromosome of Clostridium perfringens using targetron-induced mutagenesis. Plasmid. 2007;58:182–189. doi: 10.1016/j.plasmid.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cartman ST, Minton NP. A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl. Environ. Microbiol. 2010;76:1103–1109. doi: 10.1128/AEM.02525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brefort G, Magot M, Ionesco H, Sebald M. Characterization and transferability of Clostridium perfringens plasmids. Plasmid. 1977;1:52–66. doi: 10.1016/0147-619x(77)90008-7. [DOI] [PubMed] [Google Scholar]
- 31.Karlsson S, Burman LG, Akerlund T. Suppression of toxin production in Clostridium difficile VPI 10463 by amino acids. Microbiology. 1999;145:1683–1693. doi: 10.1099/13500872-145-7-1683. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.