Abstract
Recombinase-mediated cassette exchange (RMCE) is a powerful tool for unidirectional integration of DNA fragments of interest into a pre-determined genome locale. In this report, we examined how the efficiency of dual RMCE catalyzed by Flp and Cre depends on the nature of transcription units that express the recombinases. The following recombinase transcription units were analyzed: (i) Flp and Cre genes expressed as individual transcription units located on different vectors, (ii) Flp and Cre genes expressed as individual transcription units located on the same vector, (iii) Flp and Cre genes expressed from a single promoter and separated by internal ribosome entry sequence and (iv) Flp and Cre coding sequences separated by the 2A peptide and expressed as a single gene. We found that the highest level of dual RMCE (35–45% of the transfected cells) can be achieved when Flp and Cre recombinases are expressed as Flp–2A–Cre and Flp–IRES–Cre transcription units. In contrast, the lowest level of dual RMCE (∼1% of the transfected cells) is achieved when Flp and Cre are expressed as individual transcription units. The analysis shows that it is the relative Flp–to–Cre ratio that critically affects the efficiency of dual RMCE. Our results will be helpful for maximizing the efficiency of dual RMCE aimed to engineer and re-engineer genomes.
INTRODUCTION
Recombinase-mediated cassette exchange, RMCE, takes advantage of the ability of site-specific recombinases to replace a pre-determined genomic locus with a DNA fragment of interest. For RMCE to work, both the DNA fragment for replacement and the genome region to be replaced have to be flanked by recombination targets that should not recombine with each other (Figure 1A). Since RMCE is able to integrate a DNA fragment efficiently and unidirectionally, it is finding increasing usage in engineering and re-engineering of mammalian genomes (1–7). Two types of RMCE were successfully tested: with recombination targets of the same type that are sufficiently different to prevent intramolecular recombination (homotypic targets), and with recombination targets of different type (heterotypic targets). The former type of RMCE requires the use of a single recombinase, while the latter type, dual RMCE, needs two recombinases to occur (2,5). The recombinases most commonly used to perform RMCE are Flp recombinase from yeast Saccharomyces cerevisiae (1,6), Cre recombinase from coliphage P1 (8,9) and phiC31 integrase from Streptomyces phage phiC31 (3,10). Other recombinases, for example, R recombinase from yeast Zygosaccharomyces rouxii, were also shown to successfully perform RMCE, primarily in plants (11).
Figure 1.
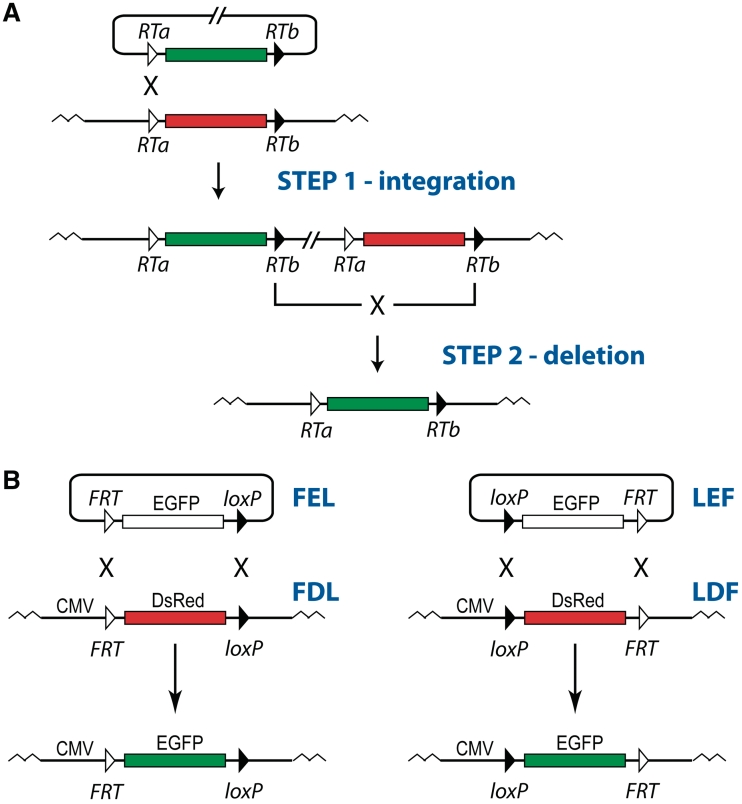
Schematics of dual RMCE (A) and FDL/FEL and LDF/LEF cassettes for testing the efficiency of dual RMCE (B). (A) In the example shown, the vector-borne EGFP-expressing cassette replaces the DsRed-expressing cassette integrated into a genome. At the first, integrative step of the reaction, the incoming vector integrates into one of the compatible recombination targets that flank the DNA fragments being swapped (the incompatible recombination targets, RTa and RTb, are depicted as open and filled triangles, respectively). At the second, deletion step of the reaction, the recombination between the other pair of compatible recombination targets excises the integrated vector and the original genomic DNA fragment leaving the vector-borne one in the genome. (B) FDL/FEL and LDF/LEF cassettes differ from each other by the nature of the recombination targets that flank the promoterless, vector-borne EGFP cassette and the DsRed-expressing cassette integrated into the genome of CHO cells: FRT–loxP or loxP–FRT, respectively. In both cassettes, dual RMCE catalyzed by Flp and Cre replaces the DsRed gene with the EGFP gene. As the result, the EGFP gene can be expressed from the genomic CMV promoter.
The mouse genome engineering programs, such as International Knockout Mouse Consortium, IKMC (12), EUCOMM (www.eucomm.org), German Gene Trap Consortium (www.genetrap.de), utilize targeting constructs that bear the recombination sites for Flp and Cre in a configuration that permits dual RMCE that can be used to modify, re-engineer the pre-integrated constructs. It was recently shown that dual RMCE is indeed applicable to re-engineering conditional mouse alleles generated by the IKMC (5). Since maximizing the efficiency of dual RMCE would benefit the field, in the present report we examined, in the model settings, how the efficiency of dual RMCE catalyzed by Flp and Cre is affected by the nature of the transcription units that express the recombinases. Two general groups of transcription units were analyzed. In one group, Flp and Cre were expressed under the control of the individual promoters with transcription units located either on the same vector or on different vectors. In the second group of the transcription units, Flp and Cre were expressed under the control of a single promoter: in one configuration, the recombinase genes were separated by the internal ribosome entry sequence, IRES, from the pIRES2-series vectors (Clontech); in the other configuration, the coding sequences of the Flp and Cre genes were separated by the self-cleaving 2A peptide TaV from the insect Thosea asigna virus (13).
The dual RMCE activity of Flp and Cre expressed from the respective transcription units was analyzed by assessing the efficiency of the replacement of a DsRed-expressing cassette, which was pre-integrated into the genome of CHO cells, with a promoterless EGFP cassette located on an incoming vector (Figure 1B). The cassettes were flanked by either the FRT/loxP pair or by the loxP/FRT pair. We found that the highest level of dual RMCE (∼45% of the transfected cells) can be achieved when Flp and Cre recombinases are expressed as one gene, in which their coding sequences are separated by the 2A peptide (Flp–2A–Cre) and the replacement cassettes are flanked by the loxP/FRT pair. If the replacement cassettes were flanked by the FRT/loxP pair, the Flp–2A–Cre transcription unit performed RMCE in ∼20% of the transfected cells. Flp–IRES–Cre transcription unit was able to perform the replacement reaction in ∼20–35% of the transfected cells, depending on the orientation of the recombination pair. Cre–2A–Flp and Cre–IRES–Flp transcription units showed intermediate dual RMCE activity (7–25% of the transfected cells). Under the same conditions, Flp and Cre recombinases expressed as individual transcription units, showed the lowest level of dual RMCE (∼0.5–1% of the transfected cells). We also found that the replacement activity of the latter group of the transcription units could be improved by either lowering the concentration of the expression vectors or by increasing the ratio of the Flp- to Cre-expressing vectors at transfection.
The results of our studies will be helpful for maximizing the efficiency of dual RMCE for genome engineering and re-engineering applications.
MATERIALS AND METHODS
Cell line and transfection
In the present study, we used Chinese Hamster ovary cells (CHO-K1; ATCC CCL-61) as model mammalian cells. CHO-K1 cells were propagated in F12–K media. Cell transfections were performed using Polyfect (Qiagen).
TD–In system
To create the TD–In system, we replaced FRT with a recombination site for TD recombinase, TDRT (14), in the original Flp–In vectors pFRT/LacZeo and pcDNA5/FRT (Invitrogen) to obtain pTDRT/LacZeo and pcTD, respectively. TDRT site used in the reporters had the following sequence (the putative recombinase binding elements are underlined): 5′-GTGCGTCAAATAA TAACGTA TTATTTGACACTT-3′. The pTDRT/LacZeo vector was integrated into the CHO genome by non-homologous recombination and the resultant cells were analyzed for the number of the copies of the integrated vector and the activity of LacZ. Several clones with singly integrated pTDRT/LacZeo and high relative activity of LacZ were expanded; of those, clone CHO–TD1 was used in all experiments described in the article. To integrate the pcTD-based reporters pFRT–DsRed–loxP and ploxP–DsRed–FRT (see next section), the CHO–TD1 cells were transfected with the respective reporter and the evolved variant of TD recombinase, TD1-40 (GenBank GU075693), that is adequately functional at 37°C. A detailed description of the evolution of TD1-40 and its activity will be published elsewhere.
Vectors
Reporter vectors
pFRT–DsRed–loxP (FDL) and ploxP–DsRed–FRT (LDF) were constructed by cloning the PCR-amplified DsRed-neo cassette from pIRES2-DsRed-Express (Clontech) into the pcTD vector (see previous section) between NheI and XhoI. The primers for amplifying the DsRed-neo cassette introduced either the FRT–loxP pair or the loxP–FRT pair, respectively, as flanking recombination targets. In the experiments described in this report, the ‘minimal’ 34-bp long FRT site was used. The minimal FRT site contains two 13-bp inverted Flp-binding elements separated by an 8-bp spacer.
pFRT–EGFP–loxP (FEL) and ploxP–EGFP–FRT (LEF) were constructed by cloning the PCR-amplified EGFP-neo cassette from pIRES2–EGFP (Clontech) into the pcTD vector between NheI and EcoRI, located in the HygroR gene. The primers for amplifying the EGFP-neo cassette introduced either the FRT–loxP pair or the loxP–FRT pair, respectively, as flanking recombination targets. The CMV promoter in the resulting plasmids was deleted by treating the plasmids with NheI and MluI, filling-in with Klenow and self-ligating.
Expression vectors
The pOG44 vector of the Flp–In system (Invitrogen) was used as a backbone for constructing the recombinase-expressing vectors. First, the gene for Flp(F70L) was deleted from pOG44 by replacing it with the NheI–BamHI linker. Then, different transcription units (Flp, Cre, Flp–2A–Cre, Cre–2A–Flp, Flp–IRES–Cre and Cre–IRES–Flp) were cloned into the resulting vector between NheI and BamHI.
All Flp-expressing vectors generated in this work utilize the Flpe variant of Flp recombinase (15). The TaV variant of the 2A self-cleaving peptides (13) was used in the Flp–2A–Cre and Cre–2A–Flp transcription units. IRES from pIRES2–EGFP (Clontech) was used in the Flp–IRES–Cre and Cre–IRES–Flp transcription units.
pDIRE (5), which expresses Flpo variant of Flp (16) and iCre variant of Cre (17), was acquired from Addgene (plasmid 26745).
Dual RMCE
Dual recombinase-mediated cassette exchange reactions were performed in 6-well plates. Under standard conditions, 0.5 µg of the reporter (FEL or LEF) and 2 µg of the expression plasmid were co-transfected into the respective cell lines (FDL or LDF) when the cells were ∼80% confluent. The efficiency of transfection was estimated separately by transfecting the cells with FEL or LEF, in which the CMV promoter was not deleted, so the number of the green cells could be counted. At 48-h post-transfection, the cells, co-transfected with the reporters and the expression vectors, were harvested and one-sixth of the cells was transferred to a 100-mm plate and expanded until confluent, after which stage the groups of green cells that formed during expansion were counted. In some experiments, the amount of the expression plasmid(s) at transfection was lowered; the specific amounts added are indicated when the respective experiments are described.
Other methods
Plasmid DNA was isolated using QIAprep Spin Miniprep Kit (Qiagen) or GeneJET Plasmid Miniprep Kit (Fermentas). Amplification of the DNA fragments used for cloning was performed using Pfu–Ultra polymerase (Agilent Technologies). PCR analysis of the mammalian genomic DNA was performed using Taq polymerase (New England Biolabs). Genomic DNA from cultured mammalian cells was isolated using DNeasy Blood and Tissue Kit (Qiagen). General genetic engineering experiments were performed as described in Sambrook and Russell (18).
RESULTS
RMCE vectors
To assess the efficiency of dual RMCE catalyzed by Flp and Cre, we created two sets of reporters (Figure 1B). Each reporter set has two vectors: one vector bears the DsRed cassette and is integrated into genome; the other vector bears the promoterless EGFP cassette that is used to replace the DsRed cassette. The reporter sets differ in the nature of the recombination sites pair that flanks the replacement cassettes (Figure 1B): FRT-replacement cassette-loxP (FDL and FEL) or loxP-replacement cassette-FRT (LDF and LEF), respectively.
For accurate comparison of the activity of different reporter sets in dual RMCE, we integrated the DsRed reporters from each set (FDL and LDF) into the same genomic location. For this, we created the TD–In system, which is analogous to the Flp–In system (Invitrogen) but relies on TD recombinase (14) and its recombination target TDRT to deliver the TDRT-bearing reporters into the TDRT site pre-integrated into the genome. We utilized the TD–In system to integrate TDRT site into the genome of CHO cells and then integrated the TDRT-bearing DsRed reporters FDL and LDF into this genomic TDRT site in the TD-dependent manner. The resultant reporter cell lines that carried either FRT–DsRed–loxP cassette or loxP–DsRed–FRT cassette were used to perform the dual RMCE experiments.
Transcription units for Flp and Cre expression
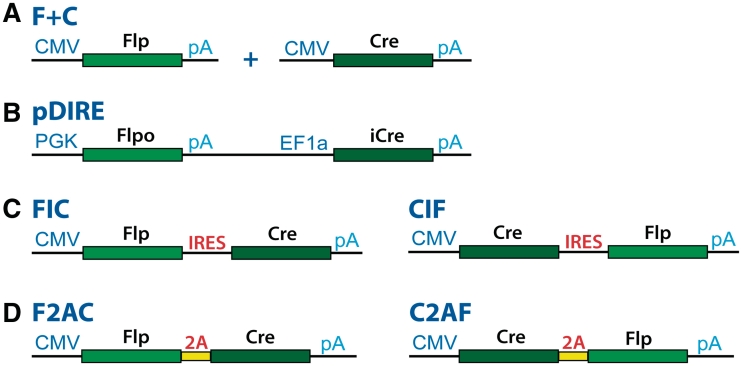
Four configurations of transcription units were used to express Flp and Cre to mediate dual RMCE (Figure 2). In the first two configurations, Flp and Cre were expressed under the control of the individual promoters and their transcription units were located either on different vectors (Figure 2A) or on the same vector (Figure 2B). As the latter vector, we used pDIRE described in Osterwalder et al. (5). In the third configuration of the transcription units, Flp and Cre were expressed under the control of a single promoter and their genes were separated by the internal ribosome entry sequence, IRES, from pIRES2–EGFP (Figure 2C). In the last configuration (Figure 2D), the coding sequences of the Flp and Cre genes were separated by the self-cleaving 2A peptide TaV from the insect T. asigna virus (13). For the latter two configurations, we generated two versions of the respective transcription units: Flp–IRES–Cre and Cre–IRES–Flp, and Flp–2A–Cre and Cre–2A–Flp.
Figure 2.
Transcription units that express Flp and Cre. (A) The Flp and Cre expressing cassettes in the (F + C) transcription unit are located on two different vectors; the recombinases are expressed from the CMV promoter. (B) in pDIRE (5), the Flpo and iCre genes are located on the same vector; the recombinases are expressed from the PGK and EF1a promoters, respectively. (C) In the transcription units FIC and CIF, the Flp and Cre genes are located on the same vector and separated by IRES from the pIRES2 series vectors. FIC and CIF are expressed from the CMV promoter. (D) In the transcription units F2AC and C2AF, the coding sequences of Flp and Cre are separated by the 2A peptide TaV (13). F2AC and C2AF are expressed from the CMV promoter.
Flp–2A–Cre and Flp–IRES–Cre are superior in performing dual RMCE
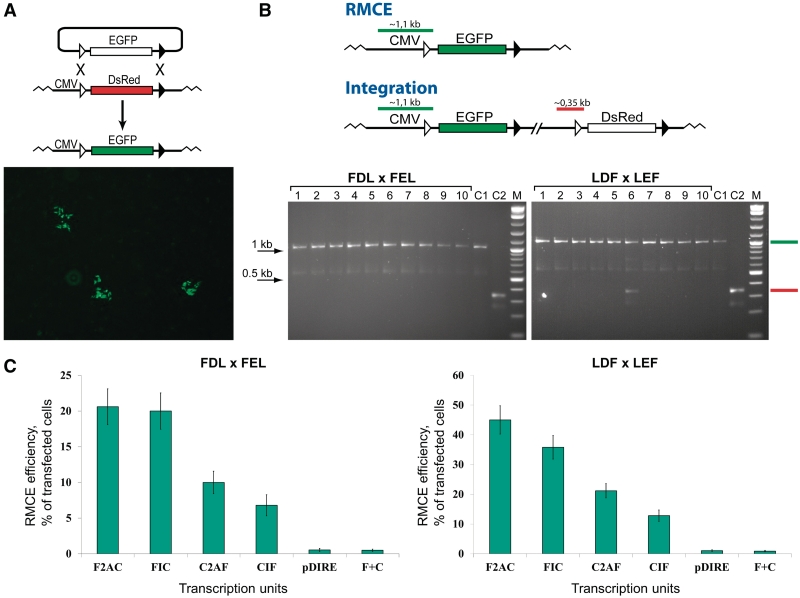
To assess the dual RMCE activity of Flp and Cre expressed from different transcription units, the reporter cell lines that carried the FDL or LDF cassettes (Figure 1B) were co-transfected with the vectors that carried the FEL or LEF cassettes, respectively, and one of the vectors that express Flp and Cre (Figure 2). At 48-h post-transfection, the cells were expanded and groups of green cells that formed during expansion counted (Figure 3A). We considered such groups of green cells, not solitary green cells, as the cells, in which a productive recombination event occurred at the target located upstream of the EGFP gene (Figure 1B). Such recombination event can indicate either simple integration of the EGFP-bearing replacement vector or dual RMCE that replaces the genomic DsRed cassette with the EGFP cassette. To distinguish between these two possibilities, we expanded several groups of green cells formed in the Flp–2A–Cre catalyzed recombination reactions (in both FDL × FEL and LDF × LEF orientations) and subjected them to the PCR analysis. As the analysis shows (Figure 3B), the majority of the green colonies are indeed the result of dual RMCE that replaced the genomic DsRed cassettes with the EGFP cassettes. Similar results were obtained in the recombination reaction catalyzed by Flp and Cre expressed from the Flp–IRES–Cre transcription unit (data not shown).
Figure 3.
Testing dual RMCE efficiency under standard conditions. (A) Typical green colonies formed as a result of LDF × LEF recombination catalyzed by F2AC after the transfected cells were expanded. (B) The PCR analysis confirms that the majority of the green colonies are indeed the result of RMCE and not just simple integration. The green and red bars schematically represent the control PCR products for the replacement and the integration reactions respectively. The control primers anneal upstream of the CMV promoter and downstream of a recombination target; the primers are not specific for the promoter or the reporter genes. The sequencing of the lower, integration-specific band in the variant #6 (LDF × LEF configuration) confirmed the nature of the band. C1, PCR product obtained by subjecting the genomic DNA of the original FDL or LDF cell line, respectively, to the PCR reaction using the control primers. C2, PCR product obtained by subjecting the FEL or LEF reporter, respectively, to the PCR reaction using the control primers. M, 2-log DNA ladder (New England Biolabs). (C) The efficiency of dual RMCE catalyzed by the transcription units F2AC, C2AF, FIC, CIF, pDIRE and (F + C) in the FDL × FEL and LDF × LEF reporter systems is represented by green bars. The concentration of the reporter and the expression plasmids at transfection was 0.4 and 2 µg, respectively. The green bars show the mean value of five experiments; the error bars indicate SD.
Having confirmed that the majority of the green colonies formed during cell expansion are indeed the result of dual RMCE, we performed a comparative analysis of the dual RMCE activity of all transcription units. As shown in Figure 3C, the efficiency of dual RMCE was dependent both on the transcription unit used to express Flp and Cre and on the relative orientation of FRT and loxP. In both orientations of the recombination targets—FDL/FEL and LDF/LEF—F2AC and FIC transcription units showed the highest activity in dual RMCE. For the FDL/FEL configuration, the efficiency of dual RMCE for both transcription units was about the same: ∼20% of the transfected cells. For the LDF/LEF orientation, the efficiency of dual RMCE for FIC was ∼35% of the transfected cells, while for F2AC—∼45% of the transfected cells.
Under the same experimental conditions, the lowest efficiency of dual RMCE (0.5–1% of the transfected cells) was observed when Flp and Cre genes were expressed from the individual promoters located either on the same plasmid, as in pDIRE, or on two plasmids (F + C). The recombination activity of these transcription units on the LDF/LEF reporter system was also higher than on the FDL/FEL reporter system.
C2AF and CIF transcription units showed intermediate activity in dual RMCE: 7–25% of the transfected cells; C2AF was more active than CIF (Figure 3C). As with the other transcription units, dual RMCE by C2AF and CIF was more efficient in the LDF/LEF reporter system.
Efficiency of dual RMCE by pDIRE can be improved by reducing its concentration at transfection
The codons in Flp and Cre genes in the transcription units F2AC, FIC, C2AF, CIF and (F + C) are essentially wild-type and are not optimized for the expression in mammalian cells. In contrast, pDIRE (5) expresses Flpo and iCre versions of Flp and Cre genes, codons of which are optimized to maximize the expression of the recombinases in mammalian cells (16,17). Moreover, Flpo and iCre in pDIRE are fused to the nuclear localization sequence, which improves the ability of the recombinases to cross the nuclear membrane and thus increases their concentration in the nucleus. Flpo and iCre were shown to be more efficient than Flp and Cre in mammalian cells in the deletion assays. However, when compared to F2AC, FIC, C2AF and CIF using similar experimental conditions, pDIRE did not show superior results in dual RMCE. To explain these results, we hypothesized that it is the high supply of Flp and Cre by pDIRE that negatively affects dual RMCE. If our hypothesis were correct than lowering the concentration of pDIRE in the transfection mixture should improve the efficiency of dual RMCE.
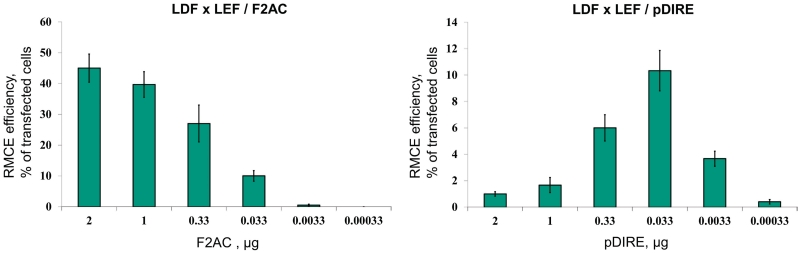
To test our hypothesis, we performed a series of dual RMCE experiments with decreasing concentration of pDIRE and F2AC (as a control) using the FDL/FEL and LEF/LDF reporter systems. We found that the efficiency of dual RMCE by pDIRE could be increased by ∼10-fold if the concentration of the vector at transfection was lowered 60-fold (2–0.033 µg). In contrast, under the same conditions, the efficiency of dual RMCE by F2AC rapidly decreased. The results in the LEF/LDF reporter system are shown in Figure 4; the results using the FDL/FEL reporter system showed similar tendencies.
Figure 4.
Lowering the concentration of F2AC and pDIRE at transfection differently affects the efficiency of dual RMCE. The results of the LDF × LEF recombination are shown. Lower amounts of F2AC added at transfection lead to the gradual drop in the replacement efficiency. In contrast, the highest efficiency of dual RMCE by pDIRE was achieved by decreasing the concentration of the plasmid at transfection. The concentration of the reporter plasmid at transfection was kept at 0.4 µg; the concentrations of F2AC and pDIRE are indicated. The green bars show the mean value of three experiments; the error bars indicate SD.
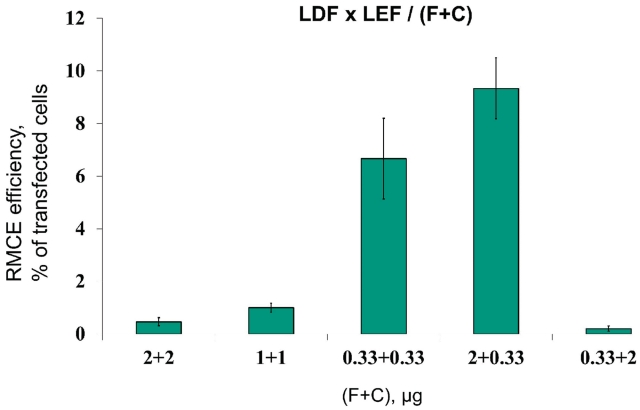
Efficiency of dual RMCE by (F + C) depends on Flp to Cre ratio
Finally, we tested whether the efficiency of dual RMCE can be affected by the Flp-to-Cre ratio. For this, we performed the replacement reaction with varying concentrations of the Flp and Cre expression vectors added at transfection while keeping the concentration of the reporter plasmids constant. The results using the LEF/LDF reporter system are shown in Figure 5; the FDL/FEL system showed similar tendencies. We found that the lower ratio of the Flp- to Cre-expressing vectors (0.33 µg:2 µg) generates fewer dual RMCE-positive colonies than under standard experimental conditions when the concentration of both Flp and Cre expression vectors are kept at 2 µg. In contrast, the higher ratio of the Flp- to Cre-expressing vectors (2 µg:0.33 µg) leads to the sharp increase in the dual RMCE efficiency. We also noted that the efficiency of dual RMCE can be increased by lowering the concentration of both expression vectors while keeping their ratio 1:1. These results suggest that both higher Flp-to-Cre ratio and lower concentration of Cre can increase the efficiency of dual RMCE.
Figure 5.
The efficiency of dual RMCE catalyzed by (F + C) is affected by the overall concentration of the recombinase expression vectors and by the relative ratios of the vectors. The results of the LDF × LEF recombination are shown. The concentration of the reporter plasmid at transfection was kept at 0.4 µg; the concentrations of (F + C) are indicated. The green bars show the mean value of three experiments; the error bars indicate SD.
DISCUSSION
RMCE is a double-reciprocal recombination reaction able to replace a genomic DNA fragment flanked with a pair of incompatible recombination targets with a vector-borne DNA fragment flanked by the same pair of recombination targets (1–3,5,8). The reaction is thought to proceed in two steps (Figure 1A). At the first step, a crossover between a pair of compatible recombination targets integrates the entire vector into the respective genomic recombination site. At the second step, a crossover between the other pair of compatible recombination targets excises the vector and the genomic DNA fragment leaving the vector-borne replacement fragment in the genome.
At dual RMCE, two recombinases may or may not be equally effective at the integration and the excision steps of the reaction. If one recombinase is more efficient than the other at the integration step then this recombinase is more likely to start the RMCE. If the other recombinase is more efficient at the excision step, then this recombinase is more likely to finish the reaction. In general, for simple tyrosine recombinases, the more efficient recombinase is likely to perform better in the excision than in the integration reaction since these two reactions are reversible and integration (bimolecular reaction) is more difficult to perform than excision (monomolecular reaction). In the case of Flp and Cre, the former recombinase is less efficient than the latter in the deletion reaction (16,19,20). In our hands, in the model settings of the CHO-TD1 cell line, Flp integrates a reporter plasmid into FRT pre-integrated into the genome with ∼0.1% efficiency when 2 µg of the Flp expressing plasmid is used in the experiments (data not shown). Under similar conditions, with loxP integrated into the same genomic location, Cre-mediated integrative recombination is barely detectable. However, when the concentrations of the recombinase expression vectors at transfection is decreased 10- to 20-fold, the efficiency of Cre-mediated integrative recombination increases to ∼0.1% of the transfected cells, while the efficiency of Flp-mediated integrative recombination drops to 0.01–0.001% of the transfected cells (data not shown). Taken together, the difference in the ability of Flp and Cre to perform integrative and excisive recombination suggests that in dual RMCE Flp primarily mediates the first, integrative, step of the reaction when the concentration of Flp is high, while Cre mediates this step when its concentration is relatively low. When the Flp-to-Cre ratio during dual RMCE is high, then both recombinases are able to mediate the integrative step of the reaction.
The above reasoning helps explain the results of the present work in which we analyzed whether the efficiency of dual RMCE can be affected by the type of transcription unit that expresses Flp and Cre. The transcription units we tested were (Figure 2): (i) Flp and Cre genes expressed from individual promoters and located either on the same plasmid [pDIRE (5)] or on different plasmids (F + C), (ii) Flp and Cre genes separated by IRES and expressed as a bicistronic unit (FIC and CIF) and (iii) Flp and Cre expressed as a single gene with their coding sequences separated by the self-cleaving 2A peptide (F2AC and C2AF). Under the standard experimental conditions used, the first group of the transcription units (pDIRE and F + C) was the least efficient in generating dual RMCE-positive colonies: ∼1% of the transfected cells (Figure 3C). Under the same conditions, the members of the second and the third groups of the transcription units were much more capable: dual RMCE-positive colonies were seen in ∼7% to ∼45% of the transfected cells (Figure 3C). Within these two groups of the transcription units, F2AC and FIC were more effective in generating dual RMCE-positive colonies than C2AF and CIF.
F2AC and FIC share two common features that help explain the observed similarity in their functional performance. First, both transcription units have a Flp gene as the first gene of the unit and a Cre gene as the second one. Second, in both transcription units the first gene of the unit is translated at a higher level than the second gene. Indeed, the analysis of the model transcription units separated by the self-cleaving peptide 2A from the insect virus TaV (this version of the 2A peptide was used in our experiments) showed that the first gene is translated about four times more efficiently than the second gene (13). Similarly, IRES that we used in the FIC and CIF constructs, is partially disabled which reduces the efficiency of the translation initiation of the second gene relative to the first gene in the bicistronic transcription unit (21). Since both C2AF and CIF were not as effective in mediating dual RMCE as F2AC and FIC, the results shown by the 2A- and IRES-based transcription units suggest that efficient dual RMCE requires higher relative expression levels of Flp than Cre.
This conclusion is also supported by the results of the analysis of the efficiency of dual RMCE using different relative ratios of Flp and Cre expression vectors, (F + C), added at transfection. As expected, based on the dependency of Flp- and Cre-mediated integration on their concentration and on the results with the 2A- and IRES-based transcription units, the efficiency of dual RMCE increased with increased Flp to Cre ratio (Figure 5). We also observed an increase in dual RMCE (albeit to a lesser extent) when the ratio of Flp to Cre expression vectors was kept 1:1 but their concentration lowered, which supports the idea that it is high concentration of Cre that is not optimal for dual RMCE.
The inability of pDIRE to be as efficient as F2AC and FIC in mediating dual RMCE was surprising at first since it was thought that high concentration of Flp and Cre expressed from pDIRE is required for reaching the highest level of dual RMCE. Based on the results discussed earlier, we reasoned that most likely it is the high expression level of Cre (and possibly Flp) that causes the problem at least because the first step of RMCE—integration—can be easily reversed in this case. Therefore, we decided to test the dual RMCE activity of pDIRE at decreasing concentrations of the plasmid at transfection. We indeed observed an increase in the dual RMCE efficiency—∼10-fold—when the concentration of pDIRE at transfection was decreased 60-fold from the level that supports the maximum dual RMCE activity of F2AC and FIC (Figures 3 and 4). However, even under these optimized conditions, pDIRE did not mediate as high level of dual RMCE as did F2AC and FIC.
We observed that the LDF/LEF reporter configuration generates more dual RMCE-positive colonies than the FDL/FEL configuration with all transcription units tested. The reasons for the phenomenon remain unclear. Since the FDL and LDF reporter vectors were integrated into the same genomic location, the position effects cannot play a role in the observed difference. On the other hand, the local environment of FRT and loxP in the LDF and FDL reporters may play a role: in these reporters the recombination targets are located either next to the CMV promoter or several kilobases away from it (Figure 1B). A potential effect of a promoter on the efficiency of site-specific recombination is not systematically studied but there is a report that suggests that the efficiency of Cre integrative recombination can be affected by a promoter located near loxP (22): a construct with stronger CMV promoter generated more integrants than constructs with weaker HSV-1 tk promoter. In our hands, in bacterial systems, the distance between FRT and the lac promoter influences the efficiency of Flp-mediated recombination: shorter distances generally lead to higher levels of recombination. Taking into account these observations, we speculate that the higher dual RMCE activity in the LDF/LEF reporter system is a result of the positive influence of the CMV promoter on the recombination and this influence is more pronounced in the Cre/loxP system than in the Flp/FRT system.
Although dual RMCE is thought to proceed in two sequential steps that are separately mediated by the respective recombinase (Figure 1A), the fact that both Flp and Cre can be present simultaneously in a cell while mediating the efficient replacement reaction, argues that the individual DNA rearranging reactions mediated by the recombinases do not interfere with each other, at least under the optimized conditions. The situation may be different for other dual RMCE systems, for example for the replacement reaction mediated by Flp and HK022 Int: when expressed simultaneously in a bacterial system, these recombinases are unable to mediate detectable RMCE (23). However, the reaction is efficient when the recombinases are expressed sequentially. Such behavior of the replacement system suggests that the individual recombination reactions mediated by Flp and HK022 Int interfere with each other, probably because of the spatial and temporal features of the synaptic complexes they form. It would be interesting to see if, in mammalian cells, the Flp/Int dual RMCE system can follow the example of the Flp/Cre system and respond positively to a simple modulation of the concentrations of the recombinase expressing vectors added simultaneously. If not, then individual approaches to optimize the replacement reaction might have to be devised for different dual RMCE systems, taking into account the complexity of the reaction complexes formed by the recombinases.
FUNDING
This work was supported by National Institutes of Health [R01GM085848 to Y.V.]; Alexander von Humboldt Foundation. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors acknowledge Vrushalee Palsule for creating the Flp-2A-Cre and Cre-2A-Flp constructs. The authors are grateful to Francis Stewart, Konstantinos Anastassiadis and Heike Petzold for helping to create CHO-TD cell lines. The authors are also grateful to Makkuni Jayaram for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Schlake T, Bode J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry. 1994;33:12746–12751. doi: 10.1021/bi00209a003. [DOI] [PubMed] [Google Scholar]
- 2.Lauth M, Spreafico F, Dethleffsen K, Meyer M. Stable and efficient cassette exchange under non-selectable conditions by combined use of two site-specific recombinases. Nucleic Acids Res. 2002;30:e115. doi: 10.1093/nar/gnf114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belteki G, Gertsenstein M, Ow DW, Nagy A. Site-specific cassette exchange and germline transmission with mouse ES cells expressing phiC31 integrase. Nat. Biotechnol. 2003;21:321–324. doi: 10.1038/nbt787. [DOI] [PubMed] [Google Scholar]
- 4.Glaser S, Anastassiadis K, Stewart AF. Current issues in mouse genome engineering. Nat. Genet. 2005;37:1187–1193. doi: 10.1038/ng1668. [DOI] [PubMed] [Google Scholar]
- 5.Osterwalder M, Galli A, Rosen B, Skarnes WC, Zeller R, Lopez-Rios J. Dual RMCE for efficient re-engineering of mouse mutant alleles. Nat. Methods. 2010;7:893–895. doi: 10.1038/nmeth.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schebelle L, Wolf C, Stribl C, Javaheri T, Schnutgen F, Ettinger A, Ivics Z, Hansen J, Ruiz P, von Melchner H, Wurst W, Floss T. Efficient conditional and promoter-specific in vivo expression of cDNAs of choice by taking advantage of recombinase-mediated cassette exchange using FlEx gene traps. Nucleic Acids Res. 2010;38:e106. doi: 10.1093/nar/gkq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turan S, Galla M, Ernst E, Qiao J, Voelkel C, Schiedlmeier B, Zehe C, Bode J. Recombinase-mediated cassette exchange (RMCE): traditional concepts and current challenges. J. Mol. Biol. 2011;407:193–221. doi: 10.1016/j.jmb.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Bethke B, Sauer B. Segmental genomic replacement by Cre-mediated recombination: genotoxic stress activation of the p53 promoter in single-copy transformants. Nucleic Acids Res. 1997;25:2828–2834. doi: 10.1093/nar/25.14.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng YQ, Seibler J, Alami R, Eisen A, Westerman KA, Leboulch P, Fiering S, Bouhassira EE. Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J. Mol. Biol. 1999;292:779–785. doi: 10.1006/jmbi.1999.3113. [DOI] [PubMed] [Google Scholar]
- 10.Bateman JR, Lee AM, Wu CT. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics. 2006;173:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nanto K, Yamada-Watanabe K, Ebinuma H. Agrobacterium-mediated RMCE approach for gene replacement. Plant Biotechnol. J. 2005;3:203–214. doi: 10.1111/j.1467-7652.2005.00118.x. [DOI] [PubMed] [Google Scholar]
- 12.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donnelly ML, Hughes LE, Luke G, Mendoza H, ten Dam E, Gani D, Ryan MD. The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J. Gen. Virol. 2001;82:1027–1041. doi: 10.1099/0022-1317-82-5-1027. [DOI] [PubMed] [Google Scholar]
- 14.Blaisonneau J, Sor F, Cheret G, Yarrow D, Fukuhara H. A circular plasmid from the yeast Torulaspora delbrueckii. Plasmid. 1997;38:202–209. doi: 10.1006/plas.1997.1315. [DOI] [PubMed] [Google Scholar]
- 15.Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 1998;16:657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- 16.Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS One. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimshek DR, Kim J, Hubner MR, Spergel DJ, Buchholz F, Casanova E, Stewart AF, Seeburg PH, Sprengel R. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32:19–26. doi: 10.1002/gene.10023. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Russell DW. Molecular Cloning : A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 19.Werdien D, Peiler G, Ryffel GU. FLP and Cre recombinase function in Xenopus embryos. Nucleic Acids Res. 2001;29:E53–E53. doi: 10.1093/nar/29.11.e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakano M, Odaka K, Ishimura M, Kondo S, Tachikawa N, Chiba J, Kanegae Y, Saito I. Efficient gene activation in cultured mammalian cells mediated by FLP recombinase-expressing recombinant adenovirus. Nucleic Acids Res. 2001;29:E40. doi: 10.1093/nar/29.7.e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rees S, Coote J, Stables J, Goodson S, Harris S, Lee MG. Bicistronic vector for the creation of stable mammalian cell lines that predisposes all antibiotic-resistant cells to express recombinant protein. Biotechniques. 1996;20:102–104. doi: 10.2144/96201st05. 106, 108–110. [DOI] [PubMed] [Google Scholar]
- 22.Baubonis W, Sauer B. Genomic targeting with purified Cre recombinase. Nucleic Acids Res. 1993;21:2025–2029. doi: 10.1093/nar/21.9.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malchin N, Molotsky T, Borovok I, Voziyanov Y, Kotlyar AB, Yagil E, Kolot M. High efficiency of a sequential recombinase-mediated cassette exchange reaction in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2010;19:117–122. doi: 10.1159/000321497. [DOI] [PMC free article] [PubMed] [Google Scholar]