Abstract
Nucleoid-associated proteins are bacterial proteins that are responsible for chromosomal DNA compaction and global gene regulation. One such protein is Escherichia coli Histone-like nucleoid structuring protein (H-NS) which functions as a global gene silencer. Whereas the DNA-binding mechanism of H-NS is well-characterized, its paralogue, StpA which is also able to silence genes is less understood. Here we show that StpA is similar to H-NS in that it is able to form a rigid filament along DNA. In contrast to H-NS, the StpA filament interacts with a naked DNA segment to cause DNA bridging which results in simultaneous stiffening and bridging of DNA. DNA accessibility is effectively blocked after the formation of StpA filament on DNA, suggesting rigid filament formation is the important step in promoting gene silencing. We also show that >1 mM magnesium promotes higher order DNA condensation, suggesting StpA may also play a role in chromosomal DNA packaging.
INTRODUCTION
The Escherichia coli chromosome is a large circular DNA molecule of several megabases in length. If it were fully stretched, this would translate to ∼1 mm in length. Since the diametre of an E. coli cell is 1–2 µm, its genomic DNA has to be highly condensed in order to fit into the cell while continuing to fulfil its biological functions. This compaction of genomic DNA is aided by DNA-binding proteins, which together with genomic DNA form the nucleoid (1–3). These DNA-binding proteins are often termed nucleoid-associated proteins (NAPs). There are about 10 major NAPs and most of them are highly expressed during exponential growth phase, suggesting their importance in cell viability and function (3).
Histone-like nucleoid structuring protein (H-NS) is one of the most studied NAPs as it is involved in many regulatory activities, in particular gene silencing and nucleoid structuring (4–9). H-NS has a molecular mass of 15.6 kDa and is heat stable with a neutral isoelectric point of 7.5 (10–12). During exponential phase there are 20 000 copies of H-NS in the cell (3). Previously, conflicting single-molecule studies demonstrated that H-NS is able to either polymerize along DNA to form a rigid nucleoprotein filament or cause DNA bridging (13–15). Recent experiments resolved this controversy by demonstrating that divalent ions such as magnesium and calcium can distinctly switch H-NS between rigid nucleoprotein filaments formation and forming DNA bridges (16). In addition, H-NS can sense changes in osmolarity, temperature and pH over a physiologically relevant range. H-NS's DNA binding is also antagonized by SsrB which leads to relieving of H-NS-mediated gene silencing and the antagonizing behaviour is only seen when H-NS forms rigid nucleoprotein filament (17,18). Antagonizing H-NS gene silencing function by displacing bound H-NS or StpA is also observed for LeuO protein (19). Structural studies demonstrated that H-NS can also form superhelical structures along DNA (20). These results suggest that in addition to DNA bridging, H-NS can play an important role in gene silencing by forming a protein filament along DNA.
StpA is an H-NS paralogue that is 58% similar to H-NS at the amino acid level (21). Due to this similarity, StpA was initially identified as a multi-copy gene suppressor that served as an H-NS substitute in H-NS-deficient cells (22). Both StpA and H-NS exhibit negative auto-regulation and are also able to suppress the promoter of the other (23,24). Studies have also shown that expression of StpA is up-regulated by high osmolarity and temperature during cell growth (25,26). This auto-regulatory relationship between H-NS and StpA suggests that H-NS is unable to suppress StpA under high osmolarity and temperature. Both H-NS and StpA are known to form concentration-dependent higher oligomers in solution (27,28) and the ability to oligomerize in solution is generally lost in gene silencing dysfunctional H-NS mutants (29–31).
We were interested in how the shared or distinct biological functions of StpA and H-NS correlate with their individual DNA-binding mechanisms. Unlike H-NS, the DNA-binding mechanism of StpA has not been extensively studied. Using atomic force microscopy (AFM) imaging, it was shown that StpA could bridge DNA at low protein concentrations, but formed globular aggregates at high protein concentrations (32). This result promoted the view that StpA was a DNA-bridging protein (33). However, the recent finding that H-NS switch between distinct DNA polymerization and bridging binding modes under the influence of divalent ions (16) raised the question as to whether StpA also has multiple DNA-binding modes and how it responds to various physiological stimuli such as monovalent salts, temperature and pH.
In the present work, we used single-molecule imaging and manipulation techniques to show that, like H-NS, StpA forms a rigid protein filament along DNA. We also showed that StpA can organize DNA into at least three major distinct DNA conformations: (i) a rigid co-filament containing a DNA backbone and a protein filament; (ii) DNA bridging between naked DNA and a DNA bound protein filament; and (iii) a magnesium-induced DNA condensation via inter-co-filament interactions. We also demonstrated that, in contrast to the DNA-H-NS co-filament (16), the DNA–StpA co-filament is largely insensitive to physiological changes in salt, temperature and pH. Importantly, once StpA forms a rigid protein filament along DNA, the DNA is inaccessible to cleavage by DNase I, indicating that DNA access was blocked after filament formation. Our results suggest that although StpA is a known paralogue of H-NS with similar biological functions, its DNA-binding mechanism varies substantially from H-NS, raising questions as to the role of StpA function in vivo.
MATERIALS AND METHODS
Over-expression and purification of StpA
pET-14b expression vector was used to express the StpA protein with N-terminal 6X-His tag. More detailed information is shown in the Supplementary Data.
AFM imaging of protein–DNA complexes
Linearized DNA of various lengths (depending on nature of experiment) was incubated with appropriate ratio of StpA before depositing on a glutaraldehyde-modified mica surface for AFM imaging experiments. More detailed information is shown in the Supplementary Data.
Magnetic tweezers single-DNA stretching experiments
A single λ-DNA (48 502 bp, New England Biolabs) was modified at both ends to attach one end on a paramagnetic bead and the other end on a modified glass edge in a transverse magnetic tweezers setup. The DNA-tethered paramagnetic bead is imaged in the focal plane to measure the DNA extension and applied force based on a home-written centroid tracking software. More detailed information is shown in the Supplementary Data.
RESULTS
StpA organizes DNA into different conformations
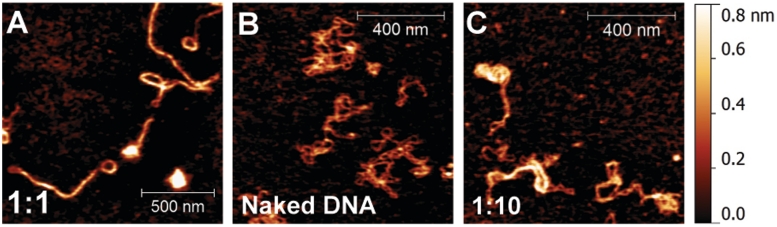
NAPs such as H-NS and HU exhibit multiple DNA-binding modes and organize DNA into different conformations depending on various conditions such as protein concentration or buffer conditions (6,16,34). Since the DNA-binding properties of StpA have not been well-characterized, we used AFM to image StpA bound to DNA at various StpA to DNA base pair (bp) ratios. For this purpose, we used glutaraldehyde-coated mica surfaces (see ‘Materials and Methods’ section), which minimally perturb DNA–protein complexes (16,35,36). Figure 1A shows the AFM image of linear DNA incubated at a 1:1 StpA:DNA ratio. The majority of DNA–StpA complexes (>80%) are a mixture of two distinct conformations: elongated co-filaments and associated large smooth loops of >250 nm in contour length. These structures are distinct from naked DNA imaged on the same mica surface, which shows random coiled conformations and has a lower height and width (Figure 1B). However, we also noted minority of the DNA–StpA complexes are small-scale aggregates (e.g. the right-bottom DNA–StpA complex in Figure 1A and in Supplementary Figure S1A). The majority of elongated DNA–StpA structures suggest an increase in DNA rigidity. The homogeneity of the DNA–StpA complexes suggests that StpA is likely evenly coated along the DNA due to the formation of a rigid protein filament along DNA. When the StpA:DNA ratio was reduced to 1 StpA to 10 bp (Figure 1C), certain regions of DNA had a greater height than others, suggesting a higher order folding of DNA and cooperativity. Further reduction of the StpA:DNA ratio to 1:100 revealed DNA bridging when StpA binding was unsaturated (Supplementary Figure S1C and D). Both 1:10 and 1:100 StpA:DNA ratio results are in agreement with previous StpA studies (32).
Figure 1.
AFM imaging demonstrates that StpA forms a rigid protein filament along DNA resulting in simultaneous stiffening and bridging of DNA. (A) Air AFM topology imaging of linearized φX174 dsDNA incubated with 1 StpA per 1 bp (1:1 StpA:DNA ratio, 300 nM StpA) shows rigid StpA-coated DNA hairpins. (B) Naked φX174 DNA on the same type of surface. Comparing with A, the naked DNA assume random coiled conformations with much thinner backbones. (C) DNA–StpA complexes at 1:10 StpA:DNA (30 nM StpA) ratio suggesting localize StpA binding and partial DNA condensation.
From the above results, it can be seen that DNA–StpA complexes form multiple conformations in a concentration-dependent manner. At low StpA concentrations, StpA causes mainly DNA condensation. At high or saturated StpA concentrations, it simultaneously organizes DNA into two distinct major conformations: elongated nucleoprotein co-filaments and large circular DNA loops. The elongated structures are likely due to formation of a rigid StpA protein filament along DNA. As StpA is able to oligomerize in solution as StpA concentration increases (28), it is possible different StpA oligomerization states at low and high concentration can be responsible for the concentration-dependent DNA organization observed here (see ‘Discussion’ section).
StpA–DNA bridging results from DNA–StpA co-filament and naked DNA interaction
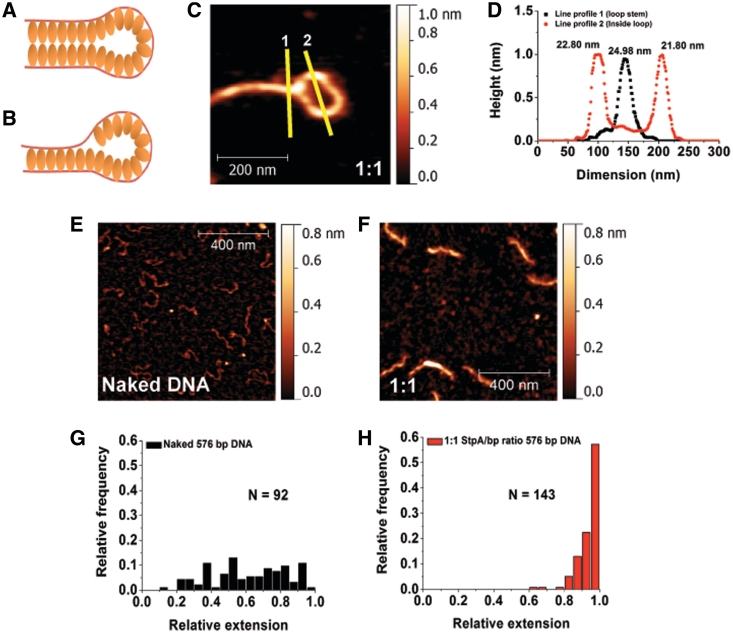
The results shown in the ‘Results’ section raise an interesting question as to how to explain the co-existence of rigid co-filament and rigid DNA end-loops that are observed in the majority of DNA–StpA complexes at saturated StpA-binding conditions. Both conformations intrinsically antagonize one another: a rigid co-filament will reduce the probability of internal loop formation due to the bending stiffness. We propose two models of mechanism that are consistent with the two conformations co-existence on the same DNA. One is that DNA bridging was caused by an attraction between two StpA protein filaments on different DNA segments, as illustrated in Figure 2A. The other mechanism can be due to StpA–DNA attraction between a naked DNA segment and a StpA filament on another DNA segment, as illustrated in Figure 2B. Our results indicate that the co-existence of rigid co-filaments and rigid DNA end-loops is likely due to the second model (see below).
Figure 2.
DNA–StpA co-filament interacts with naked DNA to form DNA bridges. (A and B) Hypothetical models of StpA-induced DNA bridging. (C) Zoomed-in StpA-coated DNA loop image. The lines are drawn to generate the width profiles in panel D. (D) AFM line profile analysis shows no significant difference between the half-height–widths of StpA-coated DNA in the loop and at the loop stem. Width values are indicated at the top of the respective peaks. (E) 1 × 1 µm image of linear 576-bp DNA with z-scale of 0–0.8 nm. (F) A 576-bp DNA substrates incubated at 1:1 StpA:DNA ratio. Comparing with E, StpA-coated 567-bp DNA is thicker and more straight, demonstrating monomeric rigid DNA–StpA co-filaments. The latter model (Figure 2B) is thus preferred over the former (Figure 2A). (G) Histogram of naked 576-bp DNA relative extension (or DNA end-to-end extension over its contour length). The distribution is widely spread, suggesting significant thermal fluctuation of DNA conformations. (H) Relative extension histogram of 576-bp DNA substrates incubated in 1:1 StpA:DNA ratio. The distribution shows that all the DNA are extended to nearly its full-contour length, suggesting significant DNA stiffening that suppresses the DNA conformational fluctuations. This is in agreement with the model in Figure 2B. Experiments were performed in 10 mM Tris, 50 mM KCl, pH 7.4 buffer.
Although we have seen minority of the structures are more compact structures of small size at StpA/bp ratio of 1:1 or above, the absence of large DNA aggregation in our AFM experiment at incubation time from 20 min to 5 h (Supplementary Figure S2A) already disfavours the first model, otherwise one should expect large globular DNA–StpA aggregates via DNA–StpA co-filaments interactions that can bring many DNA together. Additional evidence that disfavours the first model is that the apparent thickness of a segment in a rigid StpA-coated DNA loop is almost similar to that of the loop stem (Figure 2C and D). This is because the first model predicts the loop stem has a width twice as thick as the StpA-coated DNA in the loop since there will be two DNA–StpA co-filaments bridged at the loop stem (Figure 2A).
The second model also leads to a testable prediction that monomeric rigid DNA–StpA co-filaments should be observed when shorter DNA molecules are incubated with sufficiently high StpA:DNA ratios. This is because StpA–DNA bridging will have less time to occur before the DNA is fully coated by StpA. This prediction was tested by AFM imaging using shorter DNA (576 bp) at 1:1 ratio. As expected, compared with the naked DNA control (Figure 2E), the majority of DNA–StpA complexes are fully coated monomeric co-filaments and are more extended (Figure 2F). Quantitative dimension analysis (See Supplementary Method: Atomic Force Microscopy Imaging and Data Analysis and Figure S8) is shown in Figure 2G and H: without StpA, the relative extension (DNA end-to-end distance divided by its contour length) of short DNA was well distributed; while at 1:1 StpA:DNA ratio, the relative extension distribution has a sharp peak near one, suggesting highly rigid monomeric DNA that are extended nearly to its contour length. Analysis of the StpA-coated 576-bp DNA images also reveals the DNA–StpA co-filament has a thickness of ∼10–15 nm (after subtraction of AFM tip widening effect of ∼12 nm as shown in Supplementary Figure S2B) and has only a slight reduction in its contour length as compared to the naked DNA (Supplementary Figure S2C).
Finally, consistent with the second model, these fully coated monomeric 576-bp DNA–StpA co-filaments did not aggregate even when they are incubated for longer time of 4 h (Supplementary Figure S2D and E). At lower StpA:DNA ratio of 1:100, we were still able to observe StpA-DNA bridging using the same short DNA (Supplementary Figure S2F–H). At this ratio, there were always uncoated DNA segments that could interact with the coated ones. However, it could also be caused by binding of different StpA oligomerization states that exist in low StpA concentration (see ‘Discussion’ section). Overall, our results show that at high StpA/DNA ratios, StpA forms a rigid filament along DNA and StpA-induced DNA bridging can occur when this DNA–StpA co-filament segment binds to a naked DNA segment. In other words, the StpA protein filament is able to bind to two naked DNA segments simultaneously but requires just one single DNA to form (Figure 2B).
StpA-induced DNA stiffening and bridging are two kinetically competing processes
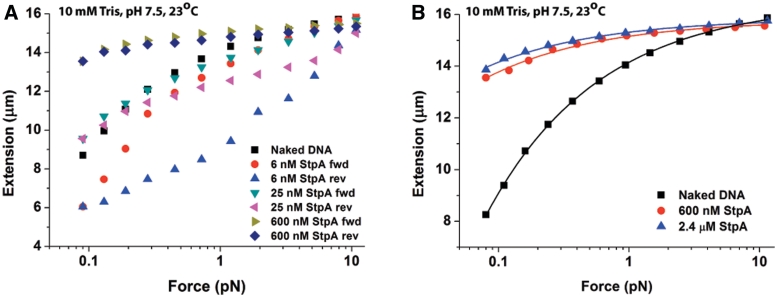
Our AFM experiments indicated that StpA could form a rigid filament on DNA at high StpA/DNA bp ratio (Figures 1A and 2F). In order to corroborate these findings, we used single-DNA stretching experiments to investigate the elastic response of DNA upon StpA binding (37). Transverse magnetic tweezers were used to probe the effects of StpA binding on the DNA force response (38). A single-DNA molecule was held at ∼11 pN when the protein was introduced into the reaction channel (Supplementary Figure S9B). The high force prevented DNA bridging before StpA was able to fully coat and stiffens the DNA. Upon addition of the protein, the force was gradually reduced to ∼0.08 pN and at each force the DNA was held for 60 s and the extension was calculated by the average in the last 30 s. To determine that StpA binding had reached a steady state, a reverse force scan was also performed by increasing the force through the same set of force values to test for hysteresis that can be caused by protein-induced DNA bridging.
Forward and reverse force-extension curves in 6, 25, 100, 300 and 600 nM StpA were recorded (Supplementary Figure S3A). For simplicity, only the curves at 6, 25 and 600 nM StpA are shown in Figure 3A. Significant hysteresis was observed at 6 nM StpA (compare 6 nM StpA forward curve; red filled circles with 6 nM StpA reverse curve; blue filled up-triangle), which was caused by DNA folding at smaller forces. This can be seen from the shorter DNA extension when compared to the naked DNA extension (see folding and unfolding time courses in Supplementary Figure S3B–D). The observed DNA folding at low StpA concentration can be explained by DNA bridging mediated by interactions between naked DNA segments and DNA–StpA co-filament segments as seen in Figure 2B. An alternative explanation is a different StpA oligomerization state which is more prevalent at lower StpA concentrations is responsible for mediating the observed DNA folding (see ‘Discussion’ section).
Figure 3.
Single-DNA stretching experiments show kinetic competition between StpA rigid protein filaments formation on DNA and DNA–StpA co-filament-dependent DNA bridging. (A) Forward and reverse force-extension curves (see panel figure legends) of 48 502 bp λ-DNA in 50 mM KCl 10 mM Tris pH 7.4 at the indicated StpA concentrations (up to saturation). At 6 nM StpA, strong hysteresis was observed, implying dominance of the StpA–DNA bridging mode. At 600 nM StpA, DNA stiffening due to protein filament formation dominated, with an increase in DNA extension and lack of hysteresis. At 25 nM StpA, a mixture of both modes is observed. (B) Force-extension curves of λ-DNA in 50 mM KCl, 10 mM Tris pH 7.4 indicate saturated binding at 600 nM StpA. This results in DNA stiffening due to protein filament formation. Only the forward curves are shown due to the absence of hysteresis. The solid lines represents fitting by the DNA worm-like-chain (WLC) model (see Supplementary Methods: Transverse Magnetic Tweezers Experimental Setup) fitting to experimental data points. The WLC model fit gives a persistence length with fitting error of 639.67 ± 34.80 nm and 909.50 ± 48.56 nm for 600 nM and 2400 nM StpA concentration, respectively. In addition, the fitted contour length with fitting error is 15 794.54 ± 32.27 nm and 15 851.05 ± 27.05 nm for 600 nM and 2400 nM StpA concentration, respectively.
Increasing the StpA concentration to 25 nM decreased the level of hysteresis and DNA folding compared with 6 nM StpA. In addition at low force (∼0.1 pN), the DNA extension increased beyond the naked DNA extension. The likely explanation is at this protein concentration, a significant portion of DNA is coated by a rigid StpA filament, resulting in an overall increase in the apparent DNA bending rigidity determined by the force-extension curve measurement (37). Thus, naked DNA segments can be depleted by higher StpA concentrations, leading to a completely stiffened DNA without any DNA bridging-dependent hysteresis. In agreement with this view, at 600 nM StpA, the DNA was stiffened over the entire force range, and the hysteresis was negligible. These results are consistent with the conclusions from the AFM imaging experiments (Figures 1A and 2F) which shows StpA can form a rigid filament along DNA at high StpA concentrations.
StpA filament formation (DNA stiffening) and the subsequent DNA–StpA co-filament and naked DNA interaction (DNA bridging) are mutually exclusive since they directly antagonizes one another—i.e. filament formation along DNA will deplete regions of naked DNA that are required for co-filament-mediated DNA bridging. This predicts a kinetic competition between them. To see this competition, another single-DNA stretching experiment was done whereby 600 nM of StpA was quickly added to the reaction channel at large force (∼6 pN) to prevent folding during protein introduction. Right after protein introduction, the DNA stretching force was immediately reduced to 0.1 pN before the filament coated the entire DNA (Supplementary Figure S3E). We observed the DNA extension initially increased due to DNA stiffening by StpA filament formation, followed by an abrupt DNA folding which is consistent with naked DNA bridging with the StpA filament-coated DNA segment. For comparison, pure stiffening was observed when the DNA was prevented from folding by holding at large force for significantly longer time (during which the naked DNA was quickly depleted by StpA filament formation) under the same protein concentration and buffer conditions (Figure 3A).
StpA protein filament increases the DNA bending rigidity more than 10-fold
We have shown StpA is able to form a rigid filament on DNA at StpA concentrations >25 nM. Since StpA has up to 25 000 copies per cell (3), the in vivo StpA concentration can be up to ∼10 µM. Therefore, our results suggest the formation of the StpA filament on DNA is highly likely a physiologically relevant DNA-binding mode (see ‘Discussion’ section). This prompts us to focus on the StpA filament formation on DNA and study its biophysical properties.
To measure the rigidity of the StpA protein filament, we analysed the force-extension response of DNA fully coated with StpA. Force-extension measurements at 600 nM and 2.4 µM StpA are shown in Figure 3B, using the same buffer conditions as in Figure 1. Addition of 600 nM StpA results in strong stiffening of the DNA, i.e. it is greatly extended (red triangles) compared to the naked DNA (black squares). Increasing the StpA concentration to 2.4 µM did not further increase DNA extension, suggesting that the DNA is saturated with StpA by 600 nM. The level of DNA stiffening is quantified by fitting the force-extension curves at saturated StpA concentration of 600 nM with the Marko–Siggia formula (39), which revealed an apparent persistence length (or bending rigidity) of 442.82 ± 161.28 nm (N = 9, see Supplementary Methods: Transverse Magnetic Tweezers Experimental Setup for curve fitting details). This is around 10-fold higher than the 50 nm persistence length of naked DNA which is due to the protein filament (see Supplementary Discussion). Since the DNA rigidity is negligible compared with the apparent bending rigidity of the DNA–StpA co-filament, we conclude the StpA filament has a bending rigidity up to 10-fold that of a naked DNA. In addition, the model fittings showed a slight reduction to DNA contour length with statistic standard deviation at 15 915.03 ± 183.41 nm (N = 9) as compared to the original λ-DNA contour length of 16 490 nm. This is in agreement with the AFM contour length analysis (Supplementary Figure S2C).
High salt disrupts the StpA protein filament on DNA
In vivo studies have shown that StpA expression is up-regulated by osmotic shock as well as increasing growth temperature (25). Single-molecule studies with H-NS demonstrated a loss of rigid filament at high salt, temperature or acidic pH (15,16). Therefore, we were interested in whether StpA responds similarly to H-NS to physiological stimuli. Using single-DNA stretching experiments, we found that the formation and structural integrity of the StpA filament is insensitive to physiological stimuli such as ionic strength in the range of 5–300 mM, temperature in 23–37°C and pH in the range 6.6–8.8 (Supplementary Figure S4A–C). In addition, low StpA concentration-induced DNA bridging by StpA is sensitive to KCl and temperature but not pH changes (Supplementary Figure S4D–F). Here we want to emphasize again that at low StpA concentration, different StpA oligomerization states may exist so the DNA folding mechanism can be different from the filament-mediated DNA bridging in high StpA concentration (see ‘Discussion’ section).
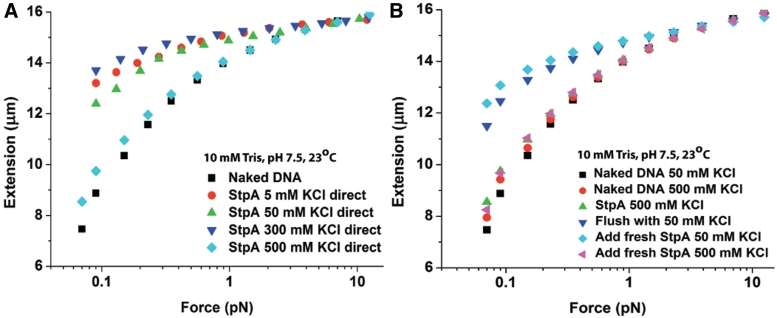
Since the formation of a rigid StpA filament was not affected by KCl concentration in the physiological range of 5–300 mM which is also confirmed with AFM imaging experiments (Supplementary Figure S4G and H), we asked whether filament formation can be prevented or disrupted at even higher salt concentration. High salt can affect the electrostatic interaction between StpA and DNA, and any possible attractive electrostatic interaction between StpA proteins can be reduced. To test this, we used magnetic tweezers at 600 nM StpA in 5–500 mM KCl, using different DNAs for each KCl concentration to eliminate a possible history dependence. As expected, for 5–300 mM KCl the force-extension curves show DNA stiffening (Figure 4A). However, at 500 mM KCl, there was a dramatic reduction in DNA extension i.e. the force-extension curve was almost similar to that of naked DNA. One explanation for the dramatic reduction in DNA stiffening was that at 500 mM KCl, the StpA DNA-binding affinity was significantly reduced, resulting in unsaturated StpA binding. Another possibility is that StpA was still bound to DNA, but the rigid filament cannot form at 500 mM KCl.
Figure 4.
The StpA protein filament was disrupted at high salt. Only the forward curves are plotted as hysteresis was not observed. (A) Force-extension curves at 600 nM StpA incubated in the KCl concentrations indicated in figure legend. As in Supplementary Figure S3, there was little change in stiffening between 5 and 300 mM KCl. However, at 500 mM KCl, almost no stiffening (filament formation) occurs. (B) Force-extension curves at 600 nM StpA during a series of buffer cycling. After StpA binds to DNA at 500 mM KCl, the remaining free protein was removed from the solution. A near saturated stiffened DNA was then obtained by changing the buffer to 50 mM KCl, implying that at 500 mM KCl StpA binding remains near saturation despite the lack of stiffening. Finally, after achieving saturation stiffening with StpA in 50 mM KCl, the stiffening was nearly eliminated by adding fresh StpA in 500 mM KCl.
To test these possibilities, we used magnetic tweezers and washed out the unbound StpA (Figure 4B). 600 nM StpA in 500 mM KCl was first added to the reaction channel, which produced the expected weak DNA stiffening shown by the green up-triangles in Figure 4B. Unbound StpA was then removed, followed by 50 mM KCl buffer and the force-extension curve was re-measured. The DNA became highly stiffened, suggesting a nearly fully coated DNA–StpA co-filament (compare with green up-triangles curve in Figure 4A). Adding fresh 600 nM StpA in 50 mM KCl further increased the extension slightly, confirming that StpA binding was almost saturated (as with 500 mM KCl). Finally, 600 nM StpA in 500 mM KCl was added again. As before, this drastically reduced the DNA extension, demonstrating that the loss of stiffening at high salt also occurred with DNA in which a rigid StpA filament had already formed. From these results, we conclude that: (i) the loss of stiffening at 500 mM KCl is not due to unsaturated binding of StpA to DNA and (ii) the drastic increase in stiffening when the buffer was switched to 50 mM KCl must be due to the re-organization of StpA protein originally bound to DNA.
The rigid StpA protein filament prevents DNA access
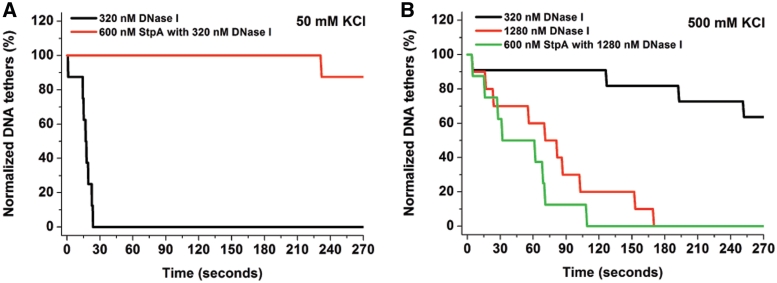
We have shown that StpA can form a rigid protein filament along DNA. An obvious question is whether this filament is related to the biological functions of StpA, in particular its ability to silence genes (23,40). We hypothesized that the formation of a rigid protein filament along DNA is able to block access to DNA by other proteins including RNA polymerase, inhibiting or suppressing transcription. To test this hypothesis, we studied the effect of StpA filament formation on the rate of DNA cleavage by DNase I, which requires accessibility to only 6 bp of exposed DNA. To achieve high throughput, we simultaneously observed 8–10 StpA-coated DNA tethers under 1–3 pN of force and then recorded the rate of DNA digestion after addition of DNase I (see Supplementary Methods: High-throughput Magnetic Tweezers for details and Figure S10). A unique advantage of this method over traditional Electrophoretic Mobility Shift Assay experiments is that a stretching force can be used to prevent DNA bridging before it is fully coated with a rigid StpA filament. Another advantage is that the kinetics of DNA breakage can be monitored in real time.
The negative control without StpA shows that all the DNA tethers were lost within 30 s after addition of 320 nM DNase I in low salt buffer (50 mM KCl, 10 mM Tris, pH 7.5). In contrast, in high salt (500 mM KCl, 10 mM Tris, pH 7.5) it took 3 min and four times as much DNase I to digest all the DNA tethers (Figure 5A and B; black and red line, respectively). This is in agreement with previous observations that DNase I activity was reduced in the presence of high monovalent salt concentrations. In 50 mM KCl, where DNA is fully coated with a rigid StpA filament by 600 nM StpA, almost all the DNA tethers remain intact after adding 320 nM DNase I (Figure 5A, red line). At 500 mM KCl, the DNA was fully bound by StpA, but a rigid filament cannot form (Figure 4A and B). Thus, all the DNA tethers are lost by ∼5 min after addition of 1280 nM DNase I (Figure 5B).
Figure 5.
StpA protein filament was able to block DNA access. (A) Normalized DNA tethers as a function of time with a tether population of 8–10. Adding 320 nM DNase I in 50 mM KCl (Black line) caused all the DNA tethers (total eight DNA tethers) to be cleaved by 30 s. Adding 600 nM StpA and then 320 nM DNase I in the same buffer condition (red line) resulted in loss of only one DNA tethers after 4 min (total eight DNA tethers). Thus, StpA protein filament protects DNA from DNase I digestion. (B) In 500 mM KCl where StpA does not form a protein filament. 320 nM DNase I in 500 mM KCl showed minimal digestion activity on naked DNA (black line). To improve the digestion efficiency, we used four times more DNase I (1280 nM) which could cut all the naked DNA tethers (total 10 DNA tethers) by 180 s (red line). Adding 600 nM StpA and then 1280 nM DNase I in 500 mM KCl, all the DNA tethers (total eight DNA tethers) were cut by 120 s (green line). Thus, the absence of filament formation in 500 mM KCl does not protect DNA from being digested.
This result indicates that formation of a rigid StpA filament along DNA, and not saturated binding, is essential for blocking DNA access. Because binding of RNA polymerase requires ∼70 bp of exposed DNA, this result suggests that the StpA filament can suppress gene transcription by blocking DNA accessibility of RNA polymerase (see ‘Discussion’ section).
Magnesium promotes StpA-induced higher order DNA compaction via inter-co-filament interactions
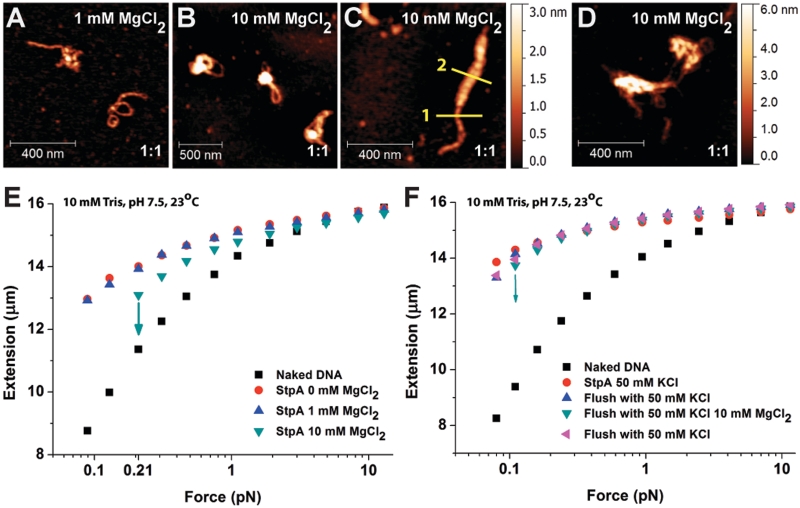
The previous experiments were performed in the absence of magnesium in order to separate the effects of magnesium from other physiological stimuli such as ionic strength, temperature and pH. In bacteria, magnesium ranges up to 4 mM (41) and it is essential for enzymatic reactions, chromosomal condensation and DNA damage repairing (41,42). Furthermore, previous studies have shown that divalent cations can affect the DNA-binding behaviour of H-NS protein (16). Previous AFM experiments reported that StpA (in the presence of magnesium) results in the formation of large aggregates (32). Such aggregates were not observed in our experiments in the absence of magnesium (Figures 1A and 2F), thus we hypothesize that magnesium can switch StpA from DNA–StpA co-filament-mediated DNA bridging to higher order DNA compaction. To test this hypothesis, we performed AFM imaging in the presence of magnesium. DNA was incubated for 20 min with 1:1 StpA:DNA ratio in 1 and 10 mM MgCl2. Figure 6A shows that in 1 mM MgCl2, the majority of DNA conformations were stiffened and exhibited simple bridging forms (comparable with the images shown in Figure 1A in the absence of magnesium). However, in 10 mM MgCl2 (Figure 6B and C), the simple bridging structure disappeared and the DNA–StpA complexes were compacted into more condensed structures. Consistently, large-scale aggregation of DNA–StpA complexes was also observed for shorter DNA, as shown in Figure 6D. This was in contrast to the monomeric configuration obtained in Figure 2F (in the absence of MgCl2), using similar DNA density. Additional experiments revealed that DNA aggregation began at ∼5 mM MgCl2, which is close to the in vivo magnesium concentration (see Supplementary Figure S5A–C for 5 mM MgCl2 images).
Figure 6.
Magnesium chloride (MgCl2) promotes DNA compaction via inter-co-filament interaction. (A–D) AFM images of 1:1 StpA:DNA complexes (300 nM StpA) incubated in buffer containing 1 or 10 mM MgCl2. The StpA/DNA ratios are at the bottom right corner of each sub panel: (A) In 1 mM MgCl2, linearized φX174 DNA substrates assumes similar rigid DNA hairpin structures to Figure 1A. (B and C) In 10 mM MgCl2, the DNA is organized into higher order aggregations. AFM width measurement showed the StpA-coated DNA has an apparent width of 20.44 nm (yellow line 1) while the thicker portion has an apparent width of 51.68 nm (yellow line 2). Considering the ∼12 nm AFM tip widening effect, the thicker portion is around four times as thick as the thinner portion, which is possibly caused by bundling of four DNA–StpA co-filaments. (D) In 10 mM MgCl2, linear 576-bp DNA substrate with the same StpA:DNA ratio and StpA concentration is organized into aggregates. (E) Force-extension curves of 600 nM StpA in varying MgCl2 buffer conditions from 0 to 10 mM MgCl2. StpA-induced DNA stiffening was observed at all non-zero MgCl2 concentrations. StpA-induced DNA folding events occurred at 10 mM MgCl2, as indicated by downward arrows. Before folding, DNA was stiffened indicated by the longer extension comparing with the naked DNA. (F) Force-extension curves of DNA fully coated with a StpA filament in the absence of surrounding free proteins in a series of buffer cycling. After the DNA folded at 10 mM MgCl2, it was completely unfolded at high force and then switched to 50 mM KCl, whereupon it was again completely stiffened with no folding. The DNA did not undergo any folding events in the absence of magnesium even when held at the lowest force (∼0.08 pN) up to 20 min (Supplementary Figure S6C). This result verifies that the observed DNA compaction by StpA was due to the presence of magnesium.
DNA condensation was also observed in magnetic tweezers experiments in the presence of magnesium. Using 600 nM StpA in 50 mM KCl with varying concentrations of MgCl2, Figure 6E shows that DNA stiffening by StpA is unaffected by 0–10 mM MgCl2. No hysteresis was observed in 1 mM MgCl2 (blue up-triangles). In 10 mM MgCl2, (green down-triangles), the DNA was still stiffened at forces >0.2 pN. However, at low force regimes (<0.2 pN), there was a dramatic decrease in its extension, indicating a DNA compaction process (indicated by the down arrow). DNA folding and unfolding dynamics are shown in Supplementary Figure S6A. Although the same DNA was used to obtain the results in Figure 6E, we confirmed that the effect of MgCl2 on StpA DNA binding was history independent (Supplementary Figure S6B). These single DNA stretching results are in agreement with the AFM imaging experiments shown in Figure 6A–D.
The StpA-induced DNA compaction at 10 mM MgCl2 cannot simply be explained by DNA bridging between DNA–StpA co-filaments and naked DNA segments as we observed at low salt and no magnesium. This is because the strong DNA stiffening effect observed at >0.2 pN in 10 mM MgCl2 indicates that the StpA protein filament was still present and thus few if any naked DNA segments were available. This was further confirmed by buffer switching experiments between 0 and 10 mM MgCl2 for a DNA fully coated with StpA filament in the absence of free protein (Figure 6F), which shows a reversible switching between pure DNA stiffening at 0 mM MgCl2 and simultaneous DNA stiffening/folding at 10 mM magnesium. Since the only varying factor in the buffer switching experiments was the presence of magnesium, the results must indicate a magnesium-dependent switching of DNA physical organization by StpA. Furthermore, since a rigid StpA filament still exists in the presence of magnesium and no naked DNA segments exist, this result also suggests that the magnesium-promoted DNA condensation is likely due to interactions between DNA–StpA co-filaments. Such a mechanism predicts that in the presence of magnesium, StpA will be able to bring DNA together through inter-co-filament interactions, which is consistent with previous AFM imaging experiments (32) and our present AFM imaging (Figure 6A–D). In addition, it was also shown the StpA filament rigidity in 10 MgCl2 buffer condition is 459.31 ± 93.76 nm (N = 6) which is generally similar to that in the absence of MgCl2 (see Supplementary Figure S6D and corresponding caption for experimental approach). Since StpA still forms a rigid filament along DNA up to 10 mM magnesium, we predict that StpA can still block DNA accessibility in 10 mM MgCl2. This was confirmed by DNase I digestion assay (Supplementary Figure S7). Taken together, in the presence of physiological magnesium, StpA binds to DNA and forms a protein filament along it, which blocks DNase I accessibility to DNA. In addition, it can simultaneously compact DNA into higher order structures through inter-co-filament interactions.
DISCUSSION
StpA binds to DNA and forms a protein filament
From AFM imaging and single-DNA stretching experiments, our results show that StpA is able to form a rigid protein filament along DNA. This protein filament forms across a range of salt, magnesium, temperature and pH which suggests formation of StpA protein filament on DNA is the fundamental mechanism of StpA–DNA binding. StpA-binding forms at least three distinct types of physical organization of the DNA: linear DNA–StpA co-filament, DNA–StpA co-filament-dependent DNA bridging (<1 mM MgCl2) and higher order DNA compaction induced by inter-co-filament interactions (>1 mM MgCl2). The StpA protein filament is a very rigid structure. Its bending persistence is ∼450 nm, around 10-fold stiffer than the DNA backbone. StpA-mediated DNA bridging and aggregation were reported previously (32,33), but the mechanism was not elucidated. For example, previous AFM experiments did not identify StpA filament formation on DNA (32). This is likely because in those experiments, >5 mM MgCl2 was used to deposit plasmid DNA onto freshly cleaved mica. According to our results, this magnesium level causes StpA to organize DNA into compact higher order structures, which makes it difficult to see the rigid filament formation. Most importantly, single-molecule manipulation measurements which can probe the existence of rigid protein structure on DNA (i.e. Figure 3A and B) were not employed. Based on our AFM imaging experiments at low StpA concentrations, StpA causes DNA condensation via DNA bridging, in agreement with previous studies (32). Due to the known concentration-dependent oligomerization states of StpA in solution (27,28), the DNA bridging at low StpA concentration is not necessarily due to co-filament-mediated DNA bridging that occurs primarily at >25 nM StpA concentration. For example, theoretical studies have shown that DNA bridging can be mediated by non-interacting H-NS dimers (43,44).
Although we have shown that StpA has multiple DNA-binding modes, we do not yet know the effective binding unit of StpA, (i.e. if StpA works as monomers or dimers or even higher oligomers) as StpA is known to oligomerize in solution as StpA concentration increases (28). This also suggests a possibility that different StpA species (i.e. monomer, dimer, higher oligomers) might be responsible for its multiple DNA-binding modes. It has been shown for both H-NS and StpA at ∼20 µM protein concentration, their oligomeric states are a broad distribution of species with majority being higher order oligomers (45). It will be important in future research to investigate the oligomerization properties of H-NS-like proteins on physical organizations of DNA.
We also want to point out that, in vivo, there are many other abundant NAPs that will compete with StpA. The total concentration of NAPs easily exceeds 100 µM. The average NAP to DNA ratio in vivo will then become 1:10 or even higher, which is comparable to our AFM imaging at 1:10 and 1:1 ratios. In our single-DNA stretching experiments, the ratio was not controlled due to the nature of single-DNA stretching experiments where only one DNA molecule is stretched. In all single-DNA stretching experiments, the StpA to DNA ratio is always in excess. In such experiments, only the concentration of the protein is meaningful. For H-NS-like proteins (such as StpA), which can form different oligomerization states in solution, the concentration is an important parameter in addition to the protein to DNA ratio. As mentioned, at different protein concentrations, the distribution of oligomerization states of the H-NS-like proteins is different. Therefore, the DNA-binding modes (not just binding affinity) may be different. Since the in vivo concentration of StpA is in the µM range, we think our single-DNA stretching experiments in 600 nM to 2.4 µM of StpA are most relevant to the in vivo condition.
Possible biological functions of StpA filament
The DNA-binding mechanism of StpA must be relevant for its biological function. A known function of StpA is to repress gene transcription (23,46). As revealed by our studies, the fundamental binding mechanism of StpA is the formation of a rigid filament structure on DNA (Figures 3B and 6 E). The StpA filament can effectively suppress DNA digestion by DNase I, which can only result from blockage of access to DNA, since DNase I requires only about 6 bp of DNA for cleavage (47). For comparison, RNA polymerase requires ∼70–80 bp of DNA for transcription in E. coli (48), which is significantly higher than that required for DNase I digestion. Thus, StpA protein filament formation likely blocks RNA polymerase from interacting with DNA, resulting in gene silencing. With a copy number of 25 000 and a 4.6 million bp chromosome (3), this translates to ∼1 StpA per 200 bp of DNA and ∼10 µM StpA concentration. At such high StpA concentration, StpA filament formation on DNA is likely the dominating DNA-binding mode in vivo. Like H-NS, StpA has DNA-binding preference to specific sites on chromosomal DNA (49) and coupled with competition with other NAPs for DNA-binding sites in vivo, StpA will localize to DNA regions with high StpA-binding affinity. Such high-affinity sites may nucleate StpA binding and direct cooperative StpA filament formation surrounding these sites. This will give rise to a sequence preference for StpA filament formation that is necessary for its selective gene silencing function.
StpA may also play an important role in chromosomal DNA packaging. Our findings have shown that StpA can organize DNA into compact higher order structures in the presence of >1 mM magnesium. Intracellular magnesium concentration is within this range (41). As such, discrete DNA–StpA co-filament islands formed on high-affinity sites will interact with another DNA–StpA co-filament islands to aid DNA compaction globally, but also able to selectively regulate genes.
Comparison between StpA and H-NS
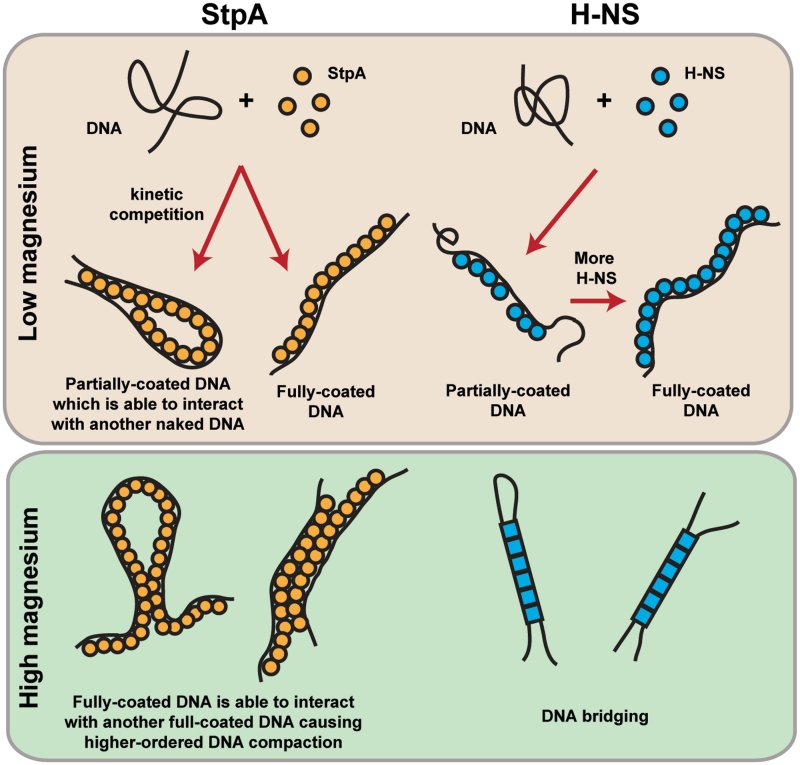
As StpA is an H-NS paralogue and they share a common function as gene silencers, it is worthwhile to compare their similarities and differences in DNA-binding behaviour (Figure 7). Both proteins can form a rigid filament along DNA (16) (see Figure 3B for StpA). However, they are distinct from each other in terms of how protein filament formation and stability responds to environmental factors. The structural integrity of the StpA filament is insensitive to ionic strength in 50–300 mM KCl, in temperature of 23–37°C and pH of 6.6–8.8 (Supplementary Figure S4A–C), while the H-NS filament is disrupted at 200 mM KCl and 37°C (16). The observation that H-NS is sensitive to environmental stimuli, whereas StpA is not might be significant in vivo. This is because the StpA protein filament is more stable as compared to H-NS and is likely to remain when there is a sudden influx in cytoplasmic potassium or when temperature is elevated, suggesting possible roles of StpA in resisting osmotic stress and heat shock. In addition, a recent in vivo study of StpA and H-NS localization in E. coli cells using super-resolution microscopy suggests a distinct difference in how StpA and H-NS are localized in cells (50). This warrants future studies on how the DNA-binding mechanism differences between H-NS and StpA might lead to their respective in vivo localization patterns.
Figure 7.
Comparison of models of StpA and H-NS DNA-binding mechanism. In low magnesium buffer conditions, StpA (orange circles) forms a rigid filament along DNA, which is able to interact with another naked DNA segment to mediate DNA bridging. The rigid filament on the DNA is unable to interact with another bound StpA filament. H-NS (blue circles) in low magnesium buffer is also able to form a rigid filament along DNA but unlike the StpA filament, the bound filament is unable to interact with another naked DNA segments (16). StpA in the presence of high magnesium buffer conditions causes inter-co-filament-mediated higher order DNA compaction. Unlike the StpA filament, formation of H-NS filament is suppressed in high magnesium concentrations. In addition, DNA bridging will occur in high magnesium concentrations (16). Blue circles and squares represent possible different H-NS-binding species in low and high magnesium conditions where two distinct binding modes were reported (16).
StpA filament can simultaneously stiffen and form DNA bridges with naked DNA at low magnesium (<1 mM) which means the rigid StpA filament can bind to two DNA segments. Such a bi-DNA-binding protein filament model was previously proposed for H-NS (20). However, this proposed protein filament model was not observed for H-NS, i.e. the H-NS protein filament is formed only at low magnesium concentrations (<2 mM) and once formed, the DNA bound H-NS filament does not interact with either a naked DNA segment or another DNA-H-NS co-filament (16). In higher magnesium concentration, H-NS is able to bridge DNA. In 10 mM magnesium, before DNA is bridged, no DNA stiffening was observed (16). This suggests that in the condition, a rigid H-NS filament does not form prior to DNA bridging, which is a distinction from the StpA filament-induced DNA bridging. Therefore, this bi-DNA-binding protein filament model seems more suitable for its StpA.
It will be interesting to compare the DNA compaction capability between H-NS and StpA. H-NS cannot compact DNA at low magnesium since it only forms rigid filament on DNA (<2 mM) (16). It can moderately compact DNA at higher magnesium (>2 mM) by forming simple DNA bridges only. In contrast, StpA filament formed at low magnesium is able to form DNA bridges. In high magnesium, StpA still maintains it filament, and it can cause higher order DNA compaction through inter-co-filament interactions. These results demonstrate that StpA has a stronger compaction capability than H-NS in both low and high magnesium concentrations, which is likely due to the higher pI value of StpA (∼8.0) than that of H-NS (∼5.4). According to these results, StpA might be a good candidate that is involved in E. coli chromosomal DNA packaging.
The H-NS and StpA filaments formed along DNA have distinct rigidity properties. Previous studies reported a bending persistence length of the H-NS-DNA co-filament to be around 130 nm, which is 2 - to 3-fold that of naked double-stranded DNA (15). In contrast, the StpA filament has an apparent persistence length with statistic standard deviation of 442.82 ± 161.28 nm (N = 9) in 50 mM KCl buffer conditions which remains more or less similarly rigid up to 300 mM KCl as shown in Figure 4A and also in the presence of 10 mM MgCl2 where the measured value with statistic standard deviation is 459.31 ± 93.76 nm (N = 6). Comparing with H-NS filament, the StpA filament is at least 3-fold higher. This difference in rigidity is likely due to a difference in the way the protein filaments are organized or formed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–10, Supplementary Methods, Supplementary Discussion and Supplementary References [51–58].
FUNDING
The Ministry of Education of Singapore (R144000251112 to J.Y.); the Mechanobiology Institute, Singapore (R714005007271 and R714015007271 to J.Y.); the Veterans Administration (101BX000372 to L.J.K.). Funding for open access charge: Mechanobiology Institute of Singapore (R144000251112 to J.Y.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs John E. Ladbury and Kin Man Suen for the kind gift of pET14b-StpA. We also thank Dr Adam Yuan and the protein expression core facility of the Mechanobiology Institute for protein purification. We also thank Dr Chen Hu for setting up the magnetic tweezers instrument. C.J.L. and Y.R.W. performed the experiments. L.J.K. and J.Y. conceived the research. C.J.L. and J.Y. designed the experiments and interpreted the data. C.J.L., Y.R.W., L.J.K. and J.Y. wrote the article.
REFERENCES
- 1.Dame RT. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol. Microbiol. 2005;56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 2.Browning DF, Grainger DC, Busby SJW. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 2010;13:773–780. doi: 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Azam TA, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auner H, Buckle M, Deufel A, Kutateladze T, Lazarus L, Mavathur R, Muskhelishvili G, Pemberton I, Schneider R, Travers A. Mechanism of transcriptional activation by FIS: role of core promoter structure and DNA topology. J. Mol. Biol. 2003;331:331–344. doi: 10.1016/s0022-2836(03)00727-7. [DOI] [PubMed] [Google Scholar]
- 5.Dame RT. Single-molecule micromanipulation studies of DNA and architectural proteins. Biochem. Soc. T. 2008;36:732–737. doi: 10.1042/BST0360732. [DOI] [PubMed] [Google Scholar]
- 6.van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc. Natl Acad. Sci. USA. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoebel DM, Free A, Dorman CJ. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology. 2008;154:2533–2545. doi: 10.1099/mic.0.2008/020693-0. [DOI] [PubMed] [Google Scholar]
- 8.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 9.Dorman CJ. H-NS, the genome sentinel. Nat. Rev. Microbiol. 2007;5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- 10.Cukier-Kahn R, Jacquet M, Gros F. Two heat-resistant, low molecular weight proteins from Escherichia coli that stimulate DNA-directed RNA synthesis. Proc. Natl Acad. Sci. USA. 1972;69:3643–3647. doi: 10.1073/pnas.69.12.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacquet M, Cukier-Kahn R, Pla J, Gros F. A thermostable protein factor acting on in vitro DNA transcription. Biochem. Biophys. Res. Commun. 1971;45:1597–1607. doi: 10.1016/0006-291x(71)90204-x. [DOI] [PubMed] [Google Scholar]
- 12.Falconi M, Gualtieri MT, La Teana A, Losso MA, Pon CL. Proteins from the prokaryotic nucleoid: primary and quaternary structure of the 15-kD Escherichia coli DNA binding protein H-NS. Mol. Microbiol. 1988;2:323–329. doi: 10.1111/j.1365-2958.1988.tb00035.x. [DOI] [PubMed] [Google Scholar]
- 13.Dame RT, Wyman C, Goosen N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 2000;28:3504–3510. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dame RT, Noom MC, Wuite GJ. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444:387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- 15.Amit R, Oppenheim AB, Stavans J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys. J. 2003;84:2467–2473. doi: 10.1016/S0006-3495(03)75051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Chen H, Kenney LJ, Yan J. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010;24:339–344. doi: 10.1101/gad.1883510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walthers D, Carroll RK, Navarre WW, Libby SJ, Fang FC, Kenney LJ. The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol. Microbiol. 2007;65:477–493. doi: 10.1111/j.1365-2958.2007.05800.x. [DOI] [PubMed] [Google Scholar]
- 18.Walthers D, Li Y, Liu YJ, Anand G, Yan J, Kenney LJ. Salmonella enterica response regulator SsrB relieves H-NS silencing by displacing H-NS bound in polymerization mode and directly activates transcription. J. Biol. Chem. 2011;286:1895–1902. doi: 10.1074/jbc.M110.164962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De la Cruz MA, Fernandez-Mora M, Guadarrama C, Flores-Valdez MA, Bustamante VH, Vazquez A, Calva E. LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Mol. Microbiol. 2007;66:727–743. doi: 10.1111/j.1365-2958.2007.05958.x. [DOI] [PubMed] [Google Scholar]
- 20.Arold ST, Leonard PG, Parkinson GN, Ladbury JE. H-NS forms a superhelical protein scaffold for DNA condensation. Proc. Natl Acad. Sci. USA. 2010;107:15728–15732. doi: 10.1073/pnas.1006966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang AX, Belfort M. Nucleotide-sequence of a newly-identified Escherichia coli gene, StpA, encoding an H-NS-like protein. Nucleic Acids Res. 1992;20:6735–6735. doi: 10.1093/nar/20.24.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi XL, Bennett GN. Plasmids bearing Hfq and the Hns-like gene StpA complement HNS mutants in modulating arginine decarboxylase gene-expression in Escherichia coli. J. Bacteriol. 1994;176:6769–6775. doi: 10.1128/jb.176.21.6769-6775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang AX, Rimsky S, Reaban ME, Buc H, Belfort M. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]
- 24.Sonden B, Uhlin BE. Coordinated and differential expression of histone-like proteins in Escherichia coli: regulation and function of the H-NS analog StpA. EMBO J. 1996;15:4970–4980. [PMC free article] [PubMed] [Google Scholar]
- 25.Free A, Dorman CJ. The Escherichia coli stpA gene is transiently expressed during growth in rich medium and is induced in minimal medium and by stress conditions. J. Bacteriol. 1997;179:909–18. doi: 10.1128/jb.179.3.909-918.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller CM, Schneider G, Dobrindt U, Emody L, Hacker J, Uhlin BE. Differential effects and interactions of endogenous and horizontally acquired H-NS-like proteins in pathogenic Escherichia coli. Mol. Microbiol. 2010;75:280–293. doi: 10.1111/j.1365-2958.2009.06995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Badaut C, Williams R, Arluison V, Bouffartigues E, Robert B, Buc H, Rimsky S. The degree of oligomerization of the H-NS nucleoid structuring protein is related to specific binding to DNA. J. Biol. Chem. 2002;277:41657–41666. doi: 10.1074/jbc.M206037200. [DOI] [PubMed] [Google Scholar]
- 28.Johansson J, Eriksson S, Sonden B, Wai SN, Uhlin BE. Heteromeric interactions among nucleoid-associated bacterial proteins: localization of StpA-stabilizing regions in H-NS of Escherichia coli. J. Bacteriol. 2001;183:2343–2347. doi: 10.1128/JB.183.7.2343-2347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueguchi C, Suzuki T, Yoshida T, Tanaka K, Mizuno T. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 1996;263:149–162. doi: 10.1006/jmbi.1996.0566. [DOI] [PubMed] [Google Scholar]
- 30.Stella S, Spurio R, Falconi M, Pon CL, Gualerzi CO. Nature and mechanism of the in vivo oligomerization of nucleoid protein H-NS. EMBO J. 2005;24:2896–2905. doi: 10.1038/sj.emboj.7600754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spurio R, Falconi M, Brandi A, Pon CL, Gualerzi CO. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 1997;16:1795–1805. doi: 10.1093/emboj/16.7.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dame RT, Luijsterburg MS, Krin E, Bertin PN, Wagner R, Wuite GJ. DNA bridging: a property shared among H-NS-like proteins. J. Bacteriol. 2005;187:1845–1848. doi: 10.1128/JB.187.5.1845-1848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keatch SA, Leonard PG, Ladbury JE, Dryden DT. StpA protein from Escherichia coli condenses supercoiled DNA in preference to linear DNA and protects it from digestion by DNase I and EcoKI. Nucleic Acids Res. 2005;33:6540–6546. doi: 10.1093/nar/gki951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao B, Johnson RC, Marko JF. Modulation of HU-DNA interactions by salt concentration and applied force. Nucleic Acids Res. 2010;38:6176–6185. doi: 10.1093/nar/gkq435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang HD, Bash R, Yodh JG, Hager GL, Lohr D, Lindsay SM. Glutaraldehyde modified mica: a new surface for atomic force microscopy of chromatin. Biophys. J. 2002;83:3619–3625. doi: 10.1016/S0006-3495(02)75362-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu H, Freedman BS, Lim CT, Heald R, Yan J. Atomic force microscope imaging of chromatin assembled in Xenopus laevis egg extract. Chromosoma. 2011;120:245–254. doi: 10.1007/s00412-010-0307-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan J, Marko JF. Effects of DNA-distorting proteins on DNA elastic response. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2003;68:011905. doi: 10.1103/PhysRevE.68.011905. [DOI] [PubMed] [Google Scholar]
- 38.Yan J, Skoko D, Marko JF. Near-field-magnetic-tweezer manipulation of single DNA molecules. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2004;70:011905. doi: 10.1103/PhysRevE.70.011905. [DOI] [PubMed] [Google Scholar]
- 39.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 40.Lucchini S, McDermott P, Thompson A, Hinton JC. The H-NS-like protein StpA represses the RpoS (sigma 38) regulon during exponential growth of Salmonella Typhimurium. Mol. Microbiol. 2009;74:1169–1186. doi: 10.1111/j.1365-2958.2009.06929.x. [DOI] [PubMed] [Google Scholar]
- 41.Lusk JE, Williams RJP, Kennedy EP. Magnesium and growth of Escherichia coli. J. Biol. Chem. 1968;243:2618. [PubMed] [Google Scholar]
- 42.Paymaster NJ. Magnesium metabolism - brief review. Ann. Roy. Coll. Surg. 1976;58:309–314. [PMC free article] [PubMed] [Google Scholar]
- 43.Wiggins PA, Dame RT, Noom MC, Wuite GJ. Protein-mediated molecular bridging: a key mechanism in biopolymer organization. Biophys. J. 2009;97:1997–2003. doi: 10.1016/j.bpj.2009.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Vries R. Influence of mobile DNA-protein-DNA bridges on DNA configurations: coarse-grained Monte-Carlo simulations. J. Chem. Phys. 2011;135:125104. doi: 10.1063/1.3636383. [DOI] [PubMed] [Google Scholar]
- 45.Leonard PG, Ono S, Gor J, Perkins SJ, Ladbury JE. Investigation of the self-association and hetero-association interactions of H-NS and StpA from Enterobacteria. Mol. Microbiol. 2009;73:165–179. doi: 10.1111/j.1365-2958.2009.06754.x. [DOI] [PubMed] [Google Scholar]
- 46.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolf E, Brukner I, Suck D. Mutational analysis of Dnase-I DNA interactions - design, expression and characterization of a Dnase-I loop insertion mutant with altered sequence selectivity. Protein Eng. 1995;8:283–291. doi: 10.1093/protein/8.3.283. [DOI] [PubMed] [Google Scholar]
- 48.Ozoline ON, Tsyganov MA. Structure of open promoter complexes with Escherichia coli Rna-polymerase as revealed by the Dnase-I footprinting technique - compilation analysis. Nucleic Acids Res. 1995;23:4533–4541. doi: 10.1093/nar/23.22.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uyar E, Kurokawa K, Yoshimura M, Ishikawa S, Ogasawara N, Oshima T. Differential binding profiles of StpA in wild-type and HNS mutant cells: a comparative analysis of cooperative partners by chromatin immunoprecipitation-microarray analysis. J. Bacteriol. 2009;191:2388–2391. doi: 10.1128/JB.01594-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang W, Li GW, Chen C, Xie XS, Zhuang X. Chromosome organization by a nucleoid-associated protein in live bacteria. Science. 2011;333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu H, Chen H, Zhang X, Qu Y, Marko JF, Yan J. Transition dynamics and selection of the distinct S-DNA and strand unpeeling modes of double helix overstretching. Nucleic Acids Res. 2011;39:3473–3481. doi: 10.1093/nar/gkq1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bao Q, Chen H, Liu Y, Yan J, Droge P, Davey CA. A divalent metal-mediated switch controlling protein-induced DNA bending. J. Mol. Biol. 2007;367:731–740. doi: 10.1016/j.jmb.2006.09.082. [DOI] [PubMed] [Google Scholar]
- 53.Rivetti C, Codeluppi S. Accurate length determination of DNA molecules visualized by atomic force microscopy: evidence for a partial B- to A-form transition on mica. Ultramicroscopy. 2001;87:55–66. doi: 10.1016/s0304-3991(00)00064-4. [DOI] [PubMed] [Google Scholar]
- 54.Vossepoel AM, Smeulders AWM. Vector code probability and metrication error in the representation of straight-lines of finite length. Comput Graphics Image Proc. 1982;20:347–364. [Google Scholar]
- 55.Allemand JF, Bensimon D, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;272:797–797 (vol 271, pg 1835, 1996). doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 56.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 57.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 58.Smith SB, Finzi L, Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.