Abstract
Lin28 is critical for stem cell maintenance and is also associated with advanced human malignancies. Our recent genome-wide studies mark Lin28 as a master post-transcriptional regulator of a subset of messenger RNAs important for cell growth and metabolism. However, the molecular basis underpinning the selective mRNA target regulation is unclear. Here, we provide evidence that Lin28 recognizes a unique motif in multiple target mRNAs, characterized by a small but critical ‘A’ bulge flanked by two G:C base pairs embedded in a complex secondary structure. This motif mediates Lin28-dependent stimulation of translation. As Lin28 is also known to inhibit the biogenesis of a cohort of miRNAs including let-7, we propose that Lin28 binding to different RNA types (precursor miRNAs versus mRNAs) may facilitate recruitment of different co-factors, leading to distinct regulatory outcomes. Our findings uncover a putative yet unexpected motif that may constitute a mechanistic base for the multitude of functions regulated by Lin28 in both stem cells and cancer cells.
INTRODUCTION
The developmentally regulated RNA-binding protein Lin28 is expressed abundantly in human and mouse embryonic stem (ES) cells and is required for their maintenance (1–4). Its aberrant expression in diverse human malignancies has been associated with advanced disease (5,6). Lin28 exerts its biological effects through at least two molecular mechanisms. First, it selectively blocks the production of a cohort of miRNAs including let-7, leading to derepression of their targets involved in cell growth and differentiation (4,5,7–11). miRNAs are short RNAs of ∼22 nt that regulate target mRNA expression through base-paired interaction (12). miRNA biogenesis involves cropping of the primary miRNA transcripts (pri-miRNAs) in the nucleus by the Drosha complex to produce ∼70-nt precursor miRNAs (pre-miRNAs), which are further cleaved in the cytoplasm by the Dicer complex to yield mature miRNAs. Second, Lin28 binds specific messenger RNAs and modulates their translation (3,13–17). Roughly, 5% of mRNAs (including that for the key pluripotency factor Oct4) in human ES cells and embryonal carcinoma (EC) cells appear to be bound and regulated by Lin28 (3).
The mechanism underlying selective inhibition of let-7 by Lin28 has been extensively studied. The common theme is that Lin28 binds to the terminal loop region of pri/pre-let-7 and blocks their processing (4,7,8,11,18–20). However, the exact sequence and structural motif that dictates Lin28 binding and inhibition remains controversial. Piskounova et al. (19) reported a conserved cytosine nucleotide in the loop region of pre-let-7g to be required for both Lin28 binding and processing inhibition. Newman et al. (8), however, showed that mutating a conserved adenosine together with two guanosine nucleotides in the loop region of pre-let-7g severely impaired Lin28-mediated let-7 inhibition. Yet, a third group reported that a conserved GGAG tetra-nucleotide sequence motif mediates Lin28-dependent miRNA uridylation (4), a process that promotes pre-let-7 degradation (21–23). Together, these studies highlight the unusual complexity of mechanisms underpinning Lin28-mediated regulation.
How Lin28 selects its mRNA targets and regulates them is currently an unexplored area and is fundamental to our understanding of the physiological functions of Lin28. Using bioinformatics approaches combined with in vitro binding and in vivo reporter analysis, we identify, in this report, a putative sequence and structural motif common to multiple Lin28 mRNA targets that confers specificity of Lin28-mediated regulation of translation. We also provide evidence that binding of Lin28 to miRNA precursors or mRNAs is associated with recruitment of different auxiliary factors, resulting in distinct regulatory outcomes.
MATERIALS AND METHODS
Antibodies and plasmids
The antibodies specific for beta-tubulin (Abcam, ab6046) and Flag (Santa Cruz, sc-807) were purchased. The FL-Lin28 and FL-Lin28ΔC expression vectors were previously described (3,17). To create FL-CCHCmut, a human Lin28 expression vector containing the indicated CCHC point mutations was used as a PCR template and the resulting PCR fragment was inserted into pFLAG-CMV-2 (Sigma, E7398) at the NotI and BamHI sites. The luciferase reporter construct Oct4-95 was previously described (3). Constructs Oct4-14T, Oct4-ΔA, RPS19-106, RPS19-75T, RPS19-3xT, HMGA1-129 and HMGA1-53T were made by cloning chemically synthesized double-stranded DNA oligos containing the indicated human gene sequences (wild-type or mutant) at the NotI and XhoI sites of the parental firefly luciferase reporter vector (3). Constructs HER2-200 and mHMGA1-133 were created by cloning PCR fragments containing the 200- and 133-nt sequences from human HER2 (NM_004448.2) and mouse HMGA1 (NM_016660.3), respectively, at the NotI and XhoI sites of the firefly luciferase reporter vector.
Cell culture and transfection
The culture and transfection of the PA-1 and HEK293 cells were carried out as preciously described (3,16).
In vitro transcription
To make a T7 promoter-containing double-stranded DNA template for in vitro transcription to generate the human pre-let-7a-1 RNA, a T7 adapter oligo (5′-gcgttaatacgactcactatagggcTGAGGTAGTAGGTTGTATAG-3′) was annealed to a longer miRNA oligo (5′-GGAAAGACAGTAGATTGTATAGTTATCTCCCAGTGGTGGGTGTGACCCTAAAACTATACAACCTACTACCTCAgccctatagtgagtcgtattaacgc-3′), followed by PCR extension using Pfu DNA polymerase. The PCR reaction was conducted at 95°C for 5 min, 59°C for 1 min and 72°C for 10 min, 1 cycle. To generate T7 DNA templates for in vitro transcription to produce Oct4-95, Oct4-14T, HMGA1-129 and HMGA1-53T RNA, PCR reactions were carried out using the following sets of primers: forward 5′-gcgttaatacgactcactatagggcTCCTGAAGCAGAAGAGGA-3′ and reverse 5′-GGCAGATGGTCGTTTGGCTGAATAC-3′ for Oct4-95, Oct4-14T and Oct4-ΔA; and forward 5′-gcgttaatacgactcactatagggcGGACGGCACTGAGAAGCG-3′ and reverse 5′-GGTCGGCCCCGAGGTCTCTTAGGTG-3′ for HMGA1-129 and HMGA1-53T, using the corresponding luciferase reporter constructs as templates. Resulting PCR fragments were gel purified and used to produce the respective RNAs using MEGAscript T7 (Ambion, AM1334) as previously described (16,17).
Electrophoretic mobility shift analysis
Binding reactions were in a total volume of 10 µl using 1 × 105 cpm of internally 32P-UTP-labeled RNA probe, together with the indicated amounts of recombinant Lin28 (rLin28) expressed and purified as described previously (8). Binding buffer contained 1.2 µg/µl yeast tRNA, 1.2 mM MgCl2, 300 mM KCl, 3.4% glycerol, 1.2 mM DTT, 24 mM HEPES, pH 7.5 and 41 mg/ml heparin. Prior to adding rLin28, samples were heated at 90°C for 3 min, followed by cooling at 25°C for 5 min. Binding reaction was conducted at 25°C for 30 min. Then, 2 µl of 50% glycerol was added and native gel electrophoresis was conducted on a 5% polyacrylamide gel at 4°C and 12 V/cm for 50 min. Band intensities of scanned films were quantified using Totallab Quant software and the data processed using the GraphPad Prism program. The total amount of RNA probe in each binding reaction was normalized against the unbound probe (in the absence of rLin28) and used to calculate the fraction bound by rLin28. The dissociation constants of Oct4-95, pre-let-7a, Oct4-14T and HMGA1-129, were calculated using the equation: fraction bound = Bmax([rLin28])/(Kd + [rLin28]), with Bmax being the observed maximum fraction of probe bound, [rLin28] the protein concentration and Kd the dissociation constant. In the cases of Oct4-95, Oct4-14T and HMGA1-129, Complex 2 (in addition to Complex 1) formed at high rLin28 concentration, and signals of Complex 1 and 2 were combined to generate probe bound signals in Kd calculation.
RNA extraction and RT-qPCR
These were carried out essentially as previously described (3,16). The PCR primers are as follows: rps19 (NM_001022.3) forward 5′-AGACGTGAACCAGCAGGAGT, rps19 reverse 5′-AGCTCGCGTGTAGAACCAGT; her2 forward 5′-AGCACTGGGGAGTCTTTGTG, her2 reverse 5′-CTGAATGGGTCGCTTTTGTT. To measure let-7a miRNA levels in HEK293 cells, total RNAs were extracted from FL-Lin28, FL-CCHCmut, FL-Lin28ΔC or vector transfected cells using Qiagen miRNeasy Mini kit (catalog number 1038703), followed by RT-qPCR using miScript reverse transcription kit (catalog number 218061) and miScript SYBR Green PCR kit (catalog number 218073) according to the manufacturers’ instructions. Let-7a miRNA levels were normalized using internal control U6 small nuclear RNA (RNU6B-2) levels.
Luciferase assays
These were carried out as previously described (3).
RESULTS
Oct4 mRNA harbors a unique motif that mediates Lin28-dependent stimulation of translation
We have previously reported a Lin28-responsive element (LRE) of 95-nt from the open reading frame (ORF) of Oct4 mRNA that elicits Lin28-dependent stimulation of translation when present in the 3′-untranslated region (3′-UTR) of a luciferase reporter (3). LREs have been mapped mostly to the ORFs (3,14,16,17), and a few to the 5′-UTR (13) and 3′-UTRs (15) of Lin28 mRNA targets. Regulation of translation by elements located in the 3′-UTRs has been extensively investigated. However, emerging evidence points to the notion that translational regulation can also be conferred by elements present in the ORFs and 5′-UTRs of mRNAs and that some elements can regulate translation from the ORFs or 5′-UTRs as efficiently as from the 3′-UTRs via a similar mechanism (24–26). Thus, it is conceivable that LREs derived from ORFs of Lin28 mRNA targets exhibit translational regulatory effects when present at the 3′-UTR of a reporter gene (3,14,16,17).
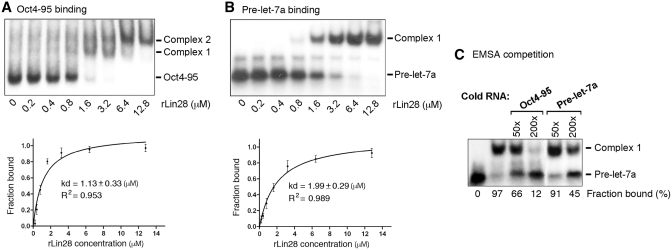
The mode of action of Lin28 in translation has been suggested: Lin28 binds to target mRNAs through recognition of LREs and subsequently recruits RNA helicase A (RHA) that plays an important role in facilitating RNP rearrangement during translation, thereby stimulating translation (3,14,16,17). To confirm a direct interaction between Lin28 and the 95-nt LRE (Oct4-95), we carried out electrophoretic mobility shift analysis (EMSA) with the Oct4-95 RNA and recombinant Lin28 protein (rLin28), using pre-let-7a RNA as a positive control. Figure 1 shows autoradiographs of non-denaturing gel electrophoresis of 32P-labeled Oct4-95 RNA (Figure 1A) or pre-let-7a RNA (Figure 1B) with increasing concentrations of rLin28. While only one protein–RNA complex (Complex 1, Figure 1B) formed with pre-let-7a, two complexes (Complex 1 and 2, Figure 1A) formed with Oct4-95 RNA, with Complex 2 forming at higher rLin28 concentrations, suggesting that more than one molecules of Lin28 may bind Oct4-95 at higher concentrations. To simplify the analysis, we combined Complex 1 and 2 as one complex in our Kd calculation. Thus, we estimated the Kd of Lin28 binding to the Oct4-95 RNA to be 1.13 µM (Figure 1A, bottom panel), and to the pre-let-7a to be 1.99 µM (Figure 1B, bottom panel). The slightly lower Kd value for Oct4-95 compared to pre-let-7 (which was reported to be 2.1 µM) (19) was also reflected by competition experiments showing that unlabeled Oct4-95 competed better for binding to Lin28 than the unlabeled pre-let-7a RNA (Figure 1C). Based on these findings we conclude that Lin28 binds Oct4-95 RNA directly and with an affinity similar to that of pre-let-7a RNA.
Figure 1.
Lin28 binds Oct4-95 in vitro. Autoradiographs of EMSA using labeled Oct4-95 (A) or pre-let-7a RNA (B) with increasing amounts of rLin28. The positions of unbound RNA– and RNA–protein complexes are marked to the right. Band intensities were quantified from three independent experiments and used to calculate the Kd values shown in the plots below. In the case of Oct4-95, Complex 1 and 2 were combined as one complex in Kd calculation. (C) Autoradiograph of EMSA performed with labeled pre-let-7a RNA and 3.2 µM of rLin28. The amounts of unlabeled competitor Oct4-95 or pre-let-7a RNA added in relative molar excess are indicated on the top. Band intensities were calculated and presented as percentage of fraction bound. Numbers are averages of two independent experiments.
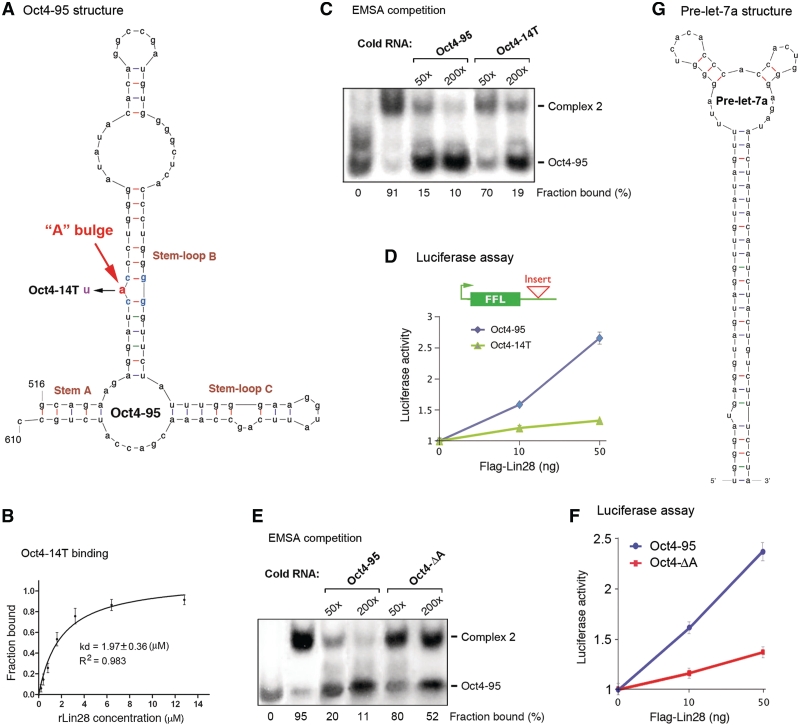
Our previous studies have suggested possible structural features involved in Lin28 recognition of its target mRNAs (3). Therefore, we employed the mFOLD program (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form) to predict the secondary structure of Oct4-95 RNA. This program has accurately predicted the secondary structures of various RNAs including the CTE (27–29), the RHA-binding elements (30) and the pre-let-7a (20). The mFOLD computation was executed at fixed conditions of 37°C with 1 M NaCl and no divalent ions. This algorithm predicted three potential structures that are highly similar to one another in that each contains a stem (Stem A) and two stem–loops (Stem–loop B and C) (Figure 2A). Importantly, this structure is maintained in a longer fragment (369 nt, previously called Oct4-R2, Supplementary Figure S1) shown to have an activity in Lin28-dependent stimulation of translation (3,16), suggesting that this structure may form and be functional in vivo in the context of full-length Oct4 mRNA. Strikingly, an additional characteristic that was initially ignored but was later found to be critical to Lin28 binding and function, is a bulged A residue (‘A’ bulge) flanked by a pair of G:C base pairs in one of the stem–loops (Figure 2A and Supplementary Figure S1). While an ‘A’ to ‘U’ mutation did not alter the secondary structure predicted by mFOLD, it did reduce the affinity for binding to Lin28 (compare Figure 2B to Figure 1A bottom panel). Also, the unlabeled Oct4-95 RNA competed better for binding to rLin28 compared to the unlabeled Oct4-14T RNA (Figure 2C). Importantly, the same point mutation resulted in the loss of Lin28-dependent stimulation of translation in vivo (Figure 2D). To further test the importance of the putative ‘A’ bulge, we deleted this unpaired nucleotide. As expected, the resulting 94-nt element (Oct4-ΔA) exhibited a reduced affinity for Lin28, as determined by the EMSA competition assay (Figure 2E). In addition, this mutant failed to induce Lin28-dependent translational stimulation (Figure 2F). Thus, the ‘A’ bulge may constitute one key recognition element for Lin28 to bind Oct4-95. The secondary structure of pre-let-7a (Figure 2G) was initially predicted by mFOLD and was subsequently validated by other methods including Footprinting (20). This secondary structure apparently differs from that of Oct4-95 (Figure 2, compare Figure 2A to G) despite their similar affinities for Lin28 (Figure 1).
Figure 2.
Structural characteristics of Oct4-95. (A) A predicted secondary structure of Oct4-95. The critical ‘A’ bulge is highlighted in red. Numbers are in nucleotides relative to the transcriptional start site of Oct4. (B) Quantification of Oct4-14T EMSA. Numbers were derived from three independent experiments. (C and E) Autoradiographs of EMSA competition experiments performed with labeled Oct4-95 RNA and 6.4 µM of rLin28. The amounts of unlabeled competitor Oct4-95, Oct4-14T or Oct4-ΔA RNA added in relative molar excess are indicated on the top. Band intensities were calculated and presented as percentage of fraction bound. Numbers are averages of two independent experiments. (D and F) Luciferase assays. Shown on top in D is a schematic of a firefly luciferase reporter construct with the green box representing its coding region and the thin line representing 3′-UTR. The position where Oct4-95, Oct4-14T or Oct4-ΔA was inserted is marked. The indicated reporter constructs were each transfected into HEK293 cells, with or without increasing amounts of FL-Lin28. Luciferase activities and mRNA levels were measured 24-h post-transfection. Relative luciferase activities were plotted after normalization against luciferase mRNA levels. Luciferase activities from cells without FL-Lin28 transfected were arbitrarily set as 1. Numbers are mean ± SD (n = 3). (G) Secondary structure of pre-let-7a predicted by mFOLD and validated by in vitro Footprint analysis (20).
The putative motif is present in multiple mRNA targets of Lin28
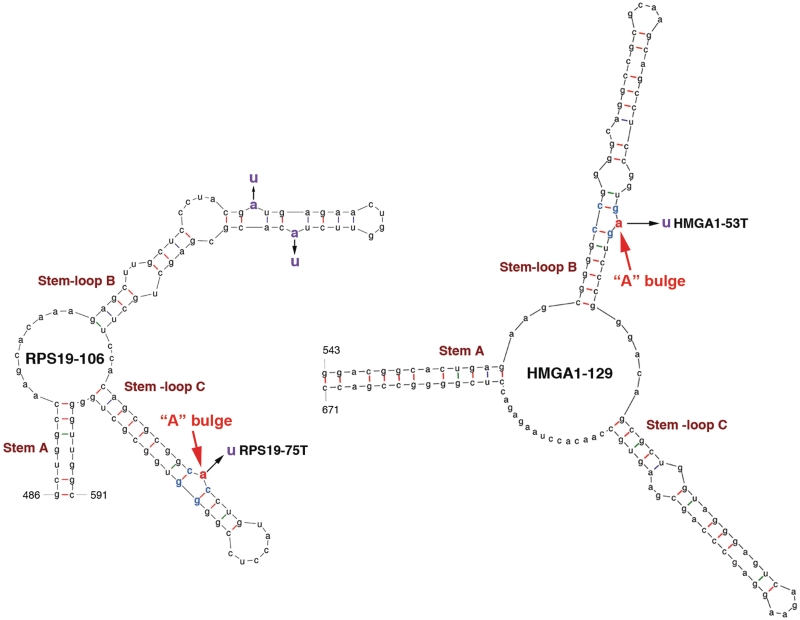
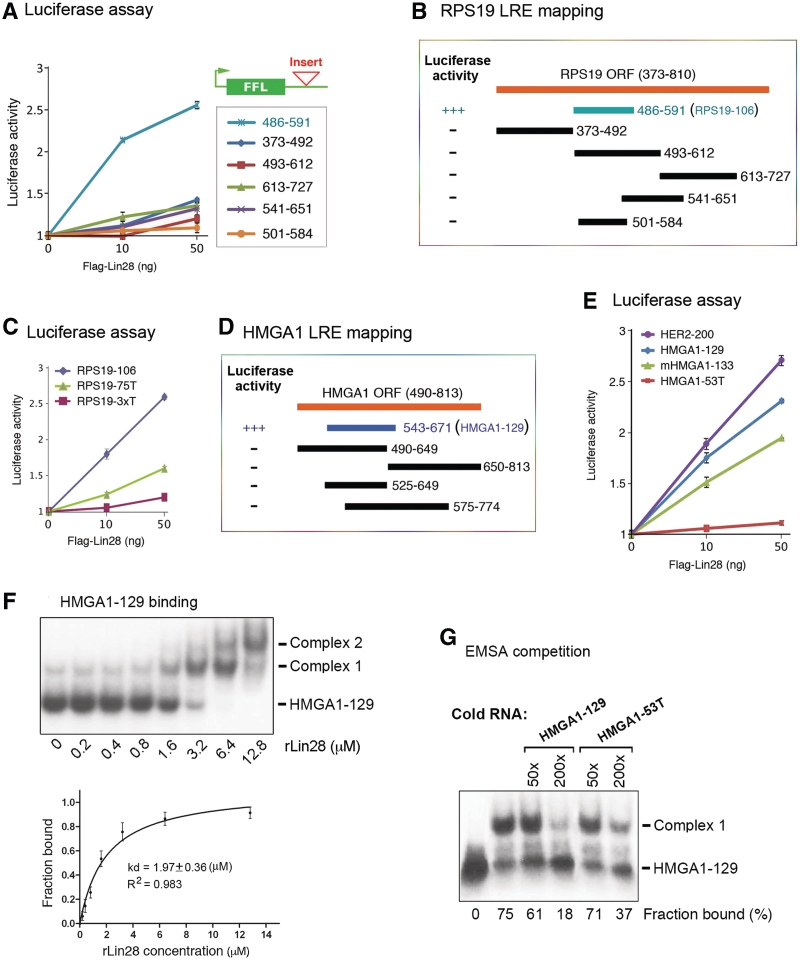
Inspired by the above observations, we decided to map LREs on other mRNA targets, with the expectation of obtaining minimal sequences in order to facilitate identification of consensus motifs. Five additional human genes (hmga1, eef1g, rps13, rps19 and her2) were chosen for further analysis. The HMGA1, EEF1G and RPS13 mRNAs have been validated as in vivo targets of Lin28 regulation at the translational level (3). Supplementary Figure S2 presents evidence that the RPS19 (ribosomal protein S19) and HER2 (human epidermal growth factor receptor 2) mRNAs are also in vivo targets of Lin28. To determine whether these mRNAs contain motifs similar to that in Oct4-95, we initially used an approach similar to that described before (3,16). Thus, luciferase reporters were constructed which incorporated RNA segments from the genes of interest. At first, long sequences were inserted, followed by stepwise shortening. This approach was next combined with sequence alignments between Oct4 mRNA and other target mRNAs and between species (bilateral animals). Despite exhaustive attempts, these approaches in every case failed to identify conserved sequence motifs essential for activity. This led us to an alternative approach in which we used mFOLD to identify common structural features, followed by reporter assays to evaluate their significance. Three criteria were used for our predictions: (i) the element contains three stem–loop structures; (ii) at least one of the stem–loops contains an ‘A’ bulge flanked by two G:C base pairs and (iii) the structure is predicted to exist in the parental full-length mRNA. First, we found 106-nt and 129-nt LREs in the ORFs of rps19 and hmga1, respectively. As shown in Figure 3, both the RPS19-106 and HMGA1-129 LREs contain the characteristic Stem A and Stem–loops B and C, with the putative ‘A’ bulge located in one of the Stem–loops. To test whether these LREs can act to mediate Lin28-dependent stimulation of translation, we inserted each of them at the 3′-UTR of the luciferase reporter (Figure 4A, top right corner) and performed luciferase analysis. While the 106-nt RPS19 element (nt 486-591) exhibited an activity of Lin28-dependent stimulation of translation, the various segments derived from other regions (based on our initial strategy) did not (Figure 4A and B). Importantly, a single ‘A’ to ‘U’ mutation at the putative ‘A’ bulge (without altering the secondary structure predicted by mFOLD) significantly reduced the activity of this element to stimulate translation (Figure 4C, compare green line with blue line), and simultaneous mutations of the three ‘A’ nucleotides further reduced the activity (compare red line with green line). Likewise, the HMGA1-129 element (Figure 4D, blue bar, nt 543-671) has a readily detectable stimulatory activity in the reporter assay, in contrast to the segments derived from other regions of the human HMGA1 coding region (Figure 4D, black bars). Again, a single ‘A’ to ‘U’ mutation (HMGA1-53T, Figure 3) without altering the secondary structure abolished its ability to stimulate translation (Figure 4E, compare red line with blue line). Like Oct4-95, HMGA1-129 forms two complexes at high rLin28 concentrations, with an apparent Kd of 1.97 (Figure 4F). As expected, the mutant HMGA1-53T exhibited slightly reduced affinity for rLin28 compared to HMGA1-129 as assessed by the EMSA competition analysis (Figure 4G).
Figure 3.
Predicted secondary structures of LREs from RPS19 and HMGA1 mRNAs. The numbers at the bases of Stem A are in nucleotides relative to the transcriptional start sites of the respective genes. The putative ‘A’ bulges are highlighted in red. The simultaneous replacements of the three ‘A’ nucleotides to ‘U’ in RPS19-106 resulted in RPS19-3xT (see Figure 4C).
Figure 4.
The predicted LREs are functional. (A) Luciferase assays with reporter genes containing the indicated segments from the RPS19 open-reading frame (ORF). Numbers are in nucleotides relative to the transcriptional start site of RPS19. (B) RPS19 LRE mapping. Luciferase activities of the corresponding fragments are presented to the left. +++ and − represent positive and negative translational stimulatory activity, respectively. The blue bar labeled with 486–591 (RPS19-106) was predicted by mFOLD, while the black bars represent fragments derived from random chopping. (C) Luciferase assays using wild-type and mutant RPS19 LREs. (D) Human HMGA1 LRE mapping. The blue bar labeled with 543–671 (HMGA1-129) was predicted by mFOLD. (E) Luciferase assays of human HER2 and HMGA1 (wild-type and mutant), and mouse HMGA1 LREs. (F) Autoradiographs of EMSA using labeled HMGA1-129 with increasing amounts of rLin28. The positions of unbound RNA– and RNA–protein complexes are marked to the right. Band intensities were quantified from three independent experiments and used to calculate the Kd values shown in the plots below. The signals of Complexes 1 and 2 were combined as one in Kd calculation. (G) Autoradiograph of EMSA performed with labeled HMGA1-129 and 3.2 µM of rLin28. The amounts of unlabeled competitor HMGA1-129 or HMGA1-53T RNA added in relative molar excess are indicated on the top. Band intensities were calculated and presented as percentage of fraction bound. Numbers are averages of two independent experiments.
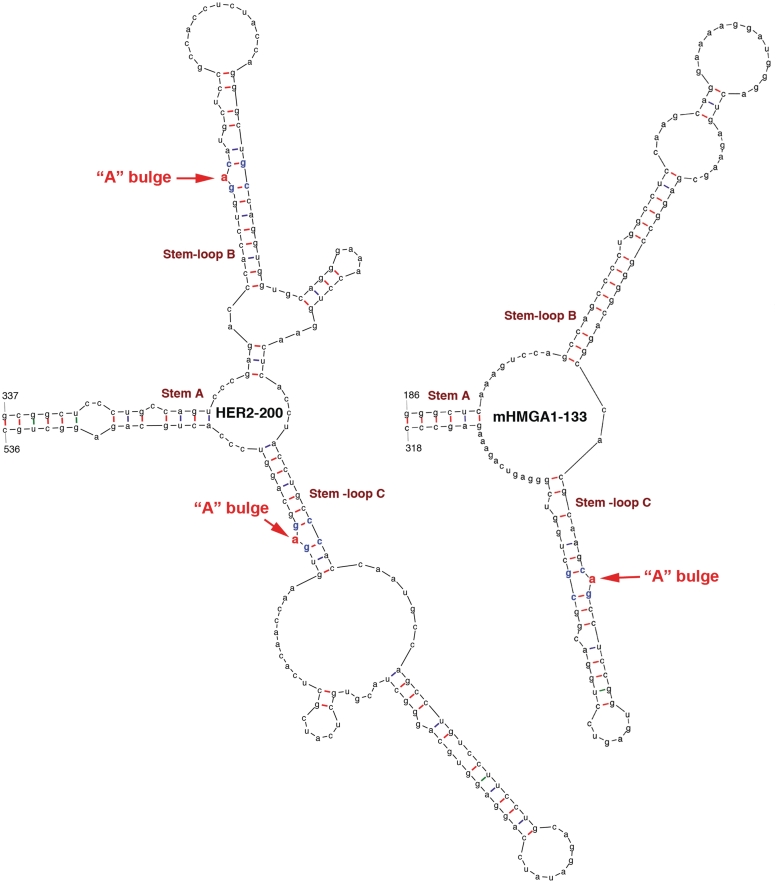
The power of this novel approach was further underscored by our ability to predict a 200-nt long LRE (HER2-200, Figure 5) from the 3767-nt long coding region of HER2 mRNA, which was tested positive in our reporter assay (Figure 4E, purple line). This structure is somewhat larger and more complex than those of Oct4-95, RPS19-106 and HMGA1-129. Moreover, the EEF1G-330 (previously called EEF1G-R3) and RPS13-456 (previously called RPS13-ORF) fragments (3) from the coding regions of EEF1G and RPS13 mRNAs, respectively, also contain the characteristic ‘A’ bulges as predicted by mFOLD (Supplementary Figures S3 and S4). Importantly, these two elements were also shown to be active in stimulating translation (3). To ask whether our prediction strategy could be extrapolated to other species, we tested the mouse HMGA1 gene. We predicted a 133-nt LRE in the coding region of this mRNA (Figure 5) that elicited a translational stimulatory activity (Figure 4E, green line). We were struck by the fact that the sequence of this LRE lies in a region that only partially overlaps with a region predicted to have the highest homology to the human HMGA1 LRE. This may explain why we could not find LREs based on sequence alignments.
Figure 5.
Predicted secondary structures of LREs from human HER2 and mouse HMGA1 mRNAs. The numbers at the bases of Stem A are in nucleotides relative to the transcriptional start sites of the respective genes. The putative ‘A’ bulges are highlighted in red.
Mutations in the CCHC domain impair the ability of Lin28 to stimulate translation
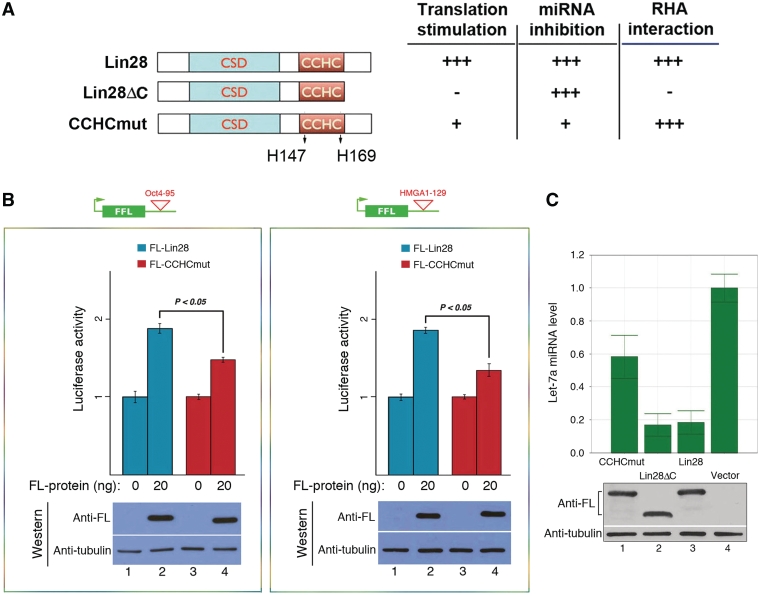
Lin28 contains a pair of retroviral-type CCHC zinc fingers shown to be important for mRNA binding (31) and inhibition of let-7 production (4,19,23). Specifically, when the two histidine residues were mutated to alanines (CCHCmut, Figure 6A, left panel), the mutant protein lost its ability to bind RNA (both mRNA and pre-let-7 miRNA) and to inhibit pre-let-7 processing/uridylation (4,23,31). We therefore postulated that the same mutations would reduce the ability of Lin28 to stimulate target mRNA translation due to impaired RNA-binding. Indeed, the CCHCmut protein exhibited a significantly reduced activity (∼50% reduction) in stimulating translation of the reporter genes containing either the Oct4-95 (Figure 6B, left panel) or the HMGA1-129 element (Figure 6B, right panel). Consistent with previous reports (4,23,31), this mutant protein also displays decreased activity in inhibiting let-7 miRNA production (Figure 6C, compare lane 1 to lane 3). Intriguingly, the C-terminal deletion of Lin28 (Lin28ΔC) does not affect its ability to inhibit let-7 production (Figure 6C, compare lane 2 to lane 3). Lin28ΔC is capable of RNA-binding but is unable to stimulate target mRNA translation, partly due to its inability to interact with RNA helicase A (RHA) (3,17). The results summarized in the right panel of Figure 6A thus suggest that the mechanisms underlying Lin28-mediated inhibition of let-7 production and stimulation of target mRNA translation are likely distinct. It is possible that binding of Lin28 to different RNA targets (i.e. precursor miRNAs versus mRNAs) is associated with recruitment of different auxiliary factors, perhaps through induced protein conformational changes following RNA binding. Such a mechanism could underlie distinct physiological functions of Lin28 (Figure 7).
Figure 6.
Differential Lin28 domain effects on translation and miRNA processing. (A) Schematics of wild-type and mutant Lin28 proteins. The two histidine residues in the CCHC domain that were replaced with alanines are marked. The effects of the mutant proteins on translational stimulation, let-7 inhibition and RHA interaction are summarized to the right. +++, 100% activity; +, ∼50% activity; −, <10% activity. (B) HEK293 cells were transfected with the reporter construct containing Oct4-95 (left panel) or HMGA1-129 (right panel), with or without co-transfection of FL-Lin28 or FL-CCHCmut. Luciferase activities were measured 24 h following transfection. Luciferase activities in the absence of the FL-protein were set as 1. Numbers are mean ± SD (n = 3). Underneath are western blot gels showing expression levels of the FL-proteins. (C) FL-tagged CCHCmut (lane1), Lin28ΔC (lane 2), wild-type Lin28 (lane 3) or empty vector (lane 4) was transfected into HEK293 cells. RNAs were isolated 48 h later and mature let-7a miRNA levels determined by RT-qPCR. The let-7a miRNA level in vector-transfected cells was set as 1. Numbers are mean ± SD (n = 3). Underneath are western blot gels showing comparable expression levels of the indicated Lin28 proteins.
Figure 7.
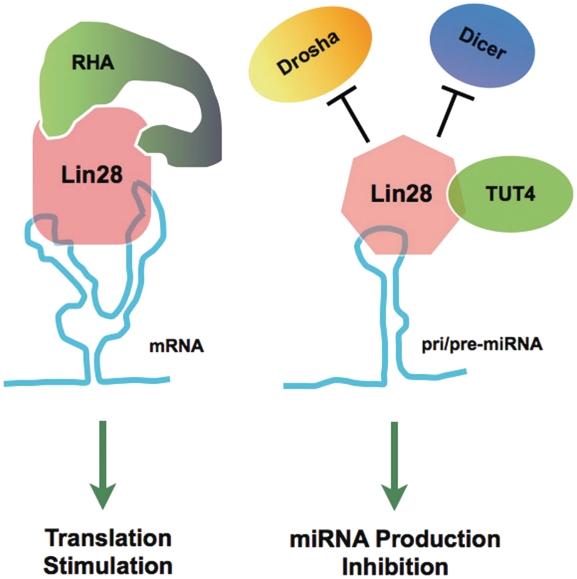
A model for differential regulation effects mediated by Lin28. Binding of Lin28 to a target mRNA through recognition of an LRE leads to the recruitment of co-factor RHA, hence stimulation of translation of the mRNA (3,13,15–17). On the other hand, binding of Lin28 to a pri/pre-miRNA through recognition of a different sequence motif prevents the association of the Drosha and Dicer complexes, and triggers the recruitment of the uridylating enzyme TUT4, which together results in a decreased production of mature miRNA (4,7,8,11,18–23).
DISCUSSION
We have identified a putative structural motif in multiple Lin28 mRNA targets that likely plays a critical role in Lin28-dependent regulation of translation. The motif is characterized by a small ‘A’ bulge flanked by two G:C base pairs in a context of a larger secondary structure. This motif was not immediately evident by inspection of sequence alignments between target genes (and LREs from target genes) and across species, or by the commonly used systematic and stepwise sequence narrowing down approach. Rather, it emerged through analysis of predicted folding patterns of sequences based on a unique set of criteria, a product of initial visual inspection at folded structures and sequences. The critical role of a single nucleotide bulge in specific RNA–protein interaction is not unprecedented. The bacteriophage R17 coat protein binds specifically to a hairpin structure in the translational operator of the replicase gene, which is required for inhibition of translation. An unpaired A residue in the hairpin stem region was found to be essential for the interaction, as deletion of this residue or substitution with a cytosine resulted in a drastic decrease in the affinity of the hairpin RNA to the coat protein (32). In the case of the iron-responsive element binding protein (IRE-BP), while formation of multiple contacts of the protein with its cognate RNA is required for high affinity binding, deletion or mutation of a single bulged C residue in the RNA hairpin stem region reduced its affinity to the protein by ∼400-fold (deletion) or ∼30- to 70-fold (mutation) (33).
While evident in all transcripts tested so far, there is possibility that this ‘A’ bulge feature may not be apparent in every Lin28 mRNA targets. This would not be surprising since Lin28 also seems to recognize discrete sequence motifs in the terminal loop (TL) of pri/pre-let-7 miRNAs that share no clear consensus with one another (4,8,19). In addition, the sequence and secondary structure of pre-let-7a (Figure 2G) differ from those of the LREs we have identified (Figures 2, 3 and 5). This is reminiscent of KSRP that acts both to regulate splicing and to mediate the degradation of AU-rich element (ARE)-containing mRNAs (34). It has also been reported to promote the biogenesis of a subset of miRNAs including let-7 (11). While this protein can bind different ARE-containing mRNAs in the context of large and structured RNA 3′-UTRs (35), it also exhibits a high affinity for G-rich stretches that are present in the TL of some, but not all, of its target precursor miRNAs (11). Likewise, hnRNP A1 participates in many aspects of mRNA metabolism but is also a negative regulator of pri-let-7 processing by competing with KSRP for binding to the TL of pri-let-7 (20). Thus, Lin28 may have evolved to recognize a variety of sequence motifs in different structural environments, constituting a mechanistic base for the multitude of functions regulated by this protein.
Lin28 has been proposed to compete with KSRP for binding to the TL, thereby blocking let-7 processing (11). However, with a Kd of ∼2 µM to the TL [this report and (8,19,36)], how could Lin28 compete in vivo with KSRP which has a Kd of ∼50 nM (11)? Also, how could the small affects of the mutations in the Lin28 LREs seen in vitro potentially have large biological effects in vivo? One possible explanation for this paradox is that many of the in vitro studies [this report and (8,19,36)] may have underestimated the affinity of Lin28 for the RNAs due to sub-optimal experimental conditions. Desjardins et al. (37) recently reported a Kd of 0.15 nM with Lin28 for pre-let-7 and they attributed this discrepancy to the absence of RNA competitor in their binding buffer and elimination of RNA contaminants during Lin28 protein purification. In addition, due to limitations in vitro (i.e. using purified proteins and short RNA elements) many biological effects may not be fully recapitulated. In vivo interactions between protein co-factors and neighboring binding sites can produce synergistic effects, leading to dramatic biological endpoints. Future studies aimed at analyzing the detailed and higher order structures of Lin28 LREs in complex with Lin28 will greatly aid in our understanding of mRNA target regulation by Lin28 in both stem cells and cancer cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4.
FUNDING
Connecticut Stem Cell Research (grant 09SCAYALE14). The contents of this material are solely the responsibility of the authors and do not necessarily represent the official views of the State of Connecticut, the Department of Public Health of the State of Connecticut or Connecticut Innovations, Incorporated. Funding for open access charge: Connecticut Innovations, Incorporated.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Richard Hochberg for critical reading of this manuscript.
REFERENCES
- 1.Darr H, Benvenisty N. Genetic analysis of the role of the reprogramming gene LIN-28 in human embryonic stem cells. Stem Cells. 2009;27:352–362. doi: 10.1634/stemcells.2008-0720. [DOI] [PubMed] [Google Scholar]
- 2.Richards M, Tan SP, Tan JH, Chan WK, Bongso A. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22:51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- 3.Peng S, Chen L-L, Lei X-X, Yang L, Lin H, Carmichael GG, Huang Y. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells. 2011;29:496–504. doi: 10.1002/stem.591. [DOI] [PubMed] [Google Scholar]
- 4.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O'Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng S, Maihle NJ, Huang Y. Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene. 2010;29:2153–2159. doi: 10.1038/onc.2009.500. [DOI] [PubMed] [Google Scholar]
- 7.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell. Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 8.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dangi-Garimella S, Yun J, Eves EM, Newman M, Erkeland SJ, Hammond SM, Minn AJ, Rosner MR. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009;28:347–358. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of MicroRNA processing by Lin-28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 13.Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu B, Huang Y. Histone H2a mRNA interacts with Lin28 and contains a Lin28-dependent posttranscriptional regulatory element. Nucleic Acids Res. 2009;37:4256–4263. doi: 10.1093/nar/gkp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu B, Zhang K, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009;15:357–361. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu C, Ma Y, Wang J, Peng S, Huang Y. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38:1240–1248. doi: 10.1093/nar/gkp1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin J, Jing W, Lei XX, Feng C, Peng S, Boris-Lawrie K, Huang Y. Evidence that Lin28 stimulates translation by recruiting RNA helicase A to polysomes. Nucleic Acids Res. 2011;39:3724–3734. doi: 10.1093/nar/gkq1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, Pasquinelli AE. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2011;18:302–308. doi: 10.1038/nsmb.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 20.Michlewski G, Caceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 24.Moretti F, Thermann R, Hentze MW. Mechanism of translational regulation by miR-2 from sites in the 5′ untranslated region or the open reading frame. RNA. 2010;16:2493–2502. doi: 10.1261/rna.2384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schnall-Levin M, Rissland OS, Johnston W, Perrimon N, Bartel DP, Berger B. Unusually effective microRNA targeting within repeat-rich coding regions of mammalian mRNAs. Genome Res. 2011;21:1395–1403. doi: 10.1101/gr.121210.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnall-Levin M, Zhao Y, Perrimon N, Berger B. Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3′-UTRs. Proc. Natl Acad. Sci. USA. 2010;107:15751–15756. doi: 10.1073/pnas.1006172107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst RK, Bray M, Rekosh D, Hammarskjöld ML. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol. Cell. Biol. 1997;17:135–144. doi: 10.1128/mcb.17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang Y, Bogerd HP, Yang H, Cullen BR. Analysis of the RNA binding specificity of the human tap protein, a constitutive transport element-specific nuclear RNA export factor. Virology. 1999;262:200–209. doi: 10.1006/viro.1999.9906. [DOI] [PubMed] [Google Scholar]
- 29.Gatfield D, Le Hir H, Schmitt C, Braun IC, Köcher T, Wilm M, Izaurralde E. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 2001;11:1716–1721. doi: 10.1016/s0960-9822(01)00532-2. [DOI] [PubMed] [Google Scholar]
- 30.Roberts TM, Boris-Lawrie K. Primary sequence and secondary structure motifs in spleen necrosis virus RU5 confer translational utilization of unspliced human immunodeficiency virus type 1 reporter RNA. J. Virol. 2003;77:11973–11984. doi: 10.1128/JVI.77.22.11973-11984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balzer E, Moss EG. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4:16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- 32.Wu H-N, Uhlenbeck OC. Role of a bulged A residue in a specific RNA-protein interaction. Biochemistry. 1987;26:8221–8227. doi: 10.1021/bi00399a030. [DOI] [PubMed] [Google Scholar]
- 33.Jaffrey SR, Haile DJ, Klausner RD, Harford JB. The interaction between the iron-responsive element binding protein and its cognate RNA is highly dependent upon both RNA sequence and structure. Nucleic Acids Res. 1993;21:4627–4631. doi: 10.1093/nar/21.19.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gherzi R, Lee K-Y, Briata P, Wegmüller D, Moroni C, Karin M, Chen C-Y. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Mayoral MF, Hollingworth D, Masino L, Díaz-Moreno I, Kelly G, Gherzi R, Chou CF, Chen CY, Ramos A. The structure of the C-terminal KH domains of KSRP reveals a noncanonical motif important for mRNA degradation. Structure. 2007;15:485–498. doi: 10.1016/j.str.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Lightfoot HL, Bugaut A, Armisen J, Lehrbach NJ, Miska EA, Balasubramanian S. A LIN28-dependent structural change in pre-let-7g directly inhibits dicer processing. Biochemistry. 2011;50:7514–7521. doi: 10.1021/bi200851d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desjardins A, Yang A, Bouvette J, Omichinski JG, Legault P. Importance of the NCp7-like domain in the recognition of pre-let-7g by the pluripotency factor Lin28. Nucleic Acids Res. 2012;40:1767–1777. doi: 10.1093/nar/gkr808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.