Abstract
Protein–RNA interactions are essential for many biological processes. However, the structural mechanisms underlying these interactions are not fully understood. Here, we analyzed the protein surface shape (dented, intermediate or protruded) and the RNA base pairing properties (paired or unpaired nucleotides) at the interfaces of 91 protein–RNA complexes derived from the Protein Data Bank. Dented protein surfaces prefer unpaired nucleotides to paired ones at the interface, and hydrogen bonds frequently occur between the protein backbone and RNA bases. In contrast, protruded protein surfaces do not show such a preference, rather, electrostatic interactions initiate the formation of hydrogen bonds between positively charged amino acids and RNA phosphate groups. Interestingly, in many protein–RNA complexes that interact via an RNA loop, an aspartic acid is favored at the interface. Moreover, in most of these complexes, nucleotide bases in the RNA loop are flipped out and form hydrogen bonds with the protein, which suggests that aspartic acid is important for RNA loop recognition through a base-flipping process. This study provides fundamental insights into the role of the shape of the protein surface and RNA secondary structures in mediating protein–RNA interactions.
INTRODUCTION
Macromolecular interactions, such as protein–protein, protein–DNA and protein–RNA interactions, are critical for many biological processes (1–4). Studies regarding the mechanisms that are involved in these interactions provide fundamental insight into the intracellular networks that regulate cellular functions. Although significant progress has been made in dissecting the mechanisms that underlie protein–protein and protein–DNA interactions, the molecular basis of protein–RNA interactions remains poorly understood.
Recently, the 3D structures of a large number of protein–RNA complexes were determined, and the resulting atomic coordinates were deposited into the Protein Data Bank (PDB) (5). These coordinates have facilitated the analysis of the structural and chemical features of protein–RNA interfaces in terms of the interacting area, composition and intermolecular bonds. Most studies consistently showed that positively charged amino acids in the protein favor the phosphate groups in the RNA due to the electrostatic interaction at the interface (6–9). However, other features at the interface, including the preferred nucleotide bases, preferred amino acid–nucleotide combinations and the least favored amino acids, were inconsistent in these studies (8). Furthermore, most of these studies did not account for the shape of the protein's surface or the RNA secondary structure, although these features have been shown to be important in specific protein–RNA interactions. Sonavane et al. (9) reported that intruded protein surfaces prefer nucleotide bases at the interface. Ray et al. (10) developed the ‘RNAcompete’ assay, a microarray-based in vitro method for estimating the binding specificities of various proteins and RNAs, and demonstrated that the RNA binding protein Vts1 specifically interacts with stem-loop RNAs, whereas the protein HuR recognizes only the non-stem–loop RNAs. Thus, it is clear that the shape of the protein surface and the secondary structure of the RNA molecule are crucial for protein–RNA interactions. Here, we report novel binding patterns that exist between specific protein surfaces and RNA nucleotides and describe the mechanisms underlying these interactions.
MATERIALS AND METHODS
Data set
As of 11 November 2010, the PDB listed 824 structures of protein–RNA complexes that had been solved using X-ray crystallography (5). From this data set, 344 complexes were selected based on the following criteria: (i) structural resolution better than 3.0 Å and (ii) polypeptides and polyribonucleotides longer than 20 amino acids and 5 nt, respectively. Ribosomal subunits were excluded from the data set because their component proteins include a large number of amino acid residues at the interface, which could lead population bias. To avoid redundancy in our data set, a standalone PISCES package was used to select a single structure with the best resolution in cases where proteins in different complexes had >30% sequence identity (with all other options set to their default) (11). After processing with PISCES, 122 non-redundant complexes were obtained. It was necessary to distinguish between the biological interactions and the crystal contacts in these non-redundant complexes because the coordinates of the X-ray structure in these complexes are formatted as an asymmetric unit, which does not always correspond to a biological unit that represents a functional form of the molecule, and because some of the asymmetric units contain crystallographic protein–RNA interfaces (i.e. crystal contacts) that can cause erroneous identification of the interaction. For each asymmetric unit, we chose a stable biological assembly that corresponded to PQS annotation (Remark 350) in PDB file using PDBePISA (12). However, for the structures having multiple copies of proteins and RNAs in the asymmetric unit, we chose a representative assembly. The resulting data set included 91 non-redundant complexes (Supplementary Table S1). Although these 91 non-redundant complexes included a few pairs of highly similar RNA structures (1DFU-1FEU, 1B23-1U0B, 2F8S-2ZI0-2ZKO) and homopolymers (poly-A: 2PO1, 2XGJ, poly-U: 2J0S, 3FHT, 3I5X), the protein component of each of these complexes was non-redundant.
Amino acid categorization based on the shape of the protein surface
The protein residues were categorized into three surface groups (dented, intermediate or protruded) using a CX algorithm (13,14) that was modified to estimate the atoms in both protruded and dented protein surfaces (Supplementary Figure S1). The modified CX value was defined as (Vext–Vint)/Vsphere, in which Vint is an occupied volume of non-hydrogen atoms within a fixed distance R, Vext is the remaining volume of the sphere and Vsphere is the total volume of the sphere. The Vint was calculated as Natom×Vatom, where Natom is the number of non-hydrogen atoms in the sphere, and Vatom is the average volume of a non-hydrogen atom. Vatom and R were set to 20.1 Å3 and 12 Å, respectively. The CX value of each atom was calculated only when the atom was located on the protein surface with an accessible surface area (ASA) that exceeded 1.0 Å2. ASA was calculated by the NACCESS program with default parameters (15). The CX value of each residue was estimated as the sum of the CX values of its component atoms, and based on this sum, the calculated CX values of the residues were categorized into the following three surface groups: dented (CX < −0.5), intermediate (−0.5 ≤ CX ≤ 0.5) or protruded (CX > 0.5).
Nucleotide categorization based on the secondary structure of the RNA molecule
Each nucleotide in the RNA structure was categorized as either a paired or an unpaired nucleotide based on its base pairing property. The paired nucleotides that form base pairs were identified by the RNAView program (16). The base pairs were categorized into 12 families depending on the types of interacting edges (Watson–Crick/Watson–Crick, Watson–Crick/Hoogsteen, Watson–Crick/Sugar, Hoogsteen/Hoogsteen, Hoogsteen/Sugar, Sugar/Sugar) and the orientation of glycosidic bonds (cis or trans) as described previously (17). The remaining nucleotides were categorized as unpaired nucleotides.
Identification of the interface and hydrogen bonds
Protein–RNA interfaces were defined when the distance between the closest atom in the amino acid–nucleotide pair was <5.0 Å. Intermolecular hydrogen bonds between the amino acid moieties (the main chain and 20 side chains) and the nucleotide moieties (the ribose, phosphate and bases) were identified using the HBplus program with default parameters (maximum donor–acceptor distance: 3.9 Å, maximum hydrogen-acceptor distance: 2.5 Å) (18). These bonds were then counted and categorized into the following six groups based on the surface shape of the protein and the RNA base pairing property: dented-unpaired, dented-paired, intermediate-unpaired, intermediate-paired, protruded-unpaired and protruded-paired.
RESULTS
Data set and statistics
Our data set comprised 91 non-redundant protein–RNA complexes that contained a total of 35 783 amino acids and 3440 nucleotides. The amino acids were first segregated according to their location in the protein molecule, and only the amino acids that were located at the surface of the protein were selected using the NACCESS program. The selected amino acids were then categorized into three groups based on the surface shape of their location (dented, intermediate or protruded). We identified 30 726 amino acids at the protein surface, of which 44, 31 and 25% belonged to the dented, intermediate and protruded groups, respectively (Supplementary Table S2). We also observed a correlation between the chemical properties of these amino acids and their location. For example, most of the hydrophobic amino acids, such as leucine and valine, were located on a dented or intermediate surface, whereas the incidence of charged amino acids, such as lysine and glutamic acid, was highest on protruded protein surfaces. Similarly, the RNA nucleotides were divided into two groups based on their base pairing properties (paired or unpaired). The paired and unpaired groups accounted for 63 and 37% of the nucleotides, respectively (Supplementary Table S3). The frequency of base pair formation appeared to correlate with the nucleotide base. Cytosine and guanine were frequently present in paired nucleotides, whereas adenine was most commonly found in unpaired nucleotides.
We next identified the amino acids and nucleotides that were located at the protein–RNA interface (defined as a distance <5 Å) in these 91 non-redundant complexes. A total of 3791 amino acids were found at the interface, among which 42% were located on a dented surface, and 30 and 28% were located on intermediate and protruded surfaces, respectively (Supplementary Table S2). Regardless of the surface shape, positively charged amino acids, such as lysine (12%) and arginine (14%), were most frequently observed at the interface. Similarly, among the 1517 nt identified at the interface, 56 and 44% were paired and unpaired nucleotides, respectively (Supplementary Table S3).
We also counted the intermolecular hydrogen bonds at each interface and identified a total of 1949 that were formed between 1323 amino acids and 894 nt. Forty percent of these amino acids formed hydrogen bonds on a dented surface, and 29 and 31% formed hydrogen bonds on intermediate and protruded surfaces, respectively (Supplementary Table S2). Similarly, the proportion of paired and unpaired nucleotides involved in hydrogen bond formation was 49 and 51%, respectively (Supplementary Table S3).
Protein surface shape in RNA recognition
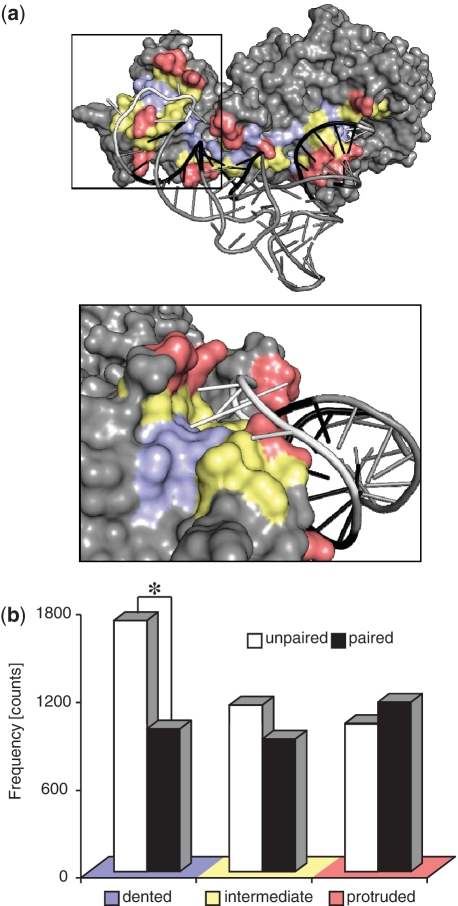
To ascertain whether a particular surface shape in the protein can identify the interacting RNA, the protein–RNA interfaces were first separated into the following three groups: protruded, intermediate and dented. Depending on the type of RNA base pairing, each of these groups was further subdivided into unpaired and paired groups, yielding in a total of six groups (Figure 1a). The frequency of amino acid–nucleotide pairs (distance <5 Å) within these six groups of protein–RNA interfaces was then calculated. The frequency of amino acid–nucleotide pairs at dented-unpaired interfaces (1714 pairs) was significantly higher than that at dented-paired interfaces (975 pairs), which suggests that a dented surface is able to distinguish unpaired and paired nucleotides at the interface. In contrast, the differences in the frequencies of amino acid nucleotide pairs at intermediate-unpaired and intermediate-paired (1138 versus 907 pairs, respectively) and protruded-unpaired and protruded-paired (1014 versus 1159 pairs, respectively) interfaces were marginal. Similar results were obtained from an analysis of the amino acid–nucleotide pairs that form hydrogen bonds at these six groups. The frequency of hydrogen bonded pair at dented-unpaired interfaces was 433 compared to 175 at dented-paired interfaces (Supplementary Figure S2).
Figure 1.
Analysis of the protein surface shapes and RNA base pairing properties at the interfaces of 91 non-redundant protein–RNA complexes. (a) The 3D structure of a representative complex (tRNA/aminoacyl tRNA synthetase complex; PDBID:1ASY) and a magnified view of the interface from the left side are shown. (b) The frequency of amino acid–nucleotide pairs at the indicated six interface groups based on the surface shape of the interacting protein and the base pairing property of the partner RNA. Dented, intermediate and protruded surfaces at the interface are shown in blue, yellow and red, respectively. Unpaired and paired nucleotides at the interface are shown in white and black, respectively. The frequency of amino acid–nucleotide pairs at six interface groups was counted for each complex. The resultant frequency distributions, obtained from all the 91 complexes, were statistically analyzed using t-test to determine the significance of the difference. Asterisk indicates P < 0.05.
Amino acid composition and RNA base pairing properties in protein–RNA recognition
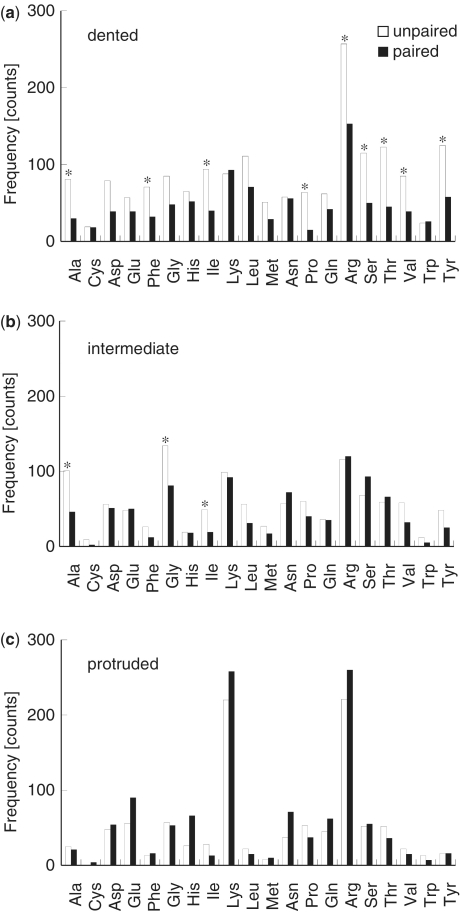
To investigate the role of the amino acids in protein–RNA binding at the interface, the frequency of each amino acid–nucleotide pair (20 amino acids and paired or unpaired nucleotides) at the three types of protein surfaces was calculated (Figure 2). At dented surfaces, a wide variety of amino acids, such as hydrophobic (Ala, Val), aromatic (Phe, Tyr) and charged (Arg) residues, showed a significant preference for unpaired nucleotides than for paired nucleotides (Figure 2a). In contrast, only a few amino acids (Ala, Gly and Ile) showed a significant preference for unpaired nucleotides at intermediate surfaces (Figure 2b), and none of the amino acids showed such preference for unpaired nucleotides at protruded surfaces (Figure 2c). These results suggest that amino acids prefer unpaired nucleotides at dented surfaces, most likely because unpaired nucleotides can easily accommodate in the dented region of the protein, whereas the base pairing between the paired nucleotides makes it difficult for them to be pushed down in such regions. Moreover, because this preference exists for a wide variety of amino acids, it is likely that the interactions at a dented surface frequently occur between the protein backbone and the RNA base.
Figure 2.
The frequency distributions of the amino acid–nucleotide pairs at dented (a), intermediate (b) and protruded (c) protein surfaces. The white and black bars represent unpaired and paired nucleotides, respectively. The frequency of amino acid–nucleotide pairs was counted for each complex. The resultant frequency distributions, obtained from all the 91 complexes, for each amino acid–nucleotide pair (amino acid-paired nucleotide versus amino acid-unpaired nucleotide) were statistically analyzed using t-test to determine the significance of the difference. Asterisk indicates P < 0.05.
At protruded surfaces, however, the interaction appeared to be dependent on the electrostatic potential of the amino acid rather than on the base pairing properties of the RNA. Positively charged amino acids, such as lysine and arginine, interacted more frequently with either unpaired or paired nucleotides when compared to other amino acids (Figure 2c). Since positively charged amino acids interact with the negatively charged phosphate groups of nucleic acids (5), our data suggest that electrostatic interactions between the positively charged side chain and the RNA phosphate groups occur more frequently at protruded surfaces.
The surface shape of the protein and RNA base pairing properties in hydrogen bond formation
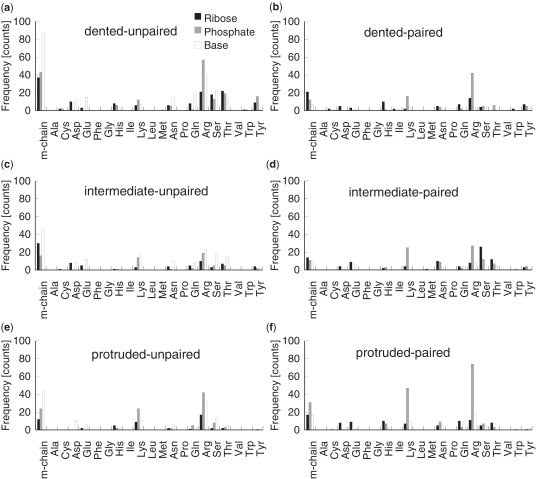
To examine whether the surface shape of protein and the RNA base pairing properties are important for the intermolecular hydrogen bonds at the interface, we measured the distribution of 1949 hydrogen bonds that were formed between the amino acid moiety (either the main chain or one of 20 side chains) and the nucleotide moiety (the ribose, phosphate or base) at the six types of protein–RNA interfaces (Figure 3). At dented-unpaired interfaces, the highest frequency of hydrogen bonds occurred between the RNA nucleotides (all three moieties) and the main chain of the protein (Figure 3a). Moreover, among the nucleotide moieties, hydrogen bonds that formed with the base were most frequent, followed by the phosphate and ribose moieties (Figure 3a). In contrast, hydrogen bonds between the protein main chain and the nucleotide bases were less frequently observed in dented-paired, intermediate-paired and protruded-paired interfaces when compared with dented-unpaired interfaces (Figure 3b, d and f, respectively). Although such hydrogen bonds were also most frequent in intermediate-unpaired and protruded-unpaired interfaces, their frequency in these interfaces was less than half than that in dented-unpaired interfaces. These results suggest that dented protein surfaces prefer unpaired nucleotides at the interface and form hydrogen bonds between the protein backbone and RNA bases.
Figure 3.
The frequency distributions of the intermolecular hydrogen bonds that are formed between the amino acid moiety (the main chain or one of 20 side chains) and the nucleotide moiety (the ribose, phosphate or base) at the following six types of interfaces: (a) dented surface and unpaired nucleotides, (b) dented surface and paired nucleotides, (c) intermediate surface and unpaired nucleotides, (d) intermediate surface and paired nucleotides, (e) protruded surface and unpaired nucleotides and (f) protruded surface and paired nucleotides. The black, gray and white bars represent the ribose, phosphate and base moieties, respectively.
Interestingly, among the six types of interfaces, protruded-paired interfaces had the highest frequency of hydrogen bonds between positively charged side chains and phosphate groups (Figure 3f). These results suggest that electrostatic interactions between positively charged side chains and the phosphate groups commonly occur at the protruded surfaces. Moreover, such interactions were more frequent at protruded-paired interfaces than at protruded-unpaired interfaces, which indicate that the base pairing properties of RNA were important in forming these interactions (Figure 3e and f). Taken together, these results suggest that both the surface shape of the protein and the base pairing properties of the RNA are critical for the formation of intermolecular hydrogen bonds at protein–RNA interfaces.
The role of amino acids in RNA loop recognition
Many RNA-binding proteins are known to preferentially recognize RNA loop structures and form intermolecular hydrogen bonds with nucleotide bases within the loop (10,19). In this study, RNA loop was considered as the region of the RNA that contained unpaired nucleotides flanked by paired nucleotides at both the 5′- and the 3′-ends. We observed that the RNA molecule interacted with the protein at a loop region that contained unpaired nucleotides (loop nucleotides) in 44 complexes. On the other hand, the RNA molecule interacted with the protein at regions that were outside of the loop and contained unpaired nucleotides (non-loop nucleotides) in 39 complexes (Supplementary Table S1).
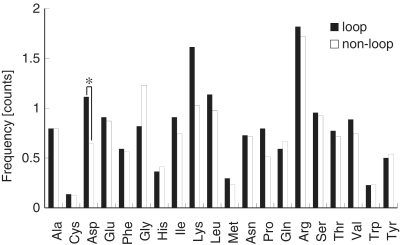
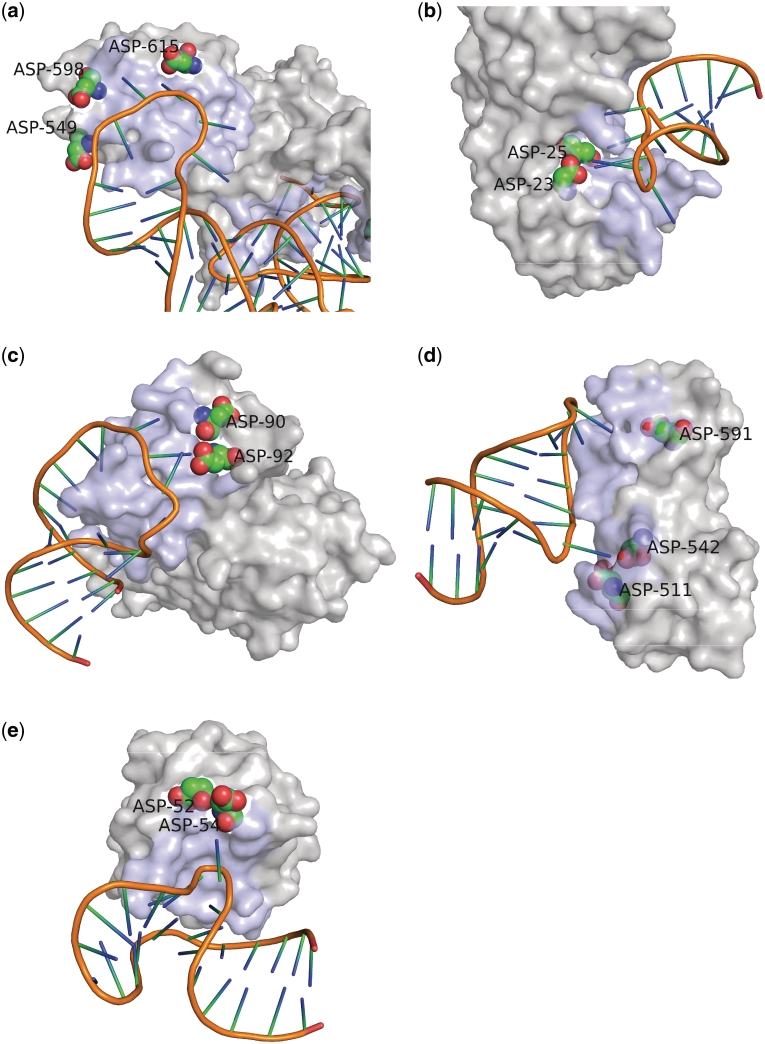
To investigate whether amino acids determine the ability of the protein to distinguish loop nucleotides from non-loop nucleotides at the interface, we compared the amino acid compositions of the proteins that interacted with RNA loop and proteins that interacted with the non-loop RNA regions (within 5 Å). Among the 20 amino acids, aspartic acid showed a significant difference in frequency between the loop and non-loop regions, whereas the frequency of other 19 amino acids did not differ significantly between these two types of regions (Figure 4). Due to the presence of large number of glycine and lysine residues in two complexes (3BOY, 3IAB), these amino acid residues also showed a greater proportional change between loop and non-loop regions; however, the difference in their proportion was statistically insignificant. This suggests that aspartic acid prefers loop nucleotides to non-loop nucleotides at the interface. Our data set contained several tRNA/aminoacyl tRNA synthetase pairs, which are structurally similar in their anticodon loops. To avoid a population bias that might have arisen from these loops, we re-analyzed the amino acid composition after excluding such protein–RNA complexes and still observed a significant difference in the frequency of aspartic acid between loop and non-loop regions (Supplementary Figure S3). Interestingly, in 27 of the 44 protein–RNA complexes that involved a loop-mediated interaction, the RNA bases in the loop region were flipped out to interact with the proteins. Some examples of aspartic acid–loop nucleotide interactions are shown in Figure 5 (20–24). Moreover, in these 27 complexes, the interacting RNAs were composed of various species, including tRNA, rRNA, mRNA, snRNA and snoRNA. These results suggest that aspartic acid plays an important role in the RNA base-flipping, and this process could be common to many protein–RNA loop interactions.
Figure 4.
Comparison of the average frequency of amino acids that interact with a loop or non-loop RNA region at the interface. The frequency of amino acid at the interface was counted for each complex. The resultant frequency distributions for each amino acid that interacted with loop or non-loop nucleotides were statistically analyzed using t-test to determine the significance of the difference. Asterisk indicates P < 0.05. The black and white bars represent loop and non-loop nucleotides, respectively.
Figure 5.
Examples of protein–RNA complexes involving aspartic acids and loop nucleotides at the interface. (a) Escherichia coli threonyl-tRNA synthetase in complex with its cognate tRNA (PDBID: 1QF6). (b) Escherichia coli 5-methyluridine methyltransferase RUMA in complex with ribosomal RNA (PDBID: 2BH2). (c) Homo sapiens spliceosomal U2B–U2A protein complex bound to the U2 snRNA fragment (PDBID: 1A9N). (d) Escherichia coli mRNA-binding domain of elongation factor SelB in complex with SECIS RNA (PDBID: 2PJP). (e) Archaeal box C/D RNA–protein complex (PDBID: 1RLG). The interacting protein surfaces are shown in light blue, and the aspartic acids at the interface are shown as spheres. The figures were generated using PyMOL software (The PyMOL Molecular Graphics System, Version 1.1r2pre, DeLano Scientific LLC.).
DISCUSSION
In this study, we characterized a data set of 91 protein–RNA complexes to identify novel mechanisms underlying protein–RNA interactions. Our data indicate that both the surface shape of the protein and the secondary structure of the RNA molecule are important in determining the binding specificity of a given protein and RNA molecule. We observed that a dented protein surface is significantly more likely to interact with unpaired nucleotides, and the hydrogen bonds at this interface are prominent between the protein backbone and RNA bases. Indeed, previous studies have shown that hydrogen bonds frequently occur between the protein backbone and RNA bases (6,7) and that a protein cavity (i.e. a dented surface) prefers nucleotide bases at the interface (9). Gupta and Gribskov (25) extensively analyzed the base pairing property in RNP region (i.e. interface) using different data set and reported the preference of unpaired nucleotides in this region. Thus, our results are consistent with these previous reports and also show that a dented protein surface can distinguish unpaired nucleotides from paired nucleotides through hydrogen bonds that form between the protein backbone and RNA bases at the interface. Consistent with previous reports (6–8), we also observed that positively charged amino acids often form electrostatic interactions with the phosphate groups of RNA. Interestingly, this type of interaction was more often observed on proteins with a protruded surface. Collectively, these data suggest that dented and protruded protein surfaces employ different recognition mechanisms for paired versus unpaired RNA nucleotides. We further hypothesize that protruded protein surface makes an initial contact with the RNA molecule through electrostatic interactions, and a dented surface determines the binding specificity through the hydrogen bonds that form with unpaired nucleotides.
A loop region is one of the major structural features of RNA and is frequently used to form an interaction with various RNA binding proteins (19). Nucleotide bases in RNA loops exhibit unique hydrogen-bonding patterns with proteins, and these patterns are key determinants of binding specificity (6, 26). In this study, we found that aspartic acids interacted more frequently with the RNA loops in which the nucleotide bases had flipped out to form hydrogen bonds with the protein. Aspartic acids are generally disfavored at protein–RNA interfaces due to electrostatic repulsion between negatively charged side chains and phosphate groups (6–8). However, aspartic acids are also known to form specific pseudo pairs with the nucleotide bases by using both their side- and main-chain atoms (27). Based on our results and previous reports, we speculate that aspartic acids are necessary for base-flipping, most likely to keep phosphate groups away from an interface and to form some specific interactions with the flipped bases.
Protein–RNA interactions are controlled by various factors, such as the composition of the amino acids and nucleotides, the shape of the macromolecules and higher order structures. Our study highlights the roles that are played by the protein surface and the secondary structure of the RNA molecule in protein–RNA interactions and also suggests a possible role of aspartic acid in RNA loop recognition. However, there are many important issues that need to be addressed for understanding the mechanism of protein–RNA interactions. For example, protein surface shapes of RNA interacting proteins should be compared with the other proteins, such as those that bind to DNA or ligands, to identify unique characteristics of such proteins. A number of prediction algorithms for protein–RNA interactions are available (28–34), and the inclusion of features such as the shape of the protein surface and the secondary structure of the RNA molecule will greatly improve the efficiency and accuracy of these algorithms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures and Supplementary Table S1–S3.
FUNDING
The Ministry of Education, Culture, Sports, Science and Technology (MEXT) Grants-in-Aid (20200070, 22370065, 22659186 and 238043); Japan Society for the Promotion of Science (JSPS). Funding for open access charge: Ministry of Education, Culture, Sports, Science and Technology.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Kei Yura (Ochanomizu University) and Gota Kawai (Chiba Institute of Technology) for their valuable advice and suggestions. They also thank Dr Maki Yoshihama (University of Miyazaki) for useful discussions.
REFERENCES
- 1.Garvie CW, Wolberger C. Recognition of specific DNA sequences. Mol. Cell. 2001;8:937–946. doi: 10.1016/s1097-2765(01)00392-6. [DOI] [PubMed] [Google Scholar]
- 2.Dixon SJ, Costanzo M, Baryshnikova A, Andrews B, Boone C. Systematic mapping of genetic interaction networks. Annu. Rev. Genet. 2009;43:601–625. doi: 10.1146/annurev.genet.39.073003.114751. [DOI] [PubMed] [Google Scholar]
- 3.Mata J, Marguerat S, Bahler J. Post-transcriptional control of gene expression: a genome-wide perspective. Trends Biochem. Sci. 2005;30:506–514. doi: 10.1016/j.tibs.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Rohs R, West S, Sosinsky A, Liu P, Mann R, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allers J, Shamoo Y. Structure-based analysis of protein-RNA interactions using the program ENTANGLE. J. Mol. Biol. 2001;311:75–86. doi: 10.1006/jmbi.2001.4857. [DOI] [PubMed] [Google Scholar]
- 7.Bahadur RP, Zacharias M, Janin J. Dissecting protein-RNA recognition sites. Nucleic Acids Res. 2008;36:2705–2716. doi: 10.1093/nar/gkn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis JJ, Broom M, Jones S. Protein-RNA interactions: structural analysis and functional classes. Proteins. 2007;66:903–911. doi: 10.1002/prot.21211. [DOI] [PubMed] [Google Scholar]
- 9.Sonavane S, Chakrabarti P. Cavities in protein-DNA and protein-RNA interfaces. Nucleic Acids Res. 2009;37:4613–4620. doi: 10.1093/nar/gkp488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ray D, Kazan H, Chan ET, Pena Castillo L, Chaudhry S, Talukder S, Blencowe BJ, Morris Q, Hughes TR. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 2009;27:667–670. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- 11.Wang G, Dunbrack RL., Jr PISCES: recent improvements to a PDB sequence culling server. Nucleic Acids Res. 2005;33:W94–W98. doi: 10.1093/nar/gki402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 13.Pintar A, Carugo O, Pongor S. CX, an algorithm that identifies protruding atoms in proteins. Bioinformatics. 2002;18:980–984. doi: 10.1093/bioinformatics/18.7.980. [DOI] [PubMed] [Google Scholar]
- 14.Yura K, Hayward S. The interwinding nature of protein-protein interfaces and its implication for protein complex formation. Bioinformatics. 2009;25:3108–3113. doi: 10.1093/bioinformatics/btp563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 16.Yang H, Jossinet F, Leontis N, Chen L, Westbrook J, Berman H, Westhof E. Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res. 2003;31:3450–3460. doi: 10.1093/nar/gkg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 19.Auweter SD, Oberstrass FC, Allain FH. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B”-U2A' protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 21.Sankaranarayanan R, Dock-Bregeon AC, Romby P, Caillet J, Springer M, Rees B, Ehresmann C, Ehresmann B, Moras D. The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999;97:371–381. doi: 10.1016/s0092-8674(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 22.Kromayer M, Neuhierl B, Friebel A, Bock A. Genetic probing of the interaction between the translation factor SelB and its mRNA binding element in Escherichia coli. Mol. Gen. Genet. 1999;262:800–806. doi: 10.1007/s004380051143. [DOI] [PubMed] [Google Scholar]
- 23.Lee TT, Agarwalla S, Stroud RM. A unique RNA Fold in the RumA-RNA-cofactor ternary complex contributes to substrate selectivity and enzymatic function. Cell. 2005;120:599–611. doi: 10.1016/j.cell.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 24.Moore T, Zhang Y, Fenley MO, Li H. Molecular basis of box C/D RNA-protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004;12:807–818. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 25.Gupta A, Gribskov M. The role of RNA sequence and structure in RNA-protein interactions. J Mol. Biol. 2011;409:574–587. doi: 10.1016/j.jmb.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 26.Morozova N, Allers J, Myers J, Shamoo Y. Protein-RNA interactions: exploring binding patterns with a three-dimensional superposition analysis of high resolution structures. Bioinformatics. 2006;22:2746–2752. doi: 10.1093/bioinformatics/btl470. [DOI] [PubMed] [Google Scholar]
- 27.Kondo J, Westhof E. Classification of pseudo pairs between nucleotide bases and amino acids by analysis of nucleotide-protein complexes. Nucleic Acids Res. 2011;39:8628–8637. doi: 10.1093/nar/gkr452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim OT, Yura K, Go N. Amino acid residue doublet propensity in the protein-RNA interface and its application to RNA interface prediction. Nucleic Acids Res. 2006;34:6450–6460. doi: 10.1093/nar/gkl819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shazman S, Mandel-Gutfreund Y. Classifying RNA-binding proteins based on electrostatic properties. PLoS Comput. Biol. 2008;4:e1000146. doi: 10.1371/journal.pcbi.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez-Cano L, Fernandez-Recio J. Optimal protein-RNA area, OPRA: a propensity-based method to identify RNA-binding sites on proteins. Proteins. 2010;78:25–35. doi: 10.1002/prot.22527. [DOI] [PubMed] [Google Scholar]
- 31.Chen YC, Lim C. Predicting RNA-binding sites from the protein structure based on electrostatics, evolution and geometry. Nucleic Acids Res. 2008;36:e29. doi: 10.1093/nar/gkn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shulman-Peleg A, Shatsky M, Nussinov R, Wolfson HJ. Prediction of interacting single-stranded RNA bases by protein-binding patterns. J. Mol. Biol. 2008;379:299–316. doi: 10.1016/j.jmb.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terribilini M, Lee JH, Yan C, Jernigan RL, Honavar V, Dobbs D. Prediction of RNA binding sites in proteins from amino acid sequence. RNA. 2006;12:1450–1462. doi: 10.1261/rna.2197306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu ZP, Wu LY, Wang Y, Zhang XS, Chen L. Prediction of protein-RNA binding sites by a random forest method with combined features. Bioinformatics. 2010;26:1616–1622. doi: 10.1093/bioinformatics/btq253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.