Abstract
In all organisms, aminoacyl tRNA synthetases covalently attach amino acids to their cognate tRNAs. Many eukaryotic tRNA synthetases have acquired appended domains, whose origin, structure and function are poorly understood. The N-terminal appended domain (NTD) of glutaminyl-tRNA synthetase (GlnRS) is intriguing since GlnRS is primarily a eukaryotic enzyme, whereas in other kingdoms Gln-tRNAGln is primarily synthesized by first forming Glu-tRNAGln, followed by conversion to Gln-tRNAGln by a tRNA-dependent amidotransferase. We report a functional and structural analysis of the NTD of Saccharomyces cerevisiae GlnRS, Gln4. Yeast mutants lacking the NTD exhibit growth defects, and Gln4 lacking the NTD has reduced complementarity for tRNAGln and glutamine. The 187-amino acid Gln4 NTD, crystallized and solved at 2.3 Å resolution, consists of two subdomains, each exhibiting an extraordinary structural resemblance to adjacent tRNA specificity-determining domains in the GatB subunit of the GatCAB amidotransferase, which forms Gln-tRNAGln. These subdomains are connected by an apparent hinge comprised of conserved residues. Mutation of these amino acids produces Gln4 variants with reduced affinity for tRNAGln, consistent with a hinge-closing mechanism proposed for GatB recognition of tRNA. Our results suggest a possible origin and function of the NTD that would link the phylogenetically diverse mechanisms of Gln-tRNAGln synthesis.
INTRODUCTION
Aminoacyl tRNA synthetases perform a critical function in conversion of the genetic code into amino acids by covalently attaching the correct amino acid to specific cognate tRNAs (1,2). These enzymes are divided into two structural classes, each arising from a common ancestor (3,4), and catalyze aminoacyl-tRNA formation by a two-step pathway: (i) an activated aminoacyl adenylate is first formed from ATP and the cognate amino acid; (ii) the amino acid is transferred to its cognate tRNA with release of AMP. Each synthetase nearly perfectly selects the correct tRNA among 20–22 different isoacceptor tRNA families (5) as well as the correct amino acid substrate; in some cases, this is achieved via the use of hydrolytic editing mechanisms to clear misactivated amino acid and/or misacylated tRNA (3,4). It is of particular interest that tRNAGln and tRNAAsn are aminoacylated by distinct mechanisms in different kingdoms. For example, whereas Gln-tRNAGln is formed in the canonical manner in the eukaryotic cytoplasm, all archaea, many bacteria and eukaryotic organelles possess an alternative two-step pathway. In this route, a non-discriminating GluRS first misaminoacylates tRNAGln; next, the Glu-tRNAGln is converted to Gln-tRNAGln by a tRNA-dependent amidotransferase belonging to either the GatCAB family (bacteria and some archaea), or the GatDE family (archaea only) (6–8). Thus, glutaminyl-tRNA synthetase (GlnRS) is primarily a eukaryotic enzyme. Synthesis of cysteinyl-tRNACys in methanogens and highly related archaea provides another example of a two-step pathway to cognate aminoacyl-tRNA, although the phylogenetic distribution of this pathway is much more limited (9).
Eukaryotic tRNA synthetases are distinctly more complex than their prokaryotic homologs because they have progressively acquired and retained additional domains throughout evolution (1,2). It is perplexing why tRNA synthetases, unlike other eukaryotic proteins, have been subject to massive progressive additions over the course of evolution (2). While some appended domains are shared among synthetase families and are similar to domains in other proteins implicated in either nucleic acid binding or protein–protein interactions (1), at least eight domains are uniquely associated with a single synthetase family, and neither their structures nor their roles are generally understood (2). An exception is the CTD of human CysRS, which is known to enhance anticodon discrimination at the expense of the aminoacylation rate, acting as a quality control step (10). This report focuses on the NTD of GlnRS, which is itself unique because GlnRS likely originated in eukaryotes, evolving directly from a progenitor eukaryotic non-discriminating GluRS (11,12). Like other eukaryotic GlnRS species, Saccharomyces cerevisiae Gln4 contains both a highly conserved C-terminal domain (CTD) with all of the known features of class I synthetases, as well as a less conserved appended N-terminal domain (NTD) with no obvious sequence homology to any known protein domain.
The origin and function of the NTD in GlnRS are of particular interest. Most eukaryotic GlnRS proteins have an appended NTD, whereas the bacterial GlnRS proteins do not, although the bacterial proteins were almost certainly acquired by horizontal transfer from eukaryotes. Saccharomyces cerevisiae GlnRS contains both a 595-amino acid CTD that contains the signature elements of a type I synthetase (4,13–15), and suffices for both catalytic function and yeast viability (16,17), and a 224-amino acid NTD that is uniquely associated with GlnRS in many eukaryotes (2). Although both Escherichia coli and Deinococcus radiodurans GlnRS proteins share extensive identity with the conserved S. cerevisiae GlnRS CTD, E. coli GlnRS entirely lacks an NTD (13) and D. radiodurans GlnRS has an unrelated domain appended to the C-terminus of the conserved domain (14). Two observations imply that the S. cerevisiae NTD contributes to synthetase function: the NTD alone exhibits a non-specific RNA binding activity (18), and the addition of the NTD to EcGlnRS results in a chimeric protein that can replace the native yeast gene (19). However, the precise role of the NTD in eukaryotic GlnRS function is unknown.
MATERIALS AND METHODS
Genetic analysis of gln4 mutants
To construct a strain (MEM70) of genotype gln4-Δ::kanR [CEN URA3 GLN4], a CEN GLN4 plasmid was transformed into yeast strain BY4741, and then the gln4-ΔKan allele was introduced by transformation, using PCR primers HWI P239 and HWI P234 (Supplementary Table S1) to amplify the fragment from the appropriate GLN4/ gln4-ΔKan heterozygous diploid (Open Biosystems ID 22424). To construct strains bearing an integrated copy of either GLN4 or gln4-Δ2–210, we used an integrating cassette (20) that carries MET15 flanked by sequences homologous to ADE2, into which we inserted GLN4 or the gln4(211–809) allele (constructed with a synthetic fragment made by Geneart). Plasmids were then digested with Stu I to release the integrating cassette and transformed into MEM70, and transformants were screened for Ade−, and plated on FOA to select for removal of the CEN URA3 GLN4 plasmid, generating the desired gln4-Δ::kanR ade2−::GLN4::MET15 (MEM133) and gln4-Δ::kanR ade2−::gln4(211–809)::MET15 (MEM141) strains. To test for growth phenotypes, MEM133 and MEM141 were transformed with a [CEN LEU2 GLN4] or a control [CEN LEU2] vector, grown overnight in SD-Leu media (21), diluted to OD600 of 1 and 2 µl of 10-fold serial dilutions were spotted onto plates containing either YPD or YP glycerol and incubated at the indicated temperatures for 1–7 days with similarly spotted control parent strains that were grown in YPD media. Oligonucleotides, yeast strains and plasmids used in these studies are reported in Supplementary Tables S1–S3.
Protein expression and purification
To express high levels of GLN4 and its derivatives in yeast, ORFs were cloned under PGAL1 control into the previously described 2 µ URA3 LIC vectors BG2483 or BG2663, in which ORFs are expressed with their C termini fused to a complex tag containing a 3C protease site, followed by an HA epitope, His6, and the ZZ domain of protein A (22), and expressed in yeast strain BCY123 (23). Gln4(1–187) was expressed in yeast strain EJG1473, which was grown in media containing selenomethionine and Ado-Methionine as described (24). Expressed proteins were purified by affinity purification on IgG sepharose, removal of GST-3C protease, concentration of samples and sizing on SuperdexHiLoad 1660 (GE Healthcare 17–1069, 10 mm× 300 mm bed dimension), as described (22).
tRNA purification and EMSA binding assay
To obtain native yeast tRNAGln(CUG), we cloned the tQ(CUG)M gene into the leu2-d URA3 vector pYEX4T (25), transformed the plasmid into BY4741, grew transformants in SD-Ura media overnight, followed by overnight growth in SD-Leu-Ura media. We then prepared low molecular weight RNA, purified the tRNAGln with biotinylated oligonucleotides oligo HWI P257 (Supplementary Table S1), and performed HPLC analysis of modified nucleotides as described (26). The ratio of modified to unmodified nucleotides was similar to that in strains with tRNAGln on a lower copy plasmid.
tRNA binding was measured, as described (27) in reaction mixtures containing Gln4 or its buffer, 2.4 nM 5′-[32P]-labeled tRNA, in buffer containing 28 mM HEPES (pH 7.5), 80 mM NaCl, 5 mM MgCl2, 0.5 mM DTT, 2.5 mM spermidine, 50 µg/ml BSA, 20 µM EDTA, 200 µg/ml polyA, 4.6 mM Tris-Cl (pH 7.5), 1 mM β-mercaptoethanol and 10% glycerol. Reactions were incubated for 20 min on ice and loaded onto prerun 5% polyacrylamide gels containing 50 mM Tris-borate, pH 8.3, 1 mM EDTA, 5 mM MgCl2 and 5% glycerol, and run at 4°C in the same buffer without glycerol.
In vitro synthesis of tRNA transcripts
Duplex DNA templates for in vitro transcription of yeast tRNAGln were synthesized from two single-stranded oligodeoxynucleotides containing a complementary overlap duplex region, as described (28). The two 3′-terminal deoxynucleotides on the non-coding strand incorporated 2′-O-methyl sugars (mU and mG in the sequences), to improve the fidelity of transcription termination by T7 RNA polymerase. Milligram quantities of each tRNA were transcribed with the Del(172–173) variant of T7 RNA polymerase, as described (28,29), and purified by denaturing polyacrylamide gel electrophoresis. tRNA was stored at 200 µM in 10 mM Tris (pH 8.0), 1 mM EDTA (TE buffer).
Steady state methods
tRNAGln transcripts were 32P-labeled at the 3′-terminal internucleotide linkage using the exchange reaction of tRNA nucleotidyltransferase (30–32), and purified again by gel electrophoresis. Steady state kinetics of tRNA aminoacylation reactions were performed in a buffer consisting of 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2 and 10 mM β-mercaptoethanol. tRNA was first refolded by heating to 85°C in TE buffer for 3 min, followed by addition of MgCl2 to 10 mM and slow-cooling to ambient temperature. Two microliters of aliquots from the reactions were added to 5 µl of a quenching solution containing 400 mM sodium acetate (pH 5.2) and 0.1% SDS, followed by addition of 3–5 ml of 0.01–0.1 mg/ml P1 nuclease (Fluka) to digest the tRNA to 5′-phosphorylated nucleosides. The digestion products were spotted on PEI-cellulose thin layer chromatography (TLC) plates and developed in a solution containing 100 mM ammonium acetate and 5% acetic acid. Raw data were quantified by phosphorimaging analysis, and corrected intensities were analyzed to obtain initial velocities. KM and Vmax were then obtained by Michaelis–Menten analysis. ATP (5 mM) was used in all reactions; saturation was confirmed for both FL-GlnRS and the NTD variant. The glutamine concentrations used to determine KM(tRNA) for FL-GlnRS and Gln4(187–809) were 10 and 60 mM, respectively; saturation was verified in each case. tRNA concentrations used were 20 nM–3 µM for FL-GlnRS and 500 nM–20 µM for Gln4(187–809). To determine KM for glutamine, the tRNA concentrations used were 1 µM for FL-GlnRS and 15 µM for Gln4(187–809). Enzyme concentrations were maintained at least 20-fold below tRNA concentrations for all experiments to ensure multiple-turnover conditions.
Crystallization and structure determination
Initial crystallization conditions were identified using a high-throughput microbatch-under-oil method (33). Crystals appeared after a 6-week incubation at 22°C in conditions containing 0.2 µl protein solution (8.9 mg/ml protein in 100 mM NaCl, 5% (v/v) glycerol, 2 mM DTT, 0.025% (w/v) NaN3, 20 mM HEPES buffer, pH 7.5) and 0.2 µl of precipitant solution (100 mM KCl, 100 mM Tris-HCl, pH 8 and 20% (w/v) PEG 4000). Crystals were extracted directly from the well and were determined suitable for X-ray data collection from initial screening. No further optimization took place.
Remote MAD data collection was carried out at 100 K on beamline 11–1 of the Stanford Synchrotron Radiation Lightsource (SSRL) (34) with a MAR 325 CCD detector. To minimize radiation effects, the data collection protocol was designed with Best (35) automated within the WebIce analysis package (36). Integration, reduction and scaling took place with XDS (37). The structure was solved with Phenix (38). Using the remote wavelength data set the structure was refined through an iterative process using Phenix with manual model building with Coot (39). Validation was carried out with Molprobity (40). The structure was deposited as PDB ID 3TL4. Experimental and refinement details are given in Supplementary Table S4. Surface charge was calculated assuming vacuum electrostatics using PyMol.
The sequences of several appended NTDs from GlnRS sequences of other organisms, listed in Table 2, were threaded to the Gln4(1–187) structure using SwissModel (41). As a control the reversed sequence was also threaded. From the models a Z-score was calculated using Prosa2003 (42) with a 20-residue moving window. The typical combined, pairwise and surface Z-scores for native proteins are (−6 to −12), (−3 to −7.5) and (−3 to −8), respectively. Alignments were performed using the ‘fit’ function of PyMOL. Due to low sequence homology, only carbon alpha atoms were included in the alignment. Loops were removed prior to RMS deviation calculation.
Table 2.
Comparison of sequences threaded to the N-term Gln(1–187) structure
| Name | Species | Residues |
Z-score |
||
|---|---|---|---|---|---|
| Combined | Pair | Surface | |||
| N-term | Saccharomyces cerevisiae | 186 | −11.23 | −7.98 | −8.71 |
| N-term reversed | Saccharomyces cerevisiae | 186 | −0.30 | −1.56 | 0.55 |
| P13188 | Saccharomyces cerevisiae | 186 | −11.28 | −7.99 | −8.75 |
| q9y7y8 | Schizosaccharomyces pombe | 190 | −6.02 | −0.79 | −7.00 |
| q9y105 | Drosophila melanogaster | 188 | −3.99 | 1.11 | −5.62 |
| q62431 | Mus musculus | 183 | −8.42 | −6.44 | −5.92 |
| p47897 | Homo sapiens | 185 | −6.55 | −3.69 | −5.20 |
| q3mhh4 | Bos taurus | 185 | −6.62 | −3.84 | −5.22 |
| p52780 | Lupinus luteus | 188 | −7.02 | −1.73 | −6.90 |
| p14325 | Dictyostelium discoideum | 185 | −7.98 | −4.06 | −6.92 |
| GatB | Thermotog maritima | 177 | −10.43 | −6.42 | −8.91 |
RESULTS
Removal of the NTD impairs Gln4 function in vivo and in vitro
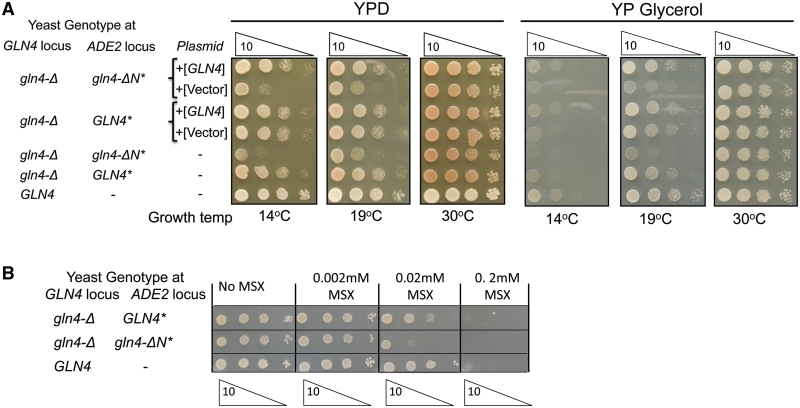
To determine if the NTD is important for the essential function of Gln4, we compared the growth of yeast strains expressing either full length GLN4 or gln4 lacking the NTD [gln4(211–809)] integrated into the chromosome under control of its own promoter, as the sole source of ScGlnRS. Growth of the gln4(211–809) mutant is impaired at 14°C and 19°C, but not at 30°C, on both YPD and YP glycerol media, and, as expected, this phenotype is complemented by full length GLN4 on a single copy plasmid but not by an empty vector (Figure 1A). In addition, the gln4(211–809) mutant is much more sensitive than wild-type to l-methionine sulfoximine, a highly specific inhibitor of glutamine synthase (43), which results in reduced concentrations of intracellular glutamine (Figure 1B). These observations demonstrate that the NTD plays an important role in the function of the native yeast enzyme in vivo.
Figure 1.
Deletion of the N-terminal domain of GLN4 impairs function. (A) Mutants bearing a gln4 mutation in which amino acids 2–210 are deleted are defective in growth at low temperature on YP media containing glucose or glycerol as a carbon source. Serial dilutions of strains with either wild-type GLN4 or gln4(211–809) (marked gln4-ΔN*) integrated at the ade2 locus in the gln4-ΔKanR mutant were grown as indicated. Indicated strains carry CEN plasmids either with or without GLN4. (B) Mutants bearing a gln4 mutation in which amino acids 2–210 are deleted are sensitive to the glutamine synthase inhibitor l-methionine sulfoximine (MSX).
Steady-state kinetic parameters were measured to directly assess the effects of the NTD on tRNAGln aminoacylation. Substantial differences between full length Gln4 and Gln4(187–809) were found. For the wild-type enzyme, similar KMtRNA (0.14 µM versus 0.19 µM) and kcat (1.7 s−1 versus 1.4 s−1) were measured for affinity-purified native tRNAGln and an unmodified transcript, suggesting that post-transcriptional modifications do not have significant effects in this system. Using unmodified tRNAGln(CUG) as substrate, we then found that Gln4(187–809) exhibits a 30-fold increase in KMtRNA (from 0.2 µM to 5.8 µM), and a 5.4-fold increase in KMGln (from 1.7 mM to 9.3 mM) although the kcat values are similar (1.4 s−1 versus 1.7 s−1) (Table 1). We infer that the NTD influences the complementarity of both the tRNA and glutamine binding sites for their respective substrates, as also suggested by the sensitivity of the Gln4(211–809) mutant to l-methionine sulfoximide.
Table 1.
Comparison of steady state kinetic parameters for Gln4 and Gln4 variants
| kcat (s−1) | KMtRNA (µM) | kcat/KMtRNA (M−1·s−1) | KMGln (mM) | kcat/KMGln (M−1·s−1) | |
|---|---|---|---|---|---|
| FL-Gln4 | 1.4 ± 0.2 | 0.19 ± 0.04 | 7.6 × 106 | 1.7 ± 0.2 | 8.5 × 102 |
| Gln4 (187–809) | 1.7 ± 0.3 | 5.85 ± 0.52 | 2.9 × 105 | 9.3 ± 0.3 | 1.8 × 102 |
| PVG-GlnRS | 2.8 ± 0.6 | 1.55 ± 0.51 | 1.8 × 106 | NA | NA |
| FL-Gln4 + native tRNA | 1.7 ± 0.1 | 0.14 ± 0.07 | 1.2 × 107 | NA | NA |
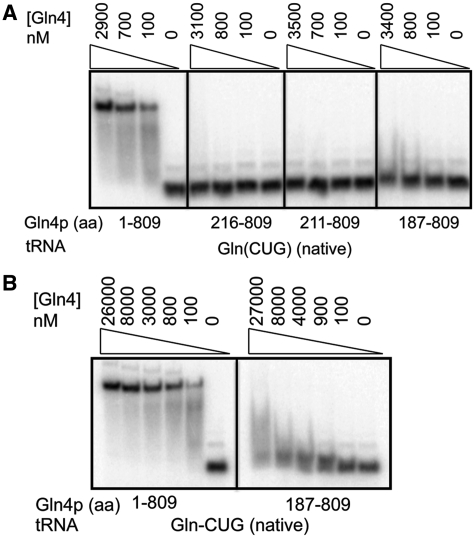
Since the kinetic analysis suggested a role for the NTD in tRNAGln binding, we developed an EMSA assay to directly measure binding. We find that yeast Gln4 binds tightly and specifically to fully modified tRNAGln(CUG) purified from S. cerevisiae, with ∼25 nM Gln4 required for 50% binding (Figure 2A and B, see Figure 5) while >800 nM Gln4 is required to bind comparably to tRNAPhe (Supplementary Figure S1). Remarkably, Gln4(187–809) binds only very weakly at 27 µM, 1000-fold above the apparent KD of wild-type Gln4 (Figure 2A and B), and other Gln4 variants Gln4(211–809) and Gln4(216–809) do not detectably bind tRNAGln(CUG) (Figure 2A). Furthermore, there was no improvement in binding of Gln4(187–809) in the presence of other Gln4 substrates including glutamine, ATP or the non-hydrolyzable ATP analog AMPPNP (Supplementary Figure S2).
Figure 2.
The N-terminal domain of Gln4 is required for specific binding to native tRNAGln(CUG). (A) Gln4 variant proteins deleted for different amounts of the NTD exhibit reduced tRNAGln(CUG) binding. (B) Gln4(187–809) protein exhibits detectable binding to tRNAGln(CUG) at high concentrations.
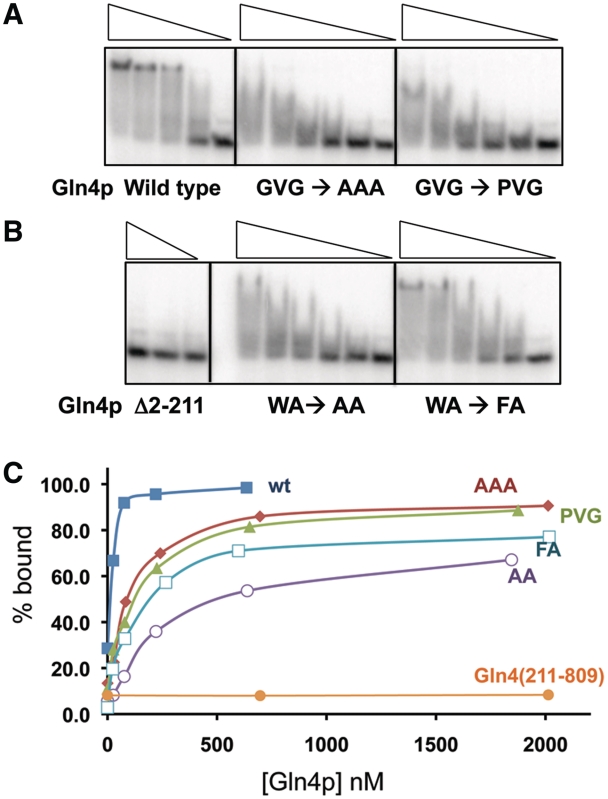
Figure 5.
Mutations in conserved amino acids in the putative hinge of the NTD affect the interaction of Gln4 with native tRNAGln(CUG). (A and B) EMSA wild-type and mutant Gln4 proteins (23–2017 nM). (C) Binding as a function of Gln4 protein concentration.
The Gln4 NTD is structurally similar to two subdomains in the amidotransferase that distinguish tRNAGln from tRNAGlu
To further discern the function of the NTD, we solved the structure of the isolated NTD, which behaves as a discrete unit to confer function when fused to the E. coli GlnRS (19). We purified three NTD variants ending at amino acids 187, which spans the region of extensive identity between the NTD of GlnRS from multiple species (see below), 215 and 228, which covers the entire region without extensive homology to E. coli GlnRS. We obtained crystals of Gln4(1–187) that diffracted to 2.3 Å, and solved the structure of a selenomethionine derivative purified from a yeast sam1-Δ sam2-Δ mutant (24) (Supplementary Table S4).
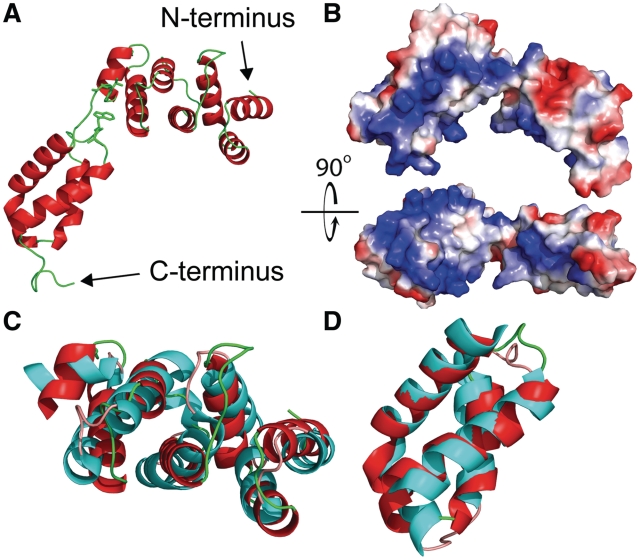
Gln4(1–187) consists of two alpha helical domains, the first from residues 1–111 containing a seven-helix bundle, and the second from residues 119–187 containing a four-helix bundle, which are connected by a seven residue G112VG114IGIT linker (Figure 3A). One face of each domain is positively charged across the length of the domain, which might facilitate interactions with the negatively charged tRNA and provide the basis for the non-specific RNA binding activity of this domain (18) (Figure 3B).
Figure 3.
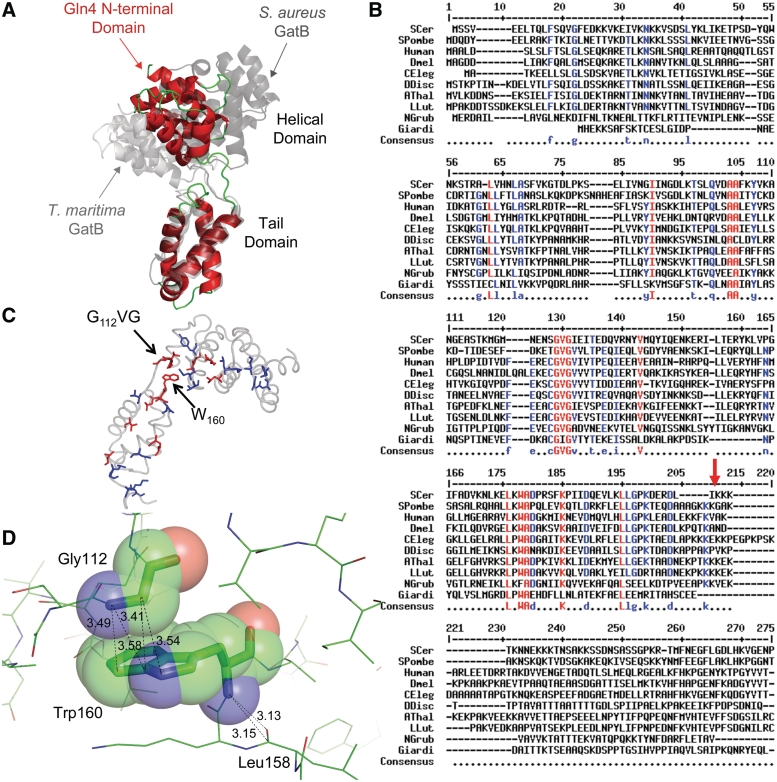
Structure of Gln4(1–187) with comparisons to domains in S. aureus GatB (PDB ID: 3IP4). (A) Crystallographic structure of Gln4 residues 1–187 in cartoon representation. The proposed hinge region (Gly112Val113Gly114) is highlighted together with the likely interacting residue Trp160, and shown in stick representation. (B) Surface electrostatic model of Gln4 residues 1–187, shown with two orientations rotated by 90° relative to each other, with positively charged residues colored blue. (C and D) Structural alignment of helical and tail domains of Gln4 NTD and S. aureus GatB (PDB ID: 3IP4) (45). (C) The crystal structure of Gln4(1–110) (red) is superposed to the helical domain of GatB(295–406) (cyan). (D) The crystal structure of Gln4(119–178) (red) is superposed on the tail domain of GatB(414–475) (cyan).
Although the NTD lacks sequence homology to any available structure, a DALI search (44) of the NTD and the individual domains revealed substantial structural homology to the helical and tail domains of the GatB subunit of GatCAB, the glutamyl-tRNA admidotransferase, from Staphylococcus aureus (PDB ID: 3IP4) (45) and Thermotoga maritima (PDB ID: 3AL0) (46) (Figure 3C and D; Supplementary Figure S3). The seven-helix bundle seen in the NTD yields an RMS deviation of 3.75 Å using carbon alpha atoms in the alpha helices of S. aureus GatB and 4.01 Å when compared with T. maritima. However, a five residue insertion between helix 4 and helix 5 appears to shift the orientation of the remaining three helices of S. aureus GatB. When aligning these three helices separately, an RMS deviation of 1.89 Å is observed. The four-helix bundle of the C-terminal subdomain of the NTD has an RMS deviation of only 1.64 Å compared with the S. aureus GatB tail domain, and 1.80 Å compared with the T. maritima GatB tail domain. Since the GatB helical and tail domains make specific and non-specific contacts with tRNAGln (46), we infer that the Gln4 NTD has similar biochemical function. Furthermore, it is likely that GlnRS NTDs from other eukaryotes adopt a similar structure, based on threading of these sequences to the Gln4(1–187) structure (47) (Table 2).
The linker between the NTD subdomains is conserved and functionally important
Three observations suggest that the linker that connects the two domains in Gln4 plays a crucial role in the tRNA binding function of this domain. First, the helical and tail domains of GatB are also connected by a linker, which appears to function as a flexible hinge that closes upon tRNA binding, based on differences in the orientation of the domains in the tRNA-bound (T. maritima) and tRNA-free (S. aureus) structures (45,46). In this regard, we note that the domains in the Gln4 NTD are oriented at an angle between that of the T. maritima tRNA-bound GatB and the S. aureus tRNA-free GatB (Figure 4A). Second, although the linker sequences in GlnRS differ from the sequences in GatB, the linker sequences in GlnRS are among the most highly conserved amino acids in the Gln4 NTD family (Figure 4B). In a comparison of highly divergent eukaryotes, although neither the length nor the sequence of the N-terminal domain is highly conserved, three of the seven amino acids in the linker region G112V113G114 are nearly 100% conserved (Figure 4B and C). Furthermore, G112 appears to interact with W160, 1 of the 10 other highly conserved residues in the NTD; the alpha carbon of G112 is in van der Waals contact with C9 of W160 (Figures 3A and 4C and D). Third, G114 is predicted to be a hinge residue, acting as a flexible connector of the two domains, based on an elastic network analysis with the program HingeProt (48).
Figure 4.
The linker between the two domains in Gln4(1–187) likely behaves as a hinge, is highly conserved and is important for tRNA binding. (A) Structure of Gln4(1–187) (red) superposed on TMGatB (light gray) and SAGatB (dark gray) by alignment of the tail domains. (B) Conservation of GlnRS NTD sequences, red-≥90%; blue-≥70%, with arrow at Gln4187., aligned using Multialin (49). (C) Conserved residues are highlighted on Gln4(1–187) according to the color code in B with the NTD backbone shown in light grey. (D) Close contacts between W160 of the Gln4 NTD and other residues.
Since the G112V113G114 residues of the linker are highly conserved, and since hinges frequently mediate conformational changes upon ligand binding (50), we considered it likely that mutations in the linker region would impair function. Thus, we purified variant proteins in which G112V113G114 was replaced with AAA and with PVG and in which W160 was replaced with F or A, and measured tRNAGln(CUG) binding. Although the variant proteins all bind tRNAGln(CUG), as measured by reduced mobility of the tRNA, all of the mutant proteins exhibit defects in binding (Figure 5A and B). Three variants (Gln4-A112A113A114, Gln4-G112P, Gln4-W160A) fail to form stable complexes with tRNAGln(CUG), as judged by lack of comigration of the complexed tRNA with that formed by wild-type Gln4, and all four variant proteins exhibit an apparently reduced affinity for tRNAGln(CUG), requiring 4–12 times more protein than the wild-type to bind comparable amounts of tRNA (Figure 5C). Moreover, the Gln4-G112P variant exhibits a 10-fold increase in the KMtRNA (from 0.19 µM to 1.6 µM) as well as a slight increase in kcat (1.4 s−1 versus 2.8 s−1) (Table 1). Thus, we conclude that the linker region is important for binding, and speculate that it acts as a hinge facilitating closure between the helical and tail domains upon tRNA binding.
DISCUSSION
The observations that the NTD of S. cerevisiae GlnRS bears a substantial structural resemblance to two domains of the bacterial GatB amidotransferase that distinguish tRNAGln from tRNAGlu, and that the NTD also participates in tRNAGln binding, imply that there is a connection between the indirect pathways for formation of Gln-tRNAGln in bacteria and archaea, and the direct pathway that evolved in eukaryotes. Since it is thought that tRNAGln was present in the last universal common ancestor, it has been puzzling that aminoacylation of this tRNA is achieved by different routes in each of the three kingdoms. Sheppard and Soll proposed that both GatCAB and GatDE were present prior to the split between archaea and bacteria (51), while the specific GlnRS evolved in eukaryotes. We propose that the tRNAGln recognition domain from an amidotransferase was most likely conscripted as an NTD to a progenitor non-discriminating GluRS, and thus played an integral part in the development of the eukaryotic GlnRS family. In particular, evolution of GlnRS from an early non-discriminating GluRS required selectivity determinants in favor of tRNAGln to evolve, while negative determinants against tRNAGlu would also appear. The proximity of the NTD to the tRNA-synthetase core domain suggests that eukaryotes may have exploited the NTD domain to provide subtle structural discrimination between tRNAGln and tRNAGlu prior to the appearance of discriminatory residues in other synthetase domains conserved between eukaryotes and bacteria.
In support of this, we find evidence that the NTD of GlnRS likely existed in the common eukaryotic ancestor, based on comparative genomic reconstruction of the Gln4 family (52). Thus, GlnRS proteins from highly diverse, free living eukaryotes, spanning lineages from the ancient JEH and POD clades through more recent clades (including Plantae, Amoebozoans and Opisthokonts) share a recognizably homologous, but diverse, NTD of 210–259 amino acids (Figure 4B and Supplementary Figure S4). Curiously, we and others (53) have also found that the appended domain is absent in some eukaryotes, including parasitic protozoa such as Trypanosoma brucei and Leishmania major, as well as the Eurotiomycetidae, Trichocomaceae fungi. There also appears to be a correlation between the presence of the appended domain and the use of U73 as the discriminator base (Supplementary Figure S5). Thus, although an appended domain is not required to construct a specific GlnRS, such a domain was likely a part of the specific GlnRS in the eukaryotic common ancestor and may have played a crucial role in the development of a specific GlnRS.
Our findings also point to a parallel between the appended domains in eukaryotic GlnRS proteins and in GlnRS in the bacterium D. radiodurans (14), even though the eukaryotic domains are located on the N terminus, upstream of the conserved core, while the appended domain of the D. radiodurans GlnRS is on the C terminus, downstream of the conserved core. Although the Gln4 NTD and the D. radiodurans GlnRS CTD have no significant sequence similarity (14), and are at opposite termini, it is likely that the D. radiodurans GlnRS CTD, like the Gln4 NTD, is structurally related to GatB, because the CTD has weak sequence homology with regions of GatB, and cross reacts with GatB antibody (14).
ACCESSION NUMBER
The structure was deposited as PDB ID 3TL4.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1–5 and Supplementary Reference [54].
FUNDING
Defense Threat Reduction Agency (Grant HDTRA1-10-C-0057 to E.H.S.) and National Institutes of Health (Grant U54 GM074899 to George DeTitta and Grant GM63713 to John Perona). Funding for open access charge: Defense Threat Reduction Agency.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, supported by DOE and NIH.
REFERENCES
- 1.Mirande M. Processivity of translation in the eukaryote cell: role of aminoacyl-tRNA synthetases. FEBS Lett. 2010;584:443–447. doi: 10.1016/j.febslet.2009.11.027. [DOI] [PubMed] [Google Scholar]
- 2.Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev. Mol. Cell. Biol. 2010;11:668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cusack S, Berthet-Colominas C, Hartlein M, Nassar N, Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990;347:249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- 4.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 5.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curnow AW, Hong K, Yuan R, Kim S, Martins O, Winkler W, Henkin TM, Soll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl Acad. Sci. USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tumbula DL, Becker HD, Chang WZ, Soll D. Domain-specific recruitment of amide amino acids for protein synthesis. Nature. 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- 8.Ibba M, Soll D. Aminoacyl-tRNAs: setting the limits of the genetic code. Genes Dev. 2004;18:731–738. doi: 10.1101/gad.1187404. [DOI] [PubMed] [Google Scholar]
- 9.Sauerwald A, Zhu W, Major TA, Roy H, Palioura S, Jahn D, Whitman WB, Yates JR, III, Ibba M, Soll D. RNA-dependent cysteine biosynthesis in archaea. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- 10.Liu C, Gamper H, Shtivelband S, Hauenstein S, Perona JJ, Hou YM. Kinetic quality control of anticodon recognition by a eukaryotic aminoacyl-tRNA synthetase. J. Mol. Biol. 2007;367:1063–1078. doi: 10.1016/j.jmb.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamour V, Quevillon S, Diriong S, N'Guyen VC, Lipinski M, Mirande M. Evolution of the Glx-tRNA synthetase family: the glutaminyl enzyme as a case of horizontal gene transfer. Proc. Natl Acad. Sci. USA. 1994;91:8670–8674. doi: 10.1073/pnas.91.18.8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nureki O, O'Donoghue P, Watanabe N, Ohmori A, Oshikane H, Araiso Y, Sheppard K, Soll D, Ishitani R. Structure of an archaeal non-discriminating glutamyl-tRNA synthetase: a missing link in the evolution of Gln-tRNAGln formation. Nucleic Acids Res. 2010;38:7286–7297. doi: 10.1093/nar/gkq605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludmerer SW, Schimmel P. Gene for yeast glutamine tRNA synthetase encodes a large amino-terminal extension and provides a strong confirmation of the signature sequence for a group of the aminoacyl-tRNA synthetases. J. Biol. Chem. 1987;262:10801–10806. [PubMed] [Google Scholar]
- 14.Deniziak M, Sauter C, Becker HD, Paulus CA, Giege R, Kern D. Deinococcus glutaminyl-tRNA synthetase is a chimer between proteins from an ancient and the modern pathways of aminoacyl-tRNA formation. Nucleic Acids Res. 2007;35:1421–1431. doi: 10.1093/nar/gkl1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rould MA, Perona JJ, Soll D, Steitz TA. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989;246:1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- 16.Ludmerer SW, Schimmel P. Construction and analysis of deletions in the amino-terminal extension of glutamine tRNA synthetase of Saccharomyces cerevisiae. J. Biol. Chem. 1987;262:10807–10813. [PubMed] [Google Scholar]
- 17.Ludmerer SW, Wright DJ, Schimmel P. Purification of glutamine tRNA synthetase from Saccharomyces cerevisiae. A monomeric aminoacyl-tRNA synthetase with a large and dispensable NH2-terminal domain. J. Biol. Chem. 1993;268:5519–5523. [PubMed] [Google Scholar]
- 18.Wang CC, Schimmel P. Species barrier to RNA recognition overcome with nonspecific RNA binding domains. J. Biol. Chem. 1999;274:16508–16512. doi: 10.1074/jbc.274.23.16508. [DOI] [PubMed] [Google Scholar]
- 19.Whelihan EF, Schimmel P. Rescuing an essential enzyme-RNA complex with a non-essential appended domain. EMBO J. 1997;16:2968–2974. doi: 10.1093/emboj/16.10.2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whipple JM, Lane EA, Chernyakov I, D'Silva S, Phizicky EM. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev. 2011;25:1173–1184. doi: 10.1101/gad.2050711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherman F, Fink G, Hicks JB. Methods in Yeast Genetics. New York: Cold Spring Harbor Laboratory Press; 1986. pp. 145–149. [Google Scholar]
- 22.Quartley E, Alexandrov A, Mikucki M, Buckner FS, Hol WG, DeTitta GT, Phizicky EM, Grayhack EJ. Heterologous expression of L. major proteins in S. cerevisiae: a test of solubility, purity, and gene recoding. J. Struct. Funct. Genomics. 2009;10:233–247. doi: 10.1007/s10969-009-9068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macbeth MR, Lingam AT, Bass BL. Evidence for auto-inhibition by the N terminus of hADAR2 and activation by dsRNA binding. RNA. 2004;10:1563–1571. doi: 10.1261/rna.7920904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malkowski MG, Quartley E, Friedman AE, Babulski J, Kon Y, Wolfley J, Said M, Luft JR, Phizicky EM, DeTitta GT, et al. Blocking S-adenosylmethionine synthesis in yeast allows selenomethionine incorporation and multiwavelength anomalous dispersion phasing. Proc. Natl Acad. Sci. USA. 2007;104:6678–6683. doi: 10.1073/pnas.0610337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martzen MR, McCraith SM, Spinelli SL, Torres FM, Fields S, Grayhack EJ, Phizicky EM. A biochemical genomics approach for identifying genes by the activity of their products. Science. 1999;286:1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- 26.Jackman JE, Montange RK, Malik HS, Phizicky EM. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA. 2003;9:574–585. doi: 10.1261/rna.5070303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkinson ML, Crary SM, Jackman JE, Grayhack EJ, Phizicky EM. The 2′-O-methyltransferase responsible for modification of yeast tRNA at position 4. RNA. 2007;13:404–413. doi: 10.1261/rna.399607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherlin LD, Bullock TL, Nissan TA, Perona JJ, Lariviere FJ, Uhlenbeck OC, Scaringe SA. Chemical and enzymatic synthesis of tRNAs for high-throughput crystallization. RNA. 2001;7:1671–1678. [PMC free article] [PubMed] [Google Scholar]
- 29.Lyakhov DL, He B, Zhang X, Studier FW, Dunn JJ, McAllister WT. Mutant bacteriophage T7 RNA polymerases with altered termination properties. J. Mol. Biol. 1997;269:28–40. doi: 10.1006/jmbi.1997.1015. [DOI] [PubMed] [Google Scholar]
- 30.Uter NT, Perona JJ. Long-range intramolecular signaling in a tRNA synthetase complex revealed by pre-steady-state kinetics. Proc. Natl Acad. Sci. USA. 2004;101:14396–14401. doi: 10.1073/pnas.0404017101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bullock TL, Uter N, Nissan TA, Perona JJ. Amino acid discrimination by a class I aminoacyl-tRNA synthetase specified by negative determinants. J. Mol. Biol. 2003;328:395–408. doi: 10.1016/s0022-2836(03)00305-x. [DOI] [PubMed] [Google Scholar]
- 32.Ibba M, Hong KW, Sherman JM, Sever S, Soll D. Interactions between tRNA identity nucleotides and their recognition sites in glutaminyl-tRNA synthetase determine the cognate amino acid affinity of the enzyme. Proc. Natl Acad. Sci. USA. 1996;93:6953–6958. doi: 10.1073/pnas.93.14.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luft JR, Collins RJ, Fehrman NA, Lauricella AM, Veatch CK, DeTitta GT. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J. Struct. Biol. 2003;142:170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 34.Soltis SM, Cohen AE, Deacon A, Eriksson T, Gonzalez A, McPhillips S, Chui H, Dunten P, Hollenbeck M, Mathews I, et al. New paradigm for macromolecular crystallography experiments at SSRL: automated crystal screening and remote data collection. Acta Crystallogr. D Biol. Crystallogr. 2008;64:1210–1221. doi: 10.1107/S0907444908030564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popov AN, Bourenkov GP. Choice of data-collection parameters based on statistic modelling. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1145–1153. doi: 10.1107/s0907444903008163. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez A, Moorhead P, McPhillips SE, Song J, Sharp K, Taylor JR, Adams PD, Sauter NK, Soltis SM. Web-Ice: integrated data collection and analysis for macromolecular crystallography. J. Appl. Crystallogr. 2008;41:176–184. [Google Scholar]
- 37.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manning JM, Moore S, Rowe WB, Meister A. Identification of L-methionine S-sulfoximine as the diastereoisomer of L-methionine SR-sulfoximine that inhibits glutamine synthetase. Biochemistry. 1969;8:2681–2685. doi: 10.1021/bi00834a066. [DOI] [PubMed] [Google Scholar]
- 44.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakamura A, Sheppard K, Yamane J, Yao M, Soll D, Tanaka I. Two distinct regions in Staphylococcus aureus GatCAB guarantee accurate tRNA recognition. Nucleic Acids Res. 2010;38:672–682. doi: 10.1093/nar/gkp955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito T, Yokoyama S. Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions. Nature. 2010;467:612–616. doi: 10.1038/nature09411. [DOI] [PubMed] [Google Scholar]
- 47.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 48.Emekli U, Schneidman-Duhovny D, Wolfson HJ, Nussinov R, Haliloglu T. HingeProt: automated prediction of hinges in protein structures. Proteins. 2008;70:1219–1227. doi: 10.1002/prot.21613. [DOI] [PubMed] [Google Scholar]
- 49.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerstein M, Lesk AM, Chothia C. Structural mechanisms for domain movements in proteins. Biochemistry. 1994;33:6739–6749. doi: 10.1021/bi00188a001. [DOI] [PubMed] [Google Scholar]
- 51.Sheppard K, Soll D. On the evolution of the tRNA-dependent amidotransferases, GatCAB and GatDE. J. Mol. Biol. 2008;377:831–844. doi: 10.1016/j.jmb.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 53.Rinehart J, Horn EK, Wei D, Soll D, Schneider A. Non-canonical eukaryotic glutaminyl- and glutamyl-tRNA synthetases form mitochondrial aminoacyl-tRNA in Trypanosoma brucei. J. Biol. Chem. 2004;279:1161–1166. doi: 10.1074/jbc.M310100200. [DOI] [PubMed] [Google Scholar]
- 54.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.