Abstract
We previously reported that miR-1 is among the most consistently down-regulated miRs in primary human prostate tumors. In this follow-up study, we further corroborated this finding in an independent data set and made the novel observation that miR-1 expression is further reduced in distant metastasis and is a candidate predictor of disease recurrence. Moreover, we performed in vitro experiments to explore the tumor suppressor function of miR-1. Cell-based assays showed that miR-1 is epigenetically silenced in human prostate cancer. Overexpression of miR-1 in these cells led to growth inhibition and down-regulation of genes in pathways regulating cell cycle progression, mitosis, DNA replication/repair and actin dynamics. This observation was further corroborated with protein expression analysis and 3′-UTR-based reporter assays, indicating that genes in these pathways are either direct or indirect targets of miR-1. A gene set enrichment analysis revealed that the miR-1-mediated tumor suppressor effects are globally similar to those of histone deacetylase inhibitors. Lastly, we obtained preliminary evidence that miR-1 alters the cellular organization of F-actin and inhibits tumor cell invasion and filipodia formation. In conclusion, our findings indicate that miR-1 acts as a tumor suppressor in prostate cancer by influencing multiple cancer-related processes and by inhibiting cell proliferation and motility.
INTRODUCTION
MicroRNAs (miRs) are small non-coding RNAs that control mRNA stability and the translation of target mRNAs by binding to regulatory sites which are mostly located in the 3′-untranslated region (UTR) of the transcript (1). Expression of these non-coding RNAs is altered in human tumors, resulting in distinct miR networks for the various tumor types (2–4). Numerous miRs have been shown to display tumor suppressor activity while others act like oncogenes (5–11). Alterations in the expression of these miRs have been linked to cancer development and metastasis and may predict disease outcome and response to therapy (6,8,12–17). Known mechanisms that cause dysregulated miR expression in tumors include genomic alterations and epigenetic promoter silencing (18–20). In addition, feedback loops between miRs and their targets are sometimes modified in cancer cells because these cells commonly express transcripts encoding growth-regulatory genes with shortened 3′-UTRs (21,22).
We and others have shown that miRs and components of the intracellular miR machinery show widespread dysregulation in prostate cancer biology (3,10,11,23–30). However, we have still an incomplete understanding of how prostate cancer-associated miRs affect disease progression because few studies have characterized miRs that are functionally linked to disease recurrence and metastasis (10,31,32). Here, we pursued the hypothesis that miR-1 is a tumor suppressor gene in prostate cancer that can serve as a prognostic marker based on our prior observation that miR-1 was commonly under-expressed in primary human prostate tumors when compared with the surrounding non-cancerous tissue (26).
miR-1 is encoded by the miR-1-133 cluster which has two copies (at 18q11 and 20q13) in the human genome producing identical mature miR sequences for miR-1 and miR-133. miR-1 is abundantly expressed in heart and skeletal muscle tissue (33–35). It was recently reported that miR-1 and miR-133, and also miR-206, which is a functional homolog of miR-1, are among the most frequently down-regulated miRs in solid human cancers (4,36). Other studies showed that miR-1 is expressed in normal human epithelial cells, albeit at low levels when compared to the heart, but silenced in cancer cells (37,38). To further define the role of miR-1 in prostate cancer progression, we analyzed a large data set consisting of primary tumors, disease metastases and patients’ recurrence status (39). We also re-expressed miR-1 in human prostate cancer cell lines. These studies revealed that miR-1 is a novel candidate marker for disease recurrence in prostate cancer and exhibits a tumor suppressor activity that affects multiple pathways, leading to higher order chromosomal and epigenetic alterations globally similar to those of histone deacetylase inhibitors.
MATERIALS AND METHODS
Cell culture and human prostate tissue samples
The human prostate cell lines, LNCaP, 22Rv1, PC-3 and RWPE-1, were obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained as indicated by the supplier. Fresh frozen human prostate tissues were collected from prostate cancer patients after prostatectomy under the IRB approved protocol ‘A case–control study of prostate cancer in the greater Baltimore area’, NCI IRB #05-C-N021. We prepared frozen serial sections from tumor tissue and adjacent non-cancerous tissue and confirmed presence/absence of tumor in these sections after review of a hematoxylin/eosin-stained section by a pathologist. Genomic DNA was extracted from these sections.
RNA isolation and expression analysis of mRNAs and miRs in cell lines
Total RNA was isolated with TRIZOL reagent (Invitrogen, Carlsbad, CA, USA). Expression profiles of protein coding genes were analyzed on the GeneChip® Human Genome U133A 2.0 array following standard protocols from Affymetrix (Santa Clara, CA, USA). Three independent experiments were performed to profile cell lines. To determine large-scale miR expression profiles, Ohio State University Comprehensive Cancer Center (OSU) arrays were used following previously established methods (40). A description of these arrays can be found at ArrayExpress under the accession numbers MEXP-258 and MEXP-1838. Normalized and raw expression data from this study for both mRNAs and miRs were deposited in GEO (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE31620. GEO also describes the V.4 microRNA platform under the accession number GPL14184.
qRT-PCR analysis of miR expression
For quantification of mature miRs, Human TaqMan® MicroRNA Assays (Applied Biosystems, Carlsbad, CA, USA) and reagents from the TaqMan® MicroRNA Reverse Transcription Kit were used, as previously reported (27). Expression levels of miRs were normalized to U6 RNA levels and presented as 2−ΔCt.
Analysis of epigenetic silenced miRs
LNCaP and 22Rv1 cells were treated with 5-Azacytidine (5-AzaC) and/or trichostatin A (TSA) (Sigma, Saint Louis, MO, USA) following similar schemes reported by others (37). Briefly, cells were plated at 1 × 106 cells per 10 cm2 for 48 h and then treated with dimethylsulfoxide (DMSO) as a control and 5-AzaC (5 µM), and/or TSA (0.3 µM). For combined treatments, TSA was added after 12 h of pre-treatment with 5-AzaC or control. After 36 h, cells were harvested for RNA extraction (n = 4–5 per treatment). OSU miRNA Version 4.0 arrays were used for profiling miR expression levels. Raw GPR data files from these experiments were imported into BRB array tools using the MAS.5 median-based normalization. Probes with low intensities (<10), minimum fold-change (i.e. <20% of expression data values with at least a 1.5-fold change in either direction from the gene's median value) and >50% missing data were excluded. Class comparisons between treatments were carried out using randomized block univariate t-tests across independent experiments.
Cloning of the miR-1-133 cluster into a lentiviral vector
Detailed information describing cloning of the miR-1-133 cluster into a lentiviral expression vector is provided in Supplementary Data.
miR oligonucleotides and transient transfections
Pre-miR mimic (Ambion, TX, USA) or lock nucleic acid (LNA) (Exiqon Inc.,Vedbaek, Denmark) oligonucleotides were used at a 30-nM final concentration in the experiments. Cells were plated in 6-well plates at 2.5 × 105 cells per well, allowed to grow for 48–72 h and then switched to antibiotic free media before being transfected with lipofectamine 2000 (Invitrogen), as directed by the manufacturer.
3′-UTR reporter construct cloning and luciferase assays
Detailed information about the cloning of the 3′-UTR reporter constructs can be found in Supplementary Data. For the luciferase assays, reporter plasmids and miR oligos were co-reverse transfected using lipofectamine 2000 as suggested by the manufacturer (Invitrogen). Briefly, 200 ng of plasmid DNA, 60 pmol of pre-miR oligos and 0.25 µl lipofectamine were complexed individually (25 µl total each) and added in various combinations to 150 µl of 4.5 × 104 cells in 96-well plates. Samples were measured after 48 h using Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA) and read on a Fluroskan Ascent FL Microplate Fluorometer (Thermo Electron, Milford, MA, USA) following the manufacturers’ procedures. Values are presented as the ratio of renilla to firefly luciferase activity (n = 6 per treatment).
Cell cycle analysis with propidium iodide
Cells were transfected as described above with miR oligos mimics and then harvested by trypsinization, washed three times with PBS, fixed with 70% cold ethanol before being stained with a 5-µg/ml propidium iodide (Sigma, MO, USA)/20 ng/ml RNase A (Roche)/PBS solution for 30 min. At least 10 000 events were analyzed for each sample using a FACSCalibur cytometer (BD Biosciences, Chicago, IL, USA), cell phase percentages were determined using the FlowJo software (Ashland, OR, USA).
Western blot analysis and antibodies
Cells were lysed in radioimmunoprecipitation assay buffer (RIPA; Sigma) and 25–50 µg protein per lane were separated on SDS–polyacrylamide gels and transferred to nitrocellulose membranes using an iBlot® Dry Blotting System (Invitrogen). Immunoblotting was performed using the following primary antibodies using the dilutions recommended by the supplier: Phospho-histone H2AX (Ser139) (Cell Signaling Technology, Danvers, MA, USA; #2577), H2A.X (#2595), histone H2A (#2578), 53BP1 (#4937), ATM (#2873), ATR (#2790), CHK2 (#2662), NOTCH3 (#2889), MCM7 (Santa Cruz Biotechnology, Santa Cruz, CA, USA; sc-9966), CHK1 (sc-8408), BRCA1 (sc-642), FN1 (sc-9068), LASP-1 (Chemicon International, Billerica, MA, USA; MAB8991), PTMA (Alexis Biochemical, San Diego, CA, USA; 4F4) and β-actin (CalbioChem, San Diego, CA, USA; #CP01). Signals were visualized after incubation with recommended secondary antibody conjugated to peroxidase and the signal was developed using Pierce ECL western blotting substrate (Thermo Fisher Scientific, Rockford, IL, USA).
Matrigel invasion assay
Invasion was measured using the BD BioCoat™ Tumor Invasion System (BD Biosciences) as indicated by the manufacturer. Briefly, 1 × 106 LNCaP, PC-3 or 22Rv1 cells were transfected with 100 nM control or pre-miR-1 oligos using Nucleofector Technology (Lonza Inc. Walkersville, MD, USA) and cell line-optimized protocols as defined by the manufacturer. After 48 h of growth in 6-well plates, cells were pre-labeled with growth media containing 10 µg/ml DilC12(3) fluorescent dye (BD Biosciences). Next, 2.5 × 104 cells in 500 µl of serum-free RPMI were seeded in triplicate into the apical cambers of both Matrigel coated and non-coated Fluroblok 24-Multiwell insert systems. Basal chambers were filled with 750 µl of RPMI including 10% FBS as a chemoattractant. After 48 h, cells that had migrated through the membrane were imaged using an inverted fluorescent microscope (20×) and quantified from at least three images per well using ImageJ software. Data is expressed as percent invasion through the Matrigel coated membrane relative to the average number of cells migrating through the non-coated membrane.
Clonogenic assay
Details can be found in Supplementary Data.
LASP1 and F-actin fluorescence labeling and determination of mitotic index
Assay details can be found in Supplementary Data.
DNA methylation analysis
Details can be found in Supplementary Data.
miR-1-133 cluster lentiviral construct and xenograft model
Lentiviral plasmid DNA was co-transfected with the Invitrogen packaging plasmids into HEK293T cells to generate VSV-g pseudotyped lentivirus particles. The cells were re-fed with DMEM media, 10% FBS, 1% Penn-Strep 24-h post-transfection and the culture supernatant was harvested 48-h post-transfection. The crude lentivirus stock was centrifuged and filtered to remove cells and other debris. Clarified lentivirus was concentrated using the Amicon filters, and virus titer was determined using flow cytometry-GFP detection. Prostate cancer cells were transduced with modified-Tween (Control) and miR-1-133 cluster lentiviral construct, expanded and sorted for GFP-positive cells. To allow for in vivo imaging of xenografts, all cells were also transduced with lentiviruses packaged with the pFerH_ffLuc2-mCherry construct (41). Xenografts were carried out by dual flank s.c. injections of 5- to 6-week-old athymic nu/nu nude mice (NCI-Animal Genetics and Production Facility, Frederick, MD, USA) with a 200 -µl mixture containing 1 × 106 of control (Tween) or miR-1-133 cluster infected cells suspended in a 1:1 solution of growth media and high-concentration Matrigel matrix (BD Biosciences). Tumor size was measured and tumor volume was calculated using the following equation: tumor volume = (W2 × L)/2 formula. For in vivo imaging, animals were intraperitoneally injected with a solution of d-Luciferin (Caliper Life Sciences, Alameda, CA, USA) and imaged with a Xenogen IVIS system (Caliper Life Sciences, Alameda, CA, USA) after 15–20 min following protocols provided by the manufacturer. All mouse studies were performed in accordance with Animal Study Protocols approved by the Animal Care and Use Committee, National Cancer Institute, National Institutes of Health.
Statistical analysis and bioinformatics
All statistical tests were two-sided, and an association was considered statistically significant with P-values <0.05. The Kaplan–Meier survival method, Cox proportional-hazards regression analysis, and the log-rank test of equality of the survival function were used for disease recurrence analysis. Affymetrix and miR array expression data from cell line-based experiments were analyzed with customized R scripts (http://www.r-project.org) after data normalization within R using robust multi-array average (RMA) for normalized intensities and MAS5 detection calls to filter out bad probes. Lists of differentially expressed genes were generated with the statistical analysis of microarray (SAM) procedure and defined using P-values from two-sided t-tests, fold changes and/or false discovery rate (FDR) cutoffs. We used BRB-ArrayTools (http://linus.nci.nih.gov/BRB-ArrayTools) to analyze mRNA and miR expression data in human prostate tumors and metastases from a publically available data set, deposited in NCBI GEO under accession number GSE21032 (39). Calculation of P-values and fold changes of differentially expressed genes were performed using the Partek Genomics Suite software (Partek Inc, St. Louis, MI, USA). Kaplan–Meier survival analysis was performed using GraphPad Prism software (La Jolla, CA, USA). Log-rank tests were performed on gene expression values that were stratified using the median as cutoff. The TargetScan database (http://www.targetscan.org) was used to define miR seed sites within miR 3′-UTRs. miR-1 targets were defined within Partek based on the TargetScan 5.0 database. Target enrichment calculations, pathway analysis and clustering, and heat maps were produced using in-house Whole Pathway Scope software, as previously described (42–44). Cluster analyses and heat map outputs of these analyses were based on ranked Fischer exact test P-values that were transformed into enrichment scores using the formula [−log10(P-value)]. Connectivity mapping (cmap) was carried out on altered gene lists using the web interface http://www.broad.mit.edu/cmap and filtered for mean connectivity scores better than ±0.6 and permutation P-values ≤0.05 for cmap query data with heat map output showing permutation P-values expressed as −log10(P-value). List Hits <2 or P-value >0.05 filtering was used to floor the scores to 0 for all heat maps. Differentially expressed gene lists were further analyzed with the Ingenuity Pathways Analysis program (Ingenuity® Systems, www.ingenuity.com) to identify miR-1-related gene networks.
RESULTS
miR-1 is under-expressed in primary and metastatic prostate cancer and is a candidate prognostic marker
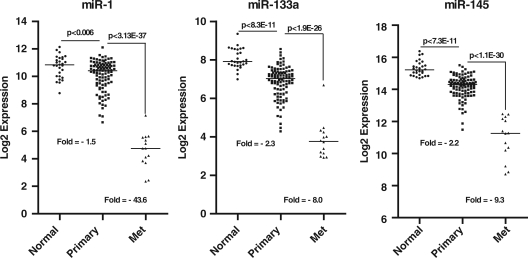
Previously, we reported that miR-1 and miR-133(a) are significantly down-regulated in prostate tumors (26). We followed up on this observation by studying mature miR-1 expression with qRT-PCR in the TRAMP mouse model of prostate cancer (45). This study showed that cancer-prone TRAMP mice at 20 week of age had only 20–30% of the miR-1 expression in their prostate when compared with miR-1 expression in the prostate of same age wild-type control animals (n = 4 each group; data not shown). To corroborate our findings, we analyzed a large publicly available data set consisting of miR and mRNA expression profiles for 99 primary tumors and 14 distant metastasis and patient data for disease recurrence (39). Consistent with our previous findings, both miR-1 and miR-133 a were found to be significantly reduced in primary tumor tissues (Figure 1). Furthermore, analysis of the profiles for distant metastases revealed an additional reduction in miR-1-133a cluster expression in these lesions (∼40-fold) when compared with primary tumor tissues. A separate genome-wide analysis of all miRs in this data set showed that miR-1 was the most reduced miR in metastatic tissue samples when compared with primary tumors (P < 1 × 10−36). For comparison, we also plotted the relative expression of the well characterized miR-145 tumor suppressor (30,46) for the same tissues (Figure 1). miR-145 followed a trend very similar to that of the miR-1-133a cluster with an incrementally reduced expression of this miR from adjacent non-cancerous tissue to primary tumor to metastasis.
Figure 1.
miR-1-133 cluster expression in primary and metastatic prostate tissues. Relative expression levels of miR-1, −133a and 145 in primary prostate tumors (n = 99) and metastatic lesions (n = 14). Data represent log2 expression values. P-values were calculated with an ANOVA-based post hoc t-test.
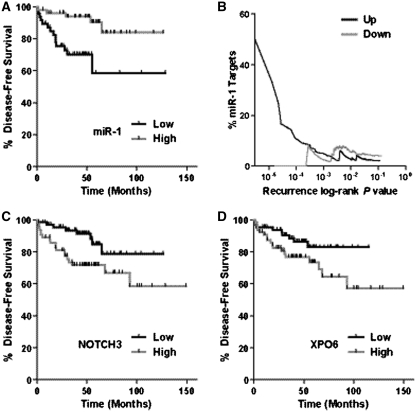
Next, we tested whether miR-1 and miR-133a are candidate prognostic markers. In a Kaplan–Meier analysis for disease recurrence, we found that miR-1 was associated with disease recurrence (Figure 2A). Tumors that had reduced miR-1 levels below the median were significantly more likely to have an early recurrence than tumors with above median miR-1 expression. The association of miR-1 with disease recurrence was independent of age and Gleason score in the adjusted Cox regression analysis (hazard ratio = 0.29; 95% confidence interval: 0.1–0.9; high- versus low-tumor miR-1). We did not find the same association with survival for miR-133a, nor was expression of the paralog miR-206-133b cluster associated with recurrence (Supplementary Figure S1). To assess to what extent predicted miR-1 target genes contribute to early disease recurrence, we plotted the enrichment of miR-1 target genes among all recurrence-associated genes (% miR-1 targets among all mRNAs at a given P-value for the association with disease recurrence) on a log-rank test-based P-value scale (Figure 2B). When the recurrence-associated mRNAs were sub-divided into up-regulated and down-regulated transcripts in the primary prostate tumors, we found a significant enrichment of miR-1 targets (‘% miR-1 target’) only among those transcripts which predict recurrence and are also up-regulated in the primary tumor, consistent with the loss of miR-1 in these tumor tissues. This enrichment increased with decreasing log-rank P-value (Figure 2B), indicating a particular enrichment for miR-1 target transcripts at the lowest false discovery for recurrence-associated transcripts. Among the recurrence-associated genes that are known miR-1 targets were NOTCH 3 (Figure 2C) and exportin-6 (Figure 2D). The findings from our miR-1 target enrichment analysis support the hypothesis that down-regulation of miR-1 in primary tumors leads to an increased risk of disease recurrence because of increased expression of oncogenic mRNAs that are otherwise down-regulated by miR-1.
Figure 2.
miR-1 is a prognostic marker. (A) Kaplan–Meier analysis of disease recurrence comparing high- versus low-miR-1 expression in the primary tumor. Tumors that had miR-1 levels below the median were significantly more likely to have an early recurrence (P = 0.008; log-rank test). (B) Enrichment of predicted miR-1 target transcripts among all recurrence-associated transcripts (‘% miR-1 target’) on a log-rank P-value scale for recurrence (P-value for the association between a mRNA and disease recurrence). Disease recurrence-associated transcripts, sub-divided into either up-regulated (‘up’) or down-regulated (‘down’) transcripts in the primary tumors, were plotted with their log-rank P-value from the recurrence analysis against enrichment of transcripts that are predicted miR-1 targets (TargetScan 5.0-based percent miR-1 targets). Among 1541 transcripts that were associated with recurrence at P ≤ 0.05 in the unstratified analysis, 61 were predicted miR-1 targets [P for enrichment of miR-1 targets (Fisher's exact-based) <0.001]. At more stringent P-value cutoffs (e.g. 10−4 or 10−5), the enrichment for miR-1 targets increased notably from 5% to over 40%. (C) Kaplan–Meier analysis of disease recurrence comparing high- versus low-NOTCH3 expression in the primary tumor. Tumors that had NOTCH3 levels above the median were significantly more likely to have an early recurrence (P = 0.004; log-rank test). (D) Kaplan–Meier analysis of disease recurrence comparing high- versus low-XPO6 (exportin-6) expression in the primary tumor. Tumors that had XPO6 levels above the median were significantly more likely to have an early recurrence (P = 0.02; log-rank test).
miR-1 family members are epigenetically regulated
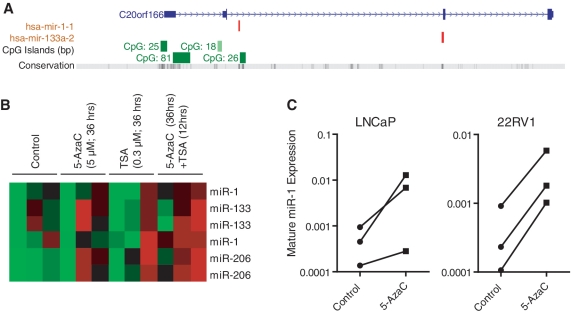
Our studies showed that the miR-1-133a cluster is under-expressed in cancerous lesions of the prostate. Previously, methylation-mediated silencing of miR-1 was found to occur in human hepatocellular carcinoma (37), consistent with the presence of regulatory CpG islands adjacent to the cluster locus (Figure 3A). To explore whether miR-1-133a is epigenetically silenced in prostate cancer, LNCaP human prostate cancer cells were treated with either the DNA hypomethylating agent, 5-azacytidine (5-AzaC) or the histone deacetylase inhibitor, trichostatin A (TSA), or with the combination of the two agents, and changes in global miR expression were examined using the OSU microarray (see methods). We observed that miR-1 and miR-133(a/b) expression was at the detection limit in untreated LNCaP cells but expression was specifically re-activated by the combination of 5-AzaC and TSA (Figure 3B). This experiment also showed that miR-206 and several other miRs (Supplementary Table S1) are epigenetically silenced in LNCaP cells. To further test, whether 5-AzaC alone can increase miR-1 expression, LNCaP and 22Rv1 human prostate cancer cells were treated in a second experiment with 5 µM 5-AzaC for 36 h, and miR-1 expression was evaluated by qRT-PCR. This experiment revealed increased mature miR-1 expression in these cells after 5-AzaC treatment (Figure 3C). To address the question whether miR-1 is promoter-methylated in the cancerous prostate, we analyzed cancer tissue from four prostate cancer patients with localized disease and the same number of surrounding non-cancerous tissues. We performed methylation analysis of the CpG island-81 within the promoter of the miRNA-1-1 gene. This CpG island was shown to be hypermethylated in the hepatocellular carcinomas (37). From our analysis using EpiTect Methyl qPCR Assay technology, we obtained evidence that this locus shows detectable DNA methylation in all analyzed prostate tissues, but hypermethylation was only present in tumors (two out of four) and not in the non-cancerous tissues (Supplementary Figure S2). The data are consistent with findings from human hepatocellular carcinoma and indicate that promoter hypermethylation may contribute to reduced miR-1 expression in at least a subset of human prostate tumors.
Figure 3.
Epigenetic regulation of miRNA-1-133 cluster miRNAs. (A) CpG islands are positioned upstream of the miR-1(-1)-133a regions. The USCS genome browser was used to identify the upstream CpG islands. (B) Heatmap showing miR microarray results for miR-1-133 family members from three independent experiments with LNCaP cells treated with either 5-AzaC, TSA or both, as indicated. Results are shown for two probesets for each miR. Red indicates increased expression of a miR (C) qRT-PCR levels showing re-expression of mature miR-1 in LNCaP and 22Rv1 human prostate cancer cells after 5-AzaC treatment (n = 3). For the microarray experiment, LNCaP cells treated with either 5 µM 5-azacytidine (5-AzaC) or 0.3 µM trichostatin A (TSA) for 36 h or the combination of the two agents; 5 µM 5-azaC for 24 h and then both 5 µM 5-AzaC and 0.3 µM TSA for an additional 12 h.
miR-1-induced gene expression alterations in LNCaP cells target cell cycle and DNA repair and show resemblance with signatures induced by histone deacetylase inhibitors
To elucidate candidate effects of miR-1 in prostate cancer, we up-regulated miR-1 in LNCaP cells using pre-miR treatment and examined miR-1-induced gene expression alterations on a global scale with Affymetrix arrays. For comparison, we also introduced into these cells miR-206, which is predicted to generate gene expression alterations very similar to miR-1, and miR-27, which is functionally unrelated to miR-1. The expression data for these miRs in the LNCaP cells are shown in Supplementary Figure S3. The expression of numerous mRNAs was found to be altered by miR-1 (Supplementary Table S2) and the other two miRs, miR-206 and miR-27, after exposure of LNCaP cells to pre-miRs for 24 h. When we queried the lists of differentially expressed mRNAs for miR seed sites within their 3′-UTR using TargetScan, a significant enrichment for these sites was observed among the down-regulated, but not up-regulated, transcripts for all 3 miRs, validating our experimental approach (Supplementary Figure S4a and Supplementary Table S3). Enrichment of predicted miR-1 and miR-206 targets among all down-regulated genes reached significant levels of 1.35 × 10−24 and 7.77 × 10−26, respectively, in the LNCaP cells, while exposure of these cells to the unrelated miR-27 did not lead to any enrichment of these miR-1/miR-206 targets among the down-regulated genes. Moreover, miR-1 and miR-206 were found to regulate abundance of many of the same transcripts (Supplementary Figure S4b), as one would expect from two homologous miRs. Collectively, the data indicate that our experiment yielded a valid list of target genes for miR-1.
Next, we queried the list of miR-1/miR-206-induced gene expression alterations for their relationship with biological processes using gene ontology annotations and also performed a cluster analysis for enrichment of differentially expressed mRNAs in these processes. This type of approach can reveal key cell functions that are influenced by a miR, either directly or indirectly. Several significant relationships between down-regulated mRNAs and specific biological processes were observed (Supplementary Figure S5a). Two main clusters defining distinct cell functions were significantly enriched for mRNAs whose expression is reduced by miR-1 and miR-206. Enriched cluster 1 included cell cycle, DNA replication and mitosis-related processes, whereas enriched cluster 2 contained DNA repair and cell cycle checkpoint-related processes. Other analysis results indicated miR-1-induced disturbances in chromosome segregation, pointing to a candidate effect of miR-1 on actin filament network organization by targeting genes like LASP1, fibronectin, exportin-6 or twinfilin-1/PTK9. Among all biological processes, cell cycle and DNA repair response had the greatest enrichment in transcripts that were down-regulated by miR-1, e.g. BRCA1 or CH(E)K1 (Supplementary Figure S5b). Notable, however, we did not observe that these cell cycle and DNA repair-related transcripts were particularly enriched for miR-1 seed sites in their 3′-UTR. A more detailed pathway analysis using the union of miR-1 and miR-206 down-regulated transcript lists identified two miR-1/miR-206 influenced networks. Numerous transcripts that encode for proteins in the origin recognition complex (ORC), which is essential for the initiation of DNA replication, were found to be repressed by miR-1/miR-206 (Supplementary Figure S5c). This included transcripts from the mini-chromosomal maintenance (MCM) family of genes; MCM2, MCM4, MCM7, MCM10 and cell division cycle-6 (CDC), CDC7, ORC6L and ORC1L. Pathway analysis also identified a miR-1-regulated network within the G2/M DNA damage checkpoint (Supplementary Figure S5d): breast cancer 1 early onset (BRCA1), CHK1, CDC25, CDC25, CDC25A, CDC25B/C and CDK1.
Having made these observations, we further explored the global effect of miR-1 in LNCaP cells by utilizing Connectivity Map (cmap) web tools (47). This analysis approach, also referred to as Gene Set Enrichment Analysis, uses the Kolmogorov–Smirnov statistics-based profile pattern matching method and can be used to compare the miR-1-induced gene expression alterations in LNCaP cells with transcriptional signatures induced by bioactive molecules that are curated in the cmap database. Our analysis discovered significant similarities between several anticancer drug-induced gene signatures and the miR-1-induced gene signature (Supplementary Figure S6). Among the top ranked molecules were two histone deacetylase inhibitors, vorinstat and trichostatin A, indicating that conferring miR-1 expression to prostate cancer cells led to gene expression alterations similar to those of histone deacetylase inhibitors.
Validation of miR-1 altered genes
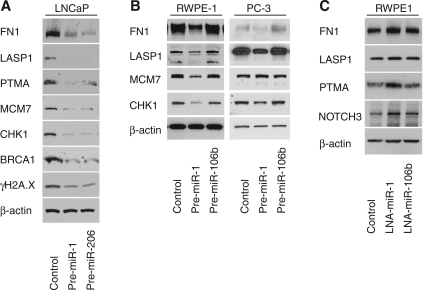
From our gene expression analysis experiments, it became evident that pathways related to cell cycle control, DNA damage response and actin filament network functions are partly controlled by miR-1. Thus, we performed Western blot analysis and 3′-UTR reporter assays for miR interference with translation, using two human prostate cancer cell lines, LNCaP and PC-3, and in the non-invasive RWPE-1 human prostate epithelial cell line to further corroborate the gene expression data (Figure 4). In this validation step, we studied several predicted targets of miR-1, such as FN1 (fibronectin), LASP1 (LIM and SH3 protein 1) and PTMA (prothymosin alpha) that were found to be significantly down-regulated in pre-miR-1 treated LNCaP cells, and also examined selected key genes in cell cycle and DNA damage control (BRCA1, CHK1, MCM7). The actin filament network-associated proteins encoded by FN1 and LASP1 were found to be down-regulated by miR-1, consistent with the array data and previous findings that another protein in this pathway, exportin-6 (XPO6), is targeted by miR-1 in human prostate tumors (26). We also confirmed that BRCA1, CHK1, MCM7 and PTMA are decreased by miR-1 at the protein level. Introduction of miR-206 caused protein level alterations very similar to miR-1, as one may expect (Figure 4A). In contrast, the introduction of miR-106b, a miR functionally unrelated to miR-1, did not cause the same protein expression alterations when examined in the RWPE-1 and PC-3 cell lines (Figure 4B). Introduction of an antisense oligo into immortalized RWPE1 cells to block miR-1 by lock-nucleic acid (LNA) technology led to a modest up-regulation of several of these proteins and also of NOTCH3, an experimentally verified miR-1 target (Figure 4C), consistent with miR-1 inhibition. Lastly, we examined whether miR-1 may affect γH2A.X levels in LNCaP cells. γH2A.X is a surrogate marker for DNA double-strand breaks and is involved in a repair pathway that includes BRCA1. Our examination of γH2A.X marker expression showed that elevated miR-1 causes a reduction of histone H2A.X phosphorylation (Figure 4A), commonly referred to as γH2A.X, suggesting that some of the miR-1 effects may signal through the γH2A.X pathway.
Figure 4.
Effects of miR-1 on protein expression in LNCaP cells. (A) Western blot with protein extracts from LNCaP cells treated with either 30 nM of pre-miR-1, pre-miR-206 or scrambled miR negative control for 48 h. (B) Western blot with protein extracts from RWPE-1 and PC-3 cells treated with either 30 nM of pre-miR-1, pre-miR-106b or scrambled miR negative control for 48 h. (C) Immunoblotting of predicted miR-1 target genes in non-invasive RWPE-1 cells transfected with inhibitory lock-nucleic acid (LNA) miR oligos targeting either miR-1 or miR-106b. Control cells were transfected with unspecific LNA oligos. miR-106b was used to show specificity for miR-1. β-actin was used as a loading control and 50 µg of protein extract was loaded per lane. Immunoblotted proteins: mini-chromosomal maintenance-7 (MCM7), breast cancer 1 early onset (BRCA1), checkpoint kinase 1 (CHK1), fibronectin 1 (FN1), LIM and SH3 protein (LASP1), Notch homolog 3 (NOTCH3) and prothymosin alpha (PTMA).
miR-1 may regulate cancer phenotypes by direct and indirect mechanisms. We could not find candidate miR-1 seed sites in the 3′-UTR of key genes in cell cycle and DNA damage control (e.g. BRCA1, CHK1 and MCM7), suggesting that miR-1 may target these genes indirectly. To further address this point, we performed analyses of the 5′-UTR and coding sequence of all miR-1/206 down-regulated genes to identify additional candidate miR-1 seed sites. This analysis showed that several cell cycle/DNA damage gene transcripts contain candidate miR-1 seed sites within their CDS or 5′-UTR region (Supplementary Table S4). For example, the BRCA-1 transcript contains a candidate miR-1 seed site in the coding sequence. On the other hand, we were able to identify potential seed sites in the 3′-UTR of FN1, LASP1 and PTMA. Therefore, we performed 3′-UTR reporter assays for miR-1 interference with translation using reporter constructs containing wild-type and mutant 3′-UTRs of FN1, LASP1 and PTMA. XPO6 and NOTCH3 have previously been shown to be a direct target by miR-1 or miR-206 (26,48). Thus, we included the 3′-UTRs of these genes in our analysis as positive controls. For all 3′-UTR constructs, transfection of LNCaP cells with miR-1 reduced luciferase activity of the wild-type construct, and this reduction was attenuated when a mutation was introduced into the predicted miR-1 seed sites (Supplementary Figure S7). The findings indicate that some of the miR-1 phenotypes in prostate cancer cells could be mediated by miR-1-induced disturbances of the actin filament network by directly targeting expression of FN1, LASP1 and XPO6, among other candidate transcripts that encode actin binding partners.
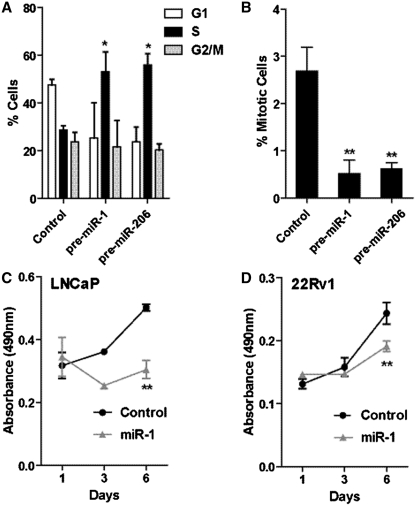
miR-1 inhibits cell cycle progression and represses mitosis and the proliferation of prostate cancer cells
Our gene expression analysis of the miR-1 effects in LNCaP cells showed that this miR targets the cell cycle. Thus, we performed a more in-depth cell cycle analysis of pre-miR-1 and pre-miR-206 transfected LNCaP cells and found that conferred expression of these miRs leads to a significant increase in the number of cells in S-phase (Figure 5A). We further tested whether the number of mitotic cells in cell culture is reduced after exposure to these miRs. Consistent with our microarray data showing that miR-1 down-regulates genes involved in DNA replication and mitosis, the fraction of mitotic LNCaP cells was significantly reduced after treatment with pre-miR-1 and pre-miR-206 oligos (Figure 5B). Moreover, these miRs also reduced cell proliferation, as shown for miR-1 in two human prostate cancer cell lines, LNCaP and 22Rv1 (Figure 5C and D). Together, the findings suggest that miR-1 may reduce proliferation of prostate cancer cells by stalling them in S-phase and preventing cell cycle progression.
Figure 5.
Expression of miR-1 alters cell cycle kinetics and suppresses mitosis and cell proliferation in prostate cancer cells. (A) FACS-based cell cycle analysis of LNCaP cells transfected with 30 nM control, pre-miR-1 or pre-miR-206 oligos. Shown is the mean percentage ± SD of propidium iodide-labeled cells in G1, S and G2/M phases of the cell cycle from three independent experiments. (B) Mitotic figures counted via DAPI staining. Three fields per miR treatment from three independent experiments were counted and presented as percentage mitotic cells. Proliferation of (C) LNCaP and (D) 22Rv1 cells transfected with 30 nM of either control or miR-1 mimic oligos. The results are given as means ± SD. n = 6 from one representative experiment. P-values were calculated using Student's t-test; *P < 0.01, **P < 0.0001.
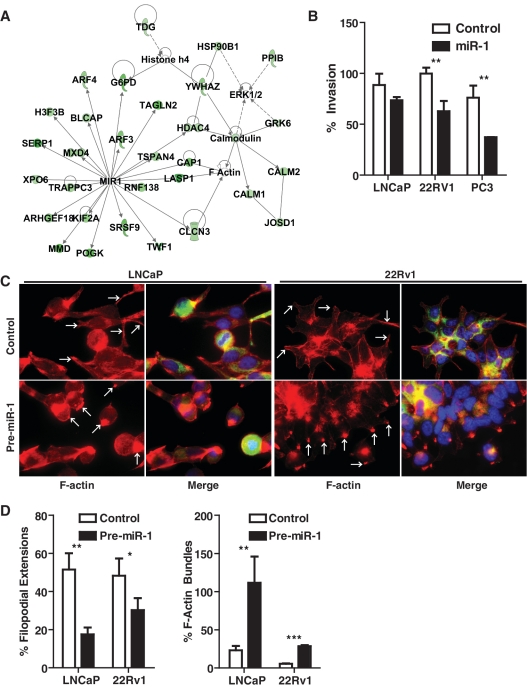
miR-1 inhibits invasion and filipodia formation
The gene expression analysis of LNCaP cells transfected with pre-miR-1 identified a group of down-regulated genes that are part of a distinct expression programming network, indicating that miR-1 may regulate the actin filament network and cell motility (Figure 6A). Within this network, several miR-1 targets are directly associated with F-actin, such as LASP1, and CLCN3, while other miR-1 targets, like G6PD and HDAC4, are indirectly linked to F-actin and histone acetylation processes. Thus, we examined whether miR-1 may affect invasiveness of human prostate cancer cells. As shown in Figure 6B, miR-1 indeed reduced matrigel invasion of prostate cancer cells, with the strongest effect in the highly invasive PC3 cells. We also examined whether the cytoskeletal organization in prostate cancer cells is altered by miR-1. Transfection of LNCaP and 22Rv1 prostate cancer cells with pre-miR-1 led to a loss of filipodia filaments, as shown in Figure 6C and D. The pre-miR-1 transfected cells had F-actin being concentrated in inactive, concentrated bundles with lacking filipodia extensions. Filipodia formation is an essential part of cell motility and is supported by actin polymerization into these filaments. The data suggest that miR-1 inhibits this process. Preliminary data summarized in Supplementary Figure S8 indicate that miR-1 function is perhaps linked to F-actin through regulation of LASP1, a gene which was found by us to be significantly down-regulated in miR-1 expressing prostate cancer cells. LASP1 is a putative oncogene and has a dual function as transcription factor and F actin binding protein which co-localizes with F-actin at peripheral cell extensions including filipodia and lamellipodia (49,50). We found that miR-1 induces re-localization of LASP1 from the nucleus into the cytoplasm (Supplementary Figure S8), which could be a compensatory process following LASP-1 down-regulation by miR-1.
Figure 6.
miR-1 targets F-actin related genes and causes changes in the cellular arrangement of F-actin. (A) Network of miR-1 targets that highlights the candidate regulation of the F-actin filament network and histone acetylation by miR-1. (B) Transwell invasion experiments show that miR-1 can inhibit the invasive potential of human prostate cancer lines. Scrambled control versus pre-miR-1 transfected cells: P < 0.05 (Student's t-test, n = 3). (C) Fluorescence stain for F-actin (red) and merged stains for F-actin, DAPI (blue) and LASP1 (green) in LNCaP and 22Rv1 prostate cancer cells transfected with either control oligos or pre-miR-1. Pre-miR-1 transfected cells showed a generally increased F-actin stain with F-actin being concentrated in inactive, concentrated bundles, as shown by the strong punctate red fluorescence and highlighted by the white arrows (in ‘Pre-miR-1’). Pre-miR transfected cells showed a significant reduction in filipodia extensions, which would arise from the base where the F-actin accumulates. For comparison, arrows highlight the filipodia filaments of the control cells (in ‘Control’). (D) Quantitative assessment of the reduction in filipodia extensions and increase in F-actin bundles by miR-1. Filipodial extensions and F-actin bundles were counted in at least four fields for an average of about 1700 cells per group and shown as a percent of total cells per field ±SD. P-values were calculated using Student's t-test; *P < 0.01, **P < 0.001,***P < 0.0001.
Altered DNA damage response to radiation in prostate cancer cells transfected with pre-miR-1 or expressing the complete miR-1-133 cluster
Our array results showed that miR-1 repressed, at least temporarily, the expression of key genes in the DNA damage response pathway. Furthermore, protein extracts from pre-miR-1 transfected LNCaP cells showed reduced γH2A.X. Thus, we were interested to see whether miR-1 would affect the response of prostate cancer cells to radiation-induced DNA damage. For this purpose, we initially transfected LNCaP and 22Rv1 cells with pre-miR-1 or control oligos and exposed these cells to 6 Gy γ-radiation, followed by 24 h of cell culture, in order to study protein expression of selected DNA damage response genes. Exposure to miR-1 in irradiated cells influenced the expression levels of some DNA damage response genes, leading to increased expression of 53BP1 and reduced levels of γH2A.X protein in both cell lines and to a modest up-regulation of other key regulators in the mammalian DNA damage response, such as ATM (ataxia telangiectasia, mutated) and ATR (ATM and Rad3-related), in the 22Rv1 cells (Supplementary Figure S9a). To further identify the kinetics that led to a reduced γH2A.X accumulation in miR-1 expressing prostate cancer cells, we either transfected 22Rv1 cells with pre-miR-1 or infected them with a lentiviral construct to express the complete miR-1-133 cluster, then irradiated the cells with 6 Gy and prepared cell extracts at various time points post-irradiation. This experiment revealed that miR-1- and miR-1-133 cluster-expressing cells initially accumulate more γH2A.X but remove γH2A.X more quickly than the control cells, leading to reduced γH2A.X levels at later time points (Supplementary Figures S9b and S9c). Lastly, we examined whether miR-1 would influence clonogenic expansion of irradiated cells. Both pre-miR-1 transfected and miR-1-133 cluster expressing 22Rv1 cells were exposed to various amounts of radiation and plated at low density to assess radiation survival, when compared with the control cells, using a traditional clonogenic survival assay as described in Supplementary Methods. 22Rv1 cells with miR-1 expression were found to have fewer radiation-surviving cells, when compared to the control cells, albeit the difference in survival between them was rather modest (Supplementary Figures S9d and S9e). Taken together, our data point to changes in the DNA repair response and to a moderately increased radiosensitivity of miR-1 expressing prostate cancer cells, consistent with the observed increased sensitization of human lung cancer cells to doxorubicin toxicity by miR-1 (38).
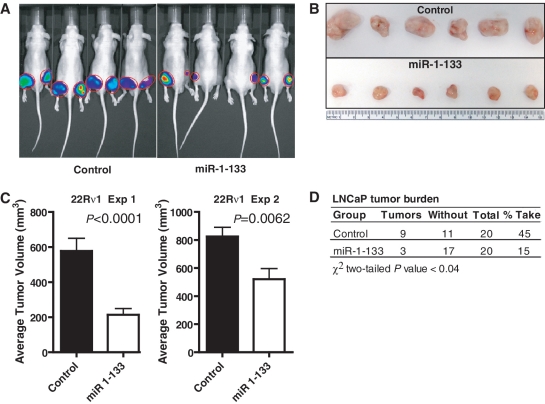
Suppression of prostate cancer xenograft growth by the miR-1-133 cluster
The influence of the complete miR-1-133 cluster on xenograft growth was tested in two subcutaneous tumor models using the 22Rv1 and LNCaP prostate cancer cell lines. 22Rv1 and LNCaP cells were infected with a lentiviral construct engineered to confer miR-1-133 expression (see Supplementary Data). Cluster expression was confirmed by qRT-PCR and showed a 10- to 100-fold up-regulation of these miRs in the infected cell lines. Overexpression of the miR cluster led to reduced tumor growth of 22Rv1 cells, as shown for two independent experiments (Figure 7A–C). In an additional xenograft experiment with LNCaP cells, we did not observed the same significant differences in the tumor volumes between the miR-1-133 and the vector control group. However, we observed a significant reduction of the number of tumors that developed in the miR-1-133 group when compared to the vector control (Figure 7D). Together, the findings are consistent with a tumor suppressor function by the miR-1-133 cluster.
Figure 7.
miR-1-133 cluster suppresses xenograft growth of prostate cancer cells. (A) Representative in vivo images of mice injected with 22Rv1 cells for 3 weeks. Cells were infected with lentiviral constructs either encoding both the complete miR-1-133 cluster and a luciferase reporter gene (miR-1-133) or encoding only the luciferase reporter gene (control). (B) Representative tumor size from this experiment (n = 6 per experimental group). Tumors were resected 3 weeks after initial cell inoculation. (C) Control tumors were significantly larger than tumors resected from animals inoculated with 22Rv1 cells that expressed the miR-1-133 cluster. Two independent experiments are shown. Average ± SEM for tumor size in the control and miR 1-133 groups (n = 20 xenografts per group) at 3-week post-inoculation. P-values calculated via Student's t-test. (D) Tumor growth of LNCaP cells expressing lentiviral constructs either encoding both the complete miR-1-133 cluster and a luciferase reporter gene (‘miR-1-133’) or encoding only the luciferase reporter gene (‘control’). Xenografts were grown for 3 weeks with n = 20 xenografts per group. There was a significant difference in tumor burden between the two groups with a reduced rate of tumor development in mice that were inoculated with miR-1-133 cluster expressing cells. Results are shown with P-values calculated using the Student's t-test.
DISCUSSION
The current study provides novel evidence that miR-1 has tumor suppressor activity in human prostate cancer and is associated with disease recurrence and metastatic spread. These findings are corroborated by a recent report in which the authors used Solexa deep sequencing of 50 organ-confined tumors and found miR-1 to be among the most down-regulated miRs in prostate tumors when compared with non-cancerous prostate tissue (51). The study by Martens-Uzunova et al. also observed, consistent with our observations, that miR-1 is further down-regulated in cancer progression, based on their analysis of lymph node metastases, and is a component of a miR-based predictor for disease outcome. However, our study is now showing, based on the analysis of a larger data set, that miR-1 alone can predict disease recurrence. Furthermore, oncogenic miR-1 target genes, such as NOTCH3, have been associated with recurrence of prostate cancer in our and other studies (32), suggesting that loss of miR-1 leads to increased expression of oncogenes, driving the association of miR-1 with disease outcome. This hypothesis is supported by our analysis of the mRNA expression profiles in the Taylor data set (39), showing that mRNA-based predictors of disease recurrence are enriched for miR-1 target transcripts. Other studies observed that the miR-1 homolog, miR-206, is a candidate tumor suppressor in both rhabdomyosarcoma development (52) and breast cancer metastasis (8), while also inducing apoptosis in a cell-based system of cervical cancer (48). The study by Tavazoie et al. (8) observed that miR-206 induces morphological changes in breast cancer cells, consistent with our observation that miR-1 inhibits formation of filipodia in prostate cancer cells. Thus, both miR-1 and miR-206 may exert similar tumor suppressor activities in cancer initiation and progression in various cancer types, even though we did not observe that miR-206 expression was a predictor of disease recurrence in our study.
miR-1 expression was thought to be largely restricted to heart and skeletal muscle tissue because miR-1-133 cluster expression is highest in these tissues but low or absent in most other tissues (33). However, more recent studies clearly demonstrated that miR-1 is expressed in non-cancerous human epithelial cells of the liver and lung (37,38). These authors also showed that miR-1 is functional in epithelial cells and has tumor suppressor-like properties, consistent with the findings in the current study. Reminiscent of findings in human liver cancer (37), we also observed that down-regulation of miR-1 in prostate cancer cells is most likely due to promoter silencing by epigenetic mechanisms. We could not detect miR-1 expression in the metastatic PC-3 and DU145 human prostate cancer cell lines by a highly sensitive qRT-PCR assay for mature miR-1, but observed a remaining low expression above the detection limit in the 22Rv1 and LNCaP cancer cell lines and a low expression in the immortalized RWPE-1 human prostate epithelial cells. Yet, introduction of antisense miR-1 oligos by LNA technology into the RWPE-1 cells led to the up-regulation of several miR-1 target genes, indicating that even a low expression of miR-1 in these cells is sufficient to partly repress oncogenic target genes like NOTCH3.
Our functional analysis of miR-1 expression in cancer cell lines pointed to a tumor suppressor function that targets processes related to cell cycle progression and cell motility. Additional mechanisms, of a more indirect nature, seem to influence DNA repair pathways and specifically the γH2A.X signaling pathway as a consequence of miR-1 expression. Other miRs have been described that target H2A.X and suppress DNA repair, rendering cells hypersensitive to genotoxic drugs (53). Thus, loss of these miRs would make a cancer cell more resistant to cancer therapy, which has been observed for miR-1 in lung cancer (38). Taken together, our data point to changes in the DNA repair response and to a moderately increased radiosensitivity of miR-1 expressing prostate cancer cells, consistent with the observed increased sensitization of human lung cancer cells to doxorubicin toxicity by miR-1 (38). A reduced sensitivity of prostate tumors with low miR-1 to radiation therapy would be one possible explanation why low-miR-1 expression in human prostate tumors is associated with early disease recurrence.
Because numerous miR-1 targets in LNCaP cells were found by us to be actin filament-associated proteins, it is plausible that some of the observed miR-1 tumor suppressor effects in solid tumors are in fact caused by a disruption of actin cytoskeleton dynamics, leading to infidelities in chromosomal segregation and aberrant mitotic events, which in turn activate cell cycle checkpoints, leading to a stalled S-phase as we observed. Future research in animal models of prostate cancer, including tissue-specific knockout of miR-1, can address these questions since we observed that the loss of miR-1 in cancer development is recapitulated in the TRAMP mouse model of prostate cancer. The inhibition of cell cycle progression in epithelial cells would also explain why miR-1 is only weakly expressed in most epithelial cells, and why members of the miR-1-133 cluster are among the most frequently down-regulated miRs in solid human cancers including prostate cancer (4,26,36,51). Thus, it is not surprising that the miR-1-133 locus is epigenetically silenced in solid tumors.
While our study showed that miR-1 is a candidate tumor suppressor, future studies are needed to further define the tumor suppressor function of miR-1 in the prostate. Currently, we do not know the relative contribution of miR-1 expression in tumor-associated stromal cells to the poor outcome signature described in this and other studies. Mir-1 is expressed in stromal cells of the prostate, though a sensitive qRT-PCR is needed to detect the expression of miR-1 in these cells (51). Thus, down-regulation of miR-1 in both the tumor epithelial and stromal cells may contribute to the association of miR-1 expression with prostate cancer outcomes. One may predict that miR-1 exerts functions in stromal cells similar to those described by our study, but miRs are known to have cell type-specific properties (54,55). An additional mechanism that may define miR expression in the prostate is exosome-mediated transfer of miRs between cells. It has been shown that cells can transfer miRs and mRNAs to neighboring cells by this mechanism, leading to detectable protein expression of mRNA-encoded transcripts after exosomal uptake (56). How efficient this mechanism is in transferring RNA from cell to cell in human tissues remains to be resolved.
The application of the cmap analysis indicated that miR-1-mediated tumor suppressor effects are globally similar to those of histone deacetylase inhibitors. The finding suggests that functional restoration of miR-1 in prostate tumors would have a therapeutic activity comparable to these compounds. This similarity in activity between miR-1 and histone deacetylase inhibitors could arise from independent mechanisms leading to common global expression changes. miR-1 may also directly target genes in the histone deacetylase pathway. Indeed, it has been shown that miR-1 targets histone deacetylase 4 (HDAC4), leading to a reduced expression of this deacetylase (37,38). Suppression of HDAC4 may contribute to some of the miR-1 tumor suppressor activity because this enzyme is a candidate oncogene in colon cancer and could be involved in the development of castration-insensitive prostate cancer (57,58). We also observed that HDAC4 is down-regulated by miR-1/miR-206 in prostate cancer cells, based on our gene expression data, consistent with our hypothesis that the underlying mechanisms for many of the miR-1-induced phenotypes in prostate cancer are higher order chromosomal and epigenetic effects by this miR.
In summary, our work describes miR-1 is a candidate tumor suppressor and potential prognostic marker in human prostate cancer. Future studies are needed to corroborate the association between miR-1 and prostate cancer outcomes and should further define the mechanisms that link this marker to disease recurrence and cancer metastasis and also examine how this knowledge can be used to target this pathway in cancer therapy.
ACCESSION NUMBER
Gene expression data from this study were deposited in GEO (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE31620. GEO also describes the V.4 microRNA platform under the accession number GPL14184.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary Tables 1–4, Supplementary Figures 1–9 and Supplementary Methods.
FUNDING
Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, Center for Cancer Research. Funding for open access charge: Center for Cancer Research, NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Barbara J. Taylor at the NCI FACS Core Laboratory and Judith A. Welsh at the Laboratory of Human Carcinogenesis for technical help. We would also like to thank staff at the University of Maryland, Departments of Pathology and Epidemiology for their support with the collection of human tissue samples.
REFERENCES
- 1.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 3.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A, Marchesini J, Mascellani N, Sana ME, Abu JR, et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589–599. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He L, He X, Lim LP, de SE, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, et al. Identification of the miR-106b∼25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci. Signal. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 14.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 15.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 16.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N. Engl. J. Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl Acad. Sci. USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl Acad. Sci. USA. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayr C, Bartel DP. Widespread shortening of 3′-UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, Dhir R. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am. J. Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 25.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 26.Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prueitt RL, Yi M, Hudson RS, Wallace TA, Howe TM, Yfantis HG, Lee DH, Stephens RM, Liu CG, Calin GA, et al. Expression of microRNAs and protein-coding genes associated with perineural invasion in prostate cancer. Prostate. 2008;68:1152–1164. doi: 10.1002/pros.20786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong AW, Fulgham P, Jay C, Chen P, Khalil I, Liu S, Senzer N, Eklund AC, Han J, Nemunaitis J. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009;16:206–216. doi: 10.1038/cgt.2008.77. [DOI] [PubMed] [Google Scholar]
- 29.Ribas J, Ni X, Haffner M, Wentzel EA, Salmasi AH, Chowdhury WH, Kudrolli TA, Yegnasubramanian S, Luo J, Rodriguez R, et al. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69:7165–7169. doi: 10.1158/0008-5472.CAN-09-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szczyrba J, Loprich E, Wach S, Jung V, Unteregger G, Barth S, Grobholz R, Wieland W, Stohr R, Hartmann A, et al. The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol. Cancer Res. 2010;8:529–538. doi: 10.1158/1541-7786.MCR-09-0443. [DOI] [PubMed] [Google Scholar]
- 31.Jung M, Schaefer A, Steiner I, Kempkensteffen C, Stephan C, Erbersdobler A, Jung K. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin. Chem. 2010;56:998–1006. doi: 10.1373/clinchem.2009.141580. [DOI] [PubMed] [Google Scholar]
- 32.Long Q, Johnson BA, Osunkoya AO, Lai YH, Zhou W, Abramovitz M, Xia M, Bouzyk MB, Nam RK, Sugar L, et al. Protein-coding and MicroRNA biomarkers of recurrence of prostate cancer following radical prostatectomy. Am. J. Pathol. 2011;179:46–54. doi: 10.1016/j.ajpath.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 34.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of Cardiogenesis, Cardiac Conduction, and Cell Cycle in Mice Lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 35.Mishima Y, breu-Goodger C, Staton AA, Stahlhut C, Shou C, Cheng C, Gerstein M, Enright AJ, Giraldez AJ. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev. 2009;23:619–632. doi: 10.1101/gad.1760209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Navon R, Wang H, Steinfeld I, Tsalenko A, Ben-Dor A, Yakhini Z. Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS ONE. 2009;4:e8003. doi: 10.1371/journal.pone.0008003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, Wang B, Suster S, Jacob ST, Ghoshal K. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J. Biol. Chem. 2008;283:33394–33405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl Acad. Sci. USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Day CP, Carter J, Bonomi C, Esposito D, Crise B, Ortiz-Conde B, Hollingshead M, Merlino G. Lentivirus-mediated bifunctional cell labeling for in vivo melanoma study. Pigment Cell Melanoma Res. 2009;22:283–295. doi: 10.1111/j.1755-148X.2009.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi M, Horton JD, Cohen JC, Hobbs HH, Stephens RM. WholePathwayScope: a comprehensive pathway-based analysis tool for high-throughput data. BMC Bioinformatics. 2006; 7:30. doi: 10.1186/1471-2105-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallace TA, Prueitt RL, Yi M, Howe TM, Gillespie JW, Yfantis HG, Stephens RM, Caporaso NE, Loffredo CA, Ambs S. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68:927–936. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 44.Yi M, Mudunuri U, Che A, Stephens RM. Seeking unique and common biological themes in multiple gene lists or datasets: pathway pattern extraction pipeline for pathway-level comparative analysis. BMC Bioinformatics. 2009;10:200. doi: 10.1186/1471-2105-10-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greenberg NM, DeMayo F, Finegold MJ, Medina D, Tilley WD, Aspinall JO, Cunha GR, Donjacour AA, Matusik RJ, Rosen JM. Prostate cancer in a transgenic mouse. Proc. Natl Acad. Sci. USA. 1995;92:3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaman MS, Chen Y, Deng G, Shahryari V, Suh SO, Saini S, Majid S, Liu J, Khatri G, Tanaka Y, et al. The functional significance of microRNA-145 in prostate cancer. Br. J. Cancer. 2010;103:256–264. doi: 10.1038/sj.bjc.6605742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 48.Song G, Zhang Y, Wang L. MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J. Biol. Chem. 2009;284:31921–31927. doi: 10.1074/jbc.M109.046862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schreiber V, Moog-Lutz C, Regnier CH, Chenard MP, Boeuf H, Vonesch JL, Tomasetto C, Rio MC. Lasp-1, a novel type of actin-binding protein accumulating in cell membrane extensions. Mol. Med. 1998;4:675–687. [PMC free article] [PubMed] [Google Scholar]
- 50.Grunewald TG, Kammerer U, Schulze E, Schindler D, Honig A, Zimmer M, Butt E. Silencing of LASP-1 influences zyxin localization, inhibits proliferation and reduces migration in breast cancer cells. Exp. Cell Res. 2006;312:974–982. doi: 10.1016/j.yexcr.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 51.Martens-Uzunova ES, Jalava SE, Dits NF, van Leenders GJ, Møller S, Trapman J, Bangma CH, Litman T, Visakorpi T, Jenster G. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene. 2012;31:978–991. doi: 10.1038/onc.2011.304. [DOI] [PubMed] [Google Scholar]
- 52.Yan D, Dong XE, Chen X, Wang L, Lu C, Wang J, Qu J, Tu L. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J. Biol. Chem. 2009;284:29596–29604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E, Bentwich Z, Lieberman J, Chowdhury D. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 2009;16:492–498. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl Acad. Sci. USA. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 56.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 57.Halkidou K, Cook S, Leung HY, Neal DE, Robson CN. Nuclear accumulation of histone deacetylase 4 (HDAC4) coincides with the loss of androgen sensitivity in hormone refractory cancer of the prostate. Eur. Urol. 2004;45:382–389. doi: 10.1016/j.eururo.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 58.Wilson AJ, Byun DS, Nasser S, Murray LB, Ayyanar K, Arango D, Figueroa M, Melnick A, Kao GD, Augenlicht LH, et al. HDAC4 promotes growth of colon cancer cells via repression of p21. Mol. Biol. Cell. 2008;19:4062–4075. doi: 10.1091/mbc.E08-02-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.