Abstract
REPs are highly repeated intergenic palindromic sequences often clustered into structures called BIMEs including two individual REPs separated by short linker of variable length. They play a variety of key roles in the cell. REPs also resemble the sub-terminal hairpins of the atypical IS200/605 family of insertion sequences which encode Y1 transposases (TnpAIS200/IS605). These belong to the HUH endonuclease family, carry a single catalytic tyrosine (Y) and promote single strand transposition. Recently, a new clade of Y1 transposases (TnpAREP) was found associated with REP/BIME in structures called REPtrons. It has been suggested that TnpAREP is responsible for REP/BIME proliferation over genomes. We analysed and compared REP distribution and REPtron structure in numerous available E. coli and Shigella strains. Phylogenetic analysis clearly indicated that tnpAREP was acquired early in the species radiation and was lost later in some strains. To understand REP/BIME behaviour within the host genome, we also studied E. coli K12 TnpAREP activity in vitro and demonstrated that it catalyses cleavage and recombination of BIMEs. While TnpAREP shared the same general organization and similar catalytic characteristics with TnpAIS200/IS605 transposases, it exhibited distinct properties potentially important in the creation of BIME variability and in their amplification. TnpAREP may therefore be one of the first examples of transposase domestication in prokaryotes.
INTRODUCTION
Repeated extragenic palindrome (REP) or Palindromic unit (PU) sequences were identified nearly 30 years ago in the intergenic regions of enterobacterial genomes (1). They play a variety of key roles in the cell. They are involved in regulating gene expression (by functioning as transcription terminators, by stabilizing mRNA and by acting as topological insulators for transcription-induced positive supercoiling (2–5), and in structuring DNA (by binding proteins such as IHF, PolI and DNA gyrase) (6–9). They are also specific target sites for several bacterial insertion sequences (10–12).
REPs are between 20- and 40-nt long, often clustered in structures called bacterial interspersed mosaic element (BIMES) as two tandem inverted copies separated by linkers, and have now been identified in a large number of bacterial genera and species where they are often found in high copy number (12–18). There are about 600 copies in Escherichia coli representing ∼1% of the genome (15,19) and over 1600 copies in Stenotrophomonas maltophilia (17). The ubiquitous nature of REPs and their multiplicity raises the important question of how they have expanded to populate their host genomes and have evolved their present multiple roles.
A clue to this may lie in members of a class of atypical bacterial insertion sequences (IS), the IS200/IS605 family, whose ends strongly resemble REPs. These ISs carry REP-like subterminal hairpins or imperfect palindromes (IP) secondary structures which are recognized and bound by the IS-specific transposase. They use a transposase, TnpA, of the HUH endonuclease family with a single catalytic tyrosine (Y1) as an attacking nucleophile and transpose using obligatory single-stranded (ss) DNA intermediates (20–23). We (ISfinder: www-is.biotoul.fr) and others (24) have identified a group of proteins, TnpAREP, closely related to IS200/IS605 transposases associated with REP sequences but forming a distinct clade defining a separate Y1 family. TnpAREP occurs in a variety of bacterial species and genera and is always flanked by REP/BIME sequences. Its presence appeared to be correlated with an increased abundance of REPs in the corresponding genomes suggesting that TnpAREP may be responsible for REP proliferation throughout their host genomes (24). The molecular mechanism generating these patterns is unknown.
In vitro and in vivo studies of two IS200/IS605 family members, IS608 and ISDra2, have provided a detailed picture of their transposition. This family differs profoundly from ‘classical’ ISs: they do not include terminal inverted repeats (IRs) and do not generate direct flanking target repeats (DRs) on insertion. Cleavage occurs at some distance from the IPs (25,26) via a transient covalent 5′-phosphotyrosine linked intermediate with the substrate DNA, leaving a free 3′-OH group on the other side of the DNA break. DNA cleavage also requires a divalent metal ion coordinated by two histidine residues, constituting the HUH motif, together with a third residue located close to the catalytic tyrosine (23,27). Transposition is strand-specific: TnpAIS608/ISDra2 recognises only the ‘top’ strand which undergoes strand cleavage and transfer to the target site. The ‘bottom’ strand is inactive. The cleavage site at both left and right ends is not recognized directly by TnpA but forms a set of hydrogen bonds with a short guide sequence located 5′ to the foot of the left and right subterminal IP (23,28,29). This recognition is essential for cleavage. Finally, excision and insertion occur preferentially at the lagging strand template in replication forks (30).
To address how BIMEs might invade and amplify within a genome, we first analysed BIME distribution and polymorphisms in the genomes of 44 assembled E. coli and Shigella strains. We also identified a single locus in a majority of the 110 available E. coli and Shigella genome sequences where a single tnpAREP gene is located. Phylogenetic analysis suggested that tnpAREP was acquired early in the radiation of the species into present-day strains. The gene is bordered by variable numbers of BIMEs in structures, similar to that of IS200/IS605 family members, called REPtrons, However, REPtrons do not appear to transpose as a unit but the BIMEs themselves are likely to be mobile and may have spread in a two-step process: transposition/recombination, which generates the observed sequence diversity of BIMEs, followed by local amplification.
To determine whether TnpAREP might be involved in this process, we analysed its cleavage and recombination activity in vitro. While TnpAREP shares similar catalytic characteristics with TnpAIS200/IS605 transposases, it exhibited distinct properties potentially important in the creation of BIME variability and in their amplification. In the light of these observations, we discuss the possible role of TnpAREP in generating variability and in proliferation of BIMEs throughout their host genomes.
To our knowledge, REP/BIME and TnpAREP probably represent the first example of bacterial transposable element domestication.
MATERIALS AND METHODS
Bioinformatic procedures
Transposase identification and analyses
The primary transposase sequence of representative elements was used as a query in a BLASTP (31) search among all complete and partial prokaryotic genome sequences available on the NCBI server. All apparently full-length transposases were retained. Recursive BLASTP searches were performed using the less conserved retrieved sequences, i.e. those with the lowest BLAST score. The procedure was terminated when the results converged to a final stable data set (no new transposase sequences were detected). BLASTP searches were performed on the NCBI BLAST online interface without the low complexity filter but with otherwise default parameters. Multiple alignments were carried out using either ClustalW (32) or MultAlin (33) and some displays were obtained using the Jalview alignment editor (34).
In a second step, we used the Markov Cluster Algorithm (MCL) (http://micans.org/mcl/) (35,36) to weigh relationships between protein clusters. An inflation factor (IF) of 1.2 was used and edges having BLASTP score values of <30 were filtered (score > 30).
REP identification
The GenBank files of the complete bacterial genomes used in this study were retrieved from the NCBI public repository (http://www.ncbi.nlm.nih.gov/genomes/ lproks.cgi).
REP identification was achieved with a combination of two methods: a method based on local alignments and a method based on sequence profiles.
For the first approach, consensus sequences were derived from Bachellier et al. (19) transformed to follow our sequence convention. They cover the three REP families y, z1 and z2 and each was used as a query for BLASTN (31) similarity searches on DNA sequences of the complete genomes. Since we are dealing with short and variable sequences, we set BLASTN parameters to permissive values to increase sensitivity (expectation value ≤ 10, word size = 4, reward for a nucleotide match = 1, penalty for a nucleotide mismatch = −1, cost to open a gap = −1). BLASTN is fast and selective but, since it produces a local alignment, the boundaries of predicted REPs can be shorter than expected. In addition, the observed nucleotide variation at each position is not included in the alignment scoring since it can decrease the sensitivity of the prediction. Thus, a second approach to predict motifs containing gaps was used based on the GLAM2 programme (37). Profiles for each REP family were built by applying the GLAM2 program on unambiguous full length REPs predicted by the previous BLASTN searches. The GLAM2SCAN program was further used to find occurrences of the GLAM2 motifs in target sequences.
To set the parameters for both approaches, we used the annotation of BIMEs in E. coli K12 (NC_000913.2) (available at: http://www.pasteur.fr/recherche/unites/pmtg/repet/index.html) as a training set. The estimated number of identified REPs corresponds to the combined results of both approaches.
Plasmid construction and TnpAREP purification
Escherichia coli MG1655 tnpAREP was cloned with a 6-His extension under control of promoter para in pBS176. Expression and purification were carried out in E. coli K12 (Rosetta, DE3) (Novagen) on Ni-agarose (Qiagen) as described for TnpAIS608 (25). Plasmid pBS180 and derivatives were constructed in several steps: the MG1655 REPtron region was first isolated by PCR directly from MG1655 genomic DNA and cloned into pBluescript, pSK. The tnpAREP gene was replaced by a Cm® cassette and the downstream BIMEs were then removed by iPCR. In pBS180mut, mutations of the conserved GTAG were introduced using the Quickchange Site-directed Mutagenesis Kit (Stratagene) and ssDNA was prepared using f1 helper phage as described by the supplier (Promega). Further details can be obtained on request.
Reactions in vitro
Oligonucleotides were 5′-end-labelled with [γ-33P] ATP (Perkin Elmer) using T4 polynucleotide kinase (NEB Inc.) or, in experiments to identify a 5′-phosphotyrosine transposase-substrate intermediate, 3′-end-labelled with [α-32P] dATP Cordycepin (Perkin Elmer) using Terminal Transferase (NEB Inc.). Labelled oligonucleotides were purified on a G25 column (GE Healthcare).
Double-stranded DNA was prepared by hybridization of complementary oligonucleotides. After 5-min denaturation at 95°C, the mixture was left to slowly cool to 25°C.
5′-Labelled oligonucleotide (0.02 µM) and unlabelled oligonucleotide (0.5 µM) were incubated with 2 and 4 µM TnpAREP (45 min, 37°C, final volume 10 µl) in 12.5 mM Tris (pH 7.5), 120 mM NaCl, 1 mM DTT, 20 µg/ml BSA, 0.5 µg of poly-dIdC and 7% glycerol in the presence or absence of 5 mM MnCl2 or MgCl2. Reactions were separated on an 8% native gel in TAE buffer, to detect retarded complexes, or on a 9% denaturing gel, to detect cleavage and recombination products, and analysed by phosphorimaging (Fuji). In reactions to detect covalent complex formation, 3′-labelled substrates were incubated with TnpAREP in the reaction mixture as described earlier and reactions were separated on 16% SDS–PAGE gel.
Cleavage sites were generally determined by comparing the band position in the sequencing gel with a sequence ladder. For certain small cleavage products, oligonucleotides of the presumed size and sequence were synthesized and used for comparison.
Primer extension
In vitro reaction mixtures were treated with Proteinase K, purified using the Promega PCR purification Kit and used as template for primer extension with 5′-end-labelled ‘a’ and ‘b’ primers with Taq polymerase (94°C 45 s, 52°C 45 s, 72°C 1 min) 35×.
-
a:
GTAAAACGACGGCCAGT.
-
b:
GCAGAACTGATCCGCTATGT.
RESULTS
REPs, BIMES and REPtrons: sequence, distribution and organization in E. coli
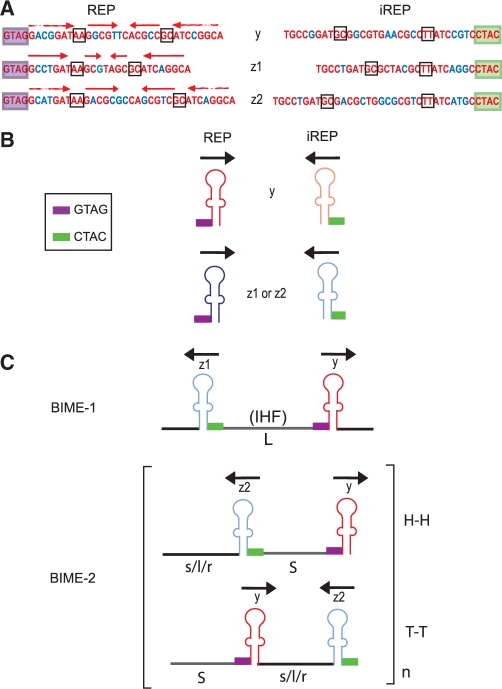
Figure 1 shows the sequence organization and previously defined nomenclature of E. coli REP and BIME elements (19): they are 30- to 40-nt long and could fold into an imperfect palindrome (IP) with a highly conserved tetranucleotide, GTAG, localized 5′ to the IP foot (Figure 1A and B). There are three major types of E. coli REP sequence, y, z1 and z2 (Figure 1A). Only 84 REPs among 584 identified in E. coli K12 are single occurrences (19). Others are organized in pairs (BIME) including two REPs in inverse orientation separated by linker sequences (Figure 1B and C): one, in the orientation including the 5′ GTAG tetranucleotide, called REP, and a second inverted sequence called iREP (Figure 1). For functional reasons (see below), the sequence convention used here is inversed compared to Bachellier et al. (19). Escherichia coli BIMEs were classified into three families (38). BIME-1 are composed of z1 and y (Figure 1) and occur as single copies in which the REP and iREP are separated by a long linker (L). BIME-2 are composed of z2 and y. They occur as multiple tandem copies with the REP and iREP components separated by a short linker, S, and with one of three types of flanking sequence, s, l or r. So-called atypical BIMEs are chimeras of BIME-1 and BIME-2, carrying different combinations of y, z1, z2, L, S, s, l and r. Like BIME-2, they also occur in multiple copies. The BIME-1 L linker is well conserved and frequently carries an IHF binding site (6) while those of BIME-2 and atypical BIMEs vary both in length and sequence. This can be seen among BIME-2 and atypical BIME copies carried by the MG1655 genome (Supplementary Figure S1). BIMEs also vary in number from locus to locus in a single strain. However, the sequence of tandem BIME copies at any one locus is well conserved (Supplementary Figure S2). Moreover, in different E. coli strains, the number of tandem BIMEs at a given locus is also variable [Supplementary Figure S3, see also (39)].
Figure 1.
Escherichia coli REPs and BIMEs. (A) Consensus sequences of E. coli y, z1 and z2 REPs. The conserved tetranucleotide GTAG is boxed in violet, conserved positions are in red. Complementary sequences (iREP) corresponding to each category are presented on the right. The CTAC tetranucleotide, complementary to the conserved GTAG sequence, is boxed in green. Two base mismatches in the hairpin stem are boxed. The red horizontal arrows indicate complementary regions able to pair. Nucleotides in blue indicate bases that differ from one REP to another but nevertheless retain complementarity. (B) Structure of REP and iREP. Violet and green boxes represent the GTAG and CTAC, respectively. Black arrows indicate REP orientation. Red indicates a y REP and blue a z1 or z2 REP. Dark and light colours indicate REP and iREP, respectively. (C) Structures of BIME-1, and BIME-2. The reader is referred to Bachellier et al. (19). BIMEs are composed of a REP and an iREP separated by long (L) or short (S) linkers, H–H and T–T represent head-to-head and tail-to-tail configurations.
Identification of tnpAREP among members of the E. coli/Shigella group
We (ISfinder: www-is.biotoul.fr) and others (24) have independently identified a group of proteins (TnpAREP), closely related to IS200/IS605 transposases, associated with REP sequences. TnpAREP occurs in a variety of bacterial species and genera and is always flanked by REP/BIME sequences.
We analysed 110 E. coli and Shigella genomes available in the PATRIC database (40) for the presence of tnpAREP and focused on its immediately surrounding region (Supplementary Figure S4 and ‘Discussion’ section). Two-thirds (74/110) carried tnpAREP located at a unique position on the circular chromosome between yafL and fhiA, even in strains (ATCC8739) (CP000946), DH1 (CP001637) and BL21 (AM946981) in which the entire region has undergone an inversion.
Figure 2A shows the distribution of REP and BIME elements in the tnpAREP-carrying region from selected E. coli strains. While the left (5′) side invariably carried a single BIME, the right (3′) included a variable number of REPs and BIMEs (e.g. MG1655, APEC01, ED1a and O157:H7). In some strains, IS-mediated rearrangements had occurred (e.g. UMN026) (Figure 2A). Albeit more complex, these structures, called REPtrons, resemble members of the IS200/IS605 family of bacterial insertion sequences. In 7 strains lacking tnpAREP, all belonging to the B2 clade of the E. coli/Shigella group (41), the surrounding REP and BIME were still present but tnpAREP had been precisely excised (e.g. CFT073). In 24 strains, all except two belonging to the B1 clade (41), no trace of tnpAREP or associated BIMEs was found but, instead, these had acquired the toxin–antitoxin genes hicA and hicB between yafL and fhiA (e.g. IAI1) (Figure 2A and Supplementary Figure S4). Mapping tnpAREP on the E. coli/Shigella phylogenetic tree (Supplementary Figure S4) suggested that the gene was acquired early in the species radiation, at least at the E. fergusoni and E. coli/Shigella separation, and was lost later in some strains by these two distinguishable events: either by replacement (together with its flanking BIMEs) with hicA and hicB or by precise deletion while retaining the flanking BIMEs.
Figure 2.
Escherichia coli phylogenetic tree, REPtron structure and TnpAREP alignment. (A) E. coli REPtron distribution and organization: the left of the figure represents, for clarity, a simplified phylogenetic tree of the Escherichia coli / Shigella group obtained by pruning the tree shown in Supplementary Figure S4 which was retrieved from the PATRIC database [http://www.patricbrc.org/portal/portal/patric/Home; (44)]. The clades (A, B1, B2, D, E, F and S) are from (41). The right of the figure shows examples of REPtrons from representative members of each clade. tnpAREP is shown in grey, the flanking genes yafL and fhiA in green and in violet, respectively. Arrows represent the direction of transcription. Flanking BIMEs are shown with the same convention as in Figure 1. The hicA and hicB genes are also indicated as black and blue arrows, respectively. (B) Alignment of TnpAIS200/IS605 and TnpAREP. TnpA from IS200/IS605 family members are boxed in red while TnpAREP derivatives are boxed in green. Conserved positions are boxed as deep blue and less well-conserved residues in lighter shades of blue. The catalytic residues of TnpAIS608 and the potential catalytic residues of E. coli MG1655 TnpAREP are shown in red above and below the alignment: histidine (H), hydrophobic (U) and tyrosine (Y) glutamine Q) and asparagine (N).
We identified REP elements in 44 complete E. coli and Shigella genomes as well as in the available genome of Escherichia fergusonii. The estimated number of REPs in E. coli and Shigella genomes varied between 286 and 574 with an average number of 422 ± 74. This large dispersion reflects the presence of two subgroups of genomes showing extreme REP frequencies: the first group with a higher REP frequency (on an average 546) corresponds to strains from a same subtree including clade A; the second group, composed of two E. coli strains (SMS-3-5 and IAI39) from clade F and Shigella dysenteriae Sd197, displays less than 310 REPs. The other genomes have an estimated number of REPs correlated to the genome size and centred around 395 ± 32. This group includes strains with and without tnpAREP. From these results, it is difficult to determine whether tnpAREP plays a role in REP amplification or maintenance or whether the loss of the gene is too recent to have had an observable effect on REP copy number. However, as only 216 REP elements have been identified in the E. fergusonii genome (which does not carry the REPtron), this distribution and the tree topology suggest that the large majority of REPs have arisen after the acquisition of the REPtron in the ancestor and before the divergence of E. coli/Shigella strains (Supplementary Figure S4).
TnpAREP and TnpAIS200/605 form two different families
Figure 2B shows an alignment of a representative group of 10 TnpAIS200/IS605 sequences (ISfinder) and a group of 10 TnpAREP sequences from the public databases. All retain a conserved tyrosine (Y) and the HUH amino acid triad (histidine–hydrophobic residue–histidine) typical of the Y1 transposase catalytic site. They also include a conserved asparagine (N), located four residues from the catalytic Y, replacing a glutamine (Q) residue of TnpAIS200/IS605 involved in divalent metal ion coordination (27). However, TnpAREP group members are generally longer, include an additional C-terminal domain compared to TnpAIS200/605 and exhibit specific conserved amino acid blocks throughout, in particular in the central and the C-terminal domains (24). MCL (Markov Clustering) analysis (35,36) of TnpAIS200/605 and TnpAREP sequences also indicated that they represent two distinct Y1 families (‘Materials and Methods’ section). Clearly, TnpAREP has sequence features suggesting it may be involved in catalysis of REP invasion and dispersal within genomes.
TnpAREP activity in vitro
To gain insight into the potential role of TnpAREP in REP proliferation, E. coli tnpAREP was cloned with a His6 carboxy-terminal tail under control of the para promoter. The resulting protein, expressed from pBS176, was purified on Ni-agarose resin (‘Materials and Methods’ section) and tested for binding and catalytic activities.
DNA binding
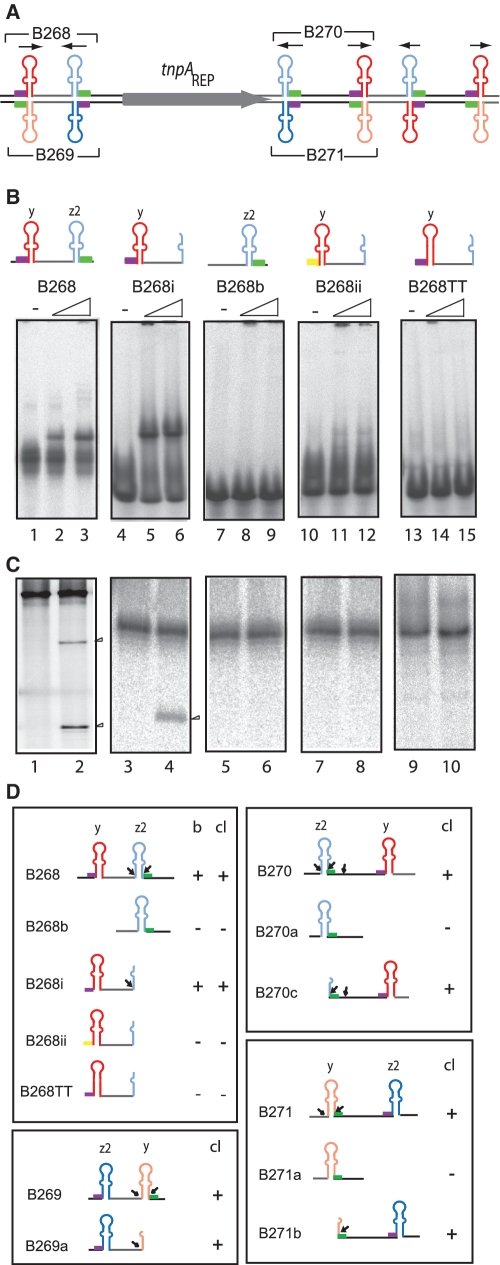
Initial DNA substrates were based on REPs and BIMEs (BIME II) from the REPtron present in MG1655 (Figure 3A and Supplementary Figure S5). Various 5′-end-labelled single- or double-strand substrates carrying REP or BIME sequences were incubated with purified TnpAREP in the absence of a divalent metal ion and analysed by EMSA (Figure 3B). No retarded band was observed with double-stranded substrates (Supplementary Figure S6A). However, B268, an ssBIME-carrying substrate from the 5′ REPtron end including a y REP and a z2 inverted REP sequence (iREP), showed a retarded band (Figure 3B, lanes 2 and 3) as did a substrate with half of the iREP (z2) (B268i; Figure 3B, lanes 5 and 6). Removal of the entire REP (y) eliminated binding (B268b, lanes 8 and 9). Mutation of the GTAG to ACGA (B268ii, lanes 11 and 12) or removal of the mismatch in the REP (y), in which mutation GC to TT should allow formation of a perfect REP palindrome sequence eliminated detectable TnpAREP binding (B268TT; lanes 14 and 15). Thus, both the GTAG tetranucleotide and the mismatch in the REP sequence are required for formation of robust TnpAREP–DNA complexes.
Figure 3.
Binding and Cleavage activity on substrates derived from REPtron. (A) The E. coli MG1655 REPtron and oligonucleotides representing ssBIMEs used in this analysis. The oligonucleotides used are indicated with numbers above and below the cartoon. They are summarized in Supplementary Table S1. (B) Binding activity observed by EMSA. 5′-end-labelled oligonucleotides were incubated in the binding buffer (‘Materials and Methods’ section) in the absence or presence of 2 or 4 µM TnpAREP (shown as a triangle above the gels). ‘−’ indicates no added TnpAREP. The yellow box represents a GTAG tetranucleotide mutated to ACGA. (C) Cleavage of ssBIME derivatives. Arrow heads show the cleavage products. 5′-end-labelled oligonucleotides in the absence or presence of 4 µM TnpAREP: B268 (116 nt), lanes 1–2; B268b (52 nt), lanes 3–4; B268i (61 nt), lanes 5–6; B268ii (61 nt, mutated for GTAG), lanes 7–8; B268TT (61 nt), lanes 9–10, respectively. (D) Structure and activity of other substrates derived from REPtron. In the right, binding and cleavage activity are summarized. (b = binding; cl = cleavage). Small black arrows indicate the position of cleavage sites.
Cleavage
To determine whether TnpAREP also has DNA cleavage activity, we incubated the 5′-end-labelled oligonucleotides used for EMSA studies with TnpAREP in reaction buffer containing Mn2+ or Mg2+ (‘Materials and Methods’ section). The products were separated in a denaturating sequencing gel. DsDNA was refractory to cleavage. TnpAREP was active only on ssDNA substrates and the reaction (using 3′-end-labelled substrates) generated a covalent DNA–protein intermediate as observed with TnpAIS608 (Our unpublished data). Cleavage was generally more efficient with Mn2+ than with Mg2+ but no significant differences in cleavage specificity were observed (Supplementary Figure S6B). Except where stated, all assays presented here were performed with ssDNA substrates in the presence of Mn2+.
B268, a 116-nt BIME-carrying substrate underwent two major cleavages at the 3′ z2 iREP to generate two labelled fragments of 85 and 55 nt (Figure 3C, lanes 1 and 2) whereas B268i, a 61-nt oligonucleotide carrying the entire y REP sequence but only part of the z2 iREP, shares the first cleavage site with B268, giving a 51-nt product (lanes 3 and 4). Even though the other B268 BIME derivatives showed no significant binding in EMSA (Figure 3B), their activity was also examined since, in the case of TnpAIS608, the fact that some DNA substrates do not form stable complexes visible by EMSA did not necessarily eliminate their capacity to undergo cleavage (18). B268b, a 52-nt substrate with only the z2 iREP was refractory to cleavage (lanes 5 and 6) as was B268ii, the 61-nt partial BIME carrying a mutated GTAG (Figure 3C, lanes 7 and 8). We confirmed this using additional GTAG mutants derived from B268 or other substrates. B268TT with a GC to TT mutation which allows formation of a perfect REP palindrome (Figure 3B, lanes 9 and 10) was also refractory to cleavage. We obtained a similar result with B268GC (mutation of AA to GC, not shown).
B269, an oligonucleotide complementary to B268 including a z2 REP and a y iREP sequence underwent two cleavages at the 3′ iREP (Figure 3D). B269a, an oligonucleotide derived from B269 carrying only part of the y iREP was also cleaved (Figure 3D). At first sight, this appears to contrast to the behaviour of the IS200/IS605 family, where only the top strand is active (21,26). However, this is clearly due to different substrate configuration: the two component REP sequences in a BIME are inverted with respect to each other. Thus, each DNA strand carries a REP and an iREP, permitting cleavage on both strands.
We also examined the cleavage properties of the BIMEs located immediately 3′ of tnpAREP. The results were similar to those obtained with the 5′ BIME. Cleavage occurred 5′ to the y REP on the top strand and to the z2 REP on the bottom strand (Figure 3D). Additional BIME variants from other E. coli chromosome regions showed similar behaviour (Supplementary Figure S5). TnpAREP catalysed cleavage of all three iREP variants, y, z1 or z2, and sometimes also cleaved sites within the linker sequence (Figure 3D, B270 and B270c) but only when the substrate also included a REP.
Thus, the data demonstrate that a REP structure is indispensable for BIME cleavage, presumably by providing a binding site for TnpAREP. Moreover, they show that TnpAREP recognises the REP with its 5′ conserved GTAG tetranucleotide and requires the non-complementary base(s) in the stem for binding and for activity but cleaves at the inverted sequence of the 5′ or 3′ REP (iREP) and at the linker sequence with the expected polarity (each cleavage resulting in a 5′ phosphotyrosine intermediate; unpublished data). The results also demonstrate that cleavage can be either 5′ or 3′ to the essential REP sequence.
Defining the cleavage sites
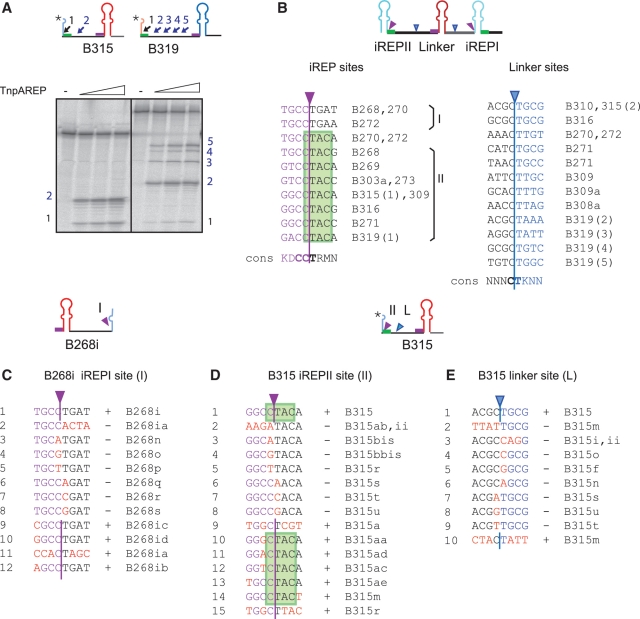
Although REP and iREP portions of BIMEs are relatively well conserved, the linkers are highly variable (Supplementary Figure S1). Like REPtron-derived substrates (Figure 3D), several of these proved to contain cleavage sites in the linker region although the efficiency of cleavage at these sites is variable. Examples are B315 a partial BIME located at araD-A in MG1655 (one site, Figure 4A) and B319, located at gltP-yjcO (four sites, Figure 4A).
Figure 4.
iREP and linker cleavage sites. (A) Examples of linker cleavage sites on B315 and B319. B315 and B319 are partial BIMEs derived from MG1655 araD-A and gltP-yjcO regions. Linker cleavage sites are indicated by blue small arrows and numbered. ‘asterisks’ indicates the position of labelling. (B) Alignment of observed in vitro ‘iREP’ and ‘linker’ sites. ‘iREP’ and ‘linker’ sites are indicated by purple or blue arrows. Their sequence is shown with the consensus sequence (cons) for each category below in bold. iREPI and iREPII sites occurred on both sides of the iREP palindrome, the second overlapped the CTAC tetranucleotide (in green), the complement of the conserved GTAG tetranucleotide. The oligonucleotides used are shown to the right of the sequences. (C) Characterization of ‘iREPI’ (I) site using B268i as an example. Wild-type bases to the left of the cleavage site are shown in purple. Those to the right are shown in black. Mutated bases are shown in red. ‘+’ indicates cleavage, ‘−’ indicates no cleavage. Nucleotide designations are shown to the right. (D and E) Characterization of ‘iREPII’ (II) and ‘linker’ (L) using B315. Colour codes are the same as for (C). [For the B315, II and L correspond to sites B315(1) and B315(2) in Figure 4A]. For the B315 linker site, nucleotides to the left of the cleavage site are shown in black and those to the right in blue. Mutated nucleotides are shown in red.
In summary, we observed two categories of cleavage site: those on either side of iREP and those in the linker regions. We refer to the first category as ‘iREP’ and to the second as ‘linker’ cleavage sites. The pattern of cleavage was similar in the presence of either Mn2+ or Mg2+ (Supplementary Figure S6).
Cleavage sites from each category obtained from a set of naturally occurring BIME sequences are aligned in Figure 4B. ‘iREP’ sites (Figure 4B left) are situated at both sides of the iREP palindrome in relatively conserved regions: the first type (I) occurred at T−4G−3C−2C−1|T1G2A3T4/A4 (where ‘|’ represents the site of cleavage) and the second (II) at G−4/T−4G−3/T−3/A−3C−2C−1|T1A2C3A4/C4/G4, within CTAC, the complement of the conserved GTAG tetranucleotide. Note that type I sites are present in z1 and z2 iREPs but are absent in y iREP sequences.
The ‘linker’ sites (Figure 4B right) appear less conserved. The deduced consensus reveals conserved CC|T (coordinates C−2C−1|T1) for ‘iREP’ and C|T (C−1|T1) sequences for ‘linker’ sites.
The importance of the C|T for cleavage
To understand the rules governing BIME processing in more detail, we analysed the cleavage pattern of a panel of mutant sites introduced at the iREPI site in the 5′ partial BIME (B268i) from the MG1655 REPtron (Figure 4C) and at the iREPII and linker sites in a second partial BIME (B315) (Figure 4D and E) present at the araD-A intergenic region. Mutations were introduced either in a block or individually in each site. Although this is not an exhaustive analysis, the results obtained show that the central C|T sequence was indispensable for cleavage as all substitutions at these positions prevented cleavage while mutations in some other positions were tolerated.
Although we have not systematically measured the efficiency of cleavage at each site, sequences flanking the CT dinucleotide may influence cleavage efficiency. For example, the weak site B319(1) includes GACClTACA compared to the stronger B315(1) with GGCClTACA: G−3 may therefore influence cleavage efficiency (Figure 4A and B). In addition, mutation of T−4 to any base (B268i, iREPI site, Figure 4C) reduced cleavage in B268ib, B268ic and B268id (Supplementary Figure S6).
Thus, ‘iREP’ and ‘linker’ sites appear to share similar sequence requirements indicating relatively limited cleavage specificity.
Cleavage of single strand circular DNA
To confirm the requirement of C|T for cleavage and to assess the distance over which TnpAREP might act, we examined cleavage of a significantly longer DNA substrate, a single strand DNA circle derived from the 4.1-kb bacteriophage f1-derived phasmid, pBluescript II SK, into which a BIME had been cloned (pBS180, ‘Materials and Methods’ section). Following cleavage with TnpAREP, DNA was deproteinized and the cleavage sites were mapped by primer extension using two different primers (‘Materials and Methods’ section). The results are presented in Figure 5. In addition to bands resulting from two natural polymerase stalling sites which depend on the presence of a BIME (lanes 1 and 5), the substrate contains many cleavage sites stretching both upstream and downstream of the resident BIME over the entire region (>400 nt) analysed (lanes 6 and 8). Moreover, cleavage is absolutely dependent on the presence of the functional BIME since no products are observed with a substrate carrying a BIME mutated for the conserved GTAG (compare lanes 2 and 4). Mapping these sites on the DNA sequence indicated that they occurred at a C|T dinucleotide.
Figure 5.
Cleavage of circular ssDNA. The substrate and primers used for analysis are shown. ssDNA circles derived from pBS180 (substrate with functional REP, lanes 1–2; 5–8) and pBS180mut (substrate with REP mutated for GTAG, lanes 3–4) were incubated in the absence or presence of TnpAREP in buffer containing MnCl2 and used for primer extension with 5′-end labelled oligonucleotide ‘a’ (lanes 1–6) and ‘b’ (7–8). Lanes 1–2 and 5–6 correspond to the same samples separated under two different migration conditions on a 6% sequencing gel. Distances in nucleotides show the distance of the complementary oligonucleotide primer from the foot of the functional REP. Cleavages revealed by primers ‘a’ and ‘b’ are shown by red and black arrowheads, respectively.
Thus, BIME-directed cleavage can occur at a considerable distance from the TnpAREP binding site.
Recombination
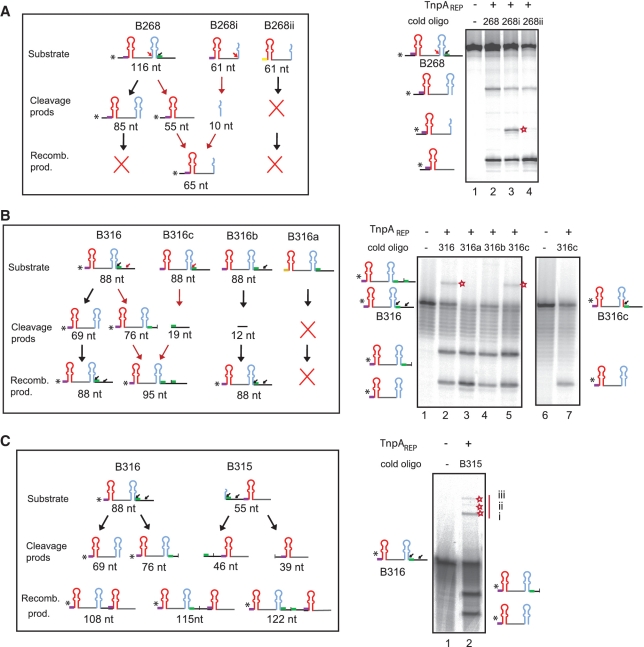
A striking characteristic of BIME-2 and atypical BIMEs is the variation in the linker sequence and in copy number at a given locus in different strains suggesting that they may undergo recombination and amplification. To investigate this in vitro, we examined the capacity of TnpAREP to promote BIME recombination with two sets of substrates. The first included ssBIMEs from the REPtron region (B268, B268i and B268ii; Figures 3 and 6A). In these experiments, we used a 20-fold molar excess of the unlabelled partner oligonucleotide to facilitate recombination. When 5′ labelled 116 nt B268 was incubated with TnpAREP, it was cleaved at two ‘iREP’ sites generating 85 and 55 nt products (Figure 6A, lanes 1 and 2). Addition of unlabelled 61 nt B268i generated a 65-nt recombination product (lane 3). This species no longer appeared in the reaction with the inactive substrate B268ii mutated for GTAG (Figure 6A, lane 4). These results demonstrate an exchange of sequences between the partner DNA molecules demonstrating recombination between two ‘iREP’ sites.
Figure 6.
TnpAREP catalysed strand exchange in vitro. DNA substrates and recombination products for each reaction are indicated, ‘*’ in the cartoons indicates radioisotope position; colour codes are as in Figure 1. Positions of recombination products are indicated by a red star, small red arrow indicate cleavage sites giving rise to detectable recombination. (A) Strand exchange between ‘iREP’ sites. 5′-end labelled oligonucleotide: B268 (116 nt): lane 1: no protein, lanes 2–4: in presence of TnpAREP and unlabelled 61 nt B268, B268i and B268ii (mutated at GTAG), respectively. (B) Recombination between ‘iREP’ and ‘linker’ sites. 5′-labelled oligonucleotide: B316 (88 nt) was incubated with: no protein (lane 1), in the presence of TnpAREP and unlabelled oligonucleotides: B316, B316a, B316b, B316c (lanes 2–5), respectively. In the reaction with 5′-labelled B316c: no protein or in the presence of protein and unlabelled B316c (lanes 6–7). (C) Recombination leading to assembly of multiple REPs. 5′-labelled B316 was incubated with: no protein (lane 1), in the presence of TnpAREP and unlabelled oligonucleotide B315 (lane 2). The products of strand transfer (marked with a red star) migrate as expected for the structures i, ii and iii shown in the left hand cartoon.
The second substrate set included derivatives of an ssBIME from the araD-A region (B316, Figure 6B). 5′-end-labelled 88 nt B316 was cleaved at a ‘iREP II’ and a linker site, generating 76 and 69 nt products (Figure 6B, lanes 2–5). In the presence of unlabelled B316, a product of 95 nt resulting from recombination between the ‘iREP’ and the ‘linker’ sites was generated (Figure 6B, lane 2). We no longer observed this species on addition of unlabelled B316a, mutated for GTAG or of unlabelled B316b, mutated at the ‘iREPII’ site, respectively (Supplementary Table S1; Figure 6B, lanes 3 and 4). The recombination product was still observed with cold B316c, mutated at the ‘linker’ site but keeping the ‘iREP’ site intact (Figure 6B, lane 5). However, this recombination product was not generated when we used 5′-end-labelled B316c in the presence of unlabelled B316c (Figure 6B, lanes 6 and 7), confirming the nature of this reaction. We did not observe recombination products from certain reactions since they reconstitute a fragment having the length of the original labelled substrate (e.g. Figure 6B). We also used the 5′-end-labelled 88 nt B316 and oligonucleotide B315 (Figure 6C). In vitro, in addition to the two cleavage products of B316, these substrates generated a series of DNA species which migrated high in the gel and had sizes consistent with recombination products which include an additional REP together with various combinations of linker sequences.
These results demonstrate recombination between iREP and linker sites which might result in BIME variability, multiplication and amplification.
DISCUSSION
tnpAREP, coding for a protein related to the Y1 transposases, was identified in association with REP/BIME sequences in structures called REPtrons found in a number of bacterial genomes Here we compared the REPtron structure and REP/BIME distribution in available E. coli and Shigella genomes. We also analysed E. coli K12 TnpAREP activity including cleavage and recombination in vitro. While TnpAREP shared the same general organization and similar catalytic characteristics as the TnpAIS200/IS605 Y1 transposases, it exhibited distinct properties potentially important in creation of BIME variability and amplification. The presumed importance of tnpAREP in REP/BIME evolution and dispersion within genomes and multiple roles assumed by REPs and BIMEs themselves in cell physiology could be interpreted as domestication. Although many cases of domestication of eukaryotic transposable elements have been documented (42,43), such domestication has not yet been described for classical bacterial elements.
REPtron evolution in E. coli and Shigella
The tnpAREP gene was present in 74 of the 110 E. coli and Shigella genomes available in the PATRIC database (http://www.patricbrc.org/portal/portal/patric/Home); (40,44) always as a single copy at the same genetic locus. This genetic conservation implies that these sequences had been acquired in a single event early in the last common ancestor of the species which gave rise to present-day strains. The phylogenetic analysis of E. coli/Shigella strains lacking tnpAREP indicates that tnpAREP had been present but had subsequently been replaced or deleted (Supplementary Figure S4).
Although REPtron organization resembles that of members of the IS200/IS605 family, we do not believe that it represents a true transposable genetic element. Its unique genetic location indicates that it has not undergone subsequent rounds of transposition. This suggests that if the spread of REPs within a genome is catalysed by TnpAREP it must occur by mobilization in trans.
We observed only minor differences in REPs/BIMEs copy number and distribution in strains with or without tnpAREP (Supplementary Figure S4), in agreement with the idea of REPtron/tnpAREP loss occurring late in the radiation of these strains. This is in contrast to the previous observation in some other bacterial species of a strong correlation between the presence of tnpAREP and the increased number of REPs/BIMEs in corresponding genomes (24), and may reflect a more recent REPtron/tnpAREP acquisition event in these strains.
The REPtron distribution therefore clearly argues for an origin which predates the radiation of E. coli/Shigella and, since E. alberti appears to carry a similar (but not identical) REPtron, it may predate the separation of this species.
TnpAREP binding, cleavage and strand transfer activity in vitro
To evaluate whether TnpAREP might be involved in the recombination events leading to REP invasion and spread, we examined its activities in vitro. We demonstrated that it binds ssREP sequences and requires the conserved 5′ GATG tetranucleotide for this. It also catalyses BIME cleavage both upstream and downstream of the REP sequence. The data suggest that the functional unit on which the enzyme acts is a (complete or partial) BIME rather than an individual REP or iREP. Cleavage requires an entire REP sequence and occurs at the iREP and in BIME linkers. Since BIMEs are composed of two inverted REP copies, this implies that TnpAREP can cleave both DNA strands, contrary to TnpAIS608/ISDra2 which functions in a strand specific manner. Moreover, the two base pair mismatch located in the REP stem is also essential for binding and activity.
Although TnpAREP forms a distinct clade within the Y1 transposase family, it appears to share with TnpAIS608 the same absolute requirement for ss substrates (Supplementary Figure S6) and the formation of a covalent protein–DNA intermediate (25) (unpublished data). However, unlike TnpAIS200/IS605, the sequence specificity for cleavage appears relatively low, requiring only the dinucleotide CT while tolerating substitutions in other surrounding positions. This limited cleavage specificity may be responsible for BIME diversification. Cleavage occurs in the presence of Mg2+ but is much more pronounced with Mn2+. However, similar cleavage patterns are observed with both metal ions (Supplementary Figure S6).
We also observed strand transfer activity in vitro. Strand transfer can therefore create sequence variability and assemble tandem BIME copies resembling the tandem amplification observed in vivo (see below).
Model of BIME variability and amplification
In light of the observed distribution and sequence variability in the collection of E. coli strains, BIME colonization and expansion throughout genomes may occur as a two-step process involving diversification followed by amplification (Figure 7). There are several ways in which this might be accomplished.
Figure 7.
Model for BIME diversification and amplification. (A) BIME excision and integration model: From a BIME array carrying several ‘linker’ cleavage sites [A(i)], cleavages at two distinct sites (‘iREP’ or ‘linker’ types) and strand exchange between Tyr-5′P (in grey) generated from the first and the 3′-OH from the second (in red) would lead to excision of a ssBIME circle [A(ii,iii)]. Cleaved again (by TnpAREP) at alternative cleavage sites and/or processed by unidentified host factors [A(iv)] before integration, this could give rise to several BIME variants [A(v)]. (B) BIME amplification model: The first cleavage at ‘iREP’ site [B(i)] would generate a 3′-OH which could act as a primer and be extended by host DNA polymerases [B(ii)]. A second cleavage occurring at a ‘linker’ site 3′ of the REP on the newly synthesized DNA strand [B(iii)] liberates another 3′-OH (in red) that attacks the first Tyr-5′P complex [B(iv), in grey]. This leads to addition of a supplementary BIME unit [B(v,vi)]. (C) Alternative model of BIME amplification [adapted from (24)]: From two BIMEs in tandem, two cleavages on the bottom strand, followed by reciprocal strand exchange [C(ii)] would lead to excision of the bottom central BIME. A 3′-OH resulting from a third cleavage on the top strand [C(iii)] would be used as primer for a ‘rolling replication amplification’ of the excised circular BIME. A fourth cleavage on the newly synthesized DNA strand [C(iv)] liberates another 3′-OH (in blue) that attacks the third Tyr-5′P complex (in pink). This leads to addition of one (or numerous) supplementary BIME(s) [C(vi)].
Diversity could be generated in an excision and insertion process by using different cleavage sites for excision and for insertion as shown in Figure 7A. From a BIME array carrying several ‘linker’ cleavage sites [Figure 7A(i)], cleavages at two distinct sites (‘iREP’ or ‘linker’ types) and joining between Tyr-5′P generated from the first and the 3′-OH from the second would lead to excision of a ssBIME circle [Figure 7A(ii,iii)] in the same way as been shown for IS608 (25). Clearly, like IS608 and ISDra2, this could involve ssDNA on the lagging strand template of a replication fork but might also use ssDNA generated during R-loop formation, triggered by transcription-induced negative supercoiling (45), repair or by supercoil-driven extrusion of the REP secondary structure element (46). If processed at different cleavage sites [Figure 7A(iv)] before integration, this intermediate could give rise to several BIME variants [Figure 7A(v)]. For BIMEs, this process can occur on both DNA strands which in principle could carry different ‘linker’ cleavage sites thus increasing the potential BIME sequence diversity. Degradation of 3′-OH end by host nucleases might also contribute to BIME variation. In this model, variation would be coupled to integration.
Amplification could occur following insertion of the excised BIME into a suitable target as that shown in Figure 7B. This uses a ‘rolling circle’ like recombination mechanism on an inserted BIME (either in the head to head or tail to tail orientation) which does not necessarily involve rolling circle replication of an excised circular product (see below). Using an example of an H–H BIME, an initial cleavage at an ‘iREP’ site [Figure 7B(i)] would generate a 3′-OH which could act as a primer and be extended by host DNA polymerases [Figure 7B(ii)]. A second cleavage occurring at a ‘linker’ site 3′ of the REP on the newly synthesized DNA strand [Figure 7B(iii)] would liberate another 3′-OH that attacks the first Tyr-5′P complex [Figure 7B(iv)]. This would lead to addition of a supplementary BIME unit [Figure 7B(v,vi)].
Alternatively, amplification could take place by a mechanism involving rolling circle replication from two BIMEs in tandem such as that proposed previously [(24); Figure 7C]. Although this model is attractive and would explain the amplification process, it requires four TnpAREP-directed cleavages and might appear complex. However, the model implies cleavages on BIMEs in both strands [Figure 7C(ii,iii)], might therefore explain the necessity to maintain BIME as unit.
Note that the recombination we observe in vitro between two ‘iREP’ sites (Figure 6A) and between a ‘iREP’ and a ‘linker’ site (Figure 6B), is equivalent to steps Aii and Biv. We believe that integration step (Aiv) corresponds to a ‘recombination’ of excised BIME with a target carrying a REP/BIME. The ability of TnpAREP to cleave at a significant distance upstream or downstream of a resident BIME (Figure 5) would indeed enable a BIME insertion/recombination at a distance from an existent BIME, therefore disseminating them on the chromosome. This capacity would also allow the unit to sequester additional flanking sequences including entire genes or gene fragments as have been observed for codA–cynR from E. coli and tnpAREP from Pseudomonas putida, Pseudomonas fluorescens, Stenotrophomonas maltophilia, Mannheimia succinicproducens (24) (Supplementary Figure S7). Acquisition of neighbouring genes is also characteristic of rolling circle (RC) transposition of the IS91, ISCR and Helitron elements (47,48)
While these data underline the potential plasticity conferred on the host genome by the tnpAREP/BIME system, neither the exact mechanism nor the inherent frequency of TnpAREP-mediated BIME recombination are at present known. Further bioinformatic approaches are expected to reveal more details concerning BIME spread through genomes. Moreover, mechanistic studies would be greatly aided by knowledge of the TnpAREP structure with and without its DNA substrate. Additionally, it will be essential in the future to develop an in vivo system to observe the activity of the tnpAREP/BIME system within its host genome since it is possible that TnpAREP activity requires host proteins and may be coupled to cell physiology such as replication, transcription or supercoiling. Such studies are underway.
This class of enzyme is widespread. It includes transposases of the IS200/IS605 and IS91/ISCR families of insertion sequences, TnpAREP, relaxases of conjugative plasmids and proteins involved in the replication of rolling circle plasmids, phage and eukaryotic viruses. These studies raise important questions concerning the evolutionary relationship between transposable elements and their domestication in cell function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–7.
FUNDING
Centre National de Recherche Scientifique (CNRS, France) (intramural). Funding for open access charge: Intramural CNRS funding.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank D. Lane, L. Lavatine, A. Hickman and F. Dyda as well as members of the Mobile Genetic Elements group for reading the manuscript and for discussions. We would also like to thank the two anonymous referees for their very constructive suggestions.
REFERENCES
- 1.Higgins CF, Ames GF, Barnes WM, Clement JM, Hofnung M. A novel intercistronic regulatory element of prokaryotic operons. Nature. 1982;298:760–762. doi: 10.1038/298760a0. [DOI] [PubMed] [Google Scholar]
- 2.Espeli O, Moulin L, Boccard F. Transcription attenuation associated with bacterial repetitive extragenic BIME elements. J. Mol. Biol. 2001;314:375–386. doi: 10.1006/jmbi.2001.5150. [DOI] [PubMed] [Google Scholar]
- 3.Khemici V, Carpousis AJ. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol. Microbiol. 2004;51:777–790. doi: 10.1046/j.1365-2958.2003.03862.x. [DOI] [PubMed] [Google Scholar]
- 4.Aguena M, Ferreira GM, Spira B. Stability of the pstS transcript of Escherichia coli. Arch. Microbiol. 2009;191:105–112. doi: 10.1007/s00203-008-0433-z. [DOI] [PubMed] [Google Scholar]
- 5.Moulin L, Rahmouni AR, Boccard F. Topological insulators inhibit diffusion of transcription-induced positive supercoils in the chromosome of Escherichia coli. Mol. Microbiol. 2005;55:601–610. doi: 10.1111/j.1365-2958.2004.04411.x. [DOI] [PubMed] [Google Scholar]
- 6.Boccard F, Prentki P. Specific interaction of IHF with RIBs, a class of bacterial repetitive DNA elements located at the 3′ end of transcription units. EMBO J. 1993;12:5019–5027. doi: 10.1002/j.1460-2075.1993.tb06195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilson E, Bachellier S, Perrin S, Perrin D, Grimont PA, Grimont F, Hofnung M. Palindromic unit highly repetitive DNA sequences exhibit species specificity within Enterobacteriaceae. Res. Microbiol. 1990;141:1103–1116. doi: 10.1016/0923-2508(90)90084-4. [DOI] [PubMed] [Google Scholar]
- 8.Gilson E, Perrin D, Hofnung M. DNA polymerase I and a protein complex bind specifically to E. coli palindromic unit highly repetitive DNA: implications for bacterial chromosome organization. Nucleic Acids Res. 1990;18:3941–3952. doi: 10.1093/nar/18.13.3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espeli O, Boccard F. In vivo cleavage of Escherichia coli BIME-2 repeats by DNA gyrase: genetic characterization of the target and identification of the cut site. Mol. Microbiol. 1997;26:767–777. doi: 10.1046/j.1365-2958.1997.6121983.x. [DOI] [PubMed] [Google Scholar]
- 10.Clement J-M, Wilde C, Bachellier S, Lambert P, Hofnung M. IS1397 is active for transposition into the chromosome of Escherichia coli K-12 and inserts specifically into palindromic units of bacterial interspersed mosaic elements. J. Bacteriol. 1999;181:6929–6936. doi: 10.1128/jb.181.22.6929-6936.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilde C, Bachellier S, Hofnung M, Clement J-M. Transposition of IS1397 in the family Enterobacteriaceae and first characterization of ISKpn1, a new insertion sequence associated with Klebsiella pneumoniae palindromic units. J. Bacteriol. 2001;183:4395–4404. doi: 10.1128/JB.183.15.4395-4404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tobes R, Pareja E. Bacterial repetitive extragenic palindromic sequences are DNA targets for insertion sequence elements. BMC Genomics. 2006;7:62. doi: 10.1186/1471-2164-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stern MJ, Ames GF-L, Smith NH, Clare Robinson E, Higgins CF. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984;37:1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- 14.Gilson E, Saurin W, Perrin D, Bachellier S, Hofnung M. Palindromic units are part of a new bacterial interspersed mosaic element (BIME) Nucl. Acids Res. 1991;19:1375–1383. doi: 10.1093/nar/19.7.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilson E, Saurin W, Perrin D, Bachellier S, Hofnung M. The BIME family of bacterial highly repetitive sequences. Res. Microbiol. 1991;142:217–222. doi: 10.1016/0923-2508(91)90033-7. [DOI] [PubMed] [Google Scholar]
- 16.Aranda-Olmedo I, Tobes R, Manzanera M, Ramos JL, Marques S. Species-specific repetitive extragenic palindromic (REP) sequences in Pseudomonas putida. Nucleic Acids Res. 2002;30:1826–1833. doi: 10.1093/nar/30.8.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rocco F, De Gregorio E, Di Nocera PP. A giant family of short palindromic sequences in Stenotrophomonas maltophilia. FEMS Microbiol. Lett. 2010;308:185–192. doi: 10.1111/j.1574-6968.2010.02010.x. [DOI] [PubMed] [Google Scholar]
- 18.Bertels F, Rainey PB. Within-genome evolution of REPINs: a new family of miniature mobile DNA in bacteria. PLoS Genet. 2011;7:e1002132. doi: 10.1371/journal.pgen.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bachellier S, Clément J-M, Hofnung M. Short palindromic repetitive DNA elements in enterobacteria: a survey. Res. Microbiol. 1999;150:627–639. doi: 10.1016/s0923-2508(99)00128-x. [DOI] [PubMed] [Google Scholar]
- 20.Kersulyte D, Velapatino B, Dailide G, Mukhopadhyay AK, Ito Y, Cahuayme L, Parkinson AJ, Gilman RH, Berg DE. Transposable element ISHp608 of Helicobacter pylori: nonrandom geographic distribution, functional organization, and insertion specificity. J. Bacteriol. 2002;184:992–1002. doi: 10.1128/jb.184.4.992-1002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ton-Hoang B, Guynet C, Ronning DR, Cointin-Marty B, Dyda F, Chandler M. Transposition of ISHp608, member of an unusual family of bacterial insertion sequences. EMBO J. 2005;24:3325–3338. doi: 10.1038/sj.emboj.7600787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ronning DR, Guynet C, Ton-Hoang B, Perez ZN, Ghirlando R, Chandler M, Dyda F. Active site sharing and subterminal hairpin recognition in a new class of DNA transposases. Mol. Cell. 2005;20:143–154. doi: 10.1016/j.molcel.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 23.Barabas O, Ronning DR, Guynet C, Hickman AB, Ton-Hoang B, Chandler M, Dyda F. Mechanism of IS200/IS605 family DNA transposases: activation and transposon-directed target site selection. Cell. 2008;132:208–220. doi: 10.1016/j.cell.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nunvar J, Huckova T, Licha I. Identification and characterization of repetitive extragenic palindromes (REP)-associated tyrosine transposases: implications for REP evolution and dynamics in bacterial genomes. BMC Genomics. 2010;11:44. doi: 10.1186/1471-2164-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guynet C, Hickman AB, Barabas O, Dyda F, Chandler M, Ton-Hoang B. In vitro reconstitution of a single-stranded transposition mechanism of IS608. Mol. Cell. 2008;29:302–312. doi: 10.1016/j.molcel.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Pasternak C, Ton-Hoang B, Coste G, Bailone A, Chandler M, Sommer S. Irradiation-induced Deinococcus radiodurans genome fragmentation triggers transposition of a single resident insertion sequence. PLoS Genet. 2010;6:e1000799. doi: 10.1371/journal.pgen.1000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hickman AB, James JA, Barabas O, Pasternak C, Ton-Hoang B, Chandler M, Sommer S, Dyda F. DNA recognition and the precleavage state during single-stranded DNA transposition in D. radiodurans. EMBO J. 2010;29:3840–3852. doi: 10.1038/emboj.2010.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guynet C, Achard A, Hoang BT, Barabas O, Hickman AB, Dyda F, Chandler M. Resetting the site: redirecting integration of an insertion sequence in a predictable way. Mol. Cell. 2009;34:612–619. doi: 10.1016/j.molcel.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He S, Hickman AB, Dyda F, Johnson NP, Chandler M, Ton-Hoang B. Reconstitution of a functional IS608 single-strand transpososome: role of non-canonical base pairing. Nucleic Acids Res. 2011;39:8503–8512. doi: 10.1093/nar/gkr566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ton-Hoang B, Pasternak C, Siguier P, Guynet C, Hickman AB, Dyda F, Sommer S, Chandler M. Single-stranded DNA transposition is coupled to host replication. Cell. 2010;142:398–408. doi: 10.1016/j.cell.2010.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Dongen S. Technical Report INS-R0010, National Research Institute for Mathematics and Computer Science. in the Netherlands. 2000. A cluster algorithm for graphs. Amsterdam. [Google Scholar]
- 36.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frith MC, Saunders NF, Kobe B, Bailey TL. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 2008;4:e1000071. doi: 10.1371/journal.pcbi.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bachellier S, Saurin W, Perrin D, Hofnung M, Gilson E. Structural and functional diversity among bacterial interspersed mosaic elements (BIMEs) Mol. Microbiol. 1994;12: 61–70. doi: 10.1111/j.1365-2958.1994.tb00995.x. [DOI] [PubMed] [Google Scholar]
- 39.Bachellier S, Clement JM, Hofnung M, Gilson E. Bacterial interspersed mosaic elements (BIMEs) are a major source of sequence polymorphism in Escherichia coli intergenic regions including specific associations with a new insertion sequence. Genetics. 1997;145:551–562. doi: 10.1093/genetics/145.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gillespie JJ, Wattam AR, Cammer SA, Gabbard JL, Shukla MP, Dalay O, Driscoll T, Hix D, Mane SP, Mao C, et al. PATRIC: the comprehensive bacterial bioinformatics resource with a focus on human pathogenic species. Infect. Immun. 2011;79:4286–4298. doi: 10.1128/IAI.00207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chalker DL, Yao MC. DNA elimination in ciliates: transposon domestication and genome surveillance. Annu. Rev. Genet. 2010;45:227–246. doi: 10.1146/annurev-genet-110410-132432. [DOI] [PubMed] [Google Scholar]
- 43.Sinzelle L, Izsvak Z, Ivics Z. Molecular domestication of transposable elements: from detrimental parasites to useful host genes. Cell. Mol. Life Sci. 2009;66:1073–1093. doi: 10.1007/s00018-009-8376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snyder EE, Kampanya N, Lu J, Nordberg EK, Karur HR, Shukla M, Soneja J, Tian Y, Xue T, Yoo H, et al. PATRIC: the VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 2007;35:D401–D406. doi: 10.1093/nar/gkl858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drolet M. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol. Microbiol. 2006;59:723–730. doi: 10.1111/j.1365-2958.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- 46.Bikard D, Loot C, Baharoglu Z, Mazel D. Folded DNA in action: hairpin formation and biological functions in prokaryotes. Microbiol. Mol. Biol. Rev. 2011;74: 570–588. doi: 10.1128/MMBR.00026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toleman MA, Bennett PM, Walsh TR. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 2006;70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kapitonov VV, Jurka J. Rolling-circle transposons in eukaryotes. Proc. Natl Acad. Sci. USA. 2001;98:8714–8719. doi: 10.1073/pnas.151269298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.