Enzymes—catalytically active proteins—are the most powerful catalysts known, endowed with exquisite selectivity for reactants and tremendous acceleration in rates (1). An ability to tailor catalysts to desired targets has been a Philosopher's Stone for chemists (2) as well as alchemists for a long time. Proteins acquire their chemical activity by folding into intricate three-dimensional structures, thousands of which are available for inspection in the Protein Data Bank (http://www.rcsb.org/pdb/). But simply looking at a structure does not tell us why a given sequence forms one structure and not another, or why so many seemingly unrelated sequences can achieve similar structures. We need to take proteins apart and tinker with their insides, as we would with any complex piece of machinery. What principles govern folding? Does a small set of key residues nucleate or lock in a structure, or does structure reflect an almost imperceptible influence of dozens or hundreds of side chains? Are local interactions more important than long-range effects or vice versa?
The work by Silverman et al. (3) presents a masterful dissection of a major protein fold, the (β/α)8 barrel structure identified first in the glycolytic enzyme triose phosphate isomerase (TIM) and since found in hundreds of enzymes. Their work investigates a total of 182 sites in the enzyme for their tolerance or intolerance to substitutions, with far-reaching implications for protein folding and design.
The work by Silverman et al. presents a masterful dissection of a major protein fold … with far-reaching implications for protein folding and design.
The most obvious feature of protein folds is their irregularity. Despite the frequent occurrence of helical and sheet substructures, there is little uniformity in the disposition of these within a native protein. There are exceptions: one is the symmetric eight-fold β/α barrel structure that is the subject of the article by Silverman et al. (3); a second is the ubiquitous α-helical coiled coil, consisting of two or more α-helical strands twined around each other (4, 5). Since Anfinsen's pioneering work (6), we know that the sequence of proteins is sufficient to dictate the folded state. It now appears that signals for structure are embedded in sequence in a highly degenerate and overdetermined fashion (7). Reading these signals has been a major challenge in many laboratories, leading inevitably to different hypotheses in efforts to decipher the folding code (8). Silverman et al. (3) analyze the effects on activity of libraries with millions of mutations in the gene of TIM, exploring the variability in the structure at a deep level.
Recognizing that burial of nonpolar side chains to form a nonpolar core structure is common to all native protein structures, the simplest “oil-droplet” model (9) proposes that the arrangement of polar and nonpolar side chains in a sequence is the dominant factor in dictating the final structure. Nonpolar or hydrophobic (H) side chains form the interior, buried from unfavorable solvent interactions, whereas polar side chains (P) face the exterior. Accordingly, patterns such as HPHPHP should read out to a β-sheet, with one nonpolar face containing H groups and one polar face with the P groups. On the other hand, patterns such as PHHPHHHP or HPPHPPP should favor helices. A striking case has been reported in which a localized change in H/P pattern is accompanied by a switch from β-sheet to α-helix structure (10).
This view is supported by the observation that there is considerable degeneracy among the nonpolar side chains that comprise the core of a globular protein, seen both in sequence alignments of proteins from different organisms (11) and in mutational studies (12–14). Conservative (H → H) substitutions are tolerated to a surprising degree. For example, when 13 positions in the hydrophobic core of the small RNase barnase were randomized, 23% of the mutants were found to be active (14). This observation is surprising because the interior of proteins tends to be well packed, more crystal like than liquid (15). Constraints can be introduced on the volume of side chains. Cavity-creating mutations can be severely destabilizing (16). But, according to the oil-droplet model, hydrophobic cores can be repacked with no great difficulty, and the main determinant is the polarity of the side chain.
In an opposing view, the van der Waals interactions in the well-packed core of a protein are thought to be crucial, so that placement of a side chain in the structure is more like fitting a key into a keyhole or a piece into a three-dimensional jigsaw puzzle (13, 15). Closer inspection of mutant proteins generated by randomizing the hydrophobic core reveals that only a small fraction of conservative hydrophobic core substitutions are equivalent to the wild type in stability and activity (17). The jigsaw-puzzle model is most obviously favored by structural and thermodynamic studies of short α-helical coiled coils. Crick's seminal analysis (18) showed that coiled coils are formed by a knob-into-holes packing of H side chains. Later studies showed clearly that the fit is a tight one: minor differences in the identity of the H residues can switch the stoichiometry from two to three or four strands (19). This result is at variance with simple H/P sequence determination, because details of the H side chains, such as the extent of branching, prove to be critical.
The oil-droplet and jigsaw-puzzle models are useful because they provide extreme but concrete targets for experimental testing. Many other factors are thought to influence the folding of a protein. The role of local structure and identity of side chains in helical regions has been considered important: Gly and Pro, for example, destabilize helical structure, whereas Ala tends to favor helix (20, 21). Signals at the ends of short helices also have been implicated: these include capping structures, in which polar side chains with the backbone near the ends of a helix (22). The corresponding factors that form H bonds determining β-sheet termini are less obvious but are likely to be relevant as well.
Silverman et al. (3) took a bold step in setting out to identify the determinants of the eight-fold (α/β) barrel structure. The motif includes about 250 residues, much larger than the model proteins studied to date. Perhaps one question to ask is, why? In addition to their remarkable frequency, β/α barrels are an attractive target for several reasons. First, the fold has a high degree of symmetry (Fig. 1A), which reduces the total sequence space. Kirschner's group has shown that individual β/α domains can be permuted with retention of activity (23). Second, the catalytic activity of (β/α) barrels resides in a series of loops that form a headpiece that rests on the core barrel structure. In a recent and elegant directed evolution experiment, the catalytic activity of a (β/α) barrel enzyme was switched from one substrate and reaction to a new one by using a combination of rational design and random mutagenesis (24). Silverman et al. (3) kept this region of the protein invariant to focus on the core itself, whereas Altamirano et al. (24) directed their mutations to the headpiece. Third, the (β/α)8 barrel structure contains more extensive hydrophobic domains than the simple globular domains explored before. In fact, there are two hydrophobic domains in TIM: one at the interface between the helices and the β-barrel and the second within the β-barrel itself (see Fig. 1).
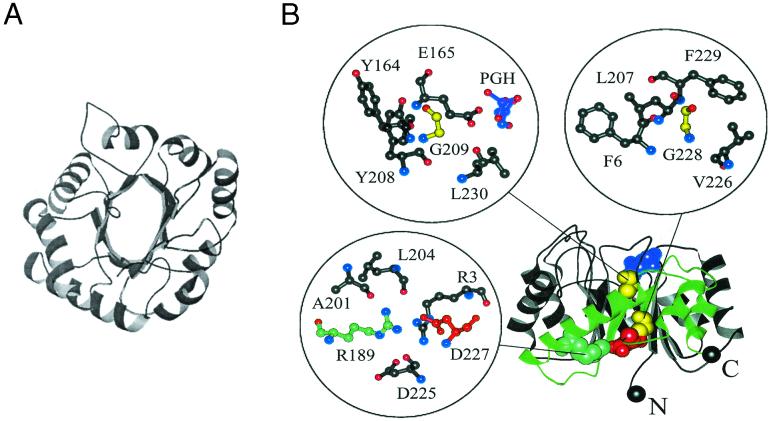
Figure 1.
Two views of the β/α barrel structure in TIM. (A) A view from the top of the structure showing the symmetry of the eight β/α domains. The N and C termini are depicted in black spheres. The active site in this orientation is facing the reader. (B) A side view showing the hydrophobic β-β-sheet interface and the four sites in the enzyme that are most sensitive to substitutions in the study by Silverman et al. (3). Insets show expanded views of the local structural environments around each residue. Phosphoglycolohydroxamate (PGH), a substrate analog included in the crystal structure, is shown in purple. Glycines 209 and 228 are colored yellow, R189 is colored cyan, and D227 is colored red. The region of the backbone proposed to be an independently folding subdomain is colored green.
Technically, the report by Silverman et al. (3) is a tour de force. Random or directed changes at multiple sites were introduced by use of mixtures of bases in desired positions in synthetic short fragments of DNA. The fragments were spliced together to generate genes or libraries of genes. Because of the vast conformational space they cover, Silverman et al. make use of technologies for generating large-scale sequence libraries and diversifying them by shuffling or recombination (25). Availability of a bacterial strain whose growth depends on the presence of an active TIM protein allowed them to screen the expressed products efficiently. Side-by-side comparison of genetically randomized libraries selected for functional activity with unselected ones is used as a probe of amino acid preferences at each site. Testing of individual enzymes was done by complementing a TIM knockout mutant or by directly measuring their rates of catalysis.
What do they find? First, the extreme oil-drop model does not fit. Although the β-sheet/α-helix hydrophobic interfaces and most surface sites can be substituted relatively freely, the authors estimate that fewer than 1 in 1010 sequences in a random library that conserves the consensus H/P pattern are active. This result contrasts starkly with the value of 1 in 4 found in barnase (14). Both internal sites and the ends of helices are also readily substituted. The β-sheet core of the TIM barrel turns out to be most intolerant to mutation, more consistent with the jigsaw-puzzle model. Interestingly, four sites in the core appear not to tolerate substitution at all (see Fig. 1B). Two of these residues are charged and form an apparently buried salt bridge in the protein, as shown in the figure. Theoretically, such interactions are thought to have a weak effect on stability (26), but the issue here may well concern specificity. Polar side chains in hydrophobic regions may in fact play a strong organizing role (27) at the expense of rapid folding and stability. As in the case of eglin c (21), other interactions postulated to play a role in folding could be tested. In particular, helix capping and stop signals seem dispensable, although several helices in the wild-type TIM enzyme are capped.
This is a landmark study, showing that large protein structures are now amenable to rigorous testing of hypotheses about folding. Practical design of barrels can now include tailoring the base as well as headpiece to generate new catalysts with greater stability and novel substrate specificity. Thermophilic barrels have already been found (28). The authors find some suggestive clues concerning the folding and assembly mechanism of barrels, too: most sensitive sites lie toward the C terminus of the protein, for example, which may define a subdomain that folds independently. Beyond this, they offer suggestive evidence that not all of the 20 natural amino acids are required to encode α/β barrels. The extent of overdetermination of structure by sequence suggests that some reduction in amino acid usage may be possible: the question is, how large a reduction? Alphabets as small as 5 amino acids have been proposed to be sufficient for folding, on the basis of experimental (29) and theoretical (30) evidence. Silverman et al. (3) point out that a reduced alphabet of 7 amino acids might be adequate to fold barrels. The set FVLAKEQ (31) produces nonperturbing single substitutions at 142 sites of the 182 they examined, although not at the invariant Gs. However, exhaustive substitutions at these sites are not reported here. Reduction to a set of about half the amino acids represents a significant simplification for protein design and has evolutionary implications as well.
It is important to appreciate that no protein fold is understood completely today. Even in coiled coils, the current paradigm, the number and orientation of strands in the final complexes are controlled by a subtle interplay of many variables in addition to specific packing interactions of interior H groups (5, 19, 32). Comparative studies of other large proteins will be needed to place this pioneering study in proper perspective. Perhaps extended hydrophobic interfaces—in α/α coiled coils and β/β structures, for example—are more sensitive to substitution than more globular structures. The achievement of Silverman et al. (3) is that they open the way to answering fundamental questions about how large and complex enzymes fold, function, achieve their stability—even evolve—and how they might be engineered for biomedical or chemical applications.
Footnotes
See companion article on page 3092.
References
- 1.Walsh C. Nature (London) 2001;409:226–231. doi: 10.1038/35051697. [DOI] [PubMed] [Google Scholar]
- 2.Koeller K M, Wang C-H. Nature (London) 2001;409:232–240. doi: 10.1038/35051706. [DOI] [PubMed] [Google Scholar]
- 3.Silverman J A, Balakrishnan R, Harbury P B. Proc Natl Acad Sci USA. 2001;98:3092–3097. doi: 10.1073/pnas.041613598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen C, Parry D. Proteins. 1990;7:1–15. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- 5.Betz S F, Bryson J W, DeGrado W F. Curr Biol. 1995;5:457–463. doi: 10.1016/0959-440x(95)80029-8. [DOI] [PubMed] [Google Scholar]
- 6.Anfinsen C B. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 7.Rose G D. Proc Natl Acad Sci USA. 2000;97:526–528. doi: 10.1073/pnas.97.2.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fersht A R. Structure and Mechanism in Protein Science. New York: Freeman; 1999. [Google Scholar]
- 9.Kamtekar S, Schiffer J M, Xiong H, Babik J M, Hecht M H. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- 10.Cordes M H, Walsh N P, McKnight C J, Sauer R T. Science. 1999;284:325–328. doi: 10.1126/science.284.5412.325. [DOI] [PubMed] [Google Scholar]
- 11.Behe M J, Lattman E E, Rose G D. Proc Natl Acad Sci USA. 1991;88:4195–4199. doi: 10.1073/pnas.88.10.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim W A, Sauer R T. Nature (London) 1989;339:31–36. doi: 10.1038/339031a0. [DOI] [PubMed] [Google Scholar]
- 13.Gassner N C, Baase W A, Matthews B W. Proc Natl Acad Sci USA. 1996;93:12155–12158. doi: 10.1073/pnas.93.22.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Axe D D, Foster NW, Fersht A R. Proc Natl Acad Sci USA. 1996;93:5590–5594. doi: 10.1073/pnas.93.11.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ponder J W, Richards F W. J Mol Biol. 1987;193:775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 16.Lim W A, Farruggio DC, Sauer R T. Biochemistry. 1992;31:4324–4333. doi: 10.1021/bi00132a025. [DOI] [PubMed] [Google Scholar]
- 17.Lim W A, Sauer R T. J Mol Biol. 1991;219:359–376. doi: 10.1016/0022-2836(91)90570-v. [DOI] [PubMed] [Google Scholar]
- 18.Crick F H C. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- 19.Harbury P B, Zhang T, Kim P S, Alber T. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 20.Pace C N, Scholtz J M. Biophys J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lahr S J, Broadwater A, Carter C W, Jr, Collier M L, Hensley L, Waldner J C, Pielak G J, Edgell M H. Proc Natl Acad Sci USA. 1999;96:14860–14865. doi: 10.1073/pnas.96.26.14860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aurora R, Rose G D. Protein Sci. 1998;7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luger K, Hommel U, Herold M, Hofsteege J, Kirschner K. Science. 1989;243:206–210. doi: 10.1126/science.2643160. [DOI] [PubMed] [Google Scholar]
- 24.Altamirano M M, Blackburn J M, Aguayo C, Fersht A R. Nature (London) 2000;403:617–622. doi: 10.1038/35001001. [DOI] [PubMed] [Google Scholar]
- 25.Arnold F H. Nature (London) 2001;409:253–257. doi: 10.1038/35051731. [DOI] [PubMed] [Google Scholar]
- 26.Hendsch Z S, Tidor B. Protein Sci. 1994;3:211–226. doi: 10.1002/pro.5560030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oakley M G, Kim P S. Biochemistry. 1998;37:12603–12610. doi: 10.1021/bi981269m. [DOI] [PubMed] [Google Scholar]
- 28.Hennig M, Sterner R, Kirschner K, Jansonius J N. Biochemistry. 1997;36:6009–6016. doi: 10.1021/bi962718q. [DOI] [PubMed] [Google Scholar]
- 29.Riddle D S, Santiago J V, Bray-Hall S T, Doshi N, Grantchorova V P, Yi Q, Baker D. Nat Struct Biol. 1997;4:805–809. doi: 10.1038/nsb1097-805. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Wang W. Nat Struct Biol. 1999;6:1022–1038. doi: 10.1038/14918. [DOI] [PubMed] [Google Scholar]
- 31.Plaxco K W, Riddle D S, Grantcharova V, Baler D. Curr Opin Struct Biol. 1998;8:80–85. doi: 10.1016/s0959-440x(98)80013-4. [DOI] [PubMed] [Google Scholar]
- 32.Hodges R S. Biochem Cell Biol. 1996;74:133–154. doi: 10.1139/o96-015. [DOI] [PubMed] [Google Scholar]