Abstract
Recent studies have shown that more than 10% of autism cases are caused by de novo structural genomic rearrangements. Given that some heritable copy number variants (CNVs) have been observed in patients as well as in healthy controls, to date little attention has been paid to the potential function of these non-de novo CNVs in causing autism. A normally intelligent patient with autism, with non-affected parents, was identified with a maternally inherited 10 Mb deletion at 13q21.2. Sequencing of the genes within the deletion identified a paternally inherited nonsynonymous amino-acid substitution at position 614 of diaphanous homolog 3 (DIAPH3) (proline to threonine; Pro614Thr). This variant, present in a highly conserved domain, was not found in 328 healthy subjects. Experiments showed a transient expression of Diaph3 in the developing murine cerebral cortex, indicating it has a function in brain development. Transfection of Pro614Thr in murine fibroblasts showed a significant reduction in the number of induced filopodia in comparison to the wild-type gene. DIAPH3 is involved in cell migration, axon guidance and neuritogenesis, and is suggested to function downstream of SHANK3. Our findings strongly suggest DIAPH3 as a novel autism susceptibility gene. Moreover, this report of a ‘double-hit’ compound heterozygote for a large, maternally inherited, genomic deletion and a paternally inherited rare missense mutation shows that not only de novo genomic variants in patients should be taken seriously in further study but that inherited CNVs may also provide valuable information.
Keywords: compound heterozygosity, structural genomic abnormality, deletion, point mutation, autism, DIAPH3
Introduction
Autism spectrum disorder (ASD) is a group of developmental disorders characterized by impairments in social interaction, communication and the presence of restrictive or repetitive patterns of behavior. The autistic spectrum (MIM 209850) ranges from severe cases (the core syndrome called autism or autistic disorder) to milder forms, which include pervasive developmental disorder, not otherwise specified and Asperger syndrome. Despite the high heritability estimated for autism (90%),1,2 identifying causative common genetic variants has proven to be challenging. Until recently, disease-causing mutations for autism had been reported in a limited number of genes, which together may account for only a small percentage (~5–10%) of autism cases.2,3 Recently, there has been a growing interest in the function of structural genomic rearrangements, including cytogenetic abnormalities and copy number variants (CNVs) associated with autism.4–6 Indeed, findings of these and other recent studies7,8 suggest that an even larger proportion than the estimated ~5–10% of autism cases may be caused by genomic rearrangements.3
When a chromosomal or genomic abnormality coincides with a disease phenotype in some patients, but unaffected carriers are also identified, a causal relation between the genetic defect and the phenotype is often considered unlikely. However, such inconsistent concurrences of a structural genomic abnormality and a disease phenotype are often encountered in neuropsychiatric disorders. For certain genetic defects a causal relationship with psychiatric disease is widely accepted, even though the concurrence of the defect and the phenotype is variable. For example, several studies have reported a high rate of schizophrenia or psychotic symptoms in subjects with the 22q11.2 deletion syndrome.9–11 Although it is highly probable that the deletion at 22q11.2 is causally related to psychosis, the majority of subjects with 22q11.2 deletion syndrome do not have psychotic symptoms. Similarly, a causal genotype–phenotype association in individuals with ASDs and with genomic duplications at chromosome 15q11–15q13 is generally accepted, even though the co-segregation is far from complete.12
Hence, such scenarios necessitate further research into other factors that could explain the variable expression of the phenotype. For instance, ‘modifier’ genes located elsewhere in the genome are likely to influence the phenotype related to the genomic variation. Alternatively, it is possible that a structural genomic alteration, such as a deletion, unmasks a mutation in an otherwise recessive gene on the noninvolved homolog.
Here, we present a male proband with an autistic disorder with a maternally inherited deletion on the long arm of chromosome 13. Closer examination of the paternally inherited homolog revealed a nonsynonymous point mutation in diaphanous homolog 3 (DIAPH3), leading to an amino-acid substitution at position 614 of DIAPH3 (proline to threonine; Pro614Thr), one of the genes located within the deleted region at 13q. Transfection of a mutant murine Diaph3 construct with the Pro614Thr point mutation showed a strongly reduced induction of filopodia in comparison to wild-type Diaph3. These findings strongly suggest that this ‘double-hit’ event, consisting of a deletion on one allele and the Pro614Thr variant point mutation in the remaining allele of DIAPH3, in the proband is functionally relevant and most likely pathogenic. Further studies showed that Diaph3 is transiently expressed in the developing murine cerebral cortex. Our results suggest that DIAPH3 may be a novel susceptibility gene for autism.
Materials and methods
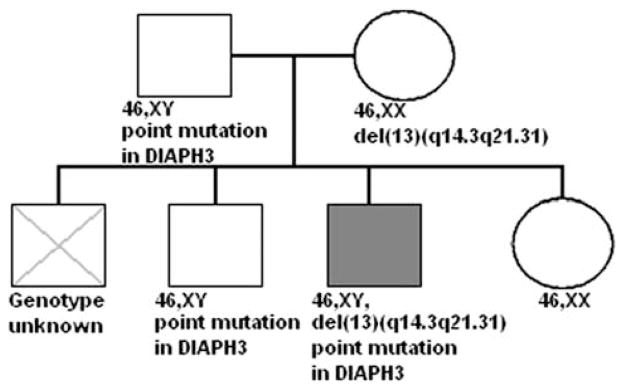
The proband was born from Caucasian parents, the third child in a family of four children (Figure 1). His eldest brother died at the age of 13 years of heart failure; his family currently comprised one elder brother ( + 3 years) and one younger sister (−8 years). At the age of 12, the proband was referred to the child psychiatry outpatient clinic at the University Medical Center Utrecht, the Netherlands.
Figure 1.
Genogram of proband’s family.
The assessment protocol, including the psychiatric, cognitive and genetic studies, was approved by the Dutch Central Committee on Research Involving Human Subjects. Additional genetic studies involving DNA from the proband, his parents and siblings was carried out at the Division of Human Genetics, Children’s Hospital in Philadelphia, USA, with the approval of the Institutional Review Board and the Committee for Protection of Human Subjects. Written informed consent was obtained from all participants in this study.
Medical, psychiatric and cognitive assessments
Phenotyping was performed at the Child and Adolescent Psychiatry Department of the University Medical Center Utrecht. Psychiatric assessment included structured and semi-structured interviews with the parents and proband, including a Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV) semi-structured interview, the sections for mood disorder and psychosis of the Schedule for Affective Disorders and Schizophrenia for School-Age Children—Present and Lifetime Version,13 the Autism Diagnostic Observation Schedule—Generic (ADOS-G)14 and the Autism Diagnostic Interview—Revised (ADI-R).15 In addition, the proband’s intelligence level was assessed using the Dutch version of the Wechsler Intelligence Scales: WISC-III.16
The parents, brother and sister were screened for psychiatric disorders with a DSM-IV semi-structured interview, whereas the possibility of ASD was examined using the ADOS and ADI-R (for the siblings), the ADI-R (for the deceased brother, parental interview items were scored relating to the presence of behaviors ‘ever’) and the DSM-IV semi-structured interview (for the parents). All psychiatric examinations were carried out by an independent clinician without earlier knowledge of the genetic status of the subjects. All the medical information available on the deceased brother was obtained from the hospital in which he had received care during his life.
Genetic studies of the subjects
In the proband and his mother, fluorescent in situ hybridization studies were carried out with peripheral blood samples, using a series of bacterial artificial chromosomes along the predicted break points based on the karyotype results. The bacterial artificial chromosomes were obtained from the clone library at the Children’s Hospital, Oakland Research Institute, Oakland, CA, USA. In the proband, the results from the fluorescent in situ hybridization studies were confirmed by comparative genomic hybridization (CGH; Illumina 550 K BeadStudio, San Diego, CA, USA). In all family members, the absence or presence of the deletion was confirmed by genotyping 17 polymorphic single-nucleotide polymorphisms (SNPs), of which 8 were located within the deleted region and 9 were outside it (a list of SNP identification numbers is available upon request).
Subsequently the involved genomic region was searched for gene content using the University of California Santa Cruz (UCSC) human genome browser, April 2004 assembly. This identified four genes with a total of 48 exons. All the exons were amplified using conventional PCR techniques and adequate primer pairs. PCR primer sequences are available on request. Sequencing was performed with a BigDye terminator cycle sequencing kit v3.1 and the ABI 3730 DNA Analyzer, using the manufacturer’s protocol (Applied Biosystems, Foster City, CA, USA). Sequences were compared to the consensus human genome sequence (UCSC genome browser).
Mutation screening in controls
After identifying the nonsynonymous mutation in DIAPH3, we screened 128 DNA control samples of mixed ethnic descent for the presence of this polymorphism. Because this sample was not formally screened for the presence of psychiatric symptoms and the reported mutation was observed in a family from the Netherlands, we screened an additional sample of 200 individuals of Dutch ancestry in whom a lifetime psychiatric diagnosis had been ruled out by the Comprehensive Assessment of Symptoms and History.17
Review of gene function and predicted effect of the mutation
The available scientific literature on DIAPH3 function was found in PubMed and reviewed. Conservation of the DNA sequence containing the mutation was examined using the PhastCons Conserved Elements, 28-way Multiz Alignment track in the UCSC human genome browser. In this track the prediction of conserved elements is calculated based on a whole-genome alignment of vertebrates, and for the placental mammalian subset of species in the alignment. Each predicted element is assigned a log-odds ‘conservation score’. For more details of this method, see Siepel et al.18
Transfection and immunohistochemistry
NIH 3T3 cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum. After overnight adherence to acid-washed glass cover slips, we transfected cells with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. After 24 h’ incubation, we fixed cells with 4% paraformaldehyde in phosphate-buffered saline and stained for F-actin using TRITC-labeled phalloidin (Sigma-Aldrich, St Louis, MO, USA). Stained cells were mounted in Prolong Gold (Invitrogen) and visualized by confocal laser scanning microscopy (Zeiss LSM510, Oberkochen, Germany). All images are shown as projected confocal stacks.
Quantification and statistical analysis
The number of filopodia per cell was quantified (n = 20 for each condition) and data were analyzed using one-way analysis of variance followed by Tukey’s post hoc test. Three independent experiments were performed for each experimental condition. Data are expressed as mean±standard error of the mean.
RNA in situ hybridization of Diaph3 during mouse development
A detailed description of this method was reported previously.19 In short, a 688 base-pair fragment of the mouse homolog of DIAPH3 (mDia2, Diaph3, NM_019670) was amplified from 14.5 days post coitum (d.p.c.) mouse brain total RNA using gene-specific primer sequences (mDia2for, TTCTCCATGAGCTGA-AAATGG; mDia2rev, TCAAGGCGATGGAAAAACTC) and the OneStep RT-PCR kit (Qiagen, Valencia, CA, USA). The reverse transcriptase–PCR product was subsequently cloned into the pGEMteasy vector (Invitrogen), and DIG-labeled cRNA probes were made using T7- or Sp6-dependent RNA polymerase (Roche, Basel, Switzerland). Sense and antisense probes were hybridized to 16μm sagittal cryosections of various mouse embryonic stages (12.5–18.5 d.p.c.) and adult mouse brain, and expression was visualized using NBT/BCIP (Roche). Images were recorded on a Zeiss Axioskop2 Plus microscope with a Sony Power HAD DXC-950P 3CCD color video camera (Tokyo, Japan). Expression was considered genuine only when the sections hybridized with the sense probe showed no significant staining.
Results
The proband was a male subject of 13 years of age. At physical examination he was found to be of small stature (height < 2.5 standard deviation (s.d.) below the norm average), of normal weight (0 s.d.) and head circumference (occipital frontal; 0 s.d.). He displayed mild dysmorphic features consisting of slightly low implanted protruding ears, long eyelashes, hypotelorism (outer canthal distance −2 s.d.), a high palate, large and irregular front teeth and a mildly triangular-shaped face. He was diagnosed with hypermetropic vision in both eyes (S + 4.0). In addition, a mild lumbar lordosis and a small difference in leg length were found, causing a slightly askew pelvis. Standard laboratory screening, including thyroid function, and additional clinical studies, including fundoscopy, an echo of the heart and kidneys, an electrocardiogram, an electroencephalogram and magnetic resonance imaging of the brain, revealed no structural abnormalities.
He fulfilled DSM-IV diagnostic criteria for autistic disorder according to clinical consensus and the results of the ADI-R and ADOS-G. He also showed problems with regard to his attention span and increased impulsivity. However, these symptoms did not meet sufficient criteria for attention deficit and hyperactivity disorder. His cognitive abilities were in the normal range (full IQ, 98; verbal IQ, 93; performance IQ, 105). Both parents and his two siblings did not display any dysmorphic features and had normal psychomotor development. None of the family members displayed autistic features.
The eldest brother was born with a congenital heart defect (single ventricle defect due to mitral valve atresia). Cardiac surgery was postoperatively complicated by renal insufficiency necessitating chronic treatment with diuretics and cardiotonic medications, and growth hormone later in life. At age 13, he died of acute heart failure, but an autopsy was not performed. The available pictures of him did not suggest facial dysmorphic features. Information from the parents indicated that he had reached psychomotor developmental milestones in time and had not displayed autistic behavior.
A karyotype performed on the proband indicated a deletion on chromosome 13q. Results of the additional serial fluorescent in situ hybridization experiments (RP11-209J19, 2n; RP11-77O2, n; RP11-67L17, n; RP11-520F9, 2n) narrowed the break points of the deletion to 52 524 189–52 955 445 (proximal) and 63 071 144–63 447 585 (distal). These findings were confirmed and further specified by the results of the 550 K Illumina SNP array, showing that the deletion spanned almost 10 Mb, starting at 52 784 996–52 805 428 (proximal), and ending at 63 062 087–63 082 454 (distal). The same deletion was found in the mother. Further, heterozygosity of the 17 SNPs was consistent with the deleted genomic region in the proband and his mother, and with the absence of the deletion in his father and his two siblings (data not shown). There were neither DNA for molecular analysis nor cells for karyotyping available from the deceased brother.
The deleted region contained only four genes: protocadherin 17 precursor, DIAPH3, tudor containing 3 and protocadherin 20 precursor. Sequencing of all the exons of these four genes revealed a non-synonymous single-nucleotide variant in exon 15 of DIAPH3 on the remaining allele. Subsequently, we found both the proband’s father and brother to be heterozygous for the same variant in DIAPH3 (see Figure 1). No other exonic variants were detected, except for one known untranslated SNP (rs339531), also in DIAPH3.
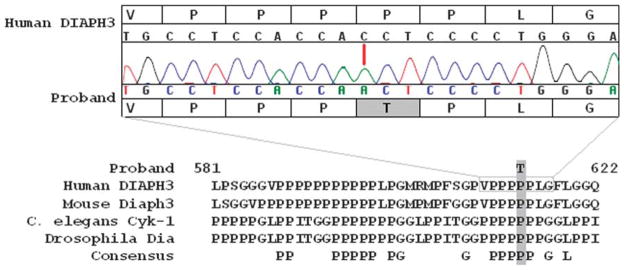
The identified point mutation (C > A) results in the substitution of proline by threonine at amino-acid position 614 (Pro614Thr) located in the formin homology 1 (FH1) domain of DIAPH3 (Figure 2). This particular polymorphism was not reported in the genome databases. In addition, we did not identify this mutation in our screening of 128 control individuals of mixed ethnic descent, nor in the additional sample of 200 Dutch individuals without a lifetime psychiatric diagnosis. The DIAPH3 Pro614Thr was not observed in these 328 control subjects (656 chromosomes), suggesting it is a very low allele frequency.
Figure 2.
Proband and consensus DNA and amino-acid sequence. Upper panel: consensus DNA and amino-acid sequence versus sequence results in the proband. Lower panel: formin homology 1 (FH1) domain in human DIAPH3 carrying a C > A point mutation that results in substitution of a conserved Pro by Thr at position 614. This amino-acid motif is conserved among species as diverse as Drosophila, Caenorhabditis elegans and mammalian species, including man.
Within DIAPH3 only one other nonsynonymous polymorphism with an allele frequency greater than 0.05 is reported by the genome databases (rs36084898, average heterozygosity: 0.148), leading to the substitution of asparagine by serine at position 363. This substitution occurs within the Rho GTPase-binding/ FH3 (GBD/FH3) domain of DIAPH3. However, this variant is unlikely to affect DIAPH3 function given that the GBD/FH3 domain of the DIAPH3 paralog DIAPH1 also contains serine at an analogous position (amino-acid position 336).
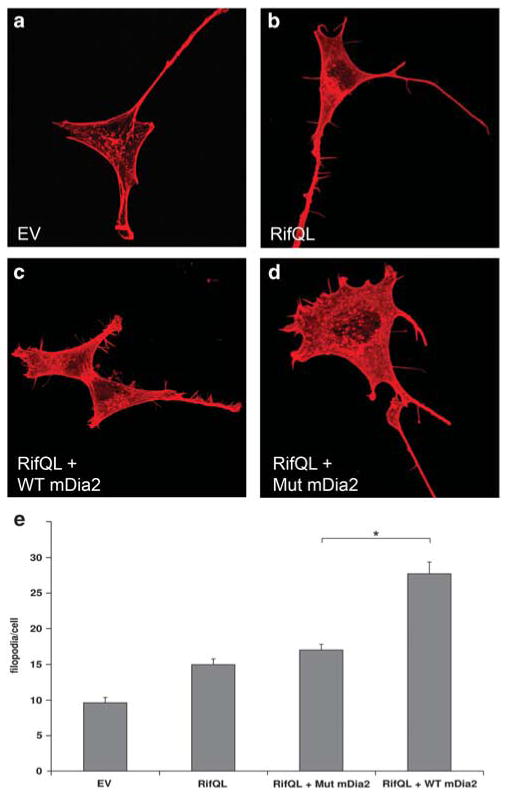
DIAPH3 is a regulator of actin dynamics, mediating assembly of unbranched actin filaments. The proline-rich FH1 domain of DIAPH3 is a binding site for the actin monomer-binding protein, profilin, thereby regulating actin nucleation.20 The probability of a functional effect of the point mutation was considered high given its location in the FH1 domain and our observation that the C > A mutation is situated in a short nucleotide sequence element that is highly conserved across vertebrates (chr13:59,443,088–59,443,116, LOD=56) and across a subset of mammalian species (Chr13:59,443,091–59,443,116, LOD = 17). Therefore, we examined whether the C > A mutation affects DIAPH3 function in the presence of an active form of the Rho family GTPase Rif (RifQL) in mouse 3T3 fibroblasts.21 In line with previous work, expression of the murine homologue of DIAPH3, mDia2 or Rif-QL alone did not lead to significant induction of filopodia formation as compared to controls (Figures 3a, b and e). In contrast, when mDia2 was co-transfected with RifQL, significantly more filopodia were observed (Figures 3c and e). Intriguingly, mutated mDia2 failed to induce filopodia formation in the presence of RifQL Figure 3d and e). Overall, these data suggest that the C > A mutation has functional consequences for DIAPH3 function and that it interferes with the effect of DIAPH3 on actin dynamics and filopodia formation.
Figure 3.
Mutant mDia2 does not induce filopodia in NIH 3T3 cells. (a and b) A small number of filopodia is found on NIH 3T3 cells transfected with empty vector (EV) or RifQL. (c) Coexpression of wild-type (WT) mDia2 and RifQL induces the formation of numerous filopodia per cell. (d) In contrast, coexpression of mutant (Mut; Pro614Thr) mDia2 and RifQL fails to induce filopodia formation as compared to transfection of RifQL alone. (e) Quantification of the number of filopodia per cell (n = 20 per experiment, three independent experiments were performed). Data are expressed as mean±s.e.m. number. *P < 0.01.
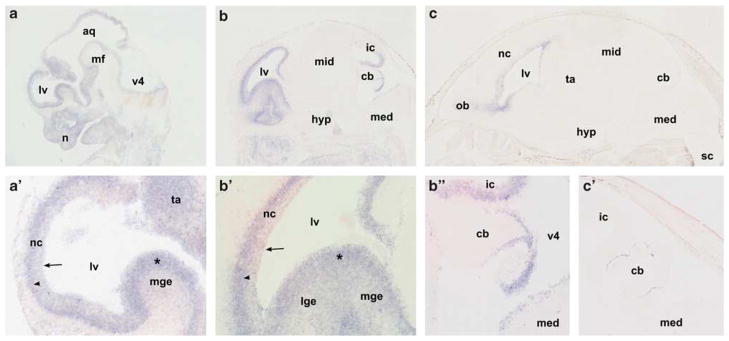
To establish whether DIAPH3 may have a function in the developing and adult brain, we performed RNA in situ hybridization of Diaph3-specific probes on cryosections of various mouse embryonic stages and adult mouse brain. During embryonic development expression was mainly observed in the subventricular zone (SVZ) of the brain (Figures 4a–c). Initially the signal was present in the ventricular zone (VZ) of the entire brain up to the spinal cord (10.5 (not shown) and 12.5 d.p.c., Figure 4a); at later stages expression became progressively restricted to the SVZ of the forebrain and roof of the midbrain (14.5 and 16.5 d.p.c., Figures 4b and c). At 18.5 d.p.c. only a low level of expression was maintained in the SVZ of the forebrain (Figure 4d). In addition, from 14.5 d.p.c. onward, staining was observed in the granular cell layer of the cerebellum (Figure 4e). No signal was detected in the adult mouse brain. Outside the brain, Diaph3 expression was detected in the neural layer of the retina, the liver, the cortical region of the kidneys and in the developing teeth (not shown). Sense probed sections did not show significant staining at any of the stages analyzed.
Figure 4.
Developmental expression of Diaph3 in the murine brain. All sections shown were hybridized with antisense probe; sense probed sections did not show significant staining. (a) At 12.5 and 10.5 days post coitum (d.p.c.) a signal was detected in the ventricular zone (VZ) of the entire central nervous system (10.5 d.p.c. not shown). (b) At 14.5 d.p.c. expression was detected in the VZ and subventricular zone (SVZ) of the forebrain and region of the inferior colliculus (a′ and b′). In the future cerebral cortex of, respectively, 12.5 and 14.5 d.p.c. embryos staining was intenser in the SVZ compared to the VZ immediately lining the ventricles (arrows indicate the border between the lateral ventricle and the neocortex, arrowheads indicate the SVZ with more intense staining. In more posterior brain regions expression was uniformly distributed over the ventricular zones, for example, the medial ganglionic eminence (*). (c) At 18.5 d.p.c. only the ventricular zone of the forebrain shows staining (b″ and c′). From 14.5 d.p.c. onward, we detected additional staining in the brain in the outer cell layer of the cerebellar primordium. Magnification: a, b, c: × 12.5; a′, b′, b″: × 50; c′: ×25. Abbreviations: aq, aqueduct; cb, cerebellar primordium; hyp, hypothalamus; ic, inferior colliculus; lge, lateral ganglionic eminence; lv, lateral ventricle; med, medulla oblongata; mf, mesencephalic flexure; mge, medial ganglionic eminence; mid, midbrain; nc, neocortex; n, nose; ob, olfactory bulb; sc, spinal cord; ta, thalamus; v4, fourth ventricle.
Discussion
Here, a normally intelligent patient with an autistic disorder is presented in whom a rare genetic ‘double hit’, consisting of a maternally inherited deletion and a paternally inherited functional single-nucleotide mutation, Pro614Thr, in a highly conserved nucleotide sequence suggest a homozygous loss- of function in the DIAPH3 gene. The results of additional experiments indicated expression of DIAPH3 during early brain development and provided evidence for a loss of function as a result of the Pro614Thr mutation. These observations strongly suggest that DIAPH3 may have a causative function in the proband’s autistic disorder. His parents and one sibling were found to be clinically unaffected carriers of either the deletion or the mutation but not both, showing the recessive nature of the respective genomic and coding variant.
In view of the recessive nature, DIAPH3 mutations resulting in ASD are likely to be rare. However, the implication of DIAPH3 in the proband provides insight into the pathological processes leading to autism.
To our knowledge, there is only one other case report of an autistic individual with a chromosomal rearrangement involving DIAPH3, XY, del (13) (q14q22).22 In addition, a large genomic deletion at 13q, such as described here, has not been reported in the database of genomic variants (HG build 36), although small CNVs within DIAPH3 have been identified in normal controls.23,24
Our case findings and the additional studies reported here provide a reasonable level of plausibility for DIAPH3 being a candidate gene for autism. The results of this study are also relevant for a better understanding of how structural genomic abnormalities may be involved in the etiology of genetic disease.
DIAPH3, a candidate gene for autism
DIAPH3 is an FH protein that has a central function in the assembly of actin filaments through the combined action of its functional FH1 (actin recruitment) and FH2 (construction of the actin filament) domains.25 DIAPH3 is closely related to its paralogs DIAPH1 and DIAPH2.
The Pro614Thr substitution occurs in the middle of a proline-rich amino-acid sequence that is highly identical with the same sequence in FH1 of DIAPH1 and DIAPH2.26
During brain development, neurons migrate from the VZs to specific cortical layers where they form synaptic connections with other neurons.27 This migratory process depends on the tightly orchestrated and highly dynamic process of cytoskeletal remodeling. As a first step in neuronal migration, the leading edge of a cell is extended by formation of membrane protrusions (filopodia). This process is mediated by two distinct actin networks, the lamellipodium and the lamella. Lamellipodia at the leading edge are involved in a repeating process of extension and retraction, but persistent leading edge advance and cell migration rely on the expansion of the lamella actin network.28 Recently, it was shown that the DIAPH3 regulates actin network dynamics within the lamella and that inhibition of DIAPH3 significantly slows cell migration.29 However, DIAPH3 also appears to have a significant function in the expansion process of lamellipodia.30
In addition, lamellipodia and filopodia are not only important for efficient axonal guidance and neuronal migration, but also have an essential function in neurite formation.31,32 Fusion of lamellipodia results in the formation of neurite extensions, which are the precursors to dendrites, dendritic spines and axons. DIAPH3 has been shown to induce filopodia formation in neurons, suggesting that DIAPH3 can also induce neurite formation.31 Moreover, a recent study showed that the inhibition of the Rif/DIAPH3 pathway in murine hippocampal neurons results in decreased synaptic activity.33 The results of our in vitro assay confirm a previous study,21 validating the evidence for the function of DIAPH3 in the induction of filopodia downstream of Rif. Importantly, the distinctive reduction of induced filopodia by mutant Diaph3 provides clear evidence for a loss of function with regard to the induction of filopodia as a result of the Pro614Thr mutation in DIAPH3.
Interestingly, SHANK3, a gene that was recently reported as a novel candidate gene for ASDs,34–36 has an important regulatory function in the process of the initiation and maturation of filopodia resulting in the formation of functional dendritic spines.37 (Although the function of SHANK3 has not been consistently replicated by all studies.38,39) In this process, SHANK3 interacts with cortactin, a protein implicated in connecting the actin network sites with receptor signaling complexes.40 On the basis of these findings, one could speculate that DIAPH3 operates downstream of SHANK3 regulation.
Spatiotemporal expression of DIAPH3 suggests a dual function in cortical development. During early corticogenesis DIAPH3 is expressed in the VZ of the whole brain, whereas in later stages expression shifts to the SVZ of the cortex only. This suggests that DIAPH3 may participate initially in proliferation of neural progenitors. The restricted expression in the cortical SVZ points to post-mitotic functions such as migration, axon outgrowth and neuritogen-esis. The function of DIAPH3 in the development of the cortex appears transient, because DIAPH3 is no longer expressed in the adult cortex (data not shown).
This also provides a neurodevelopmental window in which the pathogenesis of autism may occur. Particularly the second phase of DIAPH3 expression in the SVZ would fit with observations made in patients showing subtle cortical defects, but no gross abnormalities due to proliferation defects.
In summary, the expression profile together with the available functional data suggests that DIAPH3 may have a pivotal function in cell migration, axon guidance, neuritogenesis and synaptic activity. In addition, DIAPH3 may exert these functions in a biological pathway that includes SHANK3.
A causal relationship between DIAPH3 and autistic disorder in our proband is plausible, based on the findings of our study. First, although the deletion and the point mutation were both identified in unaffected first-degree family members, the proband is the only one carrying both alleles (that is, he is compound heterozygote). Autism is a relatively rare disorder, and both the point mutation and deletion appear to be very rare events at the population level, which makes the concurrence in one person an extremely rare event.
Second, our findings indicate that it is very likely that the function of DIAPH3 is disrupted in the proband; the point mutation in the remaining copy leads to an amino-acid substitution in a protein domain that is essential to its function, within a sequence highly conserved between the other DIAPH genes and across mammalian species. Indeed, the results of the fibroblast assay provide firm proof for a substantial loss of function with regard to the induction of filopodia due to the Pro614Thr mutation.
Third, the function and expression profile of Diaph3 is indicative of an important function in the developing cortex during embryogenesis. This is consistent with the current etiologic models of autism, which imply a disruption of normal development leading to abnormal organization and connectivity of cortical neurons.41
Although the expression pattern of Diaph3 in the mouse involves several tissues outside the brain, including retina, liver, kidney and teeth, the proband showed no major abnormalities in these organs. Laboratory analysis revealed normal liver and kidney function, and an ultrasound examination of the kidneys was unremarkable. Also, no retinal abnormalities were revealed by fundoscopy. There were, however, some minor observations: he was found to have hypermetropic eyesight and on examination his front teeth were noted to be somewhat large and irregular. The expression of Diaph3 in the murine brain appeared very restricted, both spatially and temporally, compared to a more prolonged Diaph3 expression in the tissues outside of the brain. This could suggest a different, perhaps less critical function for DIAPH3 in tissues outside the brain, consistent with our clinical observations in the proband.
Neuronal migration defects can range from very severe (no migration) to mild. Similarly, based on the function of DIAPH3 in neuronal migration one could expect to find subtle to severe cortical defects with magnetic resonance imaging, such as heterotopia or lissencephaly. It is possible that the identified point mutation affects some but not all of the functions of DIAPH3. In addition, DIAPH3 is one of three paralogs (DIAPH1, DIAPH2 and DIAPH3) with likely partially redundant functions. The loss of function of DIAPH3 may therefore be partly compensated. In summary, the function of DIAPH3 in neuronal migration may be reduced by the double-hit mutation, without being completely absent. Thus, a possible explanation is that neuronal migration defects did cause alterations in the patient’s neuroanatomy; however, these may be too subtle to be detected by the resolution of the magnetic resonance imaging technology we used. Although many neuronal migration abnormalities can indeed be detected with magnetic resonance imaging, some abnormalities remain undetectable.42
Interpretation of inherited structural abnormalities in studies on ASD
When a structural genomic abnormality is identified in a patient, establishing evidence for a causal relationship between the disease phenotype and the observed genomic abnormality is not straightforward. Naturally, causality is more likely to exist if more than one such concurrence of the phenotype and the genomic abnormality have been identified. In contrast, when other cases are observed (within or outside the index patient’s family) with the same genomic defect, but without the phenotype, it is often concluded that a causal relation is unlikely.
The interpretation of incomplete co-segregation patterns is not only clinically relevant for genetic counseling, but also is important for the interpretation of results from screens of cytogenetic abnormalities and CNVs in disease populations. CNVs and cytogenetically detectable chromosomal abnormalities are generally regarded as not causally related to a disease if the event is not de novo, that is, when one of the unaffected parents (or siblings) is also found to be a carrier.
This case shows how a causal relation between a structural genomic abnormality and a disease phenotype may be plausible, even if unaffected individuals with the same genomic abnormality are identified. This is a particularly relevant issue, given that in most hallmark studies so far, only de novo CNVs (or CNVs that were not found in unrelated normal controls) were prioritized in the analyses.4,5,7,8,43,44 Although this approach is understandable as an effort to reduce false-positive findings, one may also reason that inherited CNVs do, in fact, contribute to ASD in some cases. For instance, it has been argued that penetrance of genetic defects may be low in females.45 Alternatively, a gene within the implicated region may be affected by a common variant or a functional mutation, such as has been shown in our case report. Such a ‘double-hit’ scenario has been described for several genetic diseases; for instance, the situation of compound heterozygosity involving two different point mutations in retinal dystrophy,46 involving two different genomic rearrangements in a person with cystic fibrosis47 or involving a deletion and a mutation in an individual with 22q11.2 deletion syndrome and the Bernard–Soulier syndrome.48
Given the rarity of the deletion and the point mutation, the co-occurrence of both in one individual is likely to be an extremely rare event, which makes it almost impossible to provide statistical evidence for involvement in disease. This is a major challenge when studies reveal patients with ‘private mutations’, something that is likely to be an important cause in the etiology of autism.49 The sequencing of individual human genomes50–53 has provided evidence for the relatively frequent occurrence of ‘private’ genetic variation. For instance, approximately 18% of SNPs identified in the Watson genome and 9% of SNPs in the Venter genome are novel, that is, not previously described in the human genome databases.50,51 This finding implies that the a priori chance of detecting private, novel mutations in individuals with a disorder is likely to be high (approximately 10–20%) and emphasizes the necessity of obtaining additional evidence for a meaningful relationship between a mutation and the disorder. Our study provides a good example that, to show the relevance of private mutations suspected of being involved in disease, functional biological evidence may be essential.
Our findings suggest that assuming that only de novo genomic rearrangements are causally related to ASD may be too restrictive, and may lead to loss of valuable genetic information and biological insight. Moreover, one could argue that a ‘double-hit’ scenario, that is, the disruption of a gene by two independent mutational events affecting both alleles, is probably not a frequent cause of ASD, but it has the great advantage of pinpointing the genetic contribution to a particular gene. We therefore suggest that an effort should be made for a re-sequencing project of genes involved in non-de novo and non-unique genomic deletions patients with in ASD carrying these microdeletions. On the basis of the results of our study, we anticipate that such an effort may be an efficient approach to identify new candidate genes for autism. Recent technological advances in the field of high-throughput sequencing make it possible to pursue this worthwhile effort in the search for rare genetic variants involved in autism.
Acknowledgments
We thank Eric Strengman and Eric Rappaport for their technical support with the sequencing experiments and Harry Mellor (University of Bristol, UK) for proving us with mDia2 and RifQL expression vectors. We thank Corlinda ten Brink for help with confocal microscopy and Petra Baarendse for help with statistical analyses. We also thank Jacky Senior for the careful language corrections of the paper.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Freitag CM. The genetics of autistic disorders and its clinical relevance: a review of the literature. Mol Psychiatry. 2007;12:2–22. doi: 10.1038/sj.mp.4001896. [DOI] [PubMed] [Google Scholar]
- 2.Persico AM, Bourgeron T. Searching for ways out of the autism maze: genetic, epigenetic and environmental clues. Trends Neurosci. 2006;29:349–358. doi: 10.1016/j.tins.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Beaudet AL. Autism: highly heritable but not inherited. Nat Med. 2007;13:534–536. doi: 10.1038/nm0507-534. [DOI] [PubMed] [Google Scholar]
- 4.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vorstman JA, Staal WG, van Daalen E, van Engeland H, Hochstenbach PF, Franke L. Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism. Mol Psychiatry. 2006;11:18–28. doi: 10.1038/sj.mp.4001781. [DOI] [PubMed] [Google Scholar]
- 7.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11. 2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 9.Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- 10.Vorstman JAS, Morcus MEJ, Duijff SN, Klaassen PWJ, Heinemande Boer JA, Beemer FA, et al. The 22q11. 2 deletion in children: high rate of autistic disorders and early onset of psychotic symptoms. J Am Acad Child Adolesc Psychiatry. 2006;45:1104–1113. doi: 10.1097/01.chi.0000228131.56956.c1. [DOI] [PubMed] [Google Scholar]
- 11.Pulver AE, Nestadt G, Goldberg R, Shprintzen RJ, Lamacz M, Wolyniec PS, et al. Psychotic illness in patients diagnosed with velo-cardio-facial syndrome and their relatives. J Nerv Ment Dis. 1994;182:476–478. doi: 10.1097/00005053-199408000-00010. [DOI] [PubMed] [Google Scholar]
- 12.Veltman MW, Craig EE, Bolton PF. Autism spectrum disorders in Prader–Willi and Angelman syndromes: a systematic review. Psychiatr Genet. 2005;15:243–254. doi: 10.1097/00041444-200512000-00006. [DOI] [PubMed] [Google Scholar]
- 13.Kaufman J, Birmaher B, Brent D, Rao U, Flynn C, Moreci P, et al. Schedule for Affective Disorders and Schizophrenia for School- Age Children-Present and Lifetime Version (K-SADS-PL): initial reliability and validity data. J Am Acad Child Adolesc Psychiatry. 1997;36:980–988. doi: 10.1097/00004583-199707000-00021. [DOI] [PubMed] [Google Scholar]
- 14.Lord C, Risi S, Lambrecht L, Cook EH, Jr, Leventhal BL, DiLavore PC, et al. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord. 2000;30:205–223. [PubMed] [Google Scholar]
- 15.Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview- Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- 16.Wechsler D, Kort W, Compaan EL, Bliechrodt N, Resig WCM, Schittekatte M, et al. WISC-III NL, Handleiding. Psychological Corporation Unlimited; London, UK: 2002. [Google Scholar]
- 17.Andreasen NC, Flaum M, Arndt S. The Comprehensive Assessment of Symptoms and History (CASH). An instrument for assessing diagnosis and psychopathology. Arch Gen Psychiatry. 1992;49:615–623. doi: 10.1001/archpsyc.1992.01820080023004. [DOI] [PubMed] [Google Scholar]
- 18.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Zwaag B, Burbach JP, Scharfe C, Oefner PJ, Brunner HG, Padberg GW, et al. Identifying new candidate genes for hereditary facial paresis on chromosome 3q21-q22 by RNA in situ hybridization in mouse. Genomics. 2005;86:55–67. doi: 10.1016/j.ygeno.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 21.Pellegrin S, Mellor H. The Rho family GTPase Rif induces filopodia through mDia2. Curr Biol. 2005;15:129–133. doi: 10.1016/j.cub.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 22.Steele MM, Al Adeimi M, Siu VM, Fan YS. Brief report: a case of autism with interstitial deletion of chromosome 13. J Autism Dev Disord. 2001;31:231–234. doi: 10.1023/a:1010759401344. [DOI] [PubMed] [Google Scholar]
- 23.Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE, et al. A comprehensive analysis of common copynumber variations in the human genome. Am J Hum Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS, et al. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–1190. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evangelista M, Zigmond S, Boone C. Formins: signaling effectors for assembly and polarization of actin filaments. J Cell Sci. 2003;116:2603–2611. doi: 10.1242/jcs.00611. [DOI] [PubMed] [Google Scholar]
- 26.Katoh M, Katoh M. Identification and characterization of human DIAPH3 gene in silico. Int J Mol Med. 2004;13:473–478. [PubMed] [Google Scholar]
- 27.Sidman RL, Rakic P. Neuronal migration, with special reference to developing human brain: a review. Brain Res. 1973;62:1–35. doi: 10.1016/0006-8993(73)90617-3. [DOI] [PubMed] [Google Scholar]
- 28.Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- 29.Gupton SL, Eisenmann K, Alberts AS, Waterman-Storer CM. mDia2 regulates actin and focal adhesion dynamics and organization in the lamella for efficient epithelial cell migration. J Cell Sci. 2007;120:3475–3487. doi: 10.1242/jcs.006049. [DOI] [PubMed] [Google Scholar]
- 30.Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dent EW, Kwiatkowski AV, Mebane LM, Philippar U, Barzik M, Rubinson DA, et al. Filopodia are required for cortical neurite initiation. Nat Cell Biol. 2007;9:1347–1359. doi: 10.1038/ncb1654. [DOI] [PubMed] [Google Scholar]
- 32.Kwiatkowski AV, Rubinson DA, Dent EW, Edward van Veen J, Leslie JD, Zhang J, et al. Ena/VASP is required for neuritogenesis in the developing cortex. Neuron. 2007;56:441–455. doi: 10.1016/j.neuron.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Hotulainen P, Llano O, Smirnov S, Tanhuanpaa K, Faix J, Rivera C, et al. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol. 2009;185:323–339. doi: 10.1083/jcb.200809046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gauthier J, Spiegelman D, Piton A, Lafreniere RG, Laurent S, St- Onge J, et al. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- 37.Roussignol G, Ango F, Romorini S, Tu JC, Sala C, Worley PF, et al. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J Neurosci. 2005;25:3560–3570. doi: 10.1523/JNEUROSCI.4354-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin J, Jia M, Wang L, Lu T, Ruan Y, Liu J, et al. Association study of SHANK3 gene polymorphisms with autism in Chinese Han population. BMC Med Genet. 2009;10:61. doi: 10.1186/1471-2350-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sykes NH, Toma C, Wilson N, Volpi EV, Sousa I, Pagnamenta AT, et al. Copy number variation and association analysis of SHANK3 as a candidate gene for autism in the IMGSAC collection. Eur J Hum Genet. 2009;17:1347–1353. doi: 10.1038/ejhg.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, et al. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 41.Palmen SJ, Van EH, Hof PR, Schmitz C. Neuropathological findings in autism. Brain. 2004;127:2572–2583. doi: 10.1093/brain/awh287. [DOI] [PubMed] [Google Scholar]
- 42.Spalice A, Parisi P, Nicita F, Pizzardi G, Del BF, Iannetti P. Neuronal migration disorders: clinical, neuroradiologic and genetics aspects. Acta Paediatr. 2009;98:421–433. doi: 10.1111/j.1651-2227.2008.01160.x. [DOI] [PubMed] [Google Scholar]
- 43.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cusco I, Medrano A, Gener B, Vilardell M, Gallastegui F, Villa O, et al. Autism-specific copy number variants further implicate the phosphatidylinositol signaling pathway and the glutamatergic synapse in the etiology of the disorder. Hum Mol Genet. 2009;18:1795–1804. doi: 10.1093/hmg/ddp092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao X, Leotta A, Kustanovich V, Lajonchere C, Geschwind DH, Law K, et al. A unified genetic theory for sporadic and inherited autism. Proc Natl Acad Sci USA. 2007;104:12831– 12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janecke AR, Thompson DA, Utermann G, Becker C, Hubner CA, Schmid E, et al. Mutations in RDH12 encoding a photoreceptor cell retinol dehydrogenase cause childhood-onset severe retinal dystrophy. Nat Genet. 2004;36:850–854. doi: 10.1038/ng1394. [DOI] [PubMed] [Google Scholar]
- 47.Girardet A, Guittard C, Altieri JP, Templin C, Stremler N, Beroud C, et al. Negative genetic neonatal screening for cystic fibrosis caused by compound heterozygosity for two large CFTR rearrangements. Clin Genet. 2007;72:374–377. doi: 10.1111/j.1399-0004.2007.00850.x. [DOI] [PubMed] [Google Scholar]
- 48.Budarf ML, Konkle BA, Ludlow LB, Michaud D, Li M, Yamashiro DJ, et al. Identification of a patient with Bernard- Soulier syndrome and a deletion in the DiGeorge/velo-cardio-facial chromosomal region in 22q11. 2. Hum Mol Genet. 1995;4:763–766. doi: 10.1093/hmg/4.4.763. [DOI] [PubMed] [Google Scholar]
- 49.O’Roak BJ, State MW. Autism genetics: strategies, challenges, and opportunities. Autism Res. 2008;1:4–17. doi: 10.1002/aur.3. [DOI] [PubMed] [Google Scholar]
- 50.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 51.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]