Abstract
Nearly all free living bacteria carry toxin-antitoxin (TA) systems on their genomes, through which cell growth and death are regulated. Toxins target a variety of essential cellular functions, including DNA replication, translation, and cell division. Here we identified a novel toxin, YgfX, on the E. coli genome. The toxin, consisting of 135 residues, is composed of the N-terminal membrane domain, which encompasses two transmembrane segments, and the C-terminal cytoplasmic domain. Upon YgfX expression, the cells were initially elongated and then the middle portion of the cells became inflated to form a lemon-shape. YgfX was found to interact with MreB and FtsZ, two essential cytoskeletal proteins in E. coli. The cytoplasmic domain [YgfX(C)], was found to be responsible for the YgfX toxicity, as purified YgfX(C) was found to block polymerization of FtsZ and MreB in vitro. YgfY, located immediately upstream of YgfX, was shown to be the cognate antitoxin. Notably, YgfX is the first membrane associating toxin in bacterial TA systems. We propose to rename the toxin and the antitoxin as CptA and CptB (for Cytoskeleton Polymerization inhibiting Toxin), respectively.
Introduction
Nearly all free living bacteria contain toxin antitoxin (TA) systems on their genomes (Pandey & Gerdes, 2005). The sets of toxin and antitoxin proteins are most often encoded from a single operon. In all known cases, in normally growing cells, toxins form a stable complex with their cognate antitoxins which blocks toxin activity. Antitoxin also functions as a repressor for individual TA operons (Gerdes et al., 2005). Under stress conditions, intrinsically unstable antitoxin is lost from the cells, releasing toxin freely and inhibiting various essential cellular functions, such as DNA replication, mRNA stability, protein synthesis and cell division (Jiang et al., 2002; Zhang et al., 2003; Zhang & Inouye, 2011; Tan et al., 2011). This leads to a reversible cell growth arrest, which is implicated in the persister phenotype. TA system is also shown to be associated with pathogenicity, programmed cell death, and biofilm formation (Pandey & Gerdes, 2005; Nariya & Inouye, 2008; Wang & Wood, 2011).
E. coli have two essential bacterial cytoskeletal proteins, FtsZ and MreB. FtsZ is a highly conserved GTPase, and homologous to eukaryotic cytoskeleton protein, tubulin (Mukherjee et al., 1998). It forms a ring structure at the mid cell and functions as a scaffolding for divisome, a multi-protein complex responsible for cell division. MreB is an actin-like ATPase, essential for maintaining the typical rod shape and cell polarity in E. coli (Osborn & Rothfield, 2007). MreB is also implicated in chromosome segregation, localization of membranous organelles, and coordinating cell division with cell biosynthesis (Komeili et al., 2006; Madabhushi & Marians, 2009; Kruse et al., 2005; Garner et al., 2011; Domínguez-Escobar et al., 2011). Since Both FtsZ and MreB are involved in a number of essential cellular functions, the inhibition of their functions is detrimental to the cells. For example, the inhibition of FtsZ polymerization by SulA or MinCD results in blocking the septum formation, causing the formation of filamentous cells (Pichoff & Lutkenhaus, 2001; Mukherjee et al., 1998). The inhibition of MreB by A22 [S-(3,4-dichlorobenzyl) isothiourea] leads to loss of its rod shape, and eventual cell lysis (Bean et al., 2009; Karczmarek et al., 2007).
Here we have identified a novel TA system in E. coli genome using RASTA (Sevin & Barloy-Hubler, 2007). The putative toxin, YgfX, inhibits cell growth and causes significant changes in the cellular morphology of E. coli. Upon induction of YgfX, the cells were first elongated, and then subsequently became inflated in the middle. The YgfX toxicity was neutralized by the co-expression of YgfY, indicating that YgfY is an antitoxin of YgfX. YgfX is the first toxin of E. coli TA systems shown to be associated with membrane. We further demonstrated that YgfX physically interacts with FtsZ and MreB and inhibits their polymerization in vitro, and that the C-terminal soluble domain of the YgfX is responsible for the inhibition. On the basis of these results, we propose to rename YgfX and YgfY as CptA and CptB (for Cytoskeleton Polymerization inhibiting Toxin A and B), respectively.
Materials and Methods
Strains, Plasmids, and Growth Conditions
E. coli BW25113 (ΔaraBD) (Datsenko & Wanner, 2000) and BL21 (DE3) were grown in M9 medium supplemented with 0.2% casamino acids and 0.5% glycerol at 37°C. The primers used in this study is summarized in Table 1. The coding sequence of ygfX alone or ygfYX were PCR-amplified using primers YGFX-F and YGFX-R1, or YGFY-F and YGFX-R1, respectively. The fragments were cloned into pBAD24 vector (Guzman et al., 1995), and designated as pBAD24-ygfX and pBAD24-ygfYX, respectively. The coding 98 sequence of YgfX in a fusion with His6-tag at the C-terminal (YgfX-His) was also cloned into pBAD24 using YGFX-F and YGFX-R2. A truncated protein of YgfX (YgfX(C); cloned from V49 to Z135) was cloned into pCold-Km (unpublished results, Inouye laboratory) using YGFXs-F and YGFX-R1. His6-tagged FtsZ and MreB were constructed previously (Tan et al., 2011). FLAG-tagged FtsZ and MreB were also previously constructed in pET17b, having a tag at the C-terminal end (unpublished results, Masuda and Inouye). For examining the growth rate, 0.2% arabinose was added to the cultures during the early exponential phase.
Table 1.
Primers used for cloning of genes.
| Primer | Sequence |
|---|---|
| YGFX-F | 5′-TTTTTTTTTTCATATGGTCCTGTGGCAATCTGA-3′ |
| YGFX-R1 | 5′-TTTGAATTCTTATCTTTGCGTCTCTTGTTG-3′ |
| YGFX-R2 | 5′-TTTTGAATTCTCAGTGGTGGTGGTGGTGGTGTCTTTGCGTCTCTTGTTG-3′ |
| YGFXs-F | 5′-TTTTTTTTTTCATATGGTGGTGTTTGATTGCGTTCGC-3′ |
| YGFY-F | 5′-TTTTTTTTTTCATATGGACATTAACAACAAA-3′ |
Protein Expression, Purification, and Pull Down Assay
His6-tagged YgfX(C), FtsZ and MreB were expressed in E. coli BL21(DE3). Protein expression was induced for 2 hrs by adding 1 mM IPTG when the OD600 reached 0.8. The cells were collected by brief centrifugation at 8,000 x g and lysed by french pressure press (Thermo Fisher Scientific, MA). FtsZ and MreB was purified as described before (Tan et al., 2011). YgfX(C)-HIS was purified from the insoluble materials after dissolved in 8M urea (pH 8.0). Proteins were purified using Ni-NTA agarose according to the manufacturer’s instructions (Qiagen, CA).
Inner and outer membrane proteins were isolated following the method described previously (Hobb et al., 2009). Briefly, the total membrane were collected from the lysate by ultracentrifugation at 100,000 x g for 1 hr. The pellet was washed, then resuspended in 1% (w/v) N-lauroylsarcosine in 10 mM HEPES, pH 7.4, and incubated at 25°C for 30 min with gentle agitation. The inner and outer membrane fractions were furtherr separated by ultracentrifugation.
His6-tag pull down assays were carried out by incubating the cell lysate containing YgfX-HIS and the cell lysate containing FsZ-FLAG or MreB-FLAG (lysis buffer: 50 mM HEPES-KOH, pH 7.5, 10 mM MgCl2, 200 mM KCl, 0.1 mM EDTA, and 10% glycerol) 122 overnight at 4°C. Ni-NTA agarose (0.5 ml) was added to the lysate and the mixture was incubated at room temperature for 1 hr. The beads were washed three times with 20 ml of the same lysis buffer containing 20 mM immidazol. Protein complexes were then separated by 17.5% SDS-PAGE and visualized by western blot using monoclonal anti-FLAG antibody conjugated with horseradish peroxidase (Sigma-Aldrich, MO).
Polymerization Assays
The effect of YgfX on FtsZ and MreB polymerization was determined by a sedimentation method as previously described (Anand et al., 2004) with a few modifications. Purified FtsZ-HIS or MreB-HIS was mixed with different amounts of purified YgfX(C)-HIS in polymerization buffer P (5mM MgCl2; 50 mM NaH2PO4; 100mM NaCl; 20% glycerol, pH 6.5) and pre-incubated at room temperature for 30 min before 1 mM GTP or ATP was added to initiate polymerization. The polymerization reaction was carried out at room temperature for 30 min. FtsZ or MreB polymers were precipitated by centrifugation at 100,000 x g for 20 min, and the pellets were suspended in 50 μl of buffer P. Both the supernatant and pellet fractions were separated by a 17.5% SDS-PAGE, followed by Coomassie blue staining.
Microscopy
Cell morphology was observed using an Olympus BX40 microscope.
Results
YgfX is localized in the inner membrane
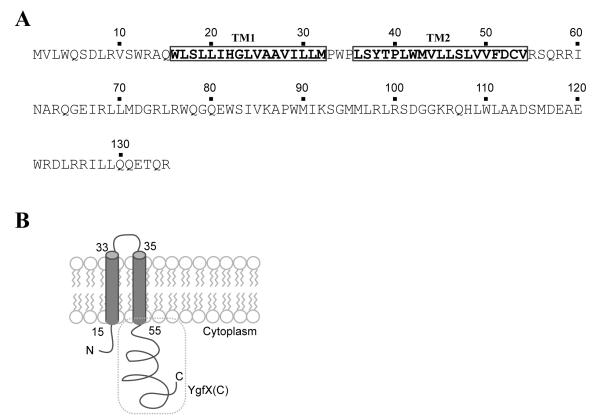
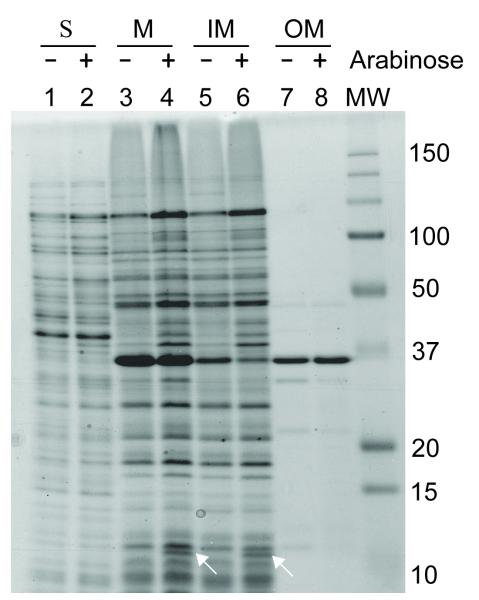
YgfX contains a long hydrophobic segment at the N-terminal region from W16 to V54 (Figure 1A). There are two Pro residues (P33 and P35) in the middle of the hydrophobic region and thus this protein likely forms a hydrophobic hair-pin structure with two transmembrane (TM) domains: TM1 from W16 to M32 and TM2 from L36 to V54. The presence 146 of positively charged residues on either side of the putative TM segments suggests that N-terminal and C-terminal soluble domain of YgfX resides in cytosol (Figure 1B). In order to experimentally determine the localization of YgfX, the full size YgfX was expressed from arabinose inducible vector, pBAD24 (pBAD24-ygfX). After YgfX expression was induced by the addition of 0.2% arabinose for 2 hrs, the total membrane was collected from the cellular lysate by ultracentrifuge. YgfX was found exclusively localized in the membrane fraction (lane 4, Figure 2). Total membrane was further separated into the inner and outer membrane fractions based on the solubility in 1% N-laurylsarcosine (Hobb et al., 2009). As predicted, YgfX was shown to be localized in the inner membrane (lane 6, Figure 2).
Figure 1. Amino acid sequence and predicted topology of YgfX.
(A) The amino acid sequence of YgfX labeled with putative transmembrane segments (TM1 and TM2).
(B) Predicted topology of YgfX. The C-terminal cytosolic domain of YgfX [YgfX(C)] is labeled with a dotted box.
Figure 2. Localization of YgfX in the inner membrane.
E. coli BL21 (DE3) cells were transformed with pBAD24-ygfX (lane 2, 4, 6, and 8) or with pBAD24 (lane 1, 3, 5, and 7), and treated with 0.2% arabinose for 1 hr. The soluble (S) fractions (lane 1 and 2) and the membrane (M) fractions (lane 3 and 4) were separated by ultracentrifugation at 100,000 x g for 1 hr. The membrane fraction was resuspended in 1% N-laurylsarcosine (Hobb et al., 2009). The detergent-soluble inner membrane (IM; lane 5 and 6) and the insoluble outer membrane (OM; lane 7 and 8) were separated by ultracentrifugation as described above. Proteins were separated by 17.5% SDS-PAGE 447 and visualized by Coommassie blue staining.
YgfX and YgfY shows toxin and antitoxin activity, respectively
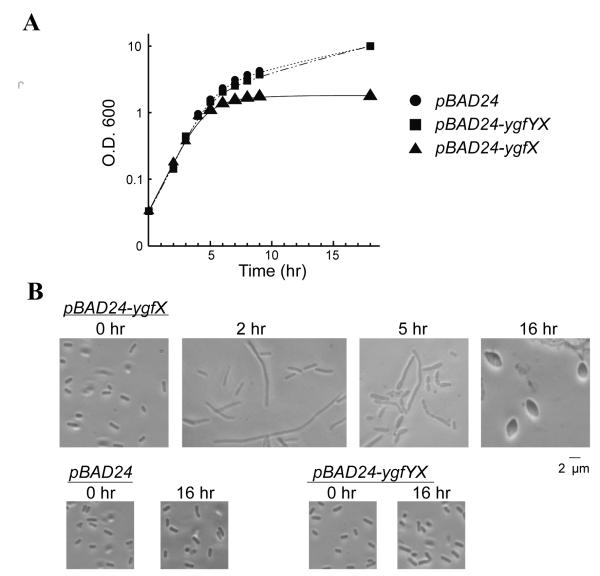
Intriguingly, the overexpression of YgfX caused growth arrest starting at 5 hrs post induction (Figure 3A). The growth arrest was accompanied by morphological change (Figure 3B). After 1 hr induction of YgfX expression from pBAD24-ygfX, some cells started to elongate. After 5 hrs, elongated cells were divided into smaller cells and simultaneously, cells became inflated in the middle or at the poles of cells. After overnight induction, cells became lemon shaped. We then examined whether YgfY can neutralize the toxicity caused by YgfX. First, the coding sequence of both ygfY and ygfX were cloned together in pBAD24. This construct did not show any growth inhibition at least for 48 hrs. The morphological change was also not observed. This result was confirmed by the expression of YgfX and YgfY separately from two independent plasmids. For this purpose, YgfY was cloned in a derivative of pCold vector (pCold-Km), and shown to be highly expressed (data not shown). In consistent with above experiments, cells expressing both YgfY and YgfX did not show any growth defect and alteration of morphology at least for 18 hrs, confirming that YgfY functions as an antitoxin for YgfX.
Figure 3. Growth inhibition and morphological change by YgfX.
E. coli BW25113 cells were transformed with pBAD24, pBAD24-ygfX or pBAD24-ygfYX. The cells were incubated in M9 medium supplemented with 0.2% casamino acids and 0.5% glycerol. The protein expression was induced at the early exponential phase with 0.2% arabinose.
(A) Growth curves of each strain.
(B) The cellular morphology at the designated period after the addition of arabinose.
YgfX interacts with FtsZ and MreB
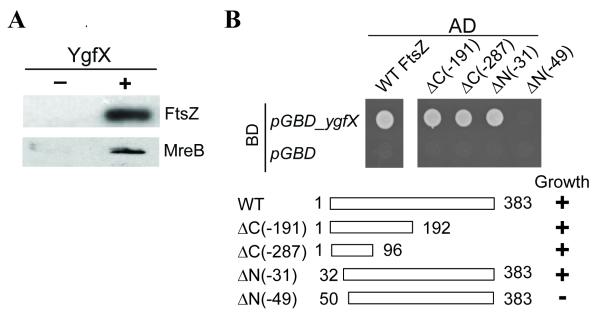
The morphological changes, observed after YgfX induction, were similar to what was observed in cells overexpressing YeeV (Tan et al., 2011). YeeV inhibits the cell division by blocking the polymerization of FtsZ and MreB. We thus examined the whether YgfX also interferes with FtsZ and MreB functions. In order to assess the physical interaction between the YgfX and FtsZ or MreB, pull down experiments were performed using the full length YgfX, which was fused to a His6-tag (YgfX-HIS). The cell lysate of E. coli BL21 cells expressing YgfX-HIS was mixed with the cell lysate containing FtsZ-FLAG or MreB-FLAG. Protein complexes were purified with affinity chromatography, using Ni-NTA beads. Eluted proteins were analyzed by SDS-PAGE, and FLAG-tagged proteins were detected by western blotting, with use of the anti-FLAG antibody (Sigma-Aldrich, MO). As a control, a lysate containing FtsZ-FLAG or MreB-FLAG was incubated with Ni-NTA beads without YgfX-HIS. As shown in Figure 4A, FtsZ-FLAG or MreB-FLAG was detected in the elution fractions only when it was mixed with YgfX-HIS, indicating that YgfX interacts with FtsZ and MreB.
Figure 4. Interactions between YgfX and cytoskeleton proteins: FtsZ and MreB.
(A) The cell lysate containing FtsZ-FLAG or MreB-FLAG was incubated with the lysate containing YgfX-HIS (+) or with that of the wild-type cells (−) before mixed with Ni-NTA column. Eluted fractions were separated by SDS-PAGE and FLAG-tagged proteins were detected using western blot with anti-FLAG antibody conjugated with horseradish peroxidase (Invitrogen, CA).
(B) Yeast two hybrid assay analyzing interaction between YgfX and FtsZ. The full length and various truncated mutants of FtsZ was fused to the activation domain (AD) of pGAD-C1, while YgfX was fused to the binding domain (BD) of pGBD-C1. The interaction was assessed by monitoring the growth on selective media (SD -trp, -leu,-his supplemented with 25 mM 3-aminotriazole).
The interaction between FtsZ and YgfX was confirmed by yeast two hybrid (Y2H) assay (James et al., 1996). The full length and various truncated mutants of FtsZ was fused to the activation domain (AD) of pGAD-C1, while YgfX was fused to the binding domain (BD) of pGBD-C1. The interaction was assessed by monitoring the growth on selective media (SD -trp, -leu,-his supplemented with 25 mM 3-aminotriazole). The growth was observed when pGBD-ygfX was co-transformed with pGAD plasmid containing the full length FtsZ as well as truncated variants of FtsZ; ΔC(−191), ΔC(−287), ΔN(−32), each lacking C-terminal 191, C192 terminal 287 and N-terminal 31 residues, respectively (Figure 4B). The interaction was lost when N-terminal 49 residues of FtsZ was deleted (ΔN(−49)). These results suggest that residues 33-96 of FtsZ are essential for the interaction with YgfX, and that the majority of C-terminal residues and the first 31 N-terminal residues are dispensable for the interaction with YgfX.
YgfX(C) inhibits FtsZ and MreB polymerization
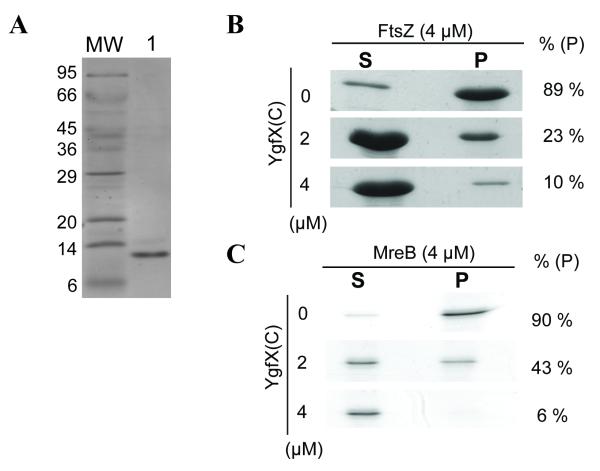
To directly assess the biological role of the interactions between YgfX and the cytoskeletal proteins, the effect of YgfX on in vitro polymerization of FtsZ and MreB was analyzed. To avoid the use of detergent to solubilize TM-containing full length YgfX for polymerization assay, the soluble C-terminal 87-residue fragment (from V49 to R135), was cloned into pCold-Km. The clone was designed to express the truncated YgfX (YgfX(C)) in the fusion with His6-tag at its N-terminal (YgfX(C)-HIS). YgfX(C)-HIS was produced at very high level in the cell, however it was entirely localized in the inclusion bodies. In order to purify YgfX(C)-HIS, the insoluble fraction was collected by centrifugation, and solubilized by 8M urea. Solubilized YgfX(C)-HIS was then purified using Ni-NTA (Qiagen, CA). YgfX(C)-HIS was highly purified by this method (Figure 5A).
Figure 5. Inhibition of in vitro polymerization of FtsZ and MreB by YgfX(C).
(A) Purified YgfX(C)-HIS from the insoluble fraction, following denaturation by 8M urea (lane 1).
(B) GTP-dependent FtsZ polymerization. FtsZ (4 μM) was polymerized with 2 mM GTP and polymers were collected by ultracentrifugation. YgfX(C)-HIS (2 or 4 μM) 471 was added to the reaction mixture prior to the addition of GTP. The monomeric FtsZ in the supernatant fraction (S) and the pellet fraction containing the polymers (P) were separated by SDS-PAGE and visualized by Commassie blue staining.
(C) ATP-dependent MreB polymerization. MreB (4 μM) was polymerized with 2 mM ATP and polymerized MreB was then collected by ultracentrifugation. YgfX(C)-HIS (2 or 4 μM) was added to the reaction mixture prior to the addition of ATP.
The GTP-dependent polymerization of FtsZ was assessed by the sedimentation assay as described previously (Anand et al., 2004). FtsZ polymer was collected in the pellet fraction by ultracentrifugation (Figure 5B). In the absence of YgfX, almost all FtsZ was polymerized and collected in the pellet fraction. However, when YgfX(C)-HIS was added to the reaction mixture, FtsZ polymer formation was decreased reciprocally to the amounts of YgfX(C)-HIS added. The polymerization of FtsZ was almost completely inhibited when YgfX(C)-HIS was added to FtsZ in the 1:1 molar ratio.
In a similar manner, the effects of YgfX on the ATP dependent polymerization of MreB was analyzed. Addition of equimolar YgfX(C)-HIS almost completely inhibited MreB polymerization (Figure 5C). These results clearly demonstrated that YgfX inhibits GTP-dependent FtsZ polymerization as well as ATP-dependent MreB polymerization, 218 and that the C-terminal 87-residue cytoplasmic domain of YgfX is responsible for the inhibition of cytoskeletal polymerization.
Discussion
YgfX is an inhibitor of FtsZ and MreB
Here, we identified a novel TA system, YgfY-YgfX, on the E. coli chromosome. The toxin, YgfX, was shown to inhibit cell division by interfering with the polymerization of essential bacterial cytoskeletal proteins, FtsZ and MreB. Unlike another recently identified soluble E. coli toxin, YeeV, which also interacts with FtsZ and MreB, YgfX is an inner membrane protein having two TM domains. This is consistent with the previous microscopic observation of GFP-YgfX, showing that YgfX is associated with the membrane (Kitagawa et al., 2005). In this study, we also demonstrated that YgfX inhibited FtsZ and MreB polymerization through its soluble C-terminal domain. The role of the TM domains of YgfX still has to be elucidated. The localization in the inner membrane may spatially limit the YgfX activity only near the membrane. For instance, Z-ring is known to be anchored to the inner membrane by ZipA (RayChaudhuri, 1999). A number of cell division proteins such as FtsW, FtsQ, FtsN, FtsL, FtsK, and FtsB, also contain a TM domain(s) (RayChaudhuri, 1999; Barondess et al., 1991; Dai et al., 2004; Buddelmeijer & Beckwith, 2002). Interestingly, spatially regulated inhibition of FtsZ polymerization by inner membrane associated MinC is responsible for the localization of Z-ring at mid-cell (Bi & Lutkenhaus, 1993). YgfX may play a similar role in temporal and spatial control of FtsZ and MreB polymerization, thus regulating cell division events in vivo.
The interaction between FtsZ and YgfX was confirmed by Y2H assay. Furthermore, using Y2H assay, region of FtsZ that are essential for the interaction with YgfX was analyzed. N-terminal 31 residues of FtsZ were not required for the interaction with YgfX. In contrast, N-terminal 31 residues are essential for the interaction with YeeV (Tan et al., 2011). This suggests that although both YeeV and YgfX target same proteins (FtsZ and MreB), and cause equivalent morphological change, they bind distinct sites of FtsZ. The predicted secondary structures differ significantly between YeeV and YgfX. This raise the possibility that a number of different protein families can bind and modulate the activity of FtsZ and/or MreB.
The interaction between YgfX and MreB, however, could not be detected by Y2H in this study. It is likely because of the presence of large activating or binding domain, fused to N-terminal of YgfX and MreB, respectively. It is equally possible that the lack of the interaction is due to low expression of YgfX in yeast. It was previously shown that the apparent interaction between YeeV and MreB was 10 fold less than the interaction between YeeV and FtsZ (Tan et al., 2011). In the case of YgfX, even the interaction with FtsZ, measured by β-galactosidase assay, was not as strong as the interaction between YeeV and FtsZ (data not shown). This apparent weaker interaction is unlikely due to a weak physical binding of YgfX with target proteins in E. coli, since the rate at which YgfX and YeeV causes morphological defects in E. coli was approximately the same.
YgfY neutralizes YgfX toxicity
Commonly, the regulation of the toxin activity occur in two different ways; one through physical sequestration of toxin by antitoxin, and the other by the autoregulatory mechanism of the toxin gene by the TA complex (Zhang et al., 2003; Motiejūnaite et al., 2007; Makarova et al., 2006). Although the toxicity of YgfX was neutralized by the co-expression of YgfY, the mechanism of how YgfY neutralizes the YgfX toxicity remains unknown. Interestingly, we could not detect the physical interaction between YgfX and YgfY, suggesting that YgfY may exert its antitoxin function at the level of transcription or by an unknown mechanism. Notably, the X-ray structure of YgfY has been determined (Lim et al., 2005), predicting 266 that YgfY is a DNA binding protein. These observations are also similar to what was observed for yeeUV; YeeU and YeeV does not physically interact. The mode of neutralization of YeeV toxicity by YeeU is also predicted to involve regulation at the level of transcription (Brown & Shaw, 2003).
Intriguingly, despite the lack of sequence similarity, YgfX and YeeV show the same mode of toxicity, and YgfY and YeeU share a similar mode of antitoxin mechanism. Interestingly, however, YeeV is a soluble protein, while YgfX is an inner membrane protein. Based on this different localization pattern, it is possible that YgfX may be able to exert its toxic function in a more specified manner than YeeV, as discussed above. More study is necessary to characterize the physiological role of ygfYX. So far, no phenotype has been shown to be associated with the deletion of ygfYX. We speculate that this TA system may be involved in cell growth regulation under stress conditions, as in other TA systems. For instance, the expression of YgfYX is affected by norfloxacin, an inhibitor of DNA gyrase (Jeong et al., 2006). It is interesting to further investigate its importance of YgfYX under such conditions.
Acknowledgements
The authors thank Dr. Peter Tupa for critical reading of the manuscript. This work was supported by grants from the National Institute of Health (RO1GM081567).
References
- Anand SP, Rajeswari H, Gupta P, Srinivasan R, Indi S, Ajitkumar P. A C-terminaldeletion mutant of Mycobacterium tuberculosis FtsZ shows fast polymerization in vitro. Microbiology. 2004;150:1119–1121. doi: 10.1099/mic.0.27038-0. [DOI] [PubMed] [Google Scholar]
- Barondess JJ, Carson M, Verduzco LM Guzman, Beckwith J. Alkaline 291 phosphatase fusions in the study of cell division genes. Res Microbiol. 1991;142:295–299. doi: 10.1016/0923-2508(91)90044-b. [DOI] [PubMed] [Google Scholar]
- Bean GJ, Flickinger ST, Westler WM, McCully ME, Sept D, Weibel DB, Amann KJ. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry. 2009;48:4852–4857. doi: 10.1021/bi900014d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol. 1993;175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot S, Corre J, Louarn JM, Cornet F, Barre FX. FtsK activities in Xer recombination, DNA mobilization and cell division involve overlapping and separate domains of the protein. Mol Microbiol. 2004;54:876–886. doi: 10.1111/j.1365-2958.2004.04335.x. [DOI] [PubMed] [Google Scholar]
- Brown JM, Shaw KJ. A novel family of Escherichia coli toxin-antitoxin gene pairs. J Bacteriol. 2003;185:6600–6608. doi: 10.1128/JB.185.22.6600-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddelmeijer N, Beckwith J. Assembly of cell division proteins at the E. coli cell center. Curr Opin Microbiol. 2002;5:553–557. doi: 10.1016/s1369-5274(02)00374-0. [DOI] [PubMed] [Google Scholar]
- Dai K, Xu Y, Lutkenhaus J. Topological characterization of the essential Escherichia coli cell division protein FtsN. J Bacteriol. 1996;178:1328–1334. doi: 10.1128/jb.178.5.1328-1334.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal 314 genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Söldner R, Carballido-López R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobb RI, Fields JA, Burns CM, Thompson SA. Evaluation of procedures for outer membrane isolation from Campylobacter jejuni. Microbiology. 2009;155:979–988. doi: 10.1099/mic.0.024539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong KS, Xie Y, Hiasa H, Khodursky AB. Analysis of pleiotropic transcriptional profiles: a case study of DNA gyrase inhibition. PLoS Genet. 2006;2:e152. doi: 10.1371/journal.pgen.0020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Pogliano J, Helinski DR, Konieczny I. ParE toxin encoded by the broad-host range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol Microbiol. 2002;44:971–979. doi: 10.1046/j.1365-2958.2002.02921.x. [DOI] [PubMed] [Google Scholar]
- Karczmarek A, Martínez-Arteaga R, Baselga RM-A, Alexeeva S, Hansen FG, Vicente M, Nanninga N, den Blaauwen T. DNA and origin region segregation are not affected by the transition from rod to sphere after inhibition of Escherichia coli MreB by A22. Mol Microbiol. 2007;65:51–63. doi: 10.1111/j.1365-2958.2007.05777.x. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- Kruse T, Bork-Jensen J, Gerdes K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol. 2005;55:78–89. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- Lim K, Doseeva V, Demirkan ES, Pullalarevu S, Krajewski W, Galkin A, Howard 359 A, Herzberg O. Crystal structure of the YgfY from Escherichia coli, a protein that may be involved in transcriptional regulation. Proteins. 2005;58:759–763. doi: 10.1002/prot.20337. [DOI] [PubMed] [Google Scholar]
- Madabhushi R, Marians KJ. Actin homolog MreB affects chromosome segregation by regulating topoisomerase IV in Escherichia coli. Mol Cell. 2009;33:171–180. doi: 10.1016/j.molcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Koonin EV. The HicAB cassette, a putative novel, RNA-targeting toxin-antitoxin system in archaea and bacteria. Bioinformatics. 2006;22:2581–2584. doi: 10.1093/bioinformatics/btl418. [DOI] [PubMed] [Google Scholar]
- Motiejūnaite R, Armalyte J, Markuckas A, Suziedeliene E. Escherichia coli dinJ-yafQ genes act as a toxin-antitoxin module. FEMS Microbiol Lett. 2007;268:112–119. doi: 10.1111/j.1574-6968.2006.00563.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Cao C, Lutkenhaus J. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2885–2890. doi: 10.1073/pnas.95.6.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nariya H, Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- Osborn MJ, Rothfield L. Cell shape determination in Escherichia coli. Curr Opin Microbiol. 2007;10:606–610. doi: 10.1016/j.mib.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J Bacteriol. 2001;183:6630–6635. doi: 10.1128/JB.183.22.6630-6635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RayChaudhuri D. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 1999;18:2372–2383. doi: 10.1093/emboj/18.9.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevin EW, Barloy-Hubler F. RASTA-Bacteria: a web-based tool for identifying toxin--antitoxin loci in prokaryotes. Genome Biol. 2007;8:R155. doi: 10.1186/gb-2007-8-8-r155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Awano N, Inouye M. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol Microbiol. 2011;79:109–118. doi: 10.1111/j.1365-2958.2010.07433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wood TK. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol. 2011;77:5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Inouye M. RatA (YfjG), an Escherichia coli toxin, inhibits 70S ribosome association to block translation initiation. Mol Microbiol. 2011;79:1418–1429. doi: 10.1111/j.1365-2958.2010.07506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell. 2003;12:913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]