INTRODUCTION
A large multinational clinical trial compared the safety and efficacy of intranasal trivalent live attenuated influenza vaccine (LAIV) with intramuscular trivalent inactivated vaccine (TIV) in children 6 to 59 months of age prior to the 2004–05 influenza season1. In the Nashville and Boston cohorts, 99 children completed the trial, six (1 with TIV, 5 with LAIV) developed medically attended wheezing within 42 days following vaccination, and eight (5 with TIV, 3 with LAIV) developed laboratory-confirmed influenza. The epidemiologic and genetic factors involved with adverse events (AEs) after vaccination and lack of vaccine efficacy are poorly understood.
Earlier studies have demonstrated that epidemiologic factors influenced the occurrence of AEs after vaccination. In one large survey evaluating injection site reactions after multiple different vaccines, significantly higher rates of pain and local reactions were seen in females when compared to males2. The pathophysiology of these differential responses was hypothesized to be multifactorial with hypersensitivity reactions, route of administration, and hormonal factors being postulated to be involved2. Another study compared the size of Bacillus Calmette-Guérin (BCG) vaccine scar between two groups of young children, one with atopy and one without, and found that children in the atopic group had significantly smaller scars than the control group3.
Recent publications have evaluated the role of genetic factors in adverse events after vaccination. We and others have shown that the systemic and local reactions after vaccinia are linked to specific genetic polymorphisms4, 5. Genetic factors are also associated with variable responses to vaccines. Twin and family studies have shown that responses to Haemophilus influenza type b (Hib) conjugate vaccine6 as well as live attenuated measles, mumps, and rubella (MMR)7 and varicella8 vaccinations have a genetic component. Furthermore, genetic studies of the HLA region suggest associations with variable response to the measles9 and rubella vaccination9–13, and candidate gene studies for the cytokines, toll-like receptors, and innate immunity response genes suggest associations with variable response to rubella vaccination14–16.
Specifically for influenza, to our knowledge, no studies have been published on the genetics or genomics of adverse reactions following the seasonal influenza vaccination. But, there is evidence that the variability in acute phase response to influenza vaccination may be in part mediated by genetic variants in HLA class II, which appear to modulate antibody responses to influenza vaccination17,18. Moreover, influenza vaccination results in a mild acute phase response in men with or without severe carotid artery disease, supporting the proposed role of genetic variants in the candidate gene NFKBIA in acute phase response to influenza vaccination19,20. Finally, at least one study suggests that altered responses to inactivated influenza vaccine may be associated with host variants in MBL and IL10 21.
Given the plausibility that both epidemiologic and genetic factors influence vaccine AEs and immunogenicity, we sought to identify the factors associated with wheezing and the occurrence of natural influenza among children who received intranasal trivalent live attenuated influenza vaccine (LAIV) or intramuscular trivalent inactivated vaccine (TIV) in the large multinational influenza trial from the Nashville and Boston cohorts.
MATERIALS AND METHODS
Study Design
Parents of children who participated in a multinational influenza vaccine efficacy trial from October 20, 2004 to August 31, 2005 at the Nashville, TN and Boston, MA sites were contacted to inquire about participation in this study. The study was approved by the Institutional Review Boards of the Centers for Disease Control and Prevention, Vanderbilt University Medical Center, and Boston University Medical Center. After consent, parents participated in a telephone interview that included demographic information and an atopy survey. The children also either came to the clinic to provide blood samples for DNA analysis or provided saliva samples that were mailed back to the study sites.
Clinical Assessments
Enrolled children were assessed for two different clinical outcomes: medically attended wheezing ≤ 42 days after vaccination and acquisition of laboratory confirmed influenza after vaccination. A medical chart review was performed to abstract demographic and exposure data such as age, sex, self-reported race, and season born.
Atopy Survey
A modified International Study of Asthma and Allergies in Childhood (ISAAC)22 was administered to parents by telephone to gather information on other exposures including premature birth (<36 weeks); ever breastfed; lived in home with pets; lived in home with smokers; patient or sibling attending daycare; and personal or family history of asthma, wheezing, or atopy, including allergies, asthma, or eczema.
DNA Collection, Extraction, and Genotyping
Either whole blood or Oragene saliva samples were collected and DNA was extracted by the Vanderbilt University DNA Resources Core. Forty-seven samples were initially genotyped on the Illumina Infinium 610-Quad, followed by an additional 40 samples (including 8 duplicates) on the 660-Quad. All genotyping was performed by the Broad Institute Center for Genotyping and Analysis. Nine samples were omitted from further analysis due to low DNA concentrations or low genotyping call rates (<95%). The majority of failed samples (7/9) came from Boston and most likely reflect the fact that all Boston DNA samples were extracted from saliva whereas Vanderbilt DNA samples were extracted from blood or saliva. In total, 70 non-duplicate samples were successfully genotyped (43 on the 610-Quad, 27 on the 660-Quad).
Statistical Analysis
Tests of association between case status and survey data were performed in STATA 10.1 using Fisher’s exact. For tests of associations with family history data, two adopted participants were excluded from the analysis.
Prior to analysis of the GWAS data, SNPs were filtered for minor allele frequency (MAF<1%) and genotyping efficiency (<99%). Tests of Hardy Weinberg Equilibrium were performed, and SNPs that deviated from expectations were flagged. Of the 598,704 SNPs genotyped across both the 610- and 660-Quad, a total 468,458 markers were analyzed.
Quality control measures and pair-wise linkage disequilibrium calculations were implemented in PLINK23. Standard allelic tests of association were performed using Fisher’s exact in PLINK. Five samples were excluded due to relatedness, and four non-European American samples (based on parent’s self-described race/ethnicity) were excluded to reduce the potential effects of population stratification. In sum, 61 samples were analyzed for genetic association. Assuming an additive genetic model in Quanto24, we had >80% power to detect large effect sizes (odds ratios >6.0) with common minor allele frequencies (>5%) at a significance level of 0.05. At more stringent significance levels (p<5.0×10−6), we were only powered to detect extremely large odds ratios (>100).
RESULTS
Study Population
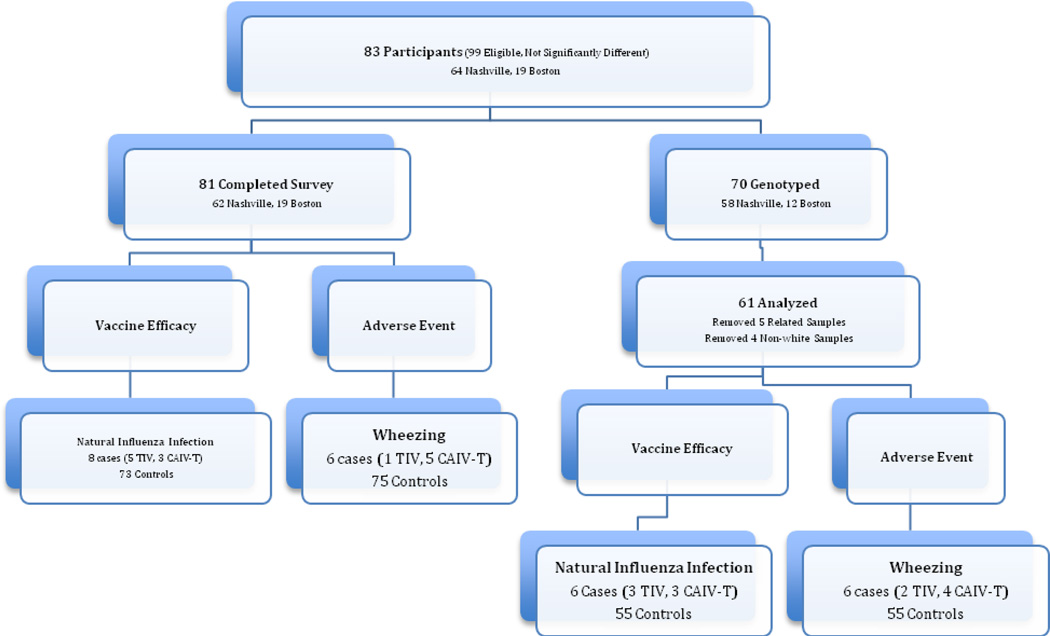
In the original influenza trial, 80 children in Nashville, and 19 children in Boston completed the study. Of the original 80 Nashville participants, 64 (80%) agreed to participate in the current study, 62 completed the atopy survey and 58 were successfully genotyped. Of the original 19 Boston participants, all 19 participants completed the survey and 12 were successfully genotyped. As mentioned above in Materials and Methods, the greater proportion of successfully genotyped samples among the Vanderbilt cohort compared with the Boston cohort (91% versus 63%) may be due to the fact that DNA was extracted from either blood or saliva whereas all the Boston DNAs were extracted from saliva only.
Of the 81 total participants who completed the survey, there were six cases with medically attended wheezing (1 with TIV, 5 with LAIV) and 75 controls without wheezing; and eight cases who developed laboratory-confirmed influenza (5 TIV, 3 LAIV) and 73 controls who did not (Figure 1). Four non-white subjects were removed from analyses because their inclusion could introduce population stratification (confounding by genetic ancestry). That is, having cases and controls of different genetic ancestry can lead to false associations. Of the samples successfully genotyped, after further quality control (see Methods), there were six cases (2 TIV, 4 LAIV) and 55 controls for medically diagnosed wheezing, and six cases (3 TIV, 3 LAIV) and 55 controls for vaccine efficacy (Figure 1).
Figure 1. Description of atopy survey and genome-wide association study sample sizes by case definition.
(Wheezing = Medically attended wheezing ≤ 42 days after vaccine)
Epidemiologic associations with medically diagnosed wheezing ≤ 42 days after vaccination
To identify epidemiologic factors associated with medically diagnosed wheezing, variables collected as part of the administered atopy questionnaire were tested for differences between the six cases with wheezing and the 75 controls without wheezing. Cases with medically diagnosed wheezing were significantly more likely to have a 1st degree relative with a history of asthma or wheezing when compared with controls (100% vs 27%, p=0.001). Both season born and daycare attendance trended toward significance (p=0.058 and p=0.11, respectively; Table 1).
Table 1. Demographic, atopy, and exposures study characteristics for cases of wheezing and controls.
Cases were defined as medically attended wheezing ≤ 42 days after vaccination.
| Cases who wheezed (n=6) | Controls who did not wheeze (n=75) | p-valuea | |
|---|---|---|---|
| Mean age – mo. | 19.0 | 23.8 | 0.26 |
| Sex – no. (%) Male Female |
4 (66.7%) 2 (33.3%) |
39 (52.0%) 36 (48.0%) |
0.68 |
| Race – no. (%) European-American African-American Mixed |
6 (100%) 0 0 |
71 (94.7%) 2 (2.7%) 2 (2.7%) |
1.00 |
| Season bornb - no. (%) Spring Summer Fall Winter |
0 1 (16.7%) 3 (50.0%) 2 (33.3%) |
27 (36.0%) 20 (26.7%) 13 (17.3%) 15 (20.0%) |
0.058 |
| No. of children in home – no. (%) One Two ≥Three |
0 3 (50%) 3 (50%) |
5 (6.9%) 31 (42.5%) 37 (50.7%) |
1.00 |
| Born premature (<36 wks) – no. (%) | 0 | 9 (12.2%) | 1.00 |
| Breastfeed – no. (%) | 5 (83.3%) | 56 (74.7%) | 1.00 |
| Live in home with pet(s)a – no. (%) | 3 (75.0%) | 33 (58.9%) | 0.64 |
| Live in home with smoker(s) – no. (%) | 0 | 6 (8.1%) | 1.00 |
| Patient or sibling attends daycare – no. (%) | 4 (57.1%) | 64 (84.2%) | 0.11 |
| Personal history of asthma/wheezing – no. (%) | 5 (71.4%) | 28 (37.3%) | 0.11 |
| Personal history of atopy – no. (%) | 2 (33.3%) | 22 (29.3%) | 1.00 |
| Maternal history of asthma/wheezing – no. (%)c | 0 | 16 (21.9%) | 0.34 |
| Maternal history of atopy – no. (%)c | 3 (50%) | 46 (63.0%) | 0.67 |
| 1st degree relative history of asthma/wheezing – no. (%)c | 6 (100%) | 20 (27.4%) | 0.001 |
| 1st degree relative history atopy – no. (%)c | 6 (100%) | 48 (65.8%) | 0.17 |
| 2nd degree relative history of asthma/wheezing – no. (%)c | 3 (50%) | 20 (27.4%) | 0.35 |
| 2nd degree relative history atopy – no. (%)c | 3 (50%) | 36 (49.3%) | 1.00 |
| Received CAIV-T vaccine | 5 (71.4%) | 31 (44.3%) | 0.24 |
Fisher’s exact test or t-test
Spring = March, April, May; Summer = June, July, August; Fall = September, October, November; Winter = December, January, February
2 adopted children were removed before analysis.
Genetic associations with medically diagnosed wheezing ≤ 42 days after vaccination
None of the common genetic variants tested for an association with medically diagnosed wheezing ≤42 days post-vaccination achieved genome-wide significance (p<5.0×10−8; Table 2). Only one association was identified at p<4.0×10−6, and ten associations were identified at p<9.0×10−5. Seven of the associations at p<9.0×10−5 were within or near genes, including myosin, heavy chain 14 (MYH14), chromosome 8 open reading frame 34 (C8orf 34), fibroblast growth factor 1 (FGF1), transcription factor 7-like 2 (T-cell specific, HMG-box) (TCF7L2), glutamate receptor interacting protein 2 (GRIP2), and xylosyltransferase I (XYLT1). Of the associations within or near genes, two involved synonymous SNPs (rs3745504 in MYH14 and rs4613440 in GRIP2) while the remaining involved intronic SNPs.
Table 2. SNPs associated with medically attended wheezing after influenza vaccination.
Cases were defined as medically diagnosed wheezing ≤ 42 days after vaccine. SNPs associated at p<9.5×10−5 for six cases and 55 controls are shown below.
| Chromoso me |
rs number |
Base position |
Allel es |
Frequency among cases |
Frequency among controls |
p- value |
Candidate gene (SNP type) |
|---|---|---|---|---|---|---|---|
| 1 | rs11265263 | 1.58E+08 | A/C | 0.58 | 0.04 | 3.34E-06 | |
| 19 | rs3745504 | 55463421 | G/A | 1.0 | 0.36 | 1.59E-05 |
MYH14 (synonymous) |
| 20 | rs4815682 | 4207113 | A/G | 1.0 | 0.36 | 1.59E-05 | |
| 4 | rs7677253 | 1.9E+08 | G/A | 0.50 | 0.036 | 4.00E-05 | |
| 19 | rs4801824 | 55448942 | G/A | 0.92 | 0.30 | 4.77E-05 |
MYH14 (intron) |
| 8 | rs11786594 | 69522698 | C/A | 0.75 | 0.16 | 5.30E-05 |
C8orf34 (intron) |
| 5 | rs249923 | 1.42E+08 | A/G | 1.0 | 0.42 | 6.87E-05 |
FGF1 (intron) |
| 10 | rs11196181 | 1.15E+08 | A/G | 0.42 | 0.02 | 7.24E-05 |
TCF7L2 (intron) |
| 6 | rs17571811 | 10443471 | G/A | 0.75 | 0.17 | 7.59E-05 | |
| 3 | rs4613440 | 14530815 | A/G | 0.92 | 0.32 | 8.11E-05 |
GRIP2 (synonymous) |
| 16 | rs12918720 | 17247437 | A/G | 0.92 | 0.32 | 8.11E-05 |
XYLT1 (intron) |
Epidemiologic associations with naturally acquired influenza infection
To identify epidemiologic factors associated with naturally acquired influenza among vaccinated children, we tested for differences between cases and controls for 18 variables including demographic factors, family history, and environmental exposures collected in the atopy survey. Mean age, sex, and race were not significantly different between cases of naturally acquired influenza and uninfected controls. Although cases were more likely to have a personal history of atopy (50%) and maternal history of atopy (75%) compared with controls (27.4% and 60.6%, respectively), none of the epidemiologic exposures tested were statistically significant at p<0.05 (Table 3).
Table 3. Demographic, atopy, and exposures study characteristics for cases of laboratory confirmed influenza and controls.
| Cases with influenza infection (n=8) | Controls with no influenza infection (n=73) | p-valuea | |
|---|---|---|---|
| Mean age – mo. | 25.3 | 23.2 | 0.43 |
| Sex – no. (%) Male Female |
4 (50%) 4 (50%) |
41 (54.7%) 34 (45.3%) |
1.00 |
| Race – no. (%) European-American African-American Mixed |
8 (100%) 0 0 |
69 (94.5%) 2 (2.7%) 2 (2.7%) |
1.00 |
| Season bornb - no. (%) Spring Summer Fall Winter |
4 (50.0%) 2 (25.0%) 1 (12.5%) 1 (12.5%) |
23 (31.5%) 19 (26.0%) 15 (20.6%) 16 (21.9%) |
0.82 |
| No. of children in home – no. (%) One Two ≥Three |
1 (12.5%) 3 (37.5%) 4 (50.0%) |
4 (5.6%) 31 (43.7%) 36 (50.7%) |
0.58 |
| Born premature (<36 wks) – no. (%) | 0 | 9 (12.5%) | 0.59 |
| Breastfeed – no. (%) | 8 (100%) | 53 (72.6%) | 0.19 |
| Live in home with pet(s)a – no. (%) | 4 (57.1%) | 32 (60.4%) | 1.00 |
| Live in home with smoker(s) – no. (%) | 2 (25.0%) | 4 (5.6%) | 0.11 |
| Patient or sibling attends daycare – no. (%) | 6 (75.0%) | 62 (82.7%) | 0.63 |
| Personal history of asthma/wheezing – no. (%) | 3 (37.5%) | 30 (40.5%) | 1.00 |
| Personal history of atopy – no. (%) | 4 (50.0%) | 20 (27.4%) | 0.29 |
| Maternal history of asthma/wheezing – no. (%)c | 1 (12.5%) | 15 (21.1%) | 1.00 |
| Maternal history of atopy – no. (%)c | 6 (75.0%) | 43 (60.6%) | 0.70 |
| 1st degree relative history of asthma/wheezing – no. (%)c | 1 (12.5%) | 25 (35.2%) | 0.26 |
| 1st degree relative history atopy – no. (%)c | 5 (62.5%) | 49 (69.0%) | 0.70 |
| 2nd degree relative history of asthma/wheezing – no. (%)c | 3 (37.5%) | 20 (28.2%) | 0.69 |
| 2nd degree relative history atopy – no. (%)c | 4 (50.0%) | 35 (49.3%) | 1.00 |
Fisher’s exact test or t-test
Spring = March, April, May; Summer = June, July, August; Fall = September, October, November; Winter = December, January, February
2 adopted children were removed before analysis.
Genetic associations with naturally acquired influenza infection
Among the 468,458 common SNPs tested for an association with the six cases that acquired natural influenza subsequent to vaccination and 55 controls who were not infected, none were associated at genome-wide significance (p<5.0×10−8). Two SNPs on chromosome 7 were associated at p<4×10−6 and a third in linkage disequilibrium (r2=0.81) was associated at p<3×10−5 (Table 4). A total of 16 associations were identified at p<9.5×10−5, and several genetic variants were located within or near genes. These candidate genes include amiloride-sensitive cation channel 1, neuronal (ACCN1), N-acetyltransferase 5 (GCN5-related, putative) (NAT5), chromosome 20 open reading frame 26 (C20orf26), PR domain containing 2, with ZNF domain (PRDM2), KIAA0232, and myosin light chain kinase (MYLK). Only one associated SNP was located in the coding region of a candidate gene (synonymous rs7263 in NAT5) while the remaining SNPs near or within genes were 5′ flanking (rs16981483) or intronic.
Table 4. SNPs associated with acquisition of natural influenza post-vaccination.
SNPs associated at p<9.5×10−5 for six cases and 55 controls are shown below.
| Chromosome | rs number | Base position | Alleles | Frequency among cases |
Frequency among controls |
p-value | Candidate gene (SNP type) |
|---|---|---|---|---|---|---|---|
| 7 | rs6593122 | 54149426 | A/G | 0.75 | 0.11 | 3.91E-06 | |
| 7 | rs2177549 | 54151555 | A/C | 0.75 | 0.11 | 3.91E-06 | |
| 7 | rs10247013 | 54156299 | G/A | 0.75 | 0.15 | 2.44E-05 | |
| 17 | rs317397 | 29200759 | G/A | 0.83 | 0.21 | 2.93E-05 |
ACCN1 (intron) |
| 17 | rs317400 | 29201307 | C/A | 0.83 | 0.21 | 2.93E-05 |
ACCN1 (intron) |
| 9 | rs4615664 | 22865745 | G/A | 0.67 | 0.11 | 4.35E-05 | |
| 8 | rs16917360 | 96196308 | A/G | 0.33 | 0 | 5.64E-05 | |
| 20 | rs7263 | 19954368 | G/A | 0.33 | 0 | 5.64E-05 |
NAT5 (synonymous) |
| 20 | rs16981483 | 19979667 | A/G | 0.33 | 0 | 5.64E-05 |
C20orf26 (5′ flanking) |
| 1 | rs4573512 | 14616559 | G/A | 0.92 | 0.31 | 6.24E-05 | |
| 1 | rs2744690 | 14010388 | A/C | 0.67 | 0.12 | 6.79E-05 |
PRDM2 (intron) |
| 8 | rs2935762 | 1.1E+08 | G/A | 0.83 | 0.24 | 7.57E-05 | |
| 8 | rs1391200 | 1.1E+08 | G/A | 0.83 | 0.24 | 7.57E-05 | |
| 7 | rs2330668 | 54156583 | A/G | 0.75 | 0.17 | 7.59E-05 | |
| 1 | rs4128690 | 14617468 | A/G | 0.92 | 0.32 | 8.11E-05 | |
| 4 | rs6848312 | 6887563 | C/A | 0.50 | 0.05 | 8.39E-05 |
KIAA0232 (intron) |
| 3 | rs11709947 | 1.25E+08 | G/A | 1.0 | 0.43 | 8.63E-05 |
MYLK (intron) |
| 9 | rs4465047 | 22862158 | C/A | 0.58 | 0.08 | 9.44E-05 | |
| 9 | rs1463014 | 22934927 | G/A | 0.58 | 0.08 | 9.44E-05 |
DISCUSSION
The epidemiologic and genetic factors involved in adverse events (AEs) after vaccination, such as wheezing, are poorly understood. Similarly, epidemiologic and genetic factors involved with vaccine effectiveness are a growing area of interest. In our pilot study of 99 children from Nashville and Boston combined who had received one of two influenza vaccines, we sought to determine any specific genetic or epidemiologic risk factors that might have predisposed a child to develop wheezing after vaccination or acquired culture-confirmed natural influenza infection after vaccination. When investigating risk factors for medically attended wheezing ≤ 42 days after vaccination, epidemiologic factors of significance included family history of asthma/wheezing. This finding is biologically plausible, since it is known that parental asthma is a risk factor for persistent wheezing in young children25–27. This finding is also potentially clinically important, since questioning parents about their asthma or asthma in the child’s siblings would be easy to do and could predict adverse events after vaccination. A larger study is needed to verify these results. In regards to the acquisition of natural influenza infection, we did not identify any findings in the atopy survey that were significantly associated with laboratory-confirmed influenza.
In our pilot study, there were several common genetic variants associated with the development of wheezing after vaccination and with acquisition of natural influenza at significance thresholds of p<10−5 and 10−6. The “top hits” for wheezing post-vaccination included several genes and genomic regions, none of which have been implicated in earlier GWAS for asthma28–34, asthma-related traits35,36, or atopy37. Likewise, the genes implicated in the cases who developed influenza after vaccination did not overlap with human leukocyte antigen genes17,18 previously reported in candidate gene studies.
Of the genes implicated here in wheezing and influenza acquisition, it is interesting to note that two are myosin-related genes: MYLK (associated here with acquisition of natural influenza) and MYH14 (associated here with wheezing post-vaccination). MYLK is a nonmuscle myosin light chain kinase isoform involved in inflammatory response and MYLK genetic variants have been implicated in susceptibility to sepsis-induced acute lung injury and asthma38–40. Also, FGF1 (associated here with wheezing post-vaccination) is a fibroblast growth factor implicated in the development of the lung41. FGF1 genetic variants have previously been associated with cord blood IgE levels42 and responsiveness to therapy for chronic hepatitis43.
It is possible that our GWAS findings represent novel associations as no GWAS has been performed for either wheezing post-vaccination or for acquisition of natural influenza post-vaccination. It is also possible that our findings represent false-positives. Indeed, none of the identified associations reached the widely accepted genome-wide significance threshold of p<5.0×10−8 44. A major limitation of our pilot GWAS was sample size. Larger studies are required to identify genetic variants associated at genome-wide significance with small to moderate effect sizes. Larger studies are also needed to determine the effects of vaccine type, natural influenza type, and other variables not examined here because of limited sample size. These limitations will continue to persist for genome-wide studies of vaccine AEs and efficacy because these events are, by design, rare. Thus, properly powered studies will most likely require large, collaborative studies such as the initial clinical trial involving a total of 249 sites1. To our knowledge, most vaccine trials do not prospectively collect DNA samples, a protocol that would greatly accelerate the pace of research in vaccine genomics.
While the novelty of our study, the use of epidemiologic and genetic data, and the application to patient populations in two geographic regions are strengths, there are also several limitations to address. First, as previously mentioned, the sample size is small, and we are inadequately powered for the GWAS unless there is a large, single genetic effect. The existence of a large, single genetic effect, however, is somewhat unlikely given that atopy is multifactorial. Second, the potential effects of case definition of wheezing should be considered. Our case definition of wheezing was limited to ≤ 42 days after vaccination, and a cutoff of 42 days is not biologically relevant if vaccine effects occur at 43 days or later. We did evaluate medically versus non-medically diagnosed wheezing within this time frame, and this expanded case definition did not alter our observations or conclusions appreciably (data not shown).
In the original parent study, a post hoc analysis for the study period through 180 days after the last dose of vaccine found that children 6–11 months of age were hospitalized for any cause at a higher rate in the LAIV group than in the TIV group (6.1% vs 2.6%, difference in rate 3.5% (95% CI 1.4, 5.8)). Rate of hospitalization for respiratory diagnosis in the LAIV group was also higher (3.2% vs 1.2%, 2.0% difference (95% CI 0.5, 3.8)). Although not statistically significant, there was a trend toward higher rate of hospitalization for any cause among children receiving LAIV who were 6–47 months of age and had a history of wheezing than among those receiving TIV also in the same age group with a history of wheezing1. These findings of possible adverse events as far as 6 months out from vaccination suggest that, should GWAS and an atopy survey be administered to a larger population, an extended duration of follow-up for adverse events might be warranted.
A third limitation is that the retrospective nature of the survey and DNA collection introduces some potential for biases such as recall bias and the bias of patients with a more positive or negative memory of their vaccine experience being more or less likely to agree to provide DNA. Despite these potential biases, we note that this study had a high participation rate for the GWAS (95.1%), and the occurrence of the AEs and acquisition of natural influenza were recorded prospectively in the original clinical trial1. Another possible bias to consider is detection bias. Families who are familiar with wheezing may be more likely to recognize it and bring it to medical attention. Alternatively, our results may have a biologic/genetic basis as discussed. One of the implications of genetic studies of wheezing and asthma is that they have the potential for sorting out whether there is, in fact, a biologic basis. If so, then the hypothesized detection bias in an epidemiologic risk factor study becomes irrelevant.
CONCLUSIONS
In summary, this pilot study demonstrates the feasibility of retrospectively obtaining atopy surveys and DNA samples from participants to identify factors associated with AE and acquisition of natural influenza infection after vaccination. Family asthma history was a risk factor for wheezing after influenza vaccination. No specific genetic polymorphisms were associated with either wheezing or laboratory-confirmed influenza infection after vaccination. This study is a paradigm for combined epidemiologic and genetic studies, and similar studies applied to larger populations should be conducted in the future.
Acknowledgements
We would like to thank all of the parents and children who participated in this study, Franklin Pediatrics in Nashville, TN, Pediatrics Associates of Fall River, MA, and all the members of the Boston University Medical Center Research team; the Vanderbilt Research Team, including Mr. Dapo Akingbade (Center for Human Genetics Research) and Cara Sutcliffe, PhD. (General Clinical Research Center and DNA Resources Core); and The Vanderbilt University Center for Human Genetics Research and Computational Genomics Core who provided computational and/or analytical support for this work. We would also like to acknowledge the ISAAC Steering Committee (isaac.auckland.ac.nz) for use of the modified atopy questionnaire. This work was supported, in part, by the Clinical Immunization and Safety Assessment (CISA) network through a subcontract with America's Health Insurance Plans (AHIP) under contract 200-2002-00732 from the Centers for Disease Control and Prevention (CDC) and the Vanderbilt Institute for Clinical and Translational Research (VICTR) CTSA grant (1UL1 RR024975-01 from NCRR/NIH), as well as the Vanderbilt Clinical and Translational Research Scholars Award (K12 RR24977-03).
Abbreviations
- LAIV
intranasal trivalent live attenuated influenza vaccine
- TIV
trivalent inactivated vaccine
- GWAS
genome-wide association study
- AEs
adverse events
- ISAAC
International Study of Asthma and Allergies in Childhood
Footnotes
Disclaimer: The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position or views of the Centers for Disease Control and Prevention.
References
- 1.Belshe RB, Edwards KM, Vesikari T, et al. Live Attenuated versus Inactivated Influenza Vaccine in Infants and Young Children. N Engl J Med. 2007 Feb 15;356(7):685–696. doi: 10.1056/NEJMoa065368. [DOI] [PubMed] [Google Scholar]
- 2.Cook IF. Sex differences in injection site reactions with human vaccines. Hum Vaccin. 2009;5(7):441–449. doi: 10.4161/hv.8476. [DOI] [PubMed] [Google Scholar]
- 3.Rehman A, Ullah I. The BCG scar size in asthmatic and non-asthmatic children. J Pak Med Assoc. 2009;59(9):625–628. [PubMed] [Google Scholar]
- 4.Stanley J, Frey SE, Taillon-Miller P, et al. The Immunogenetics of Smallpox Vaccination. The Journal of Infectious Diseases. 2007 Jul 15;196(2):212–219. doi: 10.1086/518794. [DOI] [PubMed] [Google Scholar]
- 5.Reif DM, McKinney BA, Motsinger AA, et al. Genetic Basis for Adverse Events after Smallpox Vaccination. The Journal of Infectious Diseases. 2008 Jul 1;198(1):16–22. doi: 10.1086/588670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee YC, Newport MJ, Goetghebuer T, et al. Influence of genetic and environmental factors on the immunogenicity of Hib vaccine in Gambian twins. Vaccine. 2006 Jun 19;24(25):5335–5340. doi: 10.1016/j.vaccine.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 7.Tan PL, Jacobson RM, Poland GA, Jacobsen SJ, Pankratz VS. Twin studies of immunogenicity -- determining the genetic contribution to vaccine failure. Vaccine. 2001 Mar 21;19(17–19):2434–2439. doi: 10.1016/s0264-410x(00)00468-0. [DOI] [PubMed] [Google Scholar]
- 8.Klein NP, Fireman B, Enright A, et al. A role for genetics in the immune response to the varicella vaccine. Pediatr Infect Dis J. 2007;26(4):300–305. doi: 10.1097/01.inf.0000257454.74513.07. [DOI] [PubMed] [Google Scholar]
- 9.Ovsyannikova IG, Jacobson RM, Poland GA. Variation in vaccine response in normal populations. Pharmacogenomics. 2004 Jun 1;5(4):417–427. doi: 10.1517/14622416.5.4.417. [DOI] [PubMed] [Google Scholar]
- 10.Ovsyannikova IG, Jacobson RM, Vierkant RA, Jacobsen SJ, Pankratz VS, Poland GA. Human Leukocyte Antigen Class II Alleles and Rubella-Specific Humoral and Cell-Mediated Immunity Following Measles-Mumps-Rubella II Vaccination. The Journal of Infectious Diseases. 2005 Feb 15;191(4):515–519. doi: 10.1086/427558. [DOI] [PubMed] [Google Scholar]
- 11.Ovsyannikova IG, Jacobson RM, Vierkant RA, O'Byrne MM, Poland GA. Replication of rubella vaccine population genetic studies: Validation of HLA genotype and humoral response associations. Vaccine. 2009 Nov 16;27(49):6926–6931. doi: 10.1016/j.vaccine.2009.08.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ovsyannikova IG, Jacobson RM, Vierkant RA, Jacobsen SJ, Pankratz VS, Poland GA. The contribution of HLA class I antigens in immune status following two doses of rubella vaccination. Human Immunology. 2004 Dec;65(12):1506–1515. doi: 10.1016/j.humimm.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Poland GA. Human Leukocyte Antigen Haplotypes in the Genetic Control of Immune Response to Measles-Mumps-Rubella Vaccine. The Journal of Infectious Diseases. 2006 Mar 1;193(5):655–663. doi: 10.1086/500144. [DOI] [PubMed] [Google Scholar]
- 14.Ovsyannikova I, Haralambieva I, Dhiman N, et al. Polymorphisms in the Vitamin A Receptor and Innate Immunity Genes Influence the Antibody Response to Rubella Vaccination. The Journal of Infectious Diseases. 2010 Jan 15;201(2):207–213. doi: 10.1086/649588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhiman N, Haralambieva IH, Kennedy RB, et al. SNP/haplotype associations in cytokine and cytokine receptor genes and immunity to rubella vaccine. Immunogenetics. 2010 doi: 10.1007/s00251-010-0423-6. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ovsyannikova IG, Dhiman N, Haralambieva IH, et al. Rubella vaccine-induced cellular immunity: evidence of associations with polymorphisms in the Toll-like, vitamin A and D receptors, and innate immune response genes. Hum Genet. 2010;127(2):207–221. doi: 10.1007/s00439-009-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gelder CM, Lambkin R, Hart KW, et al. Associations between human leukocyte antigens and nonresponsiveness to influenza vaccine. J Infect Dis. 2002;185(1):114–117. doi: 10.1086/338014. [DOI] [PubMed] [Google Scholar]
- 18.Poland GA, Ovsyannikova IG, Jacobson RM. Immunogenetics of seasonal influenza vaccine response. Vaccine. 2008;26(Suppl4):D35–D40. doi: 10.1016/j.vaccine.2008.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlson CS, Heagerty PJ, Nord AS, et al. TagSNP evaluation for the association of 42 inflammation loci and vascular disease: evidence of IL6, FGB, ALOX5, NFKBIA, and IL4R loci effects. Hum Genet. 2007;121(1):65–75. doi: 10.1007/s00439-006-0289-8. [DOI] [PubMed] [Google Scholar]
- 20.Carty CL, Heagerty P, Nakayama K, et al. Inflammatory Response After Influenza Vaccination in Men With and Without Carotid Artery Disease. Arterioscler Thromb Vasc Biol. 2006 Dec 1;26(12):2738–2744. doi: 10.1161/01.ATV.0000248534.30057.b5. [DOI] [PubMed] [Google Scholar]
- 21.Tang Y-W, Li H, Wu H, Shyr Y, Edwards KM. Host Single-Nucleotide Polymorphisms and Altered Responses to Inactivated Influenza Vaccine. The Journal of Infectious Diseases. 2007 Oct 1;196(7):1021–1025. doi: 10.1086/521370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellwood P, Asher MI, Beasley R, Clayton TO, Stewart AW the ISAAC Steering Committee. The International Study of Asthma and Allergies in Childhood(ISAAC): Phase Three rationale and methods. Int J Tuberc Lung Dis. 2005;9(1):10–16. [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007 Sep;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002;155:478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- 25.Crestani E, Guerra S, Wright AL, Halonen M, Martinez FD. Parental asthma as a risk factor for the development of early skin test sensitization in children. Journal of Allergy and Clinical Immunology. 2004 Feb;113(2):284–290. doi: 10.1016/j.jaci.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 26.Guilbert TW, Morgan WJ, Zeiger RS, et al. Atopic characteristics of children with recurrent wheezing at high risk for the development of childhood asthma. Journal of Allergy and Clinical Immunology. 2004 Dec;114(6):1282–1287. doi: 10.1016/j.jaci.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 27.Guerra S, Wright AL, Morgan WJ, Sherrill DL, Holberg CJ, Martinez FD. Persistence of Asthma Symptoms during Adolescence: Role of Obesity and Age at the Onset of Puberty. Am J Respir Crit Care Med. 2004 Jul 1;170(1):78–85. doi: 10.1164/rccm.200309-1224OC. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Howard TD, Zheng SL, et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. Journal of Allergy and Clinical Immunology. 2010 Feb;125(2):328–335. doi: 10.1016/j.jaci.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SH, Cho BY, Park CS, et al. Alpha-T-catenin(CTNNA3) gene was identified as a risk variant for toluene diisocyanate-induced asthma by genome-wide association analysis. Clin Exp Allergy. 2009;39(2):203–212. doi: 10.1111/j.1365-2222.2008.03117.x. [DOI] [PubMed] [Google Scholar]
- 30.Moffatt MF, Kabesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007 Jul 26;448(7152):470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 31.Himes BE, Hunninghake GM, Baurley JW, et al. Genome-wide Association Analysis Identifies PDE4D as an Asthma-Susceptibility Gene. Am J Hum Genet. 2009 May 15;84(5):581–593. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hancock DB, Romieu I, Shi M, et al. Genome-Wide Association Study Implicates Chromosome 9q21.31 as a Susceptibility Locus for Asthma in Mexican Children. PLoS Genet. 2009 Aug 28;5(8):e1000623. doi: 10.1371/journal.pgen.1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathias RA, Grant AV, Rafaels N, et al. A genome-wide association study on African-ancestry populations for asthma. Journal of Allergy and Clinical Immunology. 2010 Feb;125(2):336–346. doi: 10.1016/j.jaci.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sleiman PMA, Flory J, Imielinski M, et al. Variants of DENND1B Associated with Asthma in Children. N Engl J Med. 2010 Jan 7;362(1):36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 35.Ober C, Tan Z, Sun Y, et al. Effect of Variation in CHI3L1 on Serum YKL-40 Level, Risk of Asthma, and Lung Function. N Engl J Med. 2008 Apr 17;358(16):1682–1691. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weidinger S, Gieger C, Rodriguez E, et al. Genome-Wide Scan on Total Serum IgE Levels Identifies <italic>FCER1A</italic> as Novel Susceptibility Locus. PLoS Genet. 2008 Aug 22;4(8):e1000166. doi: 10.1371/journal.pgen.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castro-Giner F, Bustamante M, Ramon Gonzalez J, et al. A pooling-based genome-wide analysis identifies new potential candidate genes for atopy in the European Community Respiratory Health Survey(ECRHS) BMC Medical Genetics. 2009;10(1):128. doi: 10.1186/1471-2350-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao L, Grant AV, Rafaels N, et al. Polymorphisms in the myosin light chain kinase gene that confer risk of severe sepsis are associated with a lower risk of asthma. Journal of Allergy and Clinical Immunology. 2007 May;119(5):1111–1118. doi: 10.1016/j.jaci.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 39.Flores C, Ma SF, Maresso K, Ober C, Garcia JG. A variant of the myosin light chain kinase gene is associated with severe asthma in African Americans. Genet Epidemiol. 2007;31(4):296–305. doi: 10.1002/gepi.20210. [DOI] [PubMed] [Google Scholar]
- 40.Christie JD, Ma SF, Aplenc R, et al. Variation in the MYLK gene is associated with development of acute lung injury after major trauma. Crit Care Med. 2008;36(10):2794–2800. doi: 10.1097/ccm.0b013e318186b843. [DOI] [PubMed] [Google Scholar]
- 41.Kimura J, Deutsch GH. Key mechanisms of early lung development. Pediatr Dev Pathol. 2007;10(5):335–347. doi: 10.2350/07-06-0290.1. [DOI] [PubMed] [Google Scholar]
- 42.Yang KD, Chang JC, Chuang H, et al. Gene-gene and gene-environment interactions on IgE production in prenatal stage. Allergy. 2009 doi: 10.1111/j.1398-9995.2009.02260.x. In press. [DOI] [PubMed] [Google Scholar]
- 43.Hwang Y, Chen EY, Gu ZJ, et al. Genetic predisposition of responsiveness to therapy for chronic hepatitis C. Pharmacogenomics. 2006 Jul 1;7(5):697–709. doi: 10.2217/14622416.7.5.697. [DOI] [PubMed] [Google Scholar]
- 44.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008 May;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]