Abstract
We report the use of silicon chips with 16 DNA-modified electrodes (DME chips) utilizing DNA-mediated charge transport for multiplexed detection of DNA and DNA-binding protein targets. Four DNA sequences were simultaneously distinguished on a single DME chip with fourfold redundancy, including one incorporating a single base mismatch. These chips also enabled investigation of the sequence-specific activity of the restriction enzyme Alu1. DME chips supported dense DNA monolayer formation with high reproducibility, as confirmed by statistical comparison to commercially available rod electrodes. The working electrode areas on the chips were reduced to 10 µm in diameter, revealing microelectrode behavior that is beneficial for high sensitivity and rapid kinetic analysis. These results illustrate how DME chips facilitate sensitive and selective detection of DNA and DNA-binding protein targets in a robust and internally standardized multiplexed format.
INTRODUCTION
Multiplexed detection of biomarkers such as DNA, RNA, and proteins is of utility for laboratory assays as well as clinical and point-of-care disease diagnostics.1–4 Toward these ends, electrical and electrochemical devices are under development for biosensing applications, offering low cost, portability, and multiplexed capability.5–7 Carbon nanotubes,8,9 functionalized nanowires10–12 and nanoparticles,13 aptamers,14–15 and redox or impedance schemes involving DNA16–25 or other mediators26 have all served as electrical and electrochemical biosensing platforms. However, despite this proliferation of electrical biosensors, few examples of multiplexing of molecular diagnostics have been evident.12,16,18–21 Furthermore, still fewer electrical sensors have the sensitivity to distinguish single base mismatches within nucleic acid targets, and many are not suitable for sensing DNA-binding proteins. Our format offers robust, label-free, and sensitive detection that is now multiplexed.
Electrochemical detection by DNA-mediated charge transport is an emerging technology for clinical diagnostics and laboratory assays, showing great promise for sensitive and selective recognition of DNA and protein targets.27–41 Numerous studies have established that well-ordered, fully base-paired DNA facilitates electronic charge transport through the DNA π-stack over long distances, but that disruption of the base pair stack, such as by mismatched bases or bending of the duplex by proteins, greatly attenuates charge transport.27–29,41–45 Due to this sensitivity to perturbation, electrochemistry through DNA monolayers and molecular junctions has been utilized for sensitive and selective detection of DNA27–28,34–36,41 and DNA-binding proteins.29–30,32–33,38,40–41 DNA electrochemistry can be used to distinguish between targets with single base mismatches27–28 and subtle base lesions.35 DNA serves as a natural and general recognition element for DNA-binding proteins. Thus, protein sensing with DNA-mediated charge transport is a rational, sensitive, and selective platform capable of detecting unlabeled proteins.
Here we describe the fabrication and application of 16-electrode silicon chips with DNA-modified electrodes (DME chips) employing DNA-mediated electrochemistry for multiplexed detection of DNA and DNA-binding protein targets. Four DNA sequences were interrogated simultaneously on one DME chip with fourfold redundancy, demonstrating sensitivity to single-base mismatches. DME chips were used to electrochemically monitor sequence-specific DNA cleavage by the restriction enzyme Alu1. The quality of monolayer formation was investigated by statistical comparison of DME chips to commercially available rod electrodes, and the sizes of the working electrodes on the DME chips were scaled to investigate microelectrode effects. These experiments show that DME chips facilitate sensitive and selective detection of DNA and DNA-binding protein targets.
EXPERIMENTAL
Oligonucleotide synthesis
Oligonucleotides were synthesized by standard methods on solid supports using an Applied Biosystems 3400 DNA synthesizer. For thiolated strands, the 5′ end was modified with the Thiol Modifier C6 S-S phosphoramidite and standard protocols from Glen Research, Inc. DNA modified with Redmond Red on the 3′ terminus was prepared on Epoch Redmond Red CPG columns from Glen Research with ultramild phosphoramidites and reagents. For Nile Blue modified DNA, a 5-[3-acrylate NHS Ester]-deoxyuridine phosphoramidite (Glen Research) was incorporated at the 5′ terminus also using ultramild conditions. The DNA on solid support was then dried and reacted with a 10 mg/mL solution of Nile Blue perchlorate (Acros Organics) in 9:1 dichloromethane/N,N-diisopropylethylamine solution for approximately 24 hours. Excess reagents were then removed by washing three times each with dichloromethane, methanol, and acetonitrile.
Unmodified and thiolated oligonucleotides were cleaved from the solid support and deprotected by treating with concentrated ammonium hydroxide for 8 hours at 60°C. Redmond Red and Nile Blue modified DNA strands were cleaved from the support and deprotected according to ultramild conditions with 0.05 M potassium carbonate in methanol at ambient temperature for 8 hours.
Oligonucleotide purification
Oligonucleotides were purified with high performance liquid chromatography (HPLC). Following HPLC purification of the products, the oligonucleotides were treated for removal of the dimethoxytrityl (DMT) protecting group. For the thiolated oligonucleotides, the disulfide of the thiolated linker was cleaved with an excess of dithiolthreitol in concentrated ammonium hydroxide for 2 h to yield the free thiol. The DMT was removed from the unmodified, Nile Blue and Redmond Red DNA strands by treating with an 80% solution of glacial acetic acid for 20 minutes, followed by quenching of the reaction with an excess of ethanol. All of the oligonucleotides were dried and purified with a second round of HPLC. The products were characterized by HPLC, matrix assisted laser desorption ionization time-of-flight mass spectrometry, and UV-visible (UV-Vis) spectrophotometry.
The oligonucleotides were subsequently desalted and quantified by UV-Vis spectrophotometry according to their extinction coefficients (IDT Oligo Analyzer). Duplexes were formed by thermally annealing equimolar amounts of oligonucleotides at 90 °C for 5 min in deoxygenated phosphate buffer (5 mM NaPhos, 50 mM NaCl, pH 7.0) followed by slow cooling to ambient temperature.
DME chip preparation
1 mm thick Si wafers with a 10,000 Å thick oxide layer were purchased from Silicon Quest. Chips were patterned in a two-layer process. In the first layer, the gold electrodes were deposited by a lift-off technique. For the second layer, SU-8 photoresist was patterned as an insulator isolating the gold working electrode areas from the contact pads. First, wafers were cleaned thoroughly in 1165 Remover (Microchem) and vapor primed with hexamethyldisilizane (HMDS). SPR 220 3.0 photoresist (Microchem) was spin-cast at 4000 rpm and baked. The photoresist was patterned with a Karl Suss MA6 contact aligner and a chrome photomask. Following post-exposure baking, wafers were developed in AZ 300 MIF developer for one minute and rinsed thoroughly with DI water. A 15 Å Ti adhesion layer and a 1000 Å Au layer were deposited on the chips with a CHA Mark 50 electron beam evaporator. Wafers were then immersed in 1165 Remover (Microchem) overnight and sonicated as needed to complete metal lift-off. Subsequently, the wafers were thoroughly baked and cleaned by UV ozone treatment. SU-8 2002 (Microchem) was spin cast at 3000 rpm, baked and photopatterned as above. Wafers were developed in SU-8 Developer (Microchem) for one minute and baked for a permanent set of the photoresist. The wafers were subsequently diced into 1-inch by 1-inch chips by hand with a diamond scribe and stored under vacuum until use.
Preparation of DNA monolayers
Immediately prior to incubation with DNA, gold surfaces were cleaned by sonication for 15 min in acetone and 5 min in isopropanol, followed by treatment with UV-ozone for 3 min. Multi-level wells were placed over the chip, defined by a custom-made viton rubber gasket and a polypropylene clamp secured by screws to a test mount, providing a compression seal over the chip (Figure 1). This allowed for incubation with up to four distinct sequences of 25 µM duplex DNA solutions in phosphate buffer containing 100 mM MgCl2. Monolayer formation was typically allowed to proceed in a humidified environment for a period of 16–20 hours. Upon completion of film formation, the cell was backfilled with 0.5 mM 1-mercaptohexanol in a 95:5 phosphate buffer/glycerol solution for 60 minutes. The electrodes and cells were rinsed thoroughly prior to electrochemistry experiments to ensure removal of residual alkanethiols.
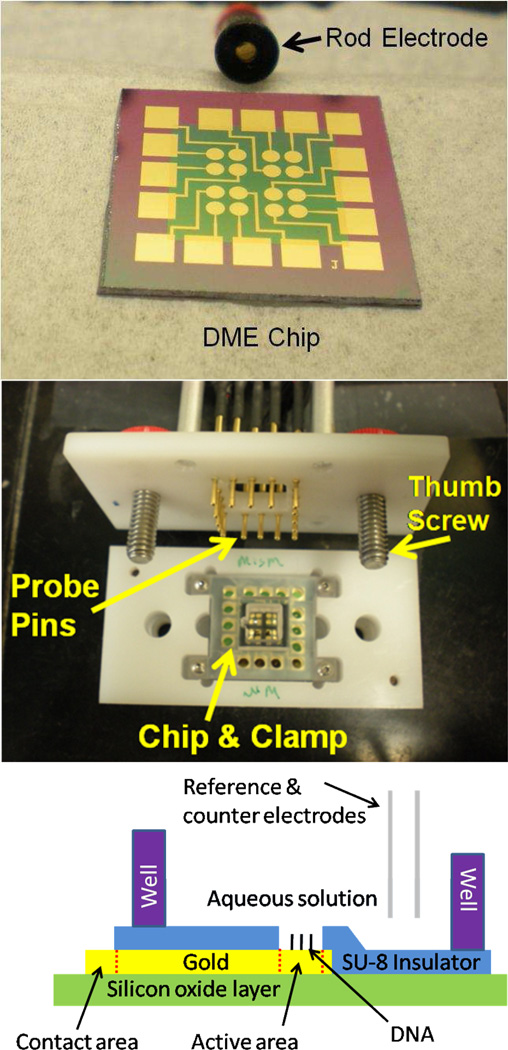
Figure 1.
Fabrication of the DME chip. (Top) A DNA-modified electrode (DME) chip with sixteen 2 mm2 gold working electrodes shown with a conventional 2 mm2 gold rod electrode. (Middle) The DME chip in a testing mount with a clamp well that splits the chip into four quadrants of four electrodes. Electrical contact is made to the chip with spring contact probe pins secured into contact with the chip by the thumb screws. (Bottom) The side-view illustration of one electrode from the fully-assembled DME chip. The working electrode area is defined and separated from the contact area by the SU-8 insulating layer. The solution of interest is confined over the chip by the well, and external reference and counter electrodes complete the circuit.
Electrochemical analysis
Cyclic voltammetry experiments were performed by automated measurement with a CH760B Electrochemical Analyzer and a 16-channel multiplexer module from CH Instruments (Austin, TX). The chips were interfaced with these instruments with a custom-built device mount bearing spring-loaded probe pins. Chips were tested with a common Pt auxiliary electrode and a common silver/silver chloride (Ag/AgCl) reference electrode. Alternatively, reference and counter electrodes can be patterned on the chip surface, though including other metals for a stable reference would increase the complexity of chip fabrication. Electrochemistry was recorded at ambient temperature in either phosphate buffer supplemented with 4 mM MgCl2, 4 mM spermidine, 50 µM ethylenediaminetetracetic acid (EDTA) and10% glycerol at pH 7.0 or tris buffer containing 50 mM Tris-HCl, 10 mM EDTA and 10 mM MgCl2, at pH 7.8. Electron transfer kinetics were obtained by Laviron analysis.46
Restriction assay
The restriction enzyme Alu1 was purchased from New England Biolabs. The shipping buffer was exchanged to tris buffer containing 50 mM Tris-HCl, 10 mM EDTA and 10 mM MgCl2, pH 7.8 using a Pierce Slide-A-Lyzer mini dialysis kit with overnight stirring. MgCl2 was not used in the phosphate buffer for DNA assembly to produce a lower density monolayer granting greater access to the restriction enzyme. The reaction was allowed to equilibrate at each point of the titration for approximately 30 minutes.
RESULTS AND DISCUSSION
Design of DME chip and testing assembly
Figure 1 shows a DME chip with 16 macroscale, 2 mm2 gold working electrodes for multiplexed analysis. The DME chips were designed as a multiplexed extension of a conventional 2 mm2 commercially available rod electrode, which is also shown for comparison in Figure 1. Each chip was patterned with four quadrants of four electrodes each so that four distinct DNA sequences could be simultaneously tested with fourfold redundancy. Isolation of the quadrants was accomplished with a gasket and clamp assembly (Figure 1) having four shallow wells surrounded by a larger well. The shallow wells, each with a maximum volume of approximately 25 µL, were used for deposition of the distinct DNA monolayers, while the larger well, with minimum and maximum working volumes of 150 µL and 600 µL, respectively, enabled all 16 electrodes to share a common analyte solution as well as common reference and counter electrodes. Each gold electrode of the DME chip was patterned with a circular working electrode with the precise active area defined by an insulating SU-8 layer. Each working electrode was connected to a square contact pad on the periphery of the DME chip. These contact pads were connected to a computer-controlled multiplexer module and electrochemical analyzer through spring contact probe pins on a testing mount secured with thumb screws. This allowed rapid electrical connection and interchange of each DME chip. In this configuration, multiplexed electrochemical testing of all 16 electrodes could be performed sequentially with common reference and counter electrodes.
Multiplexed detection of DNA sequences
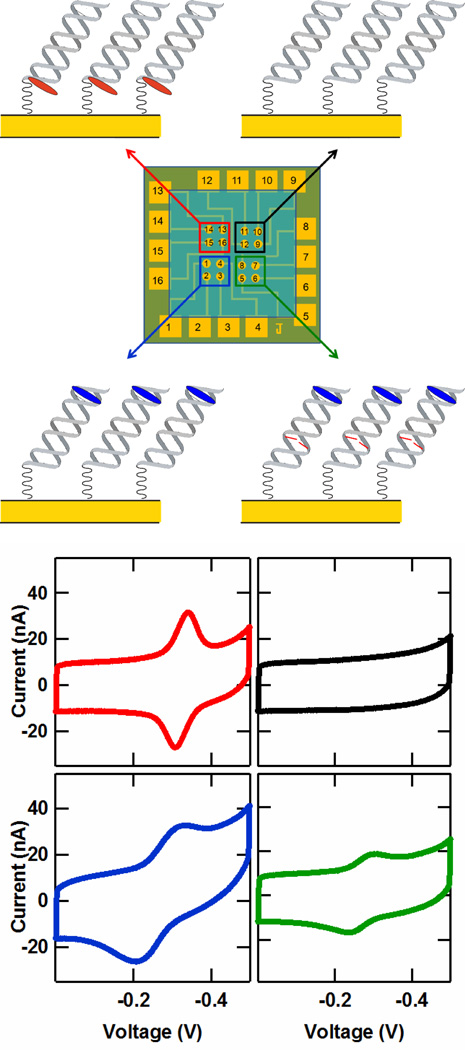
Multiplexed electrochemical analysis of four distinct DNA sequences was accomplished with the DME chip. Hybridized with identical 5′-(SH linker)-AC TTC AGC TGA GAC GCA-3′ sequences, these four 17-mer targets were distinct in either the choice of redox probe or the inclusion of a mismatch. The redox probes Nile Blue and Redmond Red were used as they have been previously demonstrated for sensitive detection of proteins40 and abasic sites47, respectively. As illustrated in Figure 2, these targets were: (i) a well matched strand with a distal 5′ Nile Blue redox probe, (ii) a well matched strand with a proximal 3′ Redmond Red probe48, (iii) a well matched strand with no redox probe, and (iv) a 5′ Nile Blue-labeled strand containing a single base-pair (CA) mismatch. The choice of these four targets illustrates the versatility of the detection technique, the generality of redox probes used, the selectivity to specific DNA sequences, and the ability to isolate the monolayers with fidelity.
Figure 2.
Multiplexed detection on the DME chip. (Upper) Illustration of a DME chip layout with four distinct DNA target complementary strands. (Lower) Cyclic voltammetry data from each of the four DNA targets depicted in the upper figure. The four sequences consisted of (i) a well matched strand with a distal 5′ Nile Blue redox probe (5′-TNBGC GTC TCA GCT GAA GT-3, blue), (ii) a well matched strand with a proximal 3′ Redmond Red probe (5′-TGC GTC TCA GCT GAA GT(RR)-3′, red), (iii) a well matched strand with no redox probe (5′-TGC GTC TCA GCT GAA GT-3′, black), and (iv) a 5′ Nile Blue-labeled strand containing a single base-pair (CA) mismatch (5′-TNBGC GTC TCA GCT AAA GT-3, green). (TNB is a thymine modified with a Nile Blue redox probe, A notes the location of a single CA mismatch, and RR denotes a Redmond Red redox probe.) The potentials are reported versus Ag/AgCl with a CV scan rate of 100 mV/s, and each curve represents an average over the four electrodes in each chip quadrant.
The cyclic voltammetry (CV) resulting from these four DNA monolayers is shown in Figure 2. The monolayer prepared with well matched, Nile Blue-labeled DNA gives a large CV peak area of 5.2 nC at -320 mV versus an Ag/AgCl reference (cathodic wave). Similarly, the well matched monolayer with a Redmond Red redox probe exhibits large CV peak area of 6.2 nC located at the distinct voltage of -340 mV (cathodic wave). These results reveal that high density DNA monolayers can be prepared on DME chips. In contrast, the monolayer prepared with a well matched complementary strand containing no redox probe shows no discernable CV peak, highlighting that no cross-contamination of the monolayers occurs between DME chip quadrants.
It has previously been demonstrated that DNA-modified electrodes can distinguish single base mismatches and other subtle lesions due to distortion imposed on the DNA base pair π-stack.27,28,35 Likewise, the CV signals on DME chips are significantly attenuated for the electrodes prepared with the complementary sequence 5′-TNBGC GTC TCA GCT AAA GT-3′, where TNB is the Nile Blue modified thymine and A indicates the position of a C-A mismatch. The CV peak area from these mismatched duplexes was 1.9 nC (cathodic wave), a factor of 2.7 lower than that found for the well matched complement. Thus, DME chips can be used for precise discernment of specific DNA and RNA targets, even distinguishing single base mismatches. Note that here DME chips provide a direct measurement of mismatch discrimination, as both well matched and mismatched films are formed under identical experimental conditions.
Overall, three DME chips were prepared identically to that of Figure 2. The ratios of integrated charge for well matched to mismatched Nile Blue strands across these 3 chips were 2.8 and 3.1 for the anodic and cathodic sweeps, respectively. The variation in signal size across a given chip can be seen in the CV data of all 16 electrodes from the chip of Figure 2 in the supporting information (Figure S-2). For this chip, the integrated cathodic peaks with standard deviations were 5.1 ± 1.6 nC for the well matched Nile Blue monolayer, 2.1 ± 0.1 nC for the mismatched Nile Blue monolayer, and 6.3 ± 0.4 for the well matched Redmond Red monolayer. Alternatively, the variation in the average integrated cathodic CV peak charge among chips was 1.5, 1.0 and 3.0 nC for well matched Nile Blue, mismatched Nile Blue, and well matched Redmond Red monolayers, respectively. Thus, in general, the variation across a chip was smaller than that between chips. Monolayers on chips were found to be relatively stable under storage at 4 °C. For example, a chip stored for 24 days at 4 °C retained over 80% of the initial integrated charge (Figure S-4), corresponding to an average loss of signal of less than 1% per day.
Electron transfer kinetics from these monolayers were also estimated from the scan rate dependence of the cyclic voltammetry by Laviron analysis.46 The electronic transfer rates were 4.2 s−1, 1.0 s−1 and 2.4 s−1, for well matched Redmond Red, well matched Nile Blue, and mismatched Nile Blue monolayers, respectively. These values are comparable to estimates on similarly prepared monolayers on rod electrodes.49 Notably, there is little difference in the transfer kinetics between matched and mismatched DNA, suggesting the same mechanism of charge transport for both sequences.
It should be noted that our platform can be extended for detection of unlabeled single stranded DNA targets. To accomplish this, single stranded DNA bearing the complementary sequence of the target is modified with a redox probe on one end and assembled on gold electrodes by a thiolated linker on the other end. Hybridization with the unlabeled target will complete the base pair π-stack and increase the DNA-mediated redox signal.
Monitoring sequence-specific enzymatic activity with DME chips
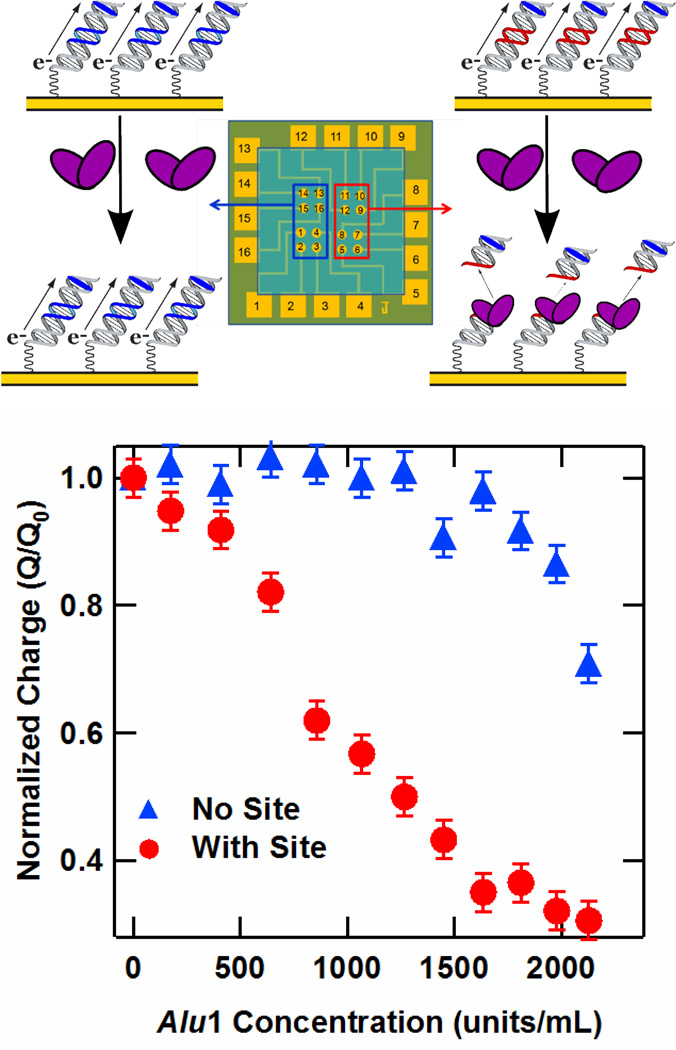
A major advantage of the multiplexed chip format over individual electrodes is the ability to measure DNA-binding protein activity with different DNA sequences on the same chip, thus exploring site-specific activity while preserving identical experimental conditions. We demonstrate this concept by measuring the sequence-specific activity of the Alu1 restriction endonuclease, which cleaves at the restriction site 5′-AGCT-3′, leaving blunt ends between the G and C bases. A DME chip was prepared with 17-mer Nile Blue-modified DNA, where half of the electrodes were assembled with a sequence containing the Alu1 recognition site, and the other half with a sequence in which this site was absent, as illustrated in Figure 3. The Alu1 restriction enzyme was titrated onto the chip, and the integrated CV peak areas were recorded at each concentration. The resulting plot of charge normalized against the initial signal versus Alu1 concentration is given in Figure 3. At low concentrations, there is a definitive drop in the integrated charge at the electrodes bearing the restriction site, while the charge from the electrodes without the site remains stable. In contrast, for the DNA-modified electrode lacking the target site there is virtually no drop in signal over this concentration range. The threshold of Alu1 restriction activity for the sequence containing the restriction site was 400 units/mL, corresponding to a concentration of approximately 10 nM.50 As the total sample volume was 250 µL, this corresponds to 2.5 picomoles of enzyme per chip, or 160 femtomoles of enzyme per electrode. Above 1600 units/mL,51 the charge at the electrodes lacking the restriction site decreases due to non-specific restriction activity, also known as star activity. In this case, the DNA without the consensus restriction site contains a pseudosite differing by only one base (5′-ATCT-3). Thus, as expected at higher enzyme concentrations, restriction cleavage at this pseudosite is apparent.
Figure 3.
Restriction assay on the DME chip. (Upper) Illustration of sequence-specific activity of the Alu1 restriction enzyme electrochemically monitored with a DME chip. (Lower) Charge versus Alu1 concentration for a DME chip for DNA with (Red) and without (Blue) the Alu1 restriction site. In particular, the sequence of the DNA containing the restriction site was the well matched 17-mer 5′-TNBGC GTC TCA GCT GAA GT-3′, where the italicized bases represent the restriction site and TNB is a thymine modified with a Nile Blue redox probe. The sequence absent this site but containing a three-base pseudosite was the well matched 17-mer 5′-TNBGC GTG CTT TAT ATC TC-3′, with the pseudosite given in italics. Charge was obtained by integrating the cathodic Nile Blue CV peaks obtained at a 50 mV/s scan rate after equilibration of the Alu1 activity at each concentration.
Several important implications arise from these observations. Cleavage by the Alu1 endonuclease requires that the DNA on these chips is in its native conformation and accessible to the protein. The observation of sequence-specific cleavage indicates that protein detection with DNA-mediated electrochemistry is highly selective. Also, by extension, incorporation of multiple DNA sequences with different protein binding characteristics on a single chip indicates that DME chips can serve as a robust platform to simultaneously monitor reactions on different oligonucleotides. Finally, this assay requires only microliter volumes of low protein concentrations, making it competitive with alternative detection methods.
Statistical comparison to rod electrodes
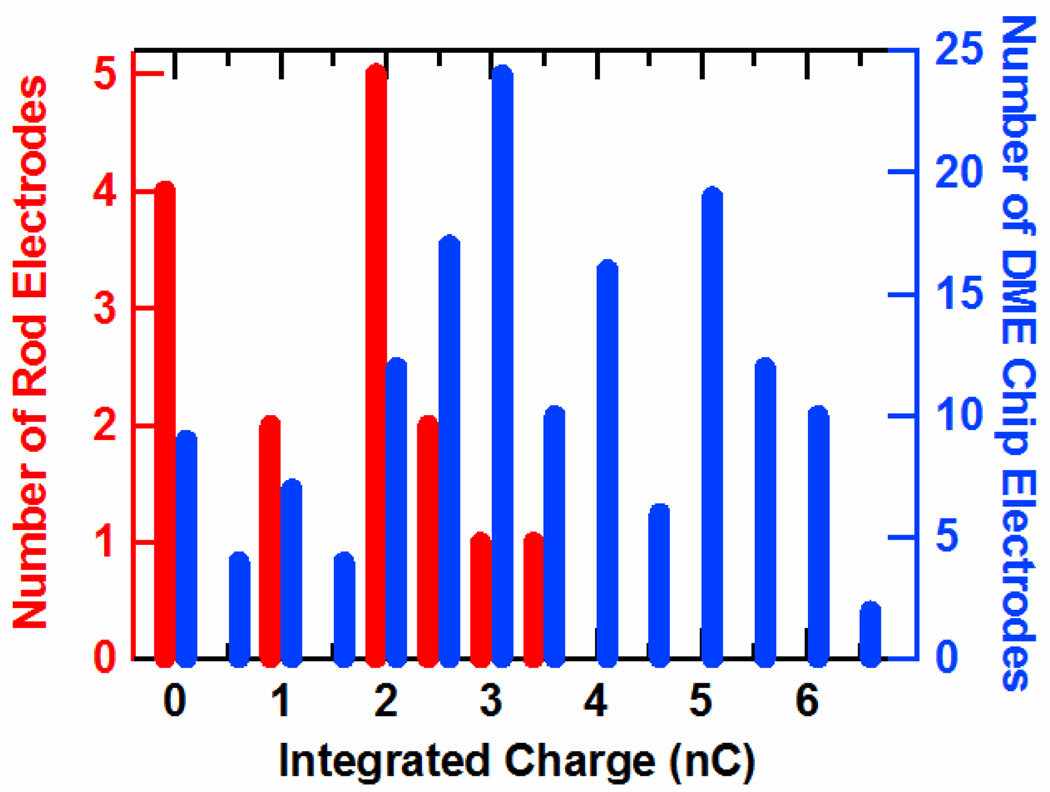
We have found that electrodes from DME chips exhibit performance superior to conventional, commercially available rod electrodes. This is clearly revealed in the histogram of Figure 4, which compares the total charge obtained by integrating the cathodic CV peaks from Nile Blue-modified, 17-mer DNA monolayers prepared on DME chips and rod electrodes. The average integrated charge value of 3.5 nC from the DME chips is nearly twice that of the 1.8 nC average obtained from rod electrodes, and the relative deviation is significantly lower, 0.5 versus 0.7. This higher average integrated charge is indicative of higher surface density of DNA at the DME electrodes. In addition, on average, fewer electrode failures (charge < 0.5 nC) were observed on the DME chips (6%) versus rod electrodes (25%). Background noise is also much smaller for the DME chips, as they display a lower capacitive current (data not shown). The higher overall signals, lower standard deviation, and better signal-to-noise ratio of the DME chips are clearly preferred for sensing and diagnostic applications.
Figure 4.
A histogram of the total charge from DNA monolayers from the electrodes of 9 DME chips (blue) and 15 rod electrodes (red). Both types of electrodes were coated with well matched DNA monolayers with the sequence 5′-TNBGC GTG CTT TAT ATC TC-3′, where TNB is a Nile Blue modified thymine. The integrated charge was obtained by integrating the cathodic peak of the cyclic voltammagram taken at a 50 mV/s scan rate. For ease of comparison, integrated charges have been sorted according to the nearest half coulomb. Note that on average a much higher signal and fewer failures (signals < 0.5 nC) are found for gold electrodes on DME chips.
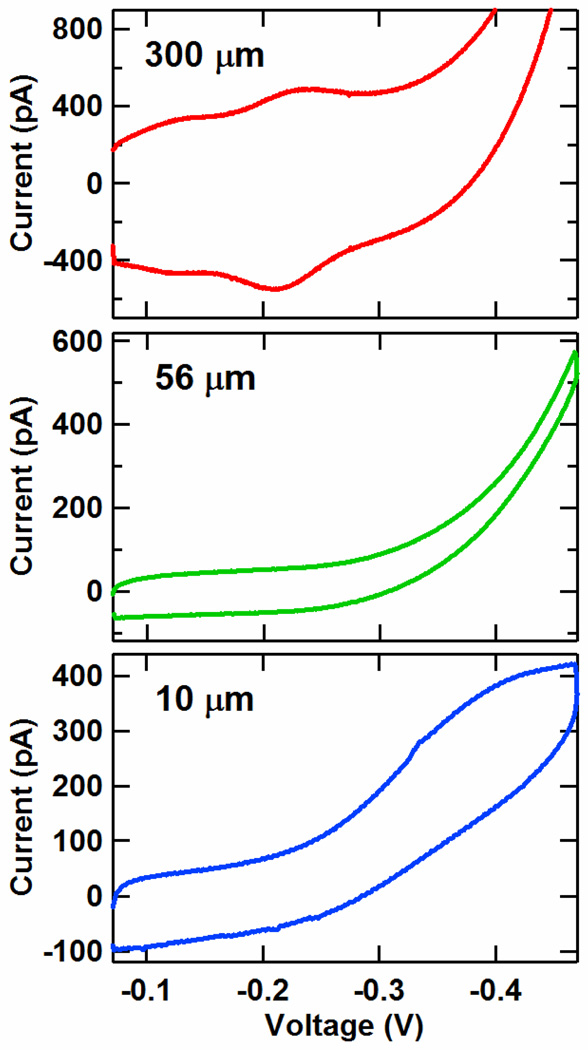
Microelectrodes
In addition to macroelectrodes, we demonstrate that DME chips can be easily prepared with microelectrodes. Microelectrodes exhibit a number of benefits for DNA and protein sensing such as high sensitivity, rapid kinetics, and lower sample volumes.18–120,40,52–53 By reducing the diameter of the opening of the SU-8 layer over each electrode, DME chips with the gold layout of Figure 1 were patterned with circular working electrodes of 300, 56 and 10 µm diameters. These electrodes were coated with double-stranded DNA monolayers of well matched, Nile Blue-modified 17-mer sequences with a distally bound Nile Blue redox probe, and the CV curves for these electrodes at a 50 mV/s scan rate are given in Figure 5. For the 300 µm diameter working electrodes, the conventional macroelectrode surface-bound redox peaks associated with Nile Blue are visible at a midpoint potential of -220 mV vs Ag/AgCl. However, for the 56 and 10 µm diameter electrodes, the voltammograms are significantly altered from this conventional shape. The 10 µm diameter electrodes exhibit the sigmoidal curves characteristic of microelectrode effects.52,53 This result is similar to our previous work with individual microelectrodes, where microelectrode effects were observed for devices of 25 µm diameter or less.40 This shape arises because ionic equilibration is achieved virtually instantaneously with the sweep of the voltage due to the small size of the electrode relative to the abundance of ions in the surrounding solution.52,53 As seen previously, the midpoint potential is shifted negatively by over 100 mV, while the limiting current and capacitive current are each higher by a factor of 5 to 6. These increases may be due to denser DNA monolayers and/or the presence of oxygen. Alternatively, the 56 µm diameter electrodes yield a voltammetry curve that is intermediate to the macroelectrode and microelectrode regimes. Microelectrode effects can thus be observed on DME chips, combining the benefits of high sensitivity and rapid equilibration to this multiplexed platform.
Figure 5.
Average cyclic voltammetry (CV) signals from Nile Blue DNA-modified 300, 56, and 10 µm diameter electrodes prepared on the same chip. The electrodes were assembled with 17-mer DNA duplexes of the sequence 5′-TNBGC GTC TCA GCT GAA GT-3′ and the well matched, thiolated complement. The potentials are reported versus Ag/AgCl with a CV scan rate of 50 mV/s, and each curve represents an average over four electrodes. The 300 µm electrodes show the conventional surface-bound macroelectrode redox peak, while the 10 µm electrodes show a sigmoidal CV curve, reflecting microelectrode effects.
CONCLUSION
We have accomplished multiplexed detection of DNA and DNA-binding protein targets with DME chips employing DNA-mediated charge transport. Four DNA sequences were simultaneously distinguished on a single DME chip with fourfold redundancy, including one incorporating a single-base mismatch, highlighting the selectivity of these detectors. These chips also enabled investigation of protein activity from the restriction enzyme Alu1, revealing sequence-specific recognition. DME chips supported high density DNA monolayer formation, as confirmed by statistical comparison to commercially available rod electrodes. The working electrode areas on the chips were reduced to 10 µm to achieve microelectrode behavior that is useful for high sensitivity and rapid kinetic detection. DME chips thus offer a new and sensitive platform for the multiplexed detection of DNA and DNA-binding protein targets.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the NIH (GM61077). J.D.S. also thanks the National Institute of Biomedical Imaging and Bioengineering for a postdoctoral fellowship (1F32EB007900). The authors thank J.Genereux, M. Buzzeo, and D. Ceres for fruitful discussions, J. DeFranco for assistance with photolithography and S. Olson and M. Roy for construction of the test mount and clamp assembly.
Footnotes
Supporting Information Available. Figure of chip test mount. DNA melting temperature data. Cyclic voltammetry data of all 16 electrodes from the multiplexed DNA detection experiment. Cyclic voltammetry peak areas versus chip position for the multiplexed DNA detection experiment. Cyclic voltammetry data for chip storage at 4°C. Cyclic voltammetry data of all 16 electrodes from the Alu1 detection experiment. Cyclic voltammetry peak areas versus chip position for the Alu1 detection experiment. Illustration of the chip layout for macroelectrode and microelectrode devices. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Darnell JE. Nat. Rev. Cancer. 2002;2:740. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 2.Libermann TA, Zerbini LF. Curr. Gene Ther. 2006;6:17. doi: 10.2174/156652306775515501. [DOI] [PubMed] [Google Scholar]
- 3.Bell J. Nature. 2004;429:453–456. doi: 10.1038/nature02624. [DOI] [PubMed] [Google Scholar]
- 4.Ludwig JA, Weinstein JN. Nat. Rev. Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 5.Drummond TG, Hill MG, Barton JK. Nat. Biotechnol. 2003;21:1192. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Cao Z, Lu Y. Chem. Rev. 2009;109:1948. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sadik OA, Aluoch AO, Zhou A. Biosens. Bioelectron. 2009;24:2749. doi: 10.1016/j.bios.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Wong SS, Joselevich E, Woolley AT, Cheung CL, Lieber CM. Nature. 1998;394:52. doi: 10.1038/27873. [DOI] [PubMed] [Google Scholar]
- 9.Chen RJ, Bangsaruntip S, Drouvalakis KA, Kam NWS, Shim M, Li YM, Kim W, Utz PJ, Dai HJ. Proc. Natl. Acad. Sci. USA. 2003;100:4984. doi: 10.1073/pnas.0837064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui Y, Wei Q, Park H, Lieber CM. Science. 2003;293:1289. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- 11.Gao Z, Agarwal A, Trigg AD, Singh N, Fang C, Tung C-H, Fan Y, Buddharaju KD, Kong J. Anal. Chem. 2007;79:3291. doi: 10.1021/ac061808q. [DOI] [PubMed] [Google Scholar]
- 12.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Nat. Biotechnol. 2005;23:1294. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 13.Park S-J, Taton TA, Mirkin CA. Science. 2002;295:1503. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 14.Zuo X, Xiao Y, Plaxco KW. J. Am. Chem. Soc. 2009;131:6944. doi: 10.1021/ja901315w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu D, Xu D, Yu X, Liu Z, He W, Ma Z. Anal. Chem. 2005;77:5107. doi: 10.1021/ac050192m. [DOI] [PubMed] [Google Scholar]
- 16.Yu CJ, Wan Y, Yowanto H, Li J, Tao C, James MD, Tan CL, Blackburn GF, Meade TJ. J. Am. Chem. Soc. 2001;123:11155. doi: 10.1021/ja010045f. [DOI] [PubMed] [Google Scholar]
- 17.Fan C, Plaxco KW, Heeger AJ. Proc. Natl. Acad. Sci. USA. 2003;100:9134. doi: 10.1073/pnas.1633515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Zhou Y, Sutherland TC, Baker B, Lee JS, Kraatz H-B. Anal Chem. 2005;77:5766. doi: 10.1021/ac050741o. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Lee JS, Kraatz H-B. Anal Chem. 2006;78:6096. doi: 10.1021/ac060533b. [DOI] [PubMed] [Google Scholar]
- 20.Elsholz B, Wörl R, Blohm L, Albers J, Feucht H, Grunwald T, Jürgen B, Schweder T, Hintsche R. Anal. Chem. 2006;78:4794. doi: 10.1021/ac0600914. [DOI] [PubMed] [Google Scholar]
- 21.Fang Z, Soleymani L, Pampalakis G, Yoshimoto M, Squire JA, Sargent EA, Kelley SO. ACS Nano. 2009;3:3207. doi: 10.1021/nn900733d. [DOI] [PubMed] [Google Scholar]
- 22.Cash KJ, Ricci F, Plaxco KW. J. Am. Chem. Soc. 2009;131:6955. doi: 10.1021/ja9011595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghindilis AL, Smith MW, Schwarzkopf KR, Roth KM, Peyvan K, Munro SB, Lodes MJ, Stӧver AG, Bernards K, Dill K, McShea A. Biosens. Bioelectron. 2007;22:1853. doi: 10.1016/j.bios.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 24.Liao JC, Mastali M, Gau V, Suchard MA, Møller AK, Bruckner DA, Babbitt JT, Li Y, Gornbein J, Landaw EM, McCabe ERB, Churchill BM, Haake DA. J. Clin. Microbiol. 2006;44:561. doi: 10.1128/JCM.44.2.561-570.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anne A, Demaille C. J. Am. Chem. Soc. 2008;130:9812. doi: 10.1021/ja801074m. [DOI] [PubMed] [Google Scholar]
- 26.G Karasinski J, Andreescu S, Sadik OA, Lavine B, Vora MN. Anal. Chem. 2005;77:7941. doi: 10.1021/ac0512150. [DOI] [PubMed] [Google Scholar]
- 27.Kelley SO, Boon EM, Barton JK, Jackson NM, Hill MG. Nucleic Acids Res. 1999;27:4830. doi: 10.1093/nar/27.24.4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boon EM, Ceres DM, Drummond TG, Hill MG, Barton JK. Nat. Biotechnol. 2000;18:1096. doi: 10.1038/80301. [DOI] [PubMed] [Google Scholar]
- 29.Boon EM, Salas JE, Barton JK. Nat. Biotechnol. 2002;20:282. doi: 10.1038/nbt0302-282. [DOI] [PubMed] [Google Scholar]
- 30.Boon EM, Livingston AL, Chmiel NH, David SS, Barton JK. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12543. doi: 10.1073/pnas.2035257100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drummond TG, Hill MG, Barton JK. J. Am. Chem. Soc. 2004;126:15010. doi: 10.1021/ja044910i. [DOI] [PubMed] [Google Scholar]
- 32.Boal AK, Yavin E, Lukianova OA, O’Shea VL, David SS, Barton JK. Biochemistry. 2005;44:8397. doi: 10.1021/bi047494n. [DOI] [PubMed] [Google Scholar]
- 33.DeRosa MC, Sancar A, Barton JK. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10788. doi: 10.1073/pnas.0503527102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inouye M, Ikeda R, Takase M, Tsuri T, Chiba J. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11606. doi: 10.1073/pnas.0502078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boal AK, Barton JK. Bioconjugate Chem. 2005;16:312. doi: 10.1021/bc0497362. [DOI] [PubMed] [Google Scholar]
- 36.E Okamoto A, Kamei T, Saito I. J. Am. Chem. Soc. 2006;128:658. doi: 10.1021/ja057040t. [DOI] [PubMed] [Google Scholar]
- 37.Gorodetsky AA, Barton JK. Langmuir. 2006;22:7917. doi: 10.1021/la0611054. [DOI] [PubMed] [Google Scholar]
- 38.Gorodetsky AA, Boal AK, Barton JK. J. Am. Chem. Soc. 2006;128:12082. doi: 10.1021/ja064784d. [DOI] [PubMed] [Google Scholar]
- 39.Wong ELS, Gooding JJ. J. Am. Chem. Soc. 2007;129:8950. doi: 10.1021/ja0723075. [DOI] [PubMed] [Google Scholar]
- 40.Gorodetsky AA, Ebrahim A, Barton JK. J. Am. Chem. Soc. 2008;130:2924. doi: 10.1021/ja7106756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo X, Gorodetsky AA, Hone J, Barton JK, Nuckolls C. Nature Nanotech. 2008;3:163. doi: 10.1038/nnano.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelley SO, Jackson NM, Hill MG, Barton JK. Angew. Chem. Int. Ed. 1998;38:941. doi: 10.1002/(SICI)1521-3773(19990401)38:7<941::AID-ANIE941>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 43.Núñez ME, Hall DB, Barton JK. Chem. Biol. 1999;6:85. doi: 10.1016/S1074-5521(99)80005-2. [DOI] [PubMed] [Google Scholar]
- 44.Kelley SO, Barton JK. Science. 1999;283:375. doi: 10.1126/science.283.5400.375. [DOI] [PubMed] [Google Scholar]
- 45.Schuster GB. Acc. Chem. Res. 2000;33:253. doi: 10.1021/ar980059z. [DOI] [PubMed] [Google Scholar]
- 46.Laviron E. J. Electroanal. Chem. 1979;101:19. [Google Scholar]
- 47.Buzzeo MC, Barton JK. Bioconjugate Chem. 2008;19:2110. doi: 10.1021/bc800339y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Redmond Red in the proximal position has been used to differentiate protein effects on charge transport when compared to a distal probe. See Gorodetsky AA, Dietrich LEP, Lee PE, Demple B, Newman DK, Barton JK. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3684. doi: 10.1073/pnas.0800093105.
- 49.Gorodetsky AA, Green O, Yavin E, Barton JK. Bioconjugate Chem. 2007;18:1434. doi: 10.1021/bc0700483. [DOI] [PubMed] [Google Scholar]
- 50.A conversion factor of 1 million units per milligram Alu1 was used New England Biolabs, personal communication.
- 51.One unit is defined as the amount of enzyme required to digest 1 µg of λ DNA in 1 hour at 37 °C in a total reaction volume of 50 µl.
- 52.Bard AJ, Faulkner LR. Electrochemical Methods. 2nd ed. New York: John Wiley & Sons; 2001. [Google Scholar]
- 53.Heinze J. Angew. Chem. Int. Ed. 1993;32:1268. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.