Abstract
The SR protein splicing factor SRSF1 is a potent proto-oncogene that is frequently upregulated in cancer. Here we show that SRSF1 is a direct target of the transcription-factor oncoprotein MYC. These two oncogenes are significantly co-expressed in lung carcinomas, and MYC knockdown downregulates SRSF1 expression in lung-cancer cell lines. MYC directly activates transcription of SRSF1 through two non-canonical E-boxes in its promoter. The resulting increase in SRSF1 protein is sufficient to modulate alternative splicing of a subset of transcripts. In particular, MYC induction leads to SRSF1-mediated alternative splicing of the signaling kinase MKNK2 and the transcription factor TEAD1. SRSF1 knockdown reduces MYC’s oncogenic activity, decreasing proliferation and anchorage-independent growth. These results suggest a mechanism for SRSF1 upregulation in tumors with elevated MYC, and identify SRSF1 as a critical MYC target that contributes to its oncogenic potential by enabling MYC to regulate the expression of specific protein isoforms through alternative splicing.
SRSF1 (formerly SF2/ASF) is a prototypical member of the SR protein family, a conserved class of splicing regulators. Besides its central roles in constitutive and alternative splicing (Ge et al., 1990; Krainer et al., 1990; Mayeda et al., 1992), SRSF1 regulates other aspects of RNA metabolism, including mRNA stability (Lemaire et al., 2002), nuclear export (Huang et al., 2003), nonsense-mediated mRNA decay (Zhang et al., 2004), translation (Sanford et al., 2004), and miRNA processing (Wu et al., 2010). The SRSF1 gene is essential, and depletion of the protein triggers genomic instability, cell-cycle arrest, and apoptosis (Xu et al., 2005; Li et al., 2005), whereas its overexpression drives transformation of immortal rodent fibroblasts (Karni et al., 2007). SRSF1 negatively autoregulates its expression through various post-transcriptional and translational mechanisms (Sun et al., 2010; Wu et al., 2010), yet despite this stringent homeostatic control, it is frequently upregulated in many different cancers (Ezponda et al, 2010; Karni et al., 2007; Thorsen et al., 2011). SRSF1 resides on Chromosome 17q23, a locus that is amplified in some tumors (Sinclair et al., 2003), accounting for some instances of SRSF1 overexpression (Karni et al., 2007).
Altered transcriptional regulation might also cause SRSF1 overexpression in tumors. MYC (alias c-Myc) is a potent oncogenic transcription factor that is frequently overexpressed or hyperactive in many cancers (Adhikary et al, 2005). Consistent with a possible role of MYC in regulating SRSF1, the SRSF1 promoter region directly or indirectly binds MYC, according to a ChIP-on-Chip analysis of CpG island arrays (Mao et al., 2003); in addition, expression microarray analyses reported SRSF1 among >100 genes upregulated in response to MYC in multiple cell lines (Coller et al., 2000; Schlosser et al., 2005).
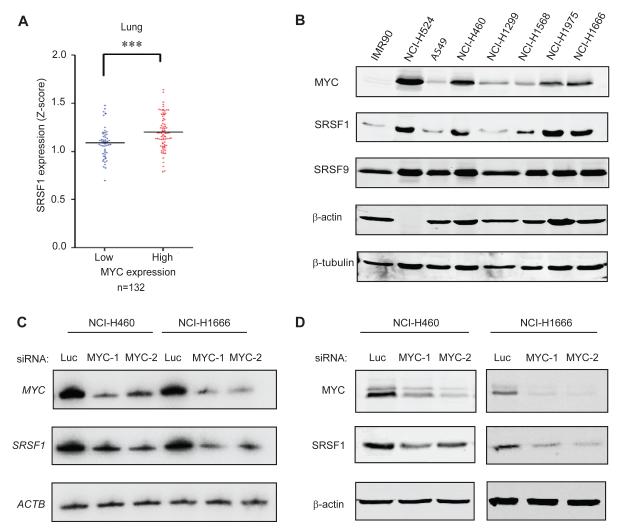
Considering that SRSF1 is markedly overexpressed in lung cancer (Ezponda et al., 2010; Karni et al., 2007), we analyzed public microarray data from a panel of 132 lung tumors, to determine whether MYC overexpression correlates with elevated SRSF1 levels in this context. Indeed, we found a strong positive correlation between MYC and SRSF1 expression at the RNA level (Figure 1A). Among eight other known or putative MYC-regulated splicing factors we analysed (David et al., 2010; Li et al., 2003; Rauch et al., 2011; Zeller et al., 2003) only hnRNPH1 and PTBP1 expression correlated significantly with MYC expression in these lung-tumor samples (Table S1). We extended this analysis to a panel of normal and tumor-derived lung cell lines, and also found a significant correlation at the protein level between MYC and SRSF1 (Figure 1B), with most cancer cell lines overexpressing both proteins, relative to IMR90 primary lung fibroblasts. In contrast, MYC expression did not correlate in these cells with that of other SR proteins, such as SRSF9 (Figure 1B) and SRSF6 (data not shown). siRNA-mediated knockdown of MYC in two of these cell lines, the large cell lung cancer cell line NCI.H460 and the bronchoalveolar adenocarcinoma cell line NCI.H1666, resulted in significant decreases in SRSF1 expression, both at the transcript and protein level, indicating that SRSF1 expression is under MYC control (Figure 1C,D). However, another bronchoalveolar adenocarcinoma cell line, A549, did not show this effect, indicating additional context-dependent levels of control (Figure S1); this may be due to threshold effects, as A549 cells have relatively low levels of both MYC and SRSF1 (Figure 1B). The imperfect correlation between SRSF1 and MYC expression in the lung cancer cell lines (Figure 1B) indicates that though MYC is an important regulator of SRSF1 expression, SRSF1 overexpression in cancer is not solely attributable to MYC expression; additional factors likely affect its expression at the transcriptional, post-transcriptional, translational, or post-translational levels.
Figure 1.
SRSF1 expression correlates with MYC levels in human lung tumors and cell lines. (A) SRSF1 expression profile from microarray analysis of 132 lung tumors (expO). The data were normalized to Z-score and divided into two categories: tumors expressing high or low MYC levels. The dot plot shows the distribution and the median (horizontal line). Mann-Whitney test ***P<0.0001. (B) Immunoblotting of MYC and SRSF1 in lung-cancer cell lines and lung primary fibroblasts, showing significant correlation between the expression of the two oncoproteins (r=0.75, one-tailed t-test *P=0.05). (C) RT-PCR and (D) Immunoblotting of MYC and SRSF1 in NCI.H460 and NCI.H1666 cells transfected with control siRNA (luciferase) or one of two siRNAs against MYC. See also Figure S1.
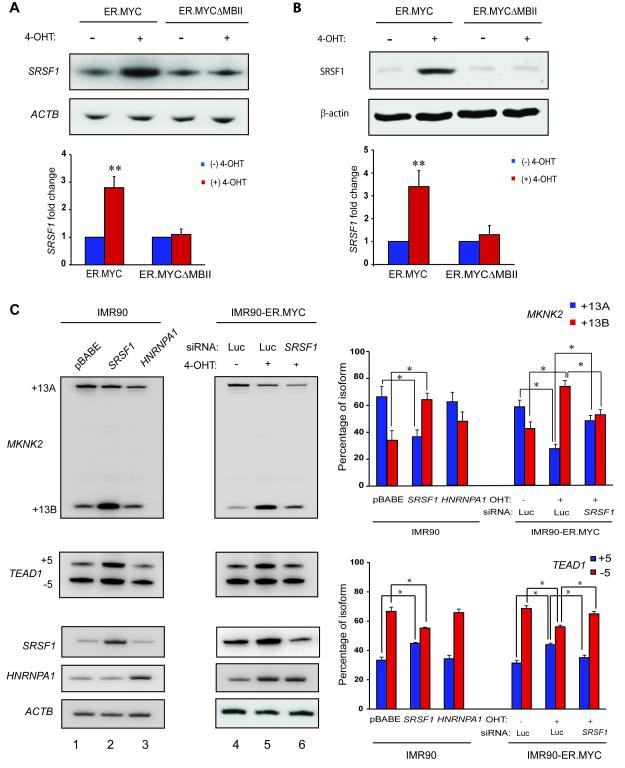
To assess more directly whether SRSF1 expression is regulated by MYC, we used an inducible MYC-Estrogen Receptor (ER) system (Eilers et al., 1989; Littlewood et al., 1995). We generated IMR90 cells stably expressing the MYC protein fused to a modified ER ligand-binding domain, which binds the synthetic estrogen analog 4-hydroxytamoxifen (4-OHT). The ER.MYC protein is held in the cytoplasm through association with the HSP-90 protein. Upon binding 4-OHT, ER.MYC translocates into the nucleus, where it regulates the expression of target genes. 4-OHT treatment of IMR90-ER.MYC cells led to significant accumulation of SRSF1 mRNA (Figure 2A) and SRSF1 protein (Figure 2B). As a control for ER.MYC induction, we verified the upregulation of a known MYC target gene, NCL (Figure S2A). Moreover, IMR90 cells transduced with empty vector did not induce SRSF1 upon 4-OHT treatment (Figure S2B). A MYC deletion mutant lacking amino acids 106-143, which comprise MYC Box II (MBII) in the transcription-activation domain (TAD) (Oster et al., 2003), failed to induce SRSF1 expression (Figure 2A,B), indicating that MYC requires an intact TAD to upregulate SRSF1 expression. We also observed increased SRSF1 levels upon MYC induction in two immortal cell lines: MCF-10A mammary epithelial cells and Rat1a fibroblasts (Figure S2C). Two additional SR protein genes, SRSF5 and SRSF11, showed no change in expression upon MYC induction, though both were predicted as MYC target genes by a genome-wide ChIP-on-Chip analysis (Li et al., 2003) (Figure S2D). In addition to showing the specificity of the effect of MYC on SRSF1, these results emphasize the need for validation to determine the true MYC targets among those predicted by genome-wide analyses.
Figure 2.
MYC regulates SRSF1 expression and alternative splicing SRSF1 Target Genes. (A) RT-PCR and (B) Immunoblotting of SRSF1 from IMR90-ER.MYC or IMR90-ER.MYCΔMBII cells induced with 4-OHT. Error bars, s.d.; n=3; t-test **P<0.01. (C) RT-PCR of MKNK2 and TEAD1 mRNA isoforms in IMR90-ER.MYC cells induced with 4-OHT, with or without SRSF1 knockdown. IMR90 cells overexpressing SRSF1 or hnRNPA1 are shown as controls. Error bars, s.d.; n=3, *P<0.05. See also Figure S2.
We next analyzed the splicing of two previously reported SRSF1 target genes, MKNK2 and TEAD1 (Karni et al., 2007). MKNK2 encodes the eIF4E-kinase MNK2 and expresses two isoforms by alternative splicing of 3′ exons 13A and 13B, whereas TEAD1 encodes the transcriptional enhancer factor protein TEF-1 and expresses two isoforms by alternative splicing of exon 5. IMR90 cells overexpressing SRSF1 predominantly expressed the +13B isoform of MKNK2 and the +5 isoform of TEAD1, as expected (Karni et al., 2007) (Figure 2C, Lanes 1-2). Another splicing factor, hnRNPA1, which is also positively regulated by MYC (Biamonti et al., 1993; David et al., 2010) and frequently antagonizes SRSF1 (Mayeda et al., 1992), did not alter MKNK2 or TEAD1 splicing (Figure 2C, lane 3). Induction of IMR90-ER.MYC cells with 4-OHT promoted a significant switch in MKNK2 splicing from the +13A to the +13B isoform and promoted inclusion of exon 5 in the TEAD1 transcript, consistent with the increase in SRSF1 (Figure 2C, lanes 4-5). Furthermore, induction of ER.MYC in cells transfected with siRNA against SRSF1 did not trigger a change in MKNK2 or TEAD1 splicing (Figure 2C, lane 6), indicating that MYC alters MKNK2 and TEAD1 splicing through upregulation of SRSF1 expression. We also observed that both SRSF1 over-expression and MYC induction led to a significant increase in the overall MKNK2 transcript level (Figure S2E), suggesting that both factors directly or indirectly regulate MKNK2 expression at the level of transcription or mRNA stability. We also measured alternative splicing of a third SRSF1 target gene, BIN1, which encodes the tumor suppressor and pro-apoptotic protein BIN1. SRSF1 overexpression promotes inclusion of the 12A exon in the BIN1 transcript (Karni et al., 2007). However, we did not observe changes in alternative splicing of BIN1 in response to MYC induction (Figure S2F), perhaps due to other splicing factors also being modulated by MYC and counteracting the SRSF1-mediated inclusion of exon 12A.
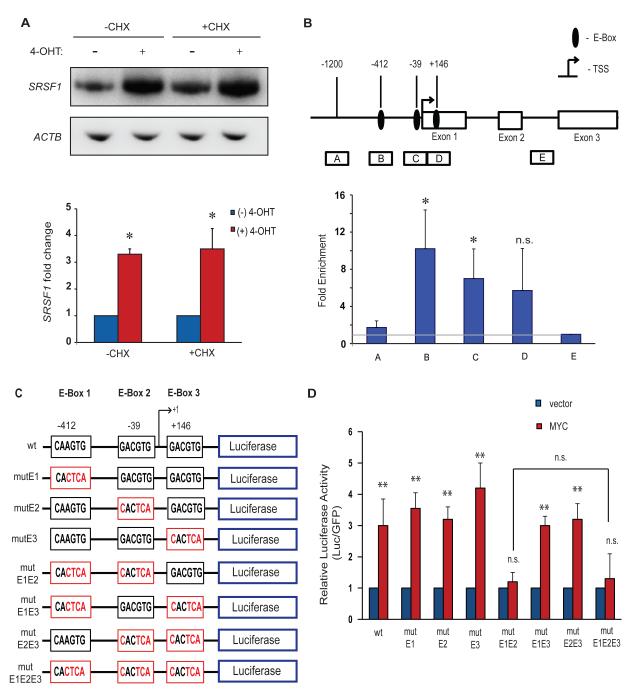
Because MYC was predicted to bind the SRSF1 promoter by ChIP-on-Chip analysis of CpG islands (Mao et al., 2003), we investigated whether SRSF1 is a direct transcriptional target of MYC. Treatment of IMR90-ER.MYC cells with the protein-synthesis inhibitor cycloheximide prior to 4-OHT induction of ER.MYC did not abrogate the upregulation of SRSF1 mRNA (Figure 3A), indicating that de novo protein synthesis is not required for MYC to activate SRSF1 expression. Moreover, analysis of the human SRSF1 promoter sequence revealed three putative non-canonical MYC binding sites (E-boxes). We therefore used ChIP to assess binding of MYC to the SRSF1 promoter locus in the lung-carcinoma cell line NCI.H460, which downregulates SRSF1 expression in response to MYC knockdown (Figure 1C,D). Our ChIP analysis revealed significant enrichment of MYC at the SRSF1 proximal promoter region comprising two E-boxes mapping at −412 and −39 (Figure 3B). We also detected MYC enrichment at a third E-box at position +146, relative to the transcription start site, but this was not significant and likely corresponds to chromatin fragments that overlap the E-box at −39 (Figure 3B). MYC binding to the SRSF1 proximal promoter region is also evident in genome-wide data from HeLa and K562 cells obtained by the ENCODE genome-wide ChIP sequencing project (UCSC genome browser, assembly NCBI36/hg18, Yale/UC Davis/Harvard study). The same study also reported the binding of MYC’s obligate hetero-dimerization partner MAX (Amati et al., 1994) to the SRSF1 proximal promoter region, suggesting MYC activity at the locus.
Figure 3.
MYC binds to and activates the human SRSF1 promoter. (A) RT-PCR of IMR90-ER.MYC cells treated with 4-OHT, with or without cycloheximide. Error bars, s.d.; n= 3; **P<0.01. (B) MYC chromatin immunoprecipitation analysis at the SRSF1 promoter locus in the lung-carcinoma NCI-H460 cell line. Diagram of the SRSF1 gene indicating the E-boxes and amplicons (A-E) used for ChIP assays. The results are expressed as DNA enrichment in fragmented chromatin immunoprecipitated with anti-MYC antibody (relative to anti-rabbit IgG immunoprecipitation) and normalized to the amplicon E signal, as measured by quantitative PCR. The horizontal gray line represents no change in MYC-specific enrichment. Error bars, s.d.; n=3; t-test *P<0.05; n.s., not significant. (C) Diagram of the wild-type SRSF1 promoter, comprising three non-canonical E-boxes, and the E-Box mutants generated for reporter assays. Mutant E-boxes and residues are indicated in red. (D) Luciferase assay of reporter constructs in (C) co-transfected with MYC cDNA or vector control into NIH3T3 cells. Luciferase activity was normalized to co-transfected GFP, and the relative activity is plotted. Error bars, s.d.; n=3; t-test **P<0.01; n.s., not significant.
To determine whether these are functional MYC binding sites, we amplified a 1500-bp genomic fragment of the SRSF1 promoter, comprising these putative E-boxes (from −1200 to +300 relative to the transcription-start site (TSS)), inserted it upstream of a luciferase reporter gene, and assayed for its MYC responsiveness in transfected NIH3T3 cells. We also generated constructs with mutations in the three E-boxes, either individually or together, to an inactive CACTCA sequence (Figure 3C). MYC overexpression resulted in ~3-fold induction of luciferase activity for the wild-type construct (Figure 3D), relative to the vector control. The double, but not the individual, mutations of E-boxes 1 and 2 abrogated this MYC-induced activation, suggesting functional redundancy between the two elements (Figure 3D). Mutation of the third putative non-canonical E-box (E-box 3), downstream of the TSS, either alone or in combination with the other E-boxes, did not abrogate, or further reduce, luciferase activity, indicating that this site is non-functional (Figure 3D). We conclude that SRSF1 is a direct transcriptional target of MYC, with two functional non-canonical E-boxes in its promoter.
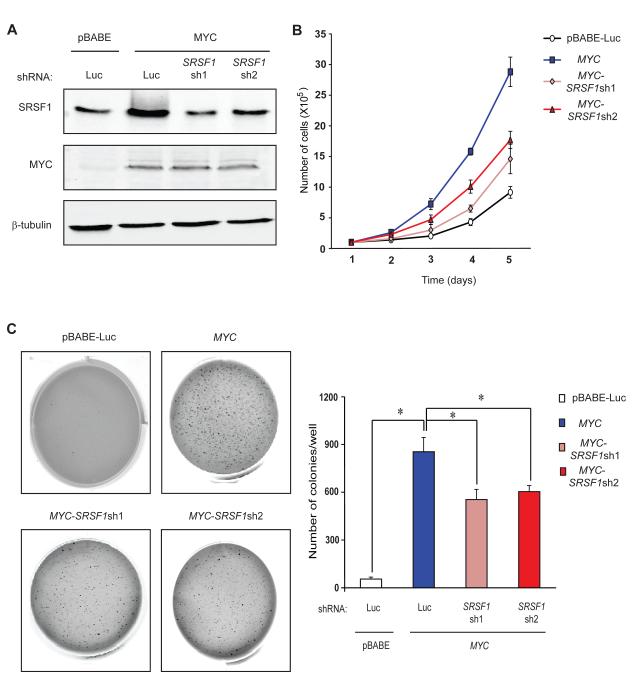
Both MYC and SRSF1 are strong oncogenes that control cell proliferation, cell-cycle progression, and apoptosis. We therefore asked whether SRSF1 induction is required for MYC-induced transformation. We generated MYC-overexpressing Rat1a fibroblasts transduced with either a control luciferase shRNA or two different shRNAs against SRSF1. SRSF1 knockdown was carefully modulated by optimizing the retroviral MOI so as to cancel out the MYC-induced increase in SRSF1 protein, but without completely depleting it from the cells (Figure 4A). As expected, Rat1a-MYC cells showed elevated SRSF1 expression and increased proliferation, compared to Rat1a-pBabe-Luc control cells (Figure 4B). SRSF1 knockdown resulted in a significant decrease in the proliferation rate of the MYC-overexpressing cells, though it remained significantly higher than the control. In accordance with this result, DNA-content analysis by flow cytometry revealed a higher percentage of the MYC-overexpressing cells in the S-G2-M phases, compared to the vector control (Figure S3). SRSF1 knockdown significantly decreased the proportion of dividing cells, with more cells accumulating in the G0-G1 phases.
Figure 4.
SRSF1 knockdown impairs anchorage-independent growth of MYC-transformed cells. (A) Immunoblotting of MYC and SRSF1 in the Rat1a-pBabe-Luc control cell line, Rat1a-MYC, and Rat1a-MYC cells transduced with one of two shRNAs against SRSF1. (B) Growth curves of the four cell lines from (A). Error bars, s.d.; n=3. (C) Anchorage-independent growth of cell lines from (A) in soft-agar colony-formation assays. Error bars, s.d.; n=3; t-test *P<0.05.
Moreover, SRSF1 knockdown did not promote cell death, ruling out apoptosis as a cause of the observed decrease in growth rate (Figure S3). Rat1a-MYC-SRSF1sh cells showed significantly decreased anchorage-independent growth, compared to Rat1a-MYC cells (Figure 4C). We conclude that SRSF1 is a critical MYC target gene, required for MYC’s full activity in tumorigenesis.
Recently, MYC was shown to regulate the expression of other splicing factors—hnRNPA1, hnRNPA2, PTB1, and hnRNPH—and through them to regulate alternative splicing of pyruvate kinase M and oncogenic A-Raf kinase pre-mRNAs (David et al., 2010; Rauch et al., 2011). In the present study we show MYC-mediated positive regulation of the oncogenic splicing factor SRSF1. MYC activates transcription from the SRSF1 promoter, and the resulting increase in SRSF1 leads to altered splicing of some but not all of its target genes. Furthermore, we found SRSF1 to be a critical MYC target, necessary for MYC’s oncogenic activity. We have also found that SRSF1 and MYC cooperate in transforming mammary epithelial cells, and their expression correlates in human breast tumors (Anczukow et al., 2011). The SRSF1 target genes that do undergo a splicing change upon MYC induction are therefore likely to be important mediators of MYC activity. Furthermore, considering the role of SRSF1 in multiple processes other than splicing, such as translation and mTOR signaling, there are likely several additional downstream effectors of SRSF1 that contribute to MYC function. The overall picture that emerges from these studies is that, in addition to regulating transcription of its target genes, MYC also indirectly regulates the expression of protein isoforms through regulation of alternative splicing of a subset of transcripts, and these changes contribute to MYC’s biological functions.
EXPERIMENTAL PROCEDURES
Plasmids
T7-tagged SRSF1 and HNRNPA1 cDNAs cloned in the pBABE-Puro retroviral vector were described previously (Karni et al., 2007). pBABE-Puro-ER.MYC (Littlewood et al., 1995) was used to generate the pBabe-Puro-ER.MYCΔMBII construct by Quick-change site-directed mutagenesis (Stratagene). The Transcriptional Regulatory Element Database (TRED) (Zhao et al., 2005) was used to obtain the SRSF1 promoter sequence (Promoter ID 18315). The SRSF1 promoter from −1200 to +300 (relative to the TSS) was amplified from human genomic DNA (Promega) and cloned into the pGL3 vector (Promega). MYC-binding sites in the wild-type SRSF1 promoter were mutated by Quick-change site-directed mutagenesis.
Cell Culture and Stable Cell Line Generation
IMR90, NIH3T3, and Rat1a cells were grown in DMEM medium (Invitrogen) supplemented with 10 % (v/v) fetal bovine serum (FBS), penicillin, and streptomycin. NCI-H524, NCI-H460, NCI-H1299, NCI-H1568, and NCI-H1975 cells were grown in RPMI 1640 medium (Invitrogen) supplemented with 10 % (v/v) FBS, penicillin, and streptomycin. A549 cells were grown in F12K medium (Invitrogen) supplemented with 10 % (v/v) FBS. NCI-H1666 cells were grown in DMEM/F12 medium (Invitrogen) supplemented with 5 % (v/v) FBS, penicillin, and streptomycin. MCF10A cells were grown in DMEM/F12 medium, supplemented with 5 % (v/v) horse serum, 20 ng/ml EGF, 100 μg/ml hydrocortisone, 10 ng/ml cholera toxin, penicillin, and streptomycin. To generate stable pools, IMR-90 and Rat1a cells were infected with pBABE-Puro or pBABE-hygro retroviral vectors expressing ER.MYC or MYC cDNAs, respectively, followed by selection with puromycin (2 μg/ml) or hygromycin (200 μg/ml) for 72 h. For MYC induction studies, ER.MYC-expressing cells were grown to confluence and treated with 2 μM 4-OHT for 8 h for RT-PCR, and 48 h for immunoblotting and splicing analysis.
RNA Interference
For inhibition of MYC or SRSF1 expression, cells were seeded (2 × 105 cells per well) in six-well plates in antibiotic-free medium. After 24 h, cells were transfected with 200 pmol short interfering RNA against MYC (Cell Signaling, Catalog No. 6553) or SRSF1 (target sequence 5′-ACGAUUGCCGCAUCUACGU-3′) using Lipofectamine RNAiMAX (Invitrogen). After a further 48 h, cells were lysed, and protein and RNA were extracted as described below. For stable knockdown of SRSF1, Rat1a cells were separately transduced with each of two SRSF1 shRNAs cloned in the retroviral vector LMP9, and selected with 2 μg/ml puromycin for 4 days.
Immunoblotting
Cells were lysed in RIPA buffer and protein was quantitated using a Bradford Protein Assay kit (Bio-Rad). 25 μg of total protein from each lysate was separated by SDS-PAGE and transferred onto a nitrocellulose membrane (Whatman), followed by blocking with 5 % (w/v) dry milk in Tris-buffered saline with 0.05 % (v/v) Tween-20, probing with the indicated antibodies, and quantitation using an Odyssey infrared-imaging system (LI-COR Biosciences). Primary antibodies used were: MYC (Cell Signaling rAb, 1:500); SRSF1 (mAb AK96 culture supernatant (Hanamura et al., 1998), 1:500); SRSF9 (mAb culture supernatant, 1:50); β-actin (Sigma mAb, 1:10,000), and β-tubulin (Genscript rAb, 1:10,000). Secondary antibodies were IRdye 800 or 680 anti-rabbit or anti-mouse (LI-COR Biosciences, 1:10,000).
RT-PCR Analysis
Cells were lysed with Trizol reagent (Invitrogen) and total RNA was extracted. Following DNAase I digestion (Promega), 2 μg of total RNA was reverse-transcribed with Improm-II reverse transcriptase (Promega). Radioactive PCR (25 cycles) with [α-32P]-dCTP was used to amplify endogenous transcripts. The products were run on a 5 % native polyacrylamide gel, visualized by autoradiography, and quantitated on a FUJIFILM FLA-5100 phosphorimager (Fuji Medical Systems) using Multi Gauge software Version 2.3 (Fujifilm). The PCR primers used were as follows:
hSRSF1F: 5′-ATGTCGGGAGGTGGTGTGATTC-3′
hSRSF1R: 5′-TGTTCCACGGCCGCTTCGAG-3′
rSRSF1F: 5′-CGCGACATCGACCTGAAGAAC-3′
rSRSF1R: 5′-CCACGACACCAGTGCCATCTCG-3′
HNRNPA1F: 5′-AAAGACCAGGTGCCCACTTA-3′
HNRNPA1R: 5′-AATCTTATCCACGGAGTCATGG-3′
MYCF: 5′-GGTACAAGCTGGAGGTGGAG-3′
MYCR: 5′-AATCTTATCCACGGAGTCATGG-3′
NCLF: 5′-TTTCTTTCCTTTGGCTGGTG-3′
NCLR: 5′-ATGGCAAGAATGCCAAGAAG-3′
MKNK2Ex11F: 5′-CCAAGTCCTGCAGCACCCCTG-3′
MKNK2Ex13aR: 5′-GATGGGAGGGTCAGGCGTGGTC-3′
MKNK2Ex13bR: 5′-GAGGAGGAAGTGACTGTCCCAC-3′
TEAD1Ex3, 4F: 5′-AGACGAAGGCAAAATGTATGG-3′
TEAD1Ex9, 8R: 5′-CGTAGGCTCAAACCCTGGAAT-3′
BIN1Ex11F: 5′-CCTCCAGATGGCTCCCCTGC-3′
BIN1Ex15R: 5′-CCCGGGGGCAGGTCCAAGCG-3′
β-actinF: 5′-GTGCCCATTTATGAGGGCTA-3′
β-actinR: 5′-CTGGCAGCTCGTAGCTCTTT-3′
Chromatin Immunoprecipitation
ChIP assays were performed as described (Steger et al., 2008). Crosslinking was performed with sequential 15 mM EGS (Pierce) and 1 % (v/v) formaldehyde treatment. Antibodies used for immunoprecipitation were rabbit anti-myc (Cell Signaling, 9402) and rabbit IgG (Cell Signaling). Immunoprecipitated DNA was analyzed by quantitative PCR using SYBR green (ABI) on an ABI 7900HT instrument. PCR primers were as follows:
Amplicon A:
F: 5′-CCCAGCCTGATTTGAATTTT-3′
R: 5′-GAAAATACCGGTCCTCTCAGG-3′
Amplicon B:
F: 5′-GGATTAGACGCACCCTACGA-3′
R: 5′-CGATTTCTCCAGGAATGAGG-3′
Amplicon C:
F: 5′-ACGTAGCCCTCGCAGCAC-3′
R: 5′-GGACTCGAGAACAGGCCTTC-3′ Amplicon D:
F: 5′-CTTTTCGTCACCGCCATGT-3′
R: 5′-GTCCTCGAACTCAACGAAGG-3′
Amplicon E:
F: 5′-GGATTGATGTGAAGGGACGA-3′
R: 5′-TGGAATCCAGAGTCCAAAAT-3′
Luciferase Reporter Assay
500 ng of MYC expression vector, 100 ng of pGL3-Luciferase reporter comprising nucleotides −1200 to +300 of the SRSF1 promoter—with or without E-box mutations—and 100 ng of pEGFP vector were co-transfected into NIH3T3 cells using Fugene 6 (Roche). 36 h after transfection, the cells were lysed, and luciferase activity was measured using a Dual Luciferase Reporter Assay kit (Promega). RNA was extracted from the remaining cell lysate, and the GFP level was measured by radioactive RT-PCR and used as a transfection control to normalize luciferase activity.
Growth Curves and Proliferation Assay
Rat1a cells transduced with pBABE-hygro, pBABE-MYC, LMP-Puro, LMP-SRSF1sh1, or LMP-SRSF1sh2 were seeded at 1 × 105 cells per 60-mm dish. At the indicated times, triplicate plates of cells were trypsinized, stained with Trypan-blue, and unstained cells were counted using a hemocytometer.
Anchorage-independent growth
Rat1a cells from each transductant pool were plated (20,000 cells per well) in triplicate in 0.35 % (w/v) agar in DMEM supplemented with 10 % (v/v) FBS on a layer of 0.7 % (w/v) agar. Cells were incubated at 37 °C and 5 % (v/v) CO2 for 14 days. Colonies were stained with 0.005 % (w/v) Crystal Violet, and whole-well images were taken using the Odyssey Imaging System. The images were analyzed using Image-J software, and the average number of colonies per well for each transductant pool was determined.
Microarray Analysis
The GEO GSE2109 dataset from the Expression Project for Oncology (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE2109) was used to obtain expression profile data from 132 clinically annotated human lung tumors. Each sample was standardized by calculating Z-scores based on the sample average and s.d. across the entire set of genes. Expression profiles of SRSF1 and MYC were extracted for all the samples. A contingency table was built showing the number of samples with high expression of both SRSF1 and MYC, only SRSF1, only MYC, or neither (Z-score>1.29, corresponding to a P-value of 0.1). A Mann-Whitney test was used to compare SRSF1 expression in lung tumors containing high versus low MYC levels (above and below the median).
Statistical Analysis
All histograms were plotted using mean ± s.d. Data points were compared using unpaired two-tailed Student’s t-tests, and P-values are indicated in the figure legends. Pearson correlation was used to evaluate the association between MYC and SRSF1 expression detected by quantitative immunoblotting.
Supplementary Material
HIGHLIGHTS.
MYC and SRSF1 are significantly co-expressed in lung and breast cancers.
SRSF1 is a direct transcriptional target of MYC.
A subset of SRSF1 targets show alternative splicing changes in response to MYC induction.
SRSF1 knockdown reduces MYC-mediated transformation.
ACKNOWLEDGMENTS
We thank John Kurland, Eric Wang, and Chris Vakoc for reagents and useful discussions; Rafaella Sordella for lung cancer cell lines; and Chaolin Zhang for SRSF1 promoter-sequence analysis. This work was supported by grant CA13106 from the NCI. The authors declare no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- Amati B, Land H. Myc-Max: a transcription factor network controlling cell cycle progression, differentiation and death. Curr. Opin. Genet. Dev. 1994;4:102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- Anczukow O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, Muthuswamy S, Krainer AR. The splicing-factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat. Struct. Mol. Biol. 2012;19:220–228. doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G, Bassi MT, Cartegni L, Mechta F, Buvoli M, Cobianchi F, Riva S. Human hnRNP protein A1 gene expression. Structural and functional characterization of the promoter. J. Mol. Biol. 1993;230:77–89. doi: 10.1006/jmbi.1993.1127. [DOI] [PubMed] [Google Scholar]
- Coller HA, et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling and adhesion. Proc. Natl. Acad. Sci. USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Picard D, Yamamoto KR, Bishop JM. Chimaraes of Myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature. 1998;340:66–68. doi: 10.1038/340066a0. [DOI] [PubMed] [Google Scholar]
- Ezponda T, et al. The oncoprotein SF2/ASF promotes non-small cell lung cancer survival by enhancing survivin expression. Clin. Cancer Res. 2010;16:4113–4125. doi: 10.1158/1078-0432.CCR-10-0076. [DOI] [PubMed] [Google Scholar]
- Ge H, Manley JL. A protein factor, ASF, controls cell-specific alternative splicing of SV-40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- Hanamura A, Cáceres JF, Mayeda A, Franza BR, Jr., Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Gattoni R, Stévenin J, Steitz JA. SR splicing factors serve as adaptor proteins for TAP-dependent mRNA export. Mol. Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Karni R, et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer AR, Conway GC, Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from Hela cells. Genes Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- Lemaire R, Prasad J, Kashima T, Gustafson J, Manley JL, Lafyatis R. Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: a novel function for SR proteins. Genes Dev. 2002;16:594–607. doi: 10.1101/gad.939502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang J, Manley JL. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 2005;19:2705–2714. doi: 10.1101/gad.1359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-myc in Burkitt’s lymphoma cells. Proc. Natl. Acad. Sci. USA. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao DY, et al. Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr. Biol. 2003;13:882–886. doi: 10.1016/s0960-9822(03)00297-5. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNPA1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Oster SK, Mao DYL, Kennedy J, Penn LZ. Functional analysis of the N-terminal domain of the Myc oncoprotein. Oncogene. 2003;22:1998–2010. doi: 10.1038/sj.onc.1206228. [DOI] [PubMed] [Google Scholar]
- Rauch J, et al. c-Myc regulates RNA splicing of the A-Raf kinase and its activation of the ERK pathway. Cancer Res. 2011;71:4664–4674. doi: 10.1158/0008-5472.CAN-10-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser I, et al. Dissection of transcriptional programmes in response to serum and c-Myc in a human B-cell line. Oncogene. 2005;13:520–524. doi: 10.1038/sj.onc.1208198. [DOI] [PubMed] [Google Scholar]
- Sinclair CS, Rowley M, Naderi A, Couch FJ. The 17q23 amplicon and breast cancer. Breast Cancer Res. Treat. 2003;78:313–322. doi: 10.1023/a:1023081624133. [DOI] [PubMed] [Google Scholar]
- Sun D, Novotny M, Bulek K, Liu C, Li X, Hamilton T. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing regulatory factor SF2 (ASF) Nat. Immun. 2011;12:853–860. doi: 10.1038/ni.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zhang Z, Sinha R, Karni R, Krainer AR. SF2/ASF autoregulation involves layers of post-transcriptional and translational control. Nat. Struct. Mol. Biol. 2010;17:306–312. doi: 10.1038/nsmb.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol. Cell. Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen K, Mansilla F, Schepeler T, Øster B, Rasmussen MH, Dyrskjøt L, Karni R, Akerman M, Krainer AR, Laurberg S, et al. Alternative splicing of SLC39A14 in colorectal cancer is regulated by the Wnt pathway. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.002998. M110.002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J. A splicing-independent function of SF2/ASF in microRNA processing. Mol. Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, et al. ASF/SF2 regulated CAMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscles. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Zeller KI, Jegga AG, Aronow BJ, O’Donell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4 doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Xuan Z, Liu L, Zhang MQ. TRED: a Transcriptional Regulatory Element Database and a platform for in silico gene regulation studies. Nucleic Acids Res. 2005;33:103–107. doi: 10.1093/nar/gki004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.