Abstract
The metabolome is the terminal downstream product of the genome and consists of the total complement of all the low molecular weight molecules (metabolites) in a cell, tissue or organism. Metabolomics aims to measure a wide breadth of small molecules in the context of physiological stimuli or in disease states. Metabolomics methodologies fall into two distinct groups; untargeted metabolomics, an intended comprehensive analysis of all the measurable analytes in a sample including chemical unknowns, and targeted metabolomics, the measurement of defined groups of chemically characterized and biochemically annotated metabolites. The methodologies considered in this unit focus on the processes of conducting targeted metabolomics experiments, and the advantages of this general approach are highlighted herein. This unit outlines the procedures for extracting nitrogenous metabolites, including the amino acids, lipids, and intermediary metabolites, including the TCA cycle oxoacids, from blood plasma. Specifically, protocols for the analysis of these metabolites using liquid chromatography-mass spectrometry-based targeted metabolomics experiments is discussed.
Keywords: Targeted Metabolomics, Liquid Chromatography-Mass Spectrometry, Multiple Reaction Monitoring
Introduction
The central biochemical and molecular biological dogma declares that information flows directionally from genomic DNA through mRNA transcripts, which are then translated to proteins. The protein products, amongst them enzymes, then influence the concentrations of their substrates and products, which are integrated in tightly controlled metabolic pathways. It is the flux of these small molecular weight metabolites within a cell, tissue or organism that generates the phenotype. While much of this holds true, it is now understood that the movement of information is far from unidirectional. Cellular communication is now known to occur via a complex system of interactions between DNA, RNA, protein and metabolite with feed-forward and negative feedback loops in play; the new methodologies of the “omics” revolution of the 1980s and 1990s provided invaluable tools for the study of these interactions on a global level and offered an alternative means of investigation to that of some of the more reductionist molecular biology approaches.
Metabolomics was defined in the 1990s to describe techniques aimed at measuring the metabolites present within a cell, tissue or organism during a genetic alteration or physiological stimulus (Oliver et al., 1998; Nicholson et al., 1998). However, the notion of cataloguing the entire set of biochemicals in organisms to extract potentially meaningful biological and clinical characteristics may go back as far as the 1950s. Measurement of time-related metabolic changes in animal models in response to genetic manipulation can be used to define the varying phenotypes observed (Lindon et al., 2003). Through this methodology metabolomics has proved complementary to the other omic techniques, identifying so-called silent phenotypes, genes that when perturbed have no apparent influence on physical characteristics or behavior (Griffin et al., 2002).
The metabolome is the terminal downstream product of the genome and consists of the total complement of all the low molecular weight molecules (metabolites) in a cell, tissue or organism required for growth, maintenance or normal function in a specific physiological state (Goodacre 2003; Goodacre et al., 2004) (Fig. 30.2.1). The potential size of the metabolome is often heavily disputed, particularly as emerging data suggest an important role for the microbiome and its metabolic products. Alongside the variety of chemical classes and physical properties that constitute metabolites and the dynamic range of metabolite concentrations across large orders of magnitude it becomes clear why it is necessary to employ an extensive array of analytical techniques in metabolomics research.
Figure 30.2.1.
The relationship between the genome, proteome and metabolome (Wang and Gerszten, Nature, 2008).
Analytical approaches used to identify changes in the concentrations and fluxes of endogenous metabolites include, but are not limited to, 1H-nuclear magnetic resonance (1H-NMR) spectroscopy, 13C-nuclear magnetic resonance (13C-NMR) spectroscopy, gas chromatography-mass spectrometry (GC-MS), gas chromatography-flame ionization detection (GC-FID), direct infusion-mass spectrometry (DI-MS) and liquid chromatography-mass spectrometry (LC-MS). Due to the wide array of metabolites and the dynamic nature of their concentrations within the cell, a complete analysis of the metabolome has thus far eluded description even when a range of analytical approaches have been employed.
Metabolomics strategies have been divided into two distinct approaches, untargeted and targeted metabolomics, each with their own inherent advantages and disadvantages. Untargeted metabolomics (UNIT 30.1) is the comprehensive analysis of all the measurable analytes in a sample, including chemical unknowns. Due to its comprehensive nature, untargeted metabolomics must be coupled to advanced chemometric techniques, such as multivariate analysis, to reduce the extensive datasets generated into a smaller set of manageable signals.
These signals then require annotation using either in silico libraries or experimental investigation and subsequent identification using analytical chemistry. Untargeted analysis offers the opportunity for novel target discovery, as coverage of the metabolome is only restricted by the methodologies of sample preparation and the inherent sensitivity and specificity of the analytical technique employed. However, the principal challenges of this approach lie in the protocols and time required to process the extensive amounts of raw data generated, the difficulties in identifying and characterizing unknown small molecules, the reliance on the intrinsic analytical coverage of the platform employed, and the bias towards detection of high-abundance molecules.
By contrast, targeted metabolomics is the measurement of defined groups of chemically characterized and biochemically annotated metabolites. Through the use of internal standards, analysis can be undertaken in a quantitative or semi-quantitative manner. This approach takes advantage of the comprehensive understanding of a vast array of metabolic enzymes, their kinetics, end products, and the known biochemical pathways to which they contribute. When utilizing targeted metabolomics sample preparation can be optimized, reducing the dominance of high-abundance molecules in the analyses; in addition, since all analyzed species are clearly defined, analytical artifacts are not carried through to downstream analysis. When predefined lists of analytes are studied, novel associations between metabolites may be illuminated in the context of specific physiological states.
This unit describes the essential methodologies for performing targeted metabolomics experiments using an LC-MS-based multiple reaction monitoring (MRM) metabolomics platform with a focus on blood plasma. Protocols for metabolite extraction and analysis of metabolites from cerebrospinal fluid, urine and tissues are also briefly discussed. The protocols outlined include sample preparation and data acquisition for the measurement of various groups of metabolites encompassing the hydrophilic amino acids employing hydrophilic interaction liquid chromatography (HILIC) (Support Protocol 1 and Basic Protocol 1) and intermediary metabolites utilizing polar reversed phase chromatography (Support Protocol 2 and Basic Protocol 2). Data analysis techniques for targeted LC-MS-based MRM of metabolites are also discussed (Basic Protocol 3).
[*Copyed: The protocols are currently presented as Support Protocol followed by Basic Protocol. This isn't the usual CP style, but it flows nicely with sample prep in the Support immediately followed by use of the sample in the Basic. I recommend keeping it as-is. Rearranging will probably annoy the author and will also cause renumbering of several figures and tables. Call me if you want to discuss. Gwen]
Strategic Planning
Due to the multiple variables analyzed during a metabolomics experiment, the intrinsic intra-individual variability, the dynamic nature of metabolite concentrations and metabolic flux, the influence of matrix and LC-MS conditions, and the need to ensure the accuracy and legitimacy of the data produced, the importance of stringent experimental design and protocol reproducibility is paramount.
General LC-MS Parameters
When using LC-MS in a targeted metabolomics experiment a number of key considerations must be taken into account that may affect the accuracy of the experimental data. Appropriately selecting the correct ionization parameters is essential. Electrospray ionization (ESI) is the most commonly used method of small molecule ionization employed in LC-MS-based metabolomics studies. ESI is a so-called “soft” ionization technique developed in the 1980s that facilitates the mass spectrometric detection of non-volatile, high-mass analytes. The principal advantages of the soft ionization technique are that it does not require chemical derivatization to improve volatility (as is the case for electron impact ionization), and that it minimizes analyte fragmentation, which can assist in the analytical interpretation of complex mixtures. However, ESI also suffers from several disadvantages, the most prominent being strong ion suppression effects when analyzing complex molecular mixtures (Petkovic et al., 2001). Ion suppression occurs when analytes compete for charge during the ionization process, with an individual analyte's ionization efficiency based on its chemical characteristics (Knochenmuss, 2003). Therefore, the observed ion count for a particular ion can change, depending on what other analytes or contaminants are being co-ionized. This issue can be particularly problematic in that ion suppression can occur even when the interfering compound is not seen in the mass spectrum. Thus, measurement must assume the mixtures are roughly the same between the groups of samples analyzed. Therefore, it is important to only compare samples of the same matrices when performing experiments. Comparing a sample of blood plasma with that of a tissue extract, for example, would be inappropriate. By separating the analytes via one or more chromatographic techniques prior to mass spectrometric analysis, the effects of ion suppression can be reduced (Petersen and Cummings, 2006). The disadvantages of ion suppression and matrix effects can be best controlled when performing a targeted metabolomics experiment through the use of isotope-labeled internal standards for absolute quantitation of metabolite concentrations (see below). In addition, the majority of LC-MS platforms utilized in targeted metabolomics are capable of both negative and positive mode ionization. Depending on the chemical and physical nature of the metabolites of interest, the correct polarity mode for efficient ionization of these molecules must be selected during the development of the LC-MS method.
Targeted Analysis and Normalization
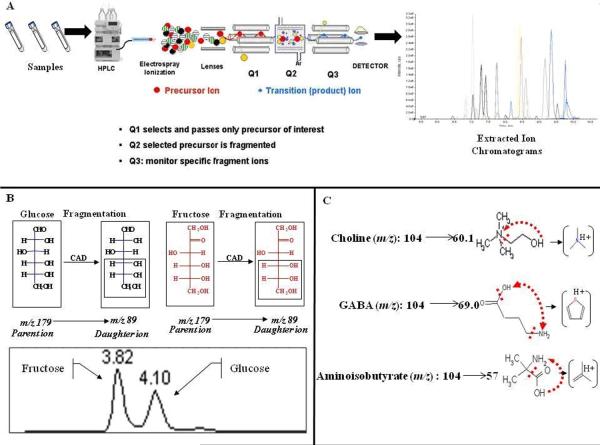
It is critical to recognize that when using a mass spectrometry-based metabolomics platform that both ion intensities and chromatographic retention times (as described below) can suffer from temporal drift. As a result, sample analysis for a particular study should be conducted in a randomized sample order and preferably the data should be acquired in the same batch on the same day, minimizing internal variation in a study set. The advantage of utilizing a targeted approach in this aspect is two-fold. First, the internal standards spiked into each sample can be used to normalize concentrations of metabolites across sample sets and batches. This is particularly useful when conducting analysis of large sample sets, where days or weeks of analysis may be required. Second, by employing MRM mass spectrometric analysis the exact identity of each metabolite can be fully defined and incorporated into the analysis. The mass spectrometric experiment is performed using a triple quadrupole instrument, as employed in the protocols outlined in this unit (Fig. 30.2.2). A metabolite precursor is ionized at the ESI source and then resolved and isolated in the first quadrupole (Q1). The second quadrupole (Q2) functions as a collision cell where, in a process known as collision-activated dissociation (CAD) or collision-induced dissociation (CID), the target ion is fragmented. Finally, the ions are accelerated into the third quadrupole, which also acts as a mass filter, selecting for a particular m/z of a fragment ion, which is then introduced to the detector. Since individual metabolites have specific precursor/product ions, known as a transition, the identity of the metabolite can be ensured, especially when combined with a known chromatographic retention time. However, the specific transitions, retention times, dynamic concentration range and collision parameters for each metabolite must be defined through experimental analysis prior to this type of targeted metabolomic experiment. This is a significant undertaking.
Figure 30.2.2.
(A) Workflow for a Liquid Chromatography Triple Quadrupole Mass Spectrometry Multiple Reaction Monitoring targeted metabolomics experiment. (B) The resolution of chromatography allows the separation of metabolites even in the case of a shared MRM transition. (C) Specific precursor/product ion transitions in MRM experiments identify individual metabolites.
Drift in instrument performance can be further controlled by monitoring a stable isotopically labeled internal standard spiked into the sample. The coefficients of variation across samples can be calculated as defined by 100 × standard deviation/mean value of the data set. Where batch-to-batch variation in the internal standard is significant, the metabolite data can be normalized to the internal standard. Furthermore, by interspacing standardized samples such as a pooled plasma sample throughout the analysis batch, temporal drift can be controlled across all analyzed metabolites. Samples can then be normalized to the standardized samples.
Biological Replicates and Confounders
The same empirical and statistical standards apply to a targeted metabolomics experiment as to any quantitative biological analysis. To obtain meaningful results, multiple biological replicates must be analyzed and subjected to appropriate statistical tests to ascertain the significant changes in metabolite concentrations between groups. In highly heterogeneous populations such as clinical studies in humans, appropriate cases and controls must be sought. The use of an appropriate number of replicates is advised as determined by a power calculation that integrates technical reproducibility, baseline biological variability, and expected differences. Corrections for multiple hypothesis testing may be required. Moreover, the appropriateness of a statistical test also depends on the type of experiment conducted. Both univariate and multivariate statistics are utilized in the field of metabolomics. Correction for multiple hypothesis testing is generally required.
Metabolite Extraction
Effective metabolite extraction from a given sample is essential to the success of any metabolomics experiment. In the case of untargeted metabolomics (UNIT 30.1) the extraction method must liberate a broad range of metabolites from the sample. By contrast, when performing a targeted metabolomics experiment the extraction protocol should focus on the efficient extraction of the metabolite class of interest, since only a subset of metabolites will be analyzed downstream. Therefore, the extraction method should be tailored to the physico-chemical properties and relative abundances of the specific subset of metabolites to be analyzed, while excluding components such as proteins and metabolites not intended for analysis. Consequentially, a range of extraction parameters must be considered. These include, but are not limited to, monophasic or biphasic, liquid-liquid, extraction, the nature, volume and ratio of the organic and aqueous solvents utilized, and the pH and temperature at which the extraction is carried out. Thus, metabolite extraction can significantly affect the total number and nature of metabolites extracted and the reproducibility of experimentation. Two protocols for the extraction of metabolites for targeted metabolomics are presented in the unit. The first focuses on the extraction of nitrogenous hydrophilic compounds, while the second outlines a biphasic methodology for the extraction of intermediary hydrophilic metabolites including the oxoacids.
Support Protocol 1
Sample Preparation for Targeted Metabolomics: Extraction of Hydrophilic Metabolites and Amino Acids from Blood Plasma, Cerebrospinal Fluid and Urine
Efficient and reproducible metabolite extraction and sample preparation are vital for ensuring reproducible and reliable results in any metabolomics experiment. The method should be carefully chosen to correspond to the groups of metabolites that are of interest. Due to the varying nature of the chemical and physical properties of metabolites, the types of solvents and protocols used affect the groups and concentrations of metabolites extracted. In addition, by reducing the steps in an extraction the metabolite yield can be increased and the inter-sample variation reduced.
The following protocol describes the sample preparation and metabolite extraction for a targeted metabolomics experiment analyzing hydrophilic metabolites including amino acids and associated amines from blood plasma, cerebrospinal fluid and urine through hydrophilic interaction liquid chromatography. The protocol is the extraction procedure for a single sample. All samples should be prepared and extracted in parallel.
Materials
Blood plasma, cerebrospinal fluid or urine experimental samples
Methanol, LCMS grade (VWR)
l-Phenylalanine-d8 (isotope labeled internal standard) (Cambridge Isotope Laboratories)
l-Valine-d8 (isotope labeled internal standard) (Sigma Aldrich Inc.)
Pipettors
Vortexer (VWR)
Refrigerated centrifuge (VWR)
Screw-top glass vials (Waters Corporation)
Deactivated low volume glass vial inserts (Waters Corporation)
Vial screw caps with bonded PTFE/Silicon Septa (Waters Corporation)
Microcentrifuge tubes (VWR)
Pipet tips (VWR)
Preparation of Internal Standard Stock Solutions
Prepare the reference standard stock solution (RS-SS) at 1000 μg/mL as follows: individually weigh out 5 mg of the L-Phenylalanine-d8 and the L-Valine-d8 into a 7 mL glass vial. Add 5 mL of HPLC-grade methanol to achieve a solution with a nominal concentration of 1000 μg/mL. The RS-SS can be stored for up to several weeks at −20°C. When required, allow the RS-SS to reach room temperature before use.
Prepare the internal standard spiking solution (IS-SS) at 0.2 μg/mL as follows: pipet 50 μL of the L-Phenylalanine-d8 RS-SS and 50 μL of the L-Valine-d8 into a 250 mL glass bottle containing 250 mL of methanol to yield a solution with a nominal concentration of 0.2 μg/mL.
Sample Preparation and Metabolite Extraction
-
3.
Allow frozen samples to equilibrate to 4°C and centrifuge for 10 minutes at 10,000 rcf to pellet insoluble materials
-
4.
Transfer the appropriate volume of sample into a 1.5 mL microcentrifuge tube, followed by the suitable amount of 0.2 μg/mL IS-SS as outlined in Table 30.2.1.
-
5.
Vortex the samples for 1 min.
-
6.
Centrifuge samples at 10000 rcf for 10 minutes at 4°C.
A protein pellet is formed at the bottom of the microcentrifuge tube and the hydrophilic metabolites are partitioned into the supernatant.
-
7.
Transfer 60 μL of supernatant into a labeled vial containing a deactivated low volume glass insert. Ensure the protein pellet is not disturbed when transferring the supernatant. Cap the vial securely.
-
8.
Proceed to Basic Protocol 1 for Targeted Metabolomic Analysis of Hydrophilic Metabolites and Amino Acids from Blood Plasma using LC-MS.
The procedure outlined in step 9 is only necessary if relative quantitation is insufficient and the goal of the metabolomics experiment is to obtain absolute quantitation of metabolites analyzed in each sample.
Table 30.2.1.
Internal Standard Spiking Solution (IS-SS) dilution factors for plasma, cerebrospinal fluid, or urine samples.
| Sample | Dilution Factor | Sample Volume | IS-SS |
|---|---|---|---|
| Blood Plasma | 10 | 10 μL | 90 μL |
| Cerebrospinal fluid | 5 | 20 μL | 80 μL |
| Urine | 5 | 20 μL | 80 μL |
Preparation of a Calibration Curve of Reference Metabolites for Absolute Quantitation
The example given in this section is of a calibration curve in pooled plasma. A calibration curve is achieved using stable isotope-labeled standards in pooled plasma at serial dilutions.
-
9.
Start with 1 mL of plasma and an isotope standard mix of labeled metabolite at a concentration of 1000 ng/μL. Follow the serial dilutions as described in Table 30.2.2.
Table 30.2.2.
Use of isotopically labeled metabolites for absolute quantitation. Serial dilutions of a 1000 ng/μL metabolite isotope mix in plasma as required to generate a calibration curve for absolute quantitation of metabolite concentrations in a plasma sample.
| Concentration (ng/μL) | 100 | 50 | 10 | 1 | 0.5 | 0.1 | 0.05 | 0 |
| Volume of Isotope Mix μL (1000 ng/μL) | 20 | - | - | - | - | - | - | - |
| Pooled Plasma μL | 180 | 50 | 90 | 180 | 50 | 180 | 50 | 100 |
| 100 ng/μL solution μL | - | 50 | 10 | - | - | - | - | - |
| 50 ng/μL solution μL | - | - | - | - | - | - | - | - |
| 10 ng/μL solution μL | - | - | - | 20 | - | - | - | - |
| 1 ng/μL solution μL | - | - | - | - | 50 | 20 | - | - |
| 0.1 ng/μL solution μL | - | - | - | - | - | - | 50 | - |
| Total Volume μL | 200 | 100 | 100 | 200 | 100 | 200 | 100 | 100 |
| Volume Remaining Post Dilution μL | 140 | 100 | 80 | 130 | 100 | 150 | 100 | 100 |
Basic Protocol 1
Targeted Metabolomic Analysis of Hydrophilic Metabolites and Amino Acids from Blood Plasma using LC-MS
The following protocol outlines the analytical conditions for a targeted metabolomics experiment analyzing hydrophilic metabolites including amino acids from blood plasma, cerebrospinal fluid and urine through hydrophilic interaction liquid chromatography. The parameters outlined are for a system comprising an Agilent 1200 liquid chromatography series pump (Agilent Technologies) coupled to a CTC-PAL HTS-xt autosampler (Leap Technologies). The mass spectrometry parameters discussed are for a 4000 QTRAP triple quadrupole mass spectrometer (AB SCIEX) coupled to an electrospray source (Turbo V).
Of note, when comparing multiple groups in a targeted metabolomics experiment the run order of samples must be randomized to ensure that comparison of metabolite concentrations in the subsequent analysis is not confounded by temporal drift in instrument performance. Therefore, differences in metabolite groups can be considered independent of any minor variations in the analytical process.
Materials
Extracted metabolite sample (Support Protocol 1)
Acetonitrile, HPLC-grade (Sigma Aldrich Inc.)
Water, HPLC-grade (VWR)
Formic acid, puriss p.a. 98% (Sigma Aldrich Inc.)
Agilent 1200 liquid chromatography series pump (Agilent Technologies)
CTC-PAL HTS-xt autosampler fitted with 100 μl syringe (Leap Technologies)
4000 QTRAP triple quadrupole mass spectrometer (AB SCIEX)
Analyst Software version 1.5.1 (AB SCIEX)
Atlantis HILIC Silica 2.1×150 mm, 3μm HPLC column (Waters Corporation)
-
1.
Prepare autosampler washes as follows: Solvent wash 1 (75% HPLC-grade water, 25% HPLC-grade acetonitrile). Solvent wash 2 (HPLC-grade acetonitrile). Set the autosampler parameters to the following.
Cycle: Analyst LC-Inj DLW Fast_Rev05_Leap
Syringe: 100 μl DLW
Loop Volume 1: 100 μl
Loop Volume 2: 100 μl
Actual Syringe: 100 μl
Injection Volume: 10 μl
Airgap Volume: 3 μl
Front Volume: 1 μl
Rear Volume: 1 μl
Filling Speed: 5 μl/sec
Pull Up Delay: 3 msec
Injection Speed: 10 μl/sec
Pre Inject Delay: 500 msec
Post Inject Delay: 500 msec
Needle Gap Valve Clean: 3 μm
Valve Clean Time Solvent 2: 3 sec
Valve Clean Time Solvent 1: 3 sec
Post Clean Time Solvent 1: 2 sec
-
2.
Perform the chromatography using an Agilent 1200 liquid chromatography series following the outlined parameters.
Column: Atlantis HILIC Silica 3μm 2.1 × 150 mm column
Prior to the use of a new HPLC column, equilibration is essential to ensure optimum chromatographic separation. In the case of the Atlantis HILIC Silica 3μm 2.1 × 150 mm column, pass 50 column volumes of 50% acetonitrile 50% water (100 min at 0.25 mL/min) followed by 20 column volumes of initial mobile phase conditions (40 min at 0.25 mL/min) through the column.
Mobile Phase A: 0.1% formic acid, 10 mM ammonium formate.
Mobile Phase B: 0.1% formic acid in acetonitrile.
Column Temperature: 30°C
Total Run Time: 32 min
Perform gradient elution at a flow rate of 250 μL/min using the chromatographic gradient outlined in Table 30.2.3.
-
3.
Perform mass spectrometric analysis using a 4000 QTRAP tandem mass spectrometer with a Turbo V electrospray source. Operate the instrument in positive MRM mode with the following mass spectrometric parameters:
Curtain Gas: 20 psi
Source Temperature: 450 °C
Ion Source Gas 1: 30 psi
Ion Source Gas 2: 40 psi
Interface Heater: ON
Collision Gas: High
IonSpray Voltage: 5000
Entrance Potential: 10
Resolution Quadrupole 1: Unit
Resolution Quadrupole 2: Unit
Multiple Reaction Monitoring (MRM) Window: 70 msec
Target Scan Time: 1 sec
Table 30.2.3.
Liquid chromatography gradient for hydrophilic metabolite elution from a HILIC column.
| Time (min) | Flow Rate (μl/min) | %B |
|---|---|---|
| 0.00 | 250 | 95 |
| 0.5 | 250 | 95 |
| 10.5 | 250 | 40 |
| 15.0 | 250 | 40 |
| 17.0 | 250 | 95 |
| 18.0 | 400 | 95 |
| 30.5 | 400 | 95 |
| 31.5 | 250 | 95 |
| 32.0 | 250 | 95 |
Set the multiple reaction monitoring (MRM) parameters for the metabolites of interest. Table 30.2.4 provides an example set of MRM parameters for a targeted metabolomics experiment.
Table 30.2.4.
Multiple Reaction Monitoring parameters for the analysis of hydrophilic metabolites.
| Metabolite | Q1 m/z | Q3 m/z | Collision Energy |
|---|---|---|---|
| L-Valine-d8 | 126.1 | 80 | 18 |
| L-Phenylalanine-d8 | 174.2 | 128 | 19 |
| Glycine | 76 | 48 | 10 |
| Alanine | 90 | 44 | 17 |
| Serine | 106 | 60 | 18 |
| Threonine | 120.1 | 74 | 18 |
| Methionine | 150.1 | 61 | 31 |
| Aspartate | 134 | 74 | 21 |
| Glutamate | 148.1 | 84 | 23 |
| Asparagine | 133.1 | 74 | 23 |
| Glutamine | 147.1 | 84 | 25 |
| Histidine | 156.1 | 110 | 21 |
| Arginine | 175.1 | 70 | 32 |
| Lysine | 147.1 | 84 | 25 |
| Valine | 118.1 | 72 | 18 |
| Leucine | 132.1 | 86.2 | 18 |
| Phenylalanine | 166.1 | 120.2 | 19 |
| Tyrosine | 182.5 | 136.1 | 19 |
| Tryptophan | 205.5 | 188.3 | 16 |
| Proline | 116.1 | 70 | 20 |
| Cis/trans hydroxyproline | 132.1 | 86.2 | 18 |
| Ornithine | 133.4 | 70 | 30 |
| Citrulline | 176 | 113.2 | 20 |
| Taurine | 126.2 | 44.1 | 31 |
| 5-Hydroxytryptophan | 221.1 | 204 | 18 |
| 5-HIAA | 192.3 | 146.2 | 18 |
| Serotonin | 177.1 | 160 | 18 |
| Cystamine | 153 | 108 | 16 |
| Cysteamine | 78 | 61 | 20 |
| Aminoisobutyric acid | 104.1 | 87 | 17 |
| Dimethylglycine | 104.1 | 58 | 20 |
| Homocysteine | 136 | 90 | 20 |
| Argininosuccinate | 291.1 | 70 | 54 |
| ADMA/SDMA | 203.1 | 70.3 | 40 |
| NMMA | 189.1 | 70 | 40 |
| Allantoin | 159 | 116 | 11 |
| GABA | 104.1 | 86 | 16 |
| Anthranilic Acid | 138 | 120 | 18 |
| Carnitine | 162.1 | 85 | 29 |
| Kynurenic Acid | 190.2 | 144 | 29 |
| 1-Methylhistamine | 126.1 | 68.1 | 29 |
| 5'-adenosylhomocysteine | 385.1 | 136.3 | 32 |
| Carnosine | 227.1 | 110 | 33 |
| Histamine | 112 | 95 | 26 |
| 3-Hydroxyanthranilic Acid | 154 | 136.2 | 18 |
| Methyl-hydroxyisobutyric Acid | 119.1 | 87 | 8 |
| N-Carbomoyl-Beta-Alanine | 133.1 | 115 | 12 |
| Cobalamin | 678.3 | 147.3 | 52 |
| Thiamine | 265.5 | 122.2 | 22 |
| Niacinamide | 123 | 80 | 30 |
| Betaine | 118.1 | 58 | 41 |
| Choline | 104.1 | 60.1 | 27 |
| Phosphocholine | 184.5 | 125 | 30 |
| Phosphoethanolamine | 142 | 44 | 22 |
| Alpha-glycerophosphocholine | 258.3 | 104.1 | 54 |
| Acetylcholine | 146.1 | 87 | 21 |
| Spermidine | 146.2 | 72 | 22 |
| Spermine | 203.2 | 129.3 | 20 |
| Creatine | 132.1 | 90 | 17 |
| Creatinine | 114.1 | 44 | 28 |
| Triiodothyronine | 651.9 | 606.1 | 35 |
| Thyroxine | 777.8 | 732 | 35 |
| Trimethylamine-N-Oxide | 76.1 | 42 | 50 |
| Glycerol | 93 | 57 | 12 |
| Glucose | 163.1 | 85 | 29 |
| Adenosine | 268.5 | 136.3 | 23 |
| Cytosine | 112 | 95.1 | 26 |
| Thymidine | 243.1 | 127 | 16 |
| Xanthosine | 285.1 | 153 | 18 |
| 2'-deoxyadenosine | 252.1 | 136.3 | 20 |
| 2'-deoxycytidine | 228.1 | 112.1 | 15 |
| cAMP | 330.3 | 136.2 | 30 |
| Isoleucine | 132.1 | 86.2 | 18 |
| Xanthine | 153 | 135 | 9 |
| 3-Hydroxykynurenine | 225 | 208 | 30 |
| Xantherenate | 206 | 160 | 30 |
| Kynurenine | 209 | 146 | 30 |
Support Protocol 2
Sample Preparation for Targeted Metabolomics: Extraction of Intermediary Metabolites and from Blood Plasma
The following protocol describes the sample preparation and metabolite extraction for a targeted metabolomics experiment analyzing intermediary metabolites including TCA cycle metabolites and additional oxoacids from blood plasma through polar reversed phase chromatography. The protocol is the extraction procedure for a single sample. All samples should be prepared and extracted in parallel.
Materials
Blood plasma experimental samples
Methanol, LCMS-grade (VWR)
Chloroform, LCMS-grade (Sigma Aldrich Inc.)
Water, LCMS-grade (VWR)
L-Phenylalanine-d8 (isotope labeled internal standard) (Cambridge Isotope Laboratories)
Pipettors (Gilson)
Vortexer (VWR)
Refrigerated Centrifuge (VWR)
SpeedVac Concentrator (Thermo Scientific)
Screw-top glass vials (Waters Corporation)
Deactivated low volume glass vial inserts (Waters Corporation)
Vial screw caps with bonded PTFE/Silicon Septa (Waters Corporation)
Microcentrifuge tubes (VWR)
Pipet tips (VWR)
Preparation of Internal Standard Stock Solutions
-
1.
Prepare a standard stock solution (RS-SS) at a concentration of 1000 μg/mL as follows: individually weigh out 5 mg of the L-Phenylalanine-d8 into a 7 mL glass vial. Add 5 mL of HPLC-grade methanol to achieve a solution with a nominal concentration of 1000 μg/mL. The RS-SS can be stored [*Author: for up to how long?] at −20°C. Allow the RS-SS to reach room temperature before use.
-
2.
Prepare an internal standard spiking solution (IS-SS) at a concentration of 0.2 μg/mL as follows: pipet 50 μL of the L-Phenylalanine-d8 RS-SS into a 250 mL glass bottle containing 250 mL of methanol to yield a solution with a nominal concentration of 0.2 μg/mL.
Sample Preparation and Metabolite Extraction
-
3.
Allow frozen plasma samples to equilibrate to ambient temperature.
-
4.
Transfer 200 μL of plasma to a 1.5 ml microcentrifuge tube.
-
5.
Add 300 μL of methanol:chloroform (2:1 v:v) to the plasma and vortex mix the samples for 1 min.
-
6.
Add 200 μl chloroform and 200 μl water to the sample and vortex mix for 1 min.
-
7.
Centrifuge the samples at 20,000 rcf for 20 min at 4°C
Centrifugation of the plasma with the addition of the organic chloroform and water results in the extraction of the metabolites from the plasma and formation of a biphasic solution with a protein pellet suspended between the phases. Metabolites are partitioned into an organic phase, containing hydrophobic metabolites, and the aqueous phase, containing hydrophilic metabolites.
-
8.
Remove the aqueous phase (top phase) containing the hydrophilic metabolites and transfer to a microcentrifuge tube. Perforate the lid of the tube.
-
9.
Evaporate the aqueous phase using a SpeedVac concentrator for 2 hours at a vacuum of 1 torr and temperature of 45°C.
The organic phase can also be removed, transferred to a microcentrifuge tube, dried using a SpeedVac concentrator and stored at −80°C for later analysis of hydrophobic metabolites, if desired.
-
10.
Reconstitute the metabolites in 50 μl IS-SS. Vortex mix for 2 min and briefly centrifuge.
-
11.
Transfer the reconstituted metabolites into a labeled vial containing a deactivated low volume glass insert. Cap the vial securely.
The sample is now ready for targeted metabolomic analysis (Basic Protocol 2).
Basic Protocol 2
Targeted Metabolomic Analysis of Intermediary Metabolites from Blood Plasma using LC-MS
The following protocol outlines the analytical conditions for a targeted metabolomics experiment analyzing intermediary metabolites including oxoacids from blood plasma through polar reversed phase chromatography. The parameters outlined are for a system comprising an Agilent 1200 liquid chromatography series pump (Agilent Technologies) coupled to a CTC-PAL HTS-xt autosampler (Leap Technologies). The mass spectrometry parameters discussed are for a 4000 QTRAP triple quadrupole mass spectrometer (AB SCIEX) coupled to an electrospray source (Turbo V).
Materials
Extracted metabolite sample (Support Protocol 2)
Acetonitrile, HPLC-grade (Sigma Aldrich Inc.)
Water, HPLC-grade (VWR)
Ammonium acetate, puriss p.a. 99% (Sigma Aldrich Inc.)
Agilent 1200 liquid chromatography series pump (Agilent Technologies)
CTC-PAL HTS-xt autosampler fitted with 100 μl syringe (Leap Technologies)
API 4000 QTRAP triple quadrupole mass spectrometer (Applied Biosystems/Sciex)
Analyst Software version 1.5.1 (Applied Biosystems/Sciex)
Synergi 4u Polar-RP 80A 4μm 4.6 × 50 mm column (Phenomenex)
-
1.
Prepare autosampler washes as follows: Solvent wash 1 (75% HPLC-grade water, 25% HPLC-grade acetonitrile). Solvent wash 2 (HPLC-grade acetonitrile). Set the autosampler parameters to the following.
Cycle: Analyst LC-Inj DLW Fast_Rev05_Leap
Syringe: 100 μl DLW
Loop Volume 1: 100 μl
Loop Volume 2: 100 μl
Actual Syringe: 100 μl
Injection Volume: 10 μl
Airgap Volume: 3 μl
Front Volume: 1 μl
Rear Volume: 1 μl
Filling Speed: 5 μl/sec
Pull Up Delay: 3 msec
Injection Speed: 10 μl/sec
Pre Inject Delay: 500 msec
Post Inject Delay: 500 msec
Needle Gap Valve Clean: 3 μm
Valve Clean Time Solvent 2: 3 sec
Valve Clean Time Solvent 1: 3 sec
Post Clean Time Solvent 1: 2 sec
-
2.
Perform the chromatography using an Agililent 1200 liquid chromatography series following the outlined parameters.
-
Column: Synergi 4u Polar-RP 80A 4μm 4.6 × 50 mm column
Prior to the use of a new HPLC column, equilibration is essential to ensure optimum chromatographic separation. Pass 50 column volumes of 50% acetonitrile 50% water (100 min at 0.25 mL/min) followed by 20 column volumes of initial mobile phase conditions (40 min at 0.25 mL/min) through the column.
Mobile Phase A: 95% water, 5% acetonitrile, 5 mM ammonium acetate
Mobile Phase B: 95% acetonitrile, 5% water, 5 mM ammonium acetate
Column Temperature: 30°C
Total Run Time: 15 min
Perform gradient elution at a flow rate of 250 μL/min using the chromatographic gradient outlined in Table 30.2.5.
-
-
3.Perform mass spectrometric analysis using an API4000 QTrap tandem mass spectrometer with a Turbo V electrospray source. Operate the instrument in negative MRM mode with the following mass spectrometric parameters:
- Curtain Gas: 20 psi
- Source Temperature: 450°C
- Ion Source Gas 1: 50 psi
- Ion Source Gas 2: 60 psi
- Interface Heater: ON
- Collision Gas: High
- IonSpray Voltage: −4500
- Entrance Potential: −10
- Resolution Quadrupole 1: Unit
- Resolution Quadrupole 2: Unit
- Multiple Reaction Monitoring (MRM) Window: 70 msec
- Target Scan Time: 1 sec
Set the multiple reaction monitoring (MRM) parameters for the metabolites of interest. Table 30.2.6 provides an example set of MRM parameters for a targeted metabolomics experiment analyzing intermediary metabolites.
Table 30.2.5.
Liquid chromatography gradient for intermediary metabolite elution from a Synergi Polar Reversed Phase column.
| Time (min) | Flow Rate (μl/min) | B% |
|---|---|---|
| 0 | 250 | 5 |
| 0.5 | 250 | 5 |
| 5 | 250 | 7 |
| 6 | 250 | 100 |
| 8 | 250 | 100 |
| 8.5 | 250 | 5 |
| 15 | 250 | 5 |
Table 30.2.6.
Multiple Reaction Monitoring parameters for the analysis of intermediary metabolites.
| Metabolite | Q1 m/z | Q3 m/z | Collision Energy |
|---|---|---|---|
| L-Phenylalanine-d8 | 172.2 | 154 | −20 |
| Glyceraldehyde | 88.8 | 59 | −10 |
| Lactic Acid | 89.2 | 43.1 | −20 |
| Acetoacetate | 101.1 | 57.1 | −15 |
| Succinate | 117.1 | 73 | −20 |
| Nicotinic Acid | 122.1 | 78.1 | −15 |
| Oxaloacetate | 130.9 | 87 | −10 |
| Malic Acid | 133 | 114.9 | −20 |
| Adenine | 134 | 107.1 | −18 |
| Hypoxanthine | 135 | 92.1 | −23 |
| α-Ketoglutarate | 144.8 | 101 | −21 |
| Orotic Acid | 154.9 | 110.6 | −15 |
| 2-Aminoadipic Acid | 160 | 116 | −18 |
| Quinollinic Acid | 166 | 122 | −13 |
| Homogentistic Acid | 166.9 | 79 | −23 |
| Uric Acid | 167 | 124 | −22 |
| Dihydroxyacetone Phosphate | 169 | 97 | −15 |
| Glycerol-3-Phosphate | 171 | 79 | −22 |
| Hippuric Acid | 177.9 | 133.9 | −16 |
| Fructose | 179 | 89 | −15 |
| Inositol | 179 | 161 | −18 |
| Hydroxyphenylpyruvate | 179 | 107.1 | −12 |
| Sorbitol | 181 | 71 | −25 |
| 4-Hydroxy-3-methoxyphenylglycol | 182.9 | 150.1 | −21 |
| 3-Phosphoglyceric Acid | 185.1 | 97.2 | −22 |
| 5-Hydroxyindole-3-acetic Acid | 190 | 146 | −16 |
| Citric Acid | 191 | 87 | −25 |
| Pantothenic Acid | 218.2 | 146.1 | −22 |
| Inosine | 267 | 135 | −30 |
| Geranyl Pyrophosphate | 313 | 79.1 | −37 |
| UMP | 323.1 | 97.1 | −60 |
| Lactose | 341.1 | 161.1 | −11 |
| cGMP | 343.8 | 150.1 | −34 |
| Glychochenodeoxycholic Acid | 448.4 | 74 | −50 |
| Glycocholic Acid | 464.4 | 74 | −50 |
| Taurochenodeoxycholic Acid | 498.7 | 79.9 | −80 |
| ATP | 506 | 159 | −45 |
| Taurocholic Acid | 514.4 | 123.8 | −85 |
| GTP | 552.2 | 158.9 | −30 |
| UDP-Glucuronic Acid | 579.2 | 403.3 | −30 |
| Billirubin | 583.2 | 285 | −36 |
| 4-Hydroxybenzoate | 137 | 93 | −20 |
| Fumarate | 115 | 71 | −15 |
| Propanoic Acid | 73 | 55 | −20 |
| Malonate | 103 | 59 | −15 |
| Mevalonate | 147.1 | 59 | −19 |
| Oxalic Acid | 89 | 61 | −18 |
| XMP | 363 | 79 | −60 |
| Aconitate | 173 | 129 | −8 |
| Phosphotyrosine | 260 | 79 | −30 |
| Pyruvate | 87 | 43 | −12 |
| NADH | 664.1 | 408 | −40 |
Basic Protocol 3
Analysis of Targeted Metabolomic LC-MS Data
Due to the specificity of the data collected in a targeted metabolomics experiment, data processing and analysis tends to be less labor intensive when compared to untargeted metabolomics (UNIT 30.1). Further, as each metabolite is defined by the MRM mass spectrometry method, the need to identify unknown metabolites, as occurs with untargeted analysis, is negated. The protocol described below outlines a method for the analysis of data generated in either of the LC-MS MRM experiments outlined above. With this methodology, a peak integration method is built in MultiQuant, a program available from AB SCIEX, for the LC-MS data generated. The data is then processed in MultiQuant, during which metabolite chromatogram peaks are automatically integrated by the MultiQuant software based on the manually generated integration method. Extracted ion chromatograms corresponding to individual metabolites in individual samples can then be visually inspected to ensure accurate peak integration.
Materials
Computer
Raw data files from LC-MS MRM experiment
Analyst Software version 1.5.1 (Applied Biosystems/Sciex)
MultiQuant Software version 2.0.2 (AB SCIEX)
-
1.
Open Analyst version 1.5.1 and then open MultiQuant version 2.0.2.
-
2.
Select the Quantitate New Data option. MultiQuant will open the Create Results Table wizard. Select the raw data to be analyzed from the available data.
-
3.
Select the Create New Method option and give the method an appropriate name.
-
4.
Select a representative sample from the list to act as a template for the analysis method.
-
5.
[*Author: This isn't really an instruction. Should it tell the reader to enter the parameters of the LC-MS method and then MultiQuant will define the components?] MultiQuant will automatically define the components for analysis based on the MRM parameters of the LC-MS method.
-
6.
MultiQuant will generate a list of the components and their extracted ion chromatograms. Scroll through the components and define the integration parameters that best fit the peak for each metabolite.
A list of integration parameters is provided on Table 30.2.7. The parameters will vary depending on the peak analyzed and care should be taken when defining these parameters as they will influence the integrated area for each peak and therefore the reliability and reproducibility of the results for the experiment.
-
8.
MultiQuant will then process and integrate the component peaks and provide integrated extracted ion chromatograms for each metabolite component in each sample. Visually inspect the integrated chromatograms to ensure the integration method has successfully and reproducibly integrated the metabolite peaks between samples. A list of results in the form of the integrated peak areas is generated; use this list to perform the required statistical analyses for the experiment.
Depending on the type of experiment conducted the results generated can be normalized to an internal standard, normalized to a standardized sample (such as pooled plasma), used for absolute quantitation with the aid of the calibration curve outlined in Support Protocol 1, or applied to a relative quantitation experiment. Univariate statistical analysis can be performed using a Student's T-Test to compare individual metabolite concentrations or integrated peak areas between groups or ANOVA between multiple groups. More complex multivariate analyses such as Principal Component Analysis (PCA) and Partial Least Squares-Disciminant Analysis (PLS-DA) can also be used to define separation between groups in binary group or multi-group analyses. A good place to start in multivariate analysis, using an excellent and freely available program, is Metaboanalyst (http://www.metaboanalyst.ca/MetaboAnalyst/faces/Home.jsp) (Xia et al., 2009).
Table 30.2.7.
Integration parameters for the analysis of Liquid Chromatography-Mass Spectrometry Multiple Reaction Monitoring data (using Multiquant 2.0.2 software). Care should be taken when altering the Gaussian Smooth Width (the smoothing between data points in the peak), the noise percentage and the peak splitting in the integration parameters.
| Integration Parameter | Setting |
|---|---|
| Gaussian Smooth Width | Recommended 0–1 points |
| Expected Retention Time (min) | Depends on MRM parameter, varies by metabolite |
| Retention Time Half Window | Varies by metabolite |
| Report Largest Peak | Dependent on the extracted ion chromatogram for metabolite |
| Minimum Peak Width | Depends on the quality of the LC-MS data and noise in the extracted ion chromatogram |
| Min Peak Height | Depends on the quality of the LC-MS data and noise in the extracted ion chromatogram |
| Noise Percentage | Recommended 20–80% to obtain optimum integration of peak |
| Peak Splitting | Alter to integrate peaks that are not fully resolved |
| Regression Parameter | Area |
| Regression Fit | Linear |
| Weighting | None to begin with but can be optimized |
Commentary
Background Information
Metabolomics arose in the 1990s following the change in thought first pioneered by functional genomics that offered a more global approach to defining and understanding biology. The omic approaches provided for the investigation of interactions across the genetic, transcriptional, protein and metabolite strata, offering an alternative to other more reductionist molecular biology techniques and leading to the generation of the genomic, transcriptomic, proteomic and metabolomic fields. Genomics aims to determine the entire genetic DNA sequence of organisms accounting for the often indefinite line between genetic and non-genetic regions of DNA, whereas transcriptomics performs functional analysis at the gene expression level by examining the expression level of mRNAs in a population of cells or tissue using high-throughput techniques often based on microarray technology. Proteomics functions at the level of protein translation and aims to identify and localize proteins and determine protein pathways within an organelle, cell, or organism (Godovac-Zimmermann and Brown, 2001; Panisko et al., 2002). Metabolomics functions to measure the small molecules (metabolites) present within a cell, tissue or organism, collectively described as the metabolome. Integrated analysis of the omics approaches carried out in parallel, and often accompanied by mathematical modeling has come to be known as Systems Biology.
Metabolomics sits at a key point in the interpretation of the biological system in question since the metabolome is the terminal downstream product of the genome. Metabolite concentrations are determined by the activity of a plethora of enzymes (which constitute part of the proteome), with the distinct components of the metabolome being the result of complex interactions between genome, transcriptome, proteome and the environment. Add to this the complexity of the interactions of metabolites themselves, where the majority of individual metabolites are involved with multiple reactions that require more than a single substrate and generate more than a single product, and it becomes clear why metabolic networks show a high degree of tightly regulated connectivity. As a result, minute perturbations or alterations in the genome, transcriptome or proteome can manifest as significant changes in metabolite concentrations. In 1971, Arthur Robinson and Linus Pauling conceived the core idea that information-rich data that reflects the functional status of a complex biological system resides in the quantitative and qualitative pattern of metabolites in body fluids (Pauling et al., 1971). The measurement of the individual components of the highly integrated metabolic network provides invaluable information about the biological function of a cell, tissue or organism in response to genetic, physiological or environmental stimuli that aid mechanistic evaluation or determination of phenotype.
In contrast to genetics and transcriptomics where relatively similar comprehensive high-throughput techniques are being rapidly developed, metabolomics has no single technique allowing for comprehensive profiling of the entire metabolome. As alluded to in the Unit Introduction, this comes about as a result of the intrinsic variation in the physico-chemical properties of different metabolite classes from small polar volatiles through to large hydrophobic lipids. Alongside this are estimates of the potential size of the metabolome ranging from ~600 metabolites in S. cerevisiae (Forster et al., 2003) to ~200,000 metabolites in plants (Fiehn, 2001). Another factor contributing to the lack of a single comprehensive technique for metabolomics is the dynamic range in metabolite concentrations that span across large orders of magnitude from femtomolar to millimolar. As a result, a range of techniques have been applied to metabolomic research in order to increase the analytical coverage of the metabolome, most of which are based on one of two distinct technologies, NMR spectroscopy and mass spectrometry. An example of some of the approaches used within metabolomics to identify the changes in the concentrations and fluxes of endogenous metabolites include 1H-NMR spectroscopy, 13C-NMR spectroscopy, 31P-NMR spectroscopy, magic angle spinning NMR spectroscopy, Fourier transform ion cyclotron resonance mass spectroscopy, GC-MS, GC-FID, DI-MS and LC-MS.
Due to the inherent complexity of lipid classes and their distinct chemical properties, focused approaches targeted to the study of these constituents of the metabolome have been developed. Although the lipidome is a sub-fraction of the metabolome, lipidomics has been separately defined as “the full characterization of lipid molecular species and of their biological roles with respect to expression of proteins involved in lipid metabolism and function, including gene regulation” (Spencer et al., 2003). Interest in lipidomics has also been driven by the widening knowledge of the role of lipid species in the cell. In the past the majority of lipids were considered to be either membrane components or an energy store. However, these molecules are now known to have diverse roles in transcriptional and translational control, cellular signaling, cell-cell interactions, and as indicators of changes to the environs of a cell or organism over time. The reader is encouraged to consult a number of reviews detailing the emergence of lipidomics (Spencer et al., 2003; Wenk, 2005; Roberts et al., 2008).
Partly as a consequence of the essential characteristics of each of the techniques employed within metabolomics, research strategies have been divided into two distinct approaches: untargeted and targeted metabolomics. These two distinct approaches are complementary and investigation utilizing both is common. Untargeted metabolomics (UNIT 30.1) is the comprehensive analysis of all the measurable analytes in a sample. This type of analysis is inclusive of chemical unknowns and so provides the opportunity for novel metabolite species, pathway and target discovery. The only limitations to metabolome coverage arise from the amount of sample biomass available, intrinsic restrictions of metabolite extraction, and the sensitivity and specificity of the analytical techniques. A result of the comprehensive coverage offered by untargeted metabolomics is the generation of large raw datasets that require complex chemometric techniques to analyze. Very often, data reduction methodologies, such as multivariate analysis using PCA or PLS-DA, are employed. Some of the major constraints of untargeted metabolomics include the reliance on the specificity and sensitivity of the analytical technique utilized, the development of protocols and time required to carry out the data analysis of the considerable datasets generated, the analytical challenges faced when attempting to identify and characterize chemical unknowns measured in the untargeted profiling, and a strong bias of detection towards higher concentration and abundance metabolites.
Targeted metabolomics aims to measure a pre-defined group of biochemically characterized and interpreted metabolites (a subset of the metabolome). This reduced coverage of the metabolome means that targeted metabolomics is reliant on a priori knowledge of metabolites and their biochemical pathways, which hinders the discovery of novel metabolic perturbations. On the other hand, through the use of isotopically labeled standards, targeted metabolomics can be made quantitative, building on the extensive understanding of a vast array of metabolic processes, enzyme kinetics and established pathways to garner a clearer representation of the physiological processes at work. This clear definition of the species measured reduces the likelihood of analytical artifacts progressed to ensuing experiments and data analysis. Specialized metabolite extraction techniques can also be exploited in targeted metabolomics, which focus on isolating the metabolites of interest, thus reducing interference by high abundance species. For a more comprehensive review of both targeted and untargeted metabolomics and the relevant techniques, see (Dunn et al., 2011; Wishart, 2011; Dudley, 2010; Griffin, 2006) Metabolomics has been used to investigate hypotheses spanning a wide range of biological models and a plethora of disciplines, including neuroscience (Griffin et al., 2004; Han, 2002), cancer biology (Odunsi et al., 2005; Griffiths et al., 2002), microbiology (Oursel et al., 2007) cellular lipid signalling (Wakelam et al., 2007) and the metabolic syndrome (Roberts et al., 2011; Wang et al., 2005; Festa et al., 2005). However, in the interest of relevance to this unit, the following applications primarily relate to instances in which LC-MS-based targeted metabolomics has been utilized. Recent studies using targeted LC-MS profiling of blood plasma have begun to yield potential new disease markers, including a study of individuals enrolled in the Framingham Heart Study who were followed for 12 years. From a screen of amino acids and amines, investigators identified [*Author: should a term be inserted here…something like “elevated plasma levels of” ?] the branched chain and aromatic amino acids isoleucine, leucine, valine, tyrosine and phenylalanine as highly significantly associated with future onset of diabetes (Wang et al., 2011). In addition, targeted LC-MS analysis of blood plasma from subjects undergoing acute exercise testing, taking part in marathon running and 302 subjects enrolled in a longitudinal exercise study provided metabolic signatures of exercise performance, the susceptibility to cardiovascular disease, and highlighted biochemical pathways of import in the benefits of exercise (Lewis et al., 2010). Targeted metabolomics has also been used to profile amino acids and acyl carnitines from the serum of 74 obese and 67 lean individuals. A metabolic signature of significantly increased branched chain amino acids and C3, C5, C6 and C8:1 acylcarnitines was characteristic of obese individuals (Newgard et al., 2009). Finally, the targeted approach has also been applied to insulin resistance studies in animal models. An et al. (2004) used metabolic profiling of acylcarnitine species by mass spectrometry to identify an increase in the concentration of the lipid-derived β-hydroxybutyrate in muscle of mice overexpressing hepatic malonyl-CoA decarboxylase and exhibiting improved insulin resistance.
The integration of genomics, transcriptomics, proteomics and metabolomics is a primary goal of systems biology. In particular, recent progress has been made towards understanding the genetic underpinnings of human metabolite levels. Geieger and colleagues (Gieger et al., 2008) integrated genome-wide analyses of common human variants with serum measurements of 363 metabolites in 284 male participants of the Cooperative Research in the Region of Augsburg (KORA) study. Even in this small proof-of-concept study, the investigators identified genetic variants in genes encoding for enzymes (FADS1, LIPC, SCAD, MCAD) that were strongly associated with the levels of specific lipid metabolites. The alignment of genetic variants, metabolite markers and pathological outcomes highlights pathways for further interrogation.
Critical Parameters and Troubleshooting
The quality of the samples obtained is paramount to the success of any metabolomics experiment before profiling is even attempted. Biological samples should be collected rapidly and in parallel to reduce variation in metabolite concentrations. Moreover, if the metabolites are not extracted immediately the samples should be stored at −80°C to prevent sample and metabolite degradation.
Amongst the most essential of parameters to be considered is the appropriate methodology for reliable and robust extraction of the metabolite classes of interest. Protocols outlined in this unit focus on the extraction of nitrogenous hydrophilic metabolites such as amino acids and intermediary metabolites such as the oxoacids. To optimize the desired extraction conditions for the metabolite classes of interest it is recommended to consult the many examples in the literature where extraction of varying metabolite classes from differing biological samples have been reviewed (Masson et al., 2010; Sellick et al., 2010; Yu Lin et al., 2007; Bligh and Dyer, 1959). Once the metabolites have been extracted and sample preparation is underway, attempts should be made to minimize the time extracted samples are left at room temperature since the solvents used are volatile and will begin to evaporate at room temperature, altering the concentration of the metabolites within the sample and introducing variation to the analysis.
Ionization of certain metabolites can be complex, especially when their chemical characteristics tend to lack readily ionizable functional groups. Attempting ionization in both negative and positive ionization mode may be of limited success in overcoming this problem. If poor ionization of metabolites in general is observed, this may be as a result of a number of complications. Firstly, ensure that the purity of solvents used for sample preparation, metabolite extraction and the LC-MS experiment is LC-MS grade or higher since contaminants, such as plasticizers and surfactants, are very often readily ionizable and can compete for charge, causing ion suppression of the metabolites and increasing the variation of ionization between samples. Ensure clean glassware is used at all times. Secondly, poor ionization may be a result of a high order of complexity of a sample resulting in extensive ion suppression; this can be overcome through fractionation of the sample, for example through the use of an upfront HPLC fractionation prior to LC-MS analysis, or by improving the chromatography, if necessary, through the use of alternative solvent systems and chromatographic columns. Of note, however, increased fractionation leads to longer data acquisition time, as well as increased variation due to additional sample handling. Finally, a lack of sensitivity may arise due to the low relative abundance of the metabolites of interest in a given sample, in which case a greater starting biomass should be injected onto the column. Failing this, an upfront concentration method such as solid phase extraction or thin layer chromatography can be employed to increase the abundance of metabolites to a detectable level; although as noted previously care should be taken to ensure reproducibility between samples in this process. These suggestions assume that the LC-MS is functioning within normal operating parameters and effort should be made prior to any experiment to ensure that this is the case.
It should be expected that a certain degree of drift in retention time and column performance will happen irrespective of the conditions used in an LC-MS experiment. To minimize the effect of this drift over time, it is important to ensure that the chromatographic columns are suitably and efficiently equilibrated prior to performing the experimental run. As differences in performance are even notable between the same type of columns produced by the same manufacturer in the same batch, where possible, all samples from a single experiment should be analyzed on the same column. In addition, it is recommended that fresh solvents are prepared prior to each LC-MS experiment. As many of these solvents are volatile at room temperature, evaporation will change the concentrations and pH of the solvent systems (especially those containing ammonium formate or ammonium acetate as given in the example protocols in this unit) resulting in drifting retention times for analytes. As mentioned in the strategic planning section of this unit, by randomizing the sample order for analysis and conducting all analyses on the same day, any observed differences in metabolite concentrations occurring as a result of this drift rather than from true differences between sample groups will be accounted for and not result in artefacts in the statistical analyses. Furthermore, normalization to the internal standards or standardized samples (such as pooled plasma) analyzed intermittently throughout the experiment can account for any drift that may occur in instrument performance.
The principal confounders to any targeted metabolomics experiment center on platform reproducibility. Contributions from three sources of variance should be considered before undertaking a metabolomics experiment: LC-MS system variance, variance introduced during sample preparation and the intra- and inter-individual biological variance. These confounders can be evaluated using a number of approaches.
As an example, to evaluate the contributions of biological and technical variance in a study, a test set of plasma samples can be collected prior and post manipulation, metabolites extracted, reconstituted and then spiked with isotope-labelled internal standard prior to LC-MS analysis (Fig. 30.2.3). In parallel, the variance in sample preparation and LC-MS analysis can be assessed by taking an aliquot of each of the test set plasma samples and pooling this to create a uniform starting solution. From this pooled solution a subset can be extracted, reconstituted and then analyzed using the LC-MS. A second subset of this pooled solution can be extracted, reconstituted and then re-pooled, yielding a single sample which can then be analyzed in multiple replicates using LC-MS to assess the contribution of the LC-MS profiling to overall variance. From these experiments coefficients of variation (CV; 100 × standard deviation/mean value of data set) can be calculated for biological variance, sample preparation variance, and LC/MS/MS variance.
Figure 30.2.3.
An example of an experiment to assess biological and technical variance (left of diagram), variance in sample preparation (centre of diagram) and contribution of the LC-MS profiling to overall variance (left of diagram).
Interestingly, when assessing the CVs for multiple mass spec analyses, the CVs for high abundance metabolites are lower than that of lower abundance metabolites. This phenomenon highlights the importance of a sensitive metabolite profiling platform (Fig. 30.2.4).
Figure 30.2.4.
Graph of coefficients of variance (CV) versus peak integrated area for metabolites analyzed from pooled plasma using Basic Protocol 1. As the integrated peak area increases the CV decreases.
Anticipated Results
The protocols outlined in this unit provide the basis for implementing targeted metabolomic experiments to define metabolic perturbations in disease states or phenotypes from physiological stimuli or genetic models. The groups and biochemical characteristics of the metabolites of interest will vary depending on the type of experiment performed. Therefore, the metabolites to be assayed in the targeted approaches used will be informed by the a priori understanding of the studies themselves and the biological systems employed. The most exigent undertaking in preparing any such experiment is likely to be defining the mass spectrometric and MRM parameters for new metabolites of interest. Determining the ideal transitions, chromatography, retention times, ionization characteristics, and dynamic/linear range for metabolites and chemical classes of interest will be laborious and taxing but ultimately provide invaluable information about specific metabolic pathways in the conditions being studied.
Time Considerations
The demand on time represented by each of the protocols outlined in this unit is dependent on several considerations, including the type of sample analyzed, the number of independent groups in the analysis and the number of technical and biological replicates assessed. A number of estimates will be provided for elements of each protocol.
The times required for extraction of metabolites in the two Support Protocols are almost wholly determined by the number of samples to be analyzed. However, the extraction of intermediary metabolites necessitates a longer period due to the 2 hr SpeedVac concentration step. For both the targeted LC-MS metabolomic analysis of hydrophilic metabolites and the intermediary metabolites, the time required to prepare the solvents, equilibrate the HPLC pumps, LC columns and mass spectrometer typically runs to 3–4 hours. For the LC-MS analysis protocols the time required is primarily dependent on the length of the chromatographic gradient. The mass spectrometry method for LC-MS analysis of hydrophilic metabolites (Basic Protocol 1) is 32 min per sample and the run time per sample for the LC-MS analysis of intermediary metabolites (Basic Protocol 2) is 15 min. Again, the time entailed for data analysis (Basic Protocol 3) of the LC-MS experiments is highly variable and depends on both the number of samples, components (metabolites) in the method and the quality of the data obtained. Individual inspection of each integrated chromatogram will be the rate-limiting step in any data analysis. As a general consideration, the period of time required for data analysis is often equivalent to the LC-MS run time.
Literature Cited
- An J, Muoio DM, Shiota M, Fujimoto Y, Cline GW, Shulman GI, Koves TR, Stevens R, Millington D, Newgard CB. Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nature Medicine. 2004;10:268–274. doi: 10.1038/nm995. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–7. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Dudley E, Yousef M, Wang Y, Griffiths WJ. Targeted metabolomics and mass spectrometry. Adv Protein Chem Struct Biol. 2010;80:45–83. doi: 10.1016/B978-0-12-381264-3.00002-3. [DOI] [PubMed] [Google Scholar]
- Dunn WB, Broadhurst DI, Atherton HJ, Goodacre R, Griffin JL. Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem Soc Rev. 2011;40:387–426. doi: 10.1039/b906712b. [DOI] [PubMed] [Google Scholar]
- Festa A, Williams K, Hanley AJ, Otvos JD, Goff DC, Wagenknecht LE, Haffner SM. Nuclear magnetic resonance lipoprotein abnormalities in prediabetic subjects in the Insulin Resistance Atherosclerosis Study. Circulation. 2005;111:3465–3472. doi: 10.1161/CIRCULATIONAHA.104.512079. [DOI] [PubMed] [Google Scholar]
- Fiehn O. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comp. Funct. Genomics. 2001;2:155–168. doi: 10.1002/cfg.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster J, Famili I, Fu P, Palsson BO, Nielsen J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003;13:244–253. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieger C, Geistlinger L, Altmaier E, Hrabé de Angelis M, Kronenberg F, Meitinger T, Mewes HW, Wichmann H-E, Weinberger KM, Adamski J, Illig T, Suhre K. Genetics Meets Metabolomics: A Genome-Wide Association Study of Metabolite Profiles in Human Serum. PLOS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godovac-Zimmermann J, Brown LR. Perspectives for mass spectrometry and functional proteomics. Mass Spectrom. Rev. 2001;20:1–57. doi: 10.1002/1098-2787(2001)20:1<1::AID-MAS1001>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Goodacre R. Metabolic Profiling: Its role in Biomarker Discovery and Gene Function Analysis. Kluwer Academic Publishers; London: 2003. [Google Scholar]
- Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22:245–252. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Griffin JL. The Cinderella story of metabolic profiling: does metabolomics get to go to the functional genomics ball? Philos Trans R Soc Lond B Biol Sci. 2006;361:147–61. doi: 10.1098/rstb.2005.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JL, Cemal CK, Pook MA. Defining a metabolic phenotype in the brain of a transgenic mouse model of spinocerebellar ataxia 3. Physiol. Genomics. 2004;16:334–340. doi: 10.1152/physiolgenomics.00149.2003. [DOI] [PubMed] [Google Scholar]
- Griffin JL, Nicholls AW, Keun HC, Mortishire-Smith RJ, Nicholson JK, Kuehn T. Metabolic profiling of rodent biological fluids via 1H NMR spectroscopy using a 1 mm microlitre probe. Analyst. 2002;127:582–584. doi: 10.1039/b201207c. [DOI] [PubMed] [Google Scholar]
- Giffiths JR, McSheehy PM, Robinson SP, Troy H, Chung YL, Leek RD, Williams KJ, Stratford IJ, Harris AL, Stubbs M. Metabolic changes detected by in vivo magnetic resonance studies of HEPA-1 wild-type tumours deficient in hypoxia-inducible factor-1 beta(HIF-1beta) evidence of an anabolic role for the HIF-1 pathway. Cancer Res. 2002;62:688–95. [PubMed] [Google Scholar]
- Han X, Holtzman D,M, McKeel DW, Jr, Kelley J, Morris JC. Substantial sulfatide deficiency and ceramide elevation in very early Alzheimer's disease: Potential role in disease pathogenesis. J. Neurochem. 2002;82:809–818. doi: 10.1046/j.1471-4159.2002.00997.x. [DOI] [PubMed] [Google Scholar]
- Knochenmuss R. A quantitative model of ultraviolet matrix-assisted laser desorption/ionization including analyte ion generation. Anal. Chem. 2003;75:2199–207. doi: 10.1021/ac034032r. [DOI] [PubMed] [Google Scholar]
- Lewis GD, Farrell L, Wood MJ, Martinovic M, Arany Z, Rowe GC, Souza A, Cheng S, McCabe EL, Yang E, Shi X, Deo R, Roth FP, Asnani A, Rhee EP, Systrom DM, Semigran MJ, Vasan RS, Carr SA, Wang TJ, Sabatine MS, Clish CB, Gerszten RE. Metabolic signatures of exercise in human plasma. Sci Transl Med. 2010;2:33ra37. doi: 10.1126/scitranslmed.3001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindon JC, Holmes E, Nicholson JK. So what's the deal with metabonomics? Anal Chem. 2003;75:384A–391 A. doi: 10.1021/ac031386+. [DOI] [PubMed] [Google Scholar]
- Masson P, Couto Alves A, Ebbels TMD, Nicholson JK, Want EJ. Optimization and Evaluation of Metabolite Extraction Protocols for Untargeted Metabolic Profiling of Liver Samples by UPLC-MS. Anal Chem. 2010;82:7779–7786. doi: 10.1021/ac101722e. [DOI] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, Rochon J, Gallup D, Ilkayeva O, Wenner BR, Yancy WS, Jr., Eisenson H, Musante G, Surwit RS, Millington DS, Butler MD, Svetkey LP. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–26. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Lindon JC, Holmes E. `Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1998;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- Odunsi K, Wollman RM, Ambrosone CB, Hutson A, McCann SE, Tammela J, Geisler JP, Miller G, Sellers T, Cliby W, Qian F, Keitz B, Intengan M, Lele S, Alderfer JL. Detection of epithelial ovarian cancer using 1H-NMR-based metabonomics. Int J Cancer. 2005;113:782–8. doi: 10.1002/ijc.20651. [DOI] [PubMed] [Google Scholar]
- Oliver SG, Winson MK, Kell DB, Baganz F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998;16:373–378. doi: 10.1016/s0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- Oursel D, Loutelier-Bourhis C, Orange N, Chevalier S, Norris V, Lange CM. Identification and relative quantification of fatty acids in Escherichia coli membranes by gas chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 2007;21:3229–3233. doi: 10.1002/rcm.3177. [DOI] [PubMed] [Google Scholar]
- Panisko EA, Conrads TP, Goshe MB, Veenstra TB. The postgenomic age: characterization of proteomes. Exp Hematol. 2002;30:97–107. doi: 10.1016/s0301-472x(01)00771-8. [DOI] [PubMed] [Google Scholar]
- Pauling L, Robinson AB, Teranishi R, Cary P. Quantitative analysis of urine vapor and breath by gas-liquid partition chromatography. Proc Natl Acad Sci U S A. 1971;68:2374–6. doi: 10.1073/pnas.68.10.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BL, Cummings BS. A review of chromatographic methods for the assessment of phospholipids in biological samples. Biomed. Chromatogr. 2006;20:227–243. doi: 10.1002/bmc.563. [DOI] [PubMed] [Google Scholar]
- Petkovic M, Schiller J, Muller M, Benard S, Reichl S, Arnold K, Arnhold J. Detection of individual phospholipids in lipid mixtures by matrix-assisted laser desorption/ionization timeof-flight mass spectrometry: phosphatidylcholine prevents the detection of further species. Anal. Biochem. 2001;289:202–216. doi: 10.1006/abio.2000.4926. [DOI] [PubMed] [Google Scholar]
- Roberts LD, McCombie G, Titman CM, Griffin JL. A matter of fat: an introduction to lipidomic profiling methods. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2008;871:174–81. doi: 10.1016/j.jchromb.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Roberts LD, Murray AJ, Menassa D, Ashmore T, Nicholls AW, Griffin JL. The contrasting roles of PPARδ and PPARγ in regulating the metabolic switch between oxidation and storage of fats in white adipose tissue. Genome Biol. 2011;12:R75. doi: 10.1186/gb-2011-12-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick CA, Knight D, Croxford AS, Maqsood AR, Stephens GM, Goodacre R, Dickson AJ. Evaluation of extraction processed for intracellular metabolite profiling of mammalian cells: matching extraction approaches to cell type and metabolite targets. Metabolomics. 2010;6:427–438. [Google Scholar]
- Spencer F, Lagarde M, Geloen A, Record M. What is lipi- domics? Eur. J. Lipid Sci. Technol. 2003;105:481–482. [Google Scholar]
- Wang C, Kong H, Guan Y, Yang J, Gu J, Yang S, Xu G. Plasma phospholipid metabolic profiling and biomarkers of type 2 diabetes mellitus based on high-performance liquid chromatography/electrospray mass spectrometry and multivariate statistical analysis. Anal. Chem. 2005;77:4108–16. doi: 10.1021/ac0481001. [DOI] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O'Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–53. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakelam MJ, Pettitt TR, Postle AD. Lipidomic analysis of signaling pathways. Methods Enzymol. 2007;432:233–46. doi: 10.1016/S0076-6879(07)32010-7. [DOI] [PubMed] [Google Scholar]
- Wenk MR. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- Wishart DS. Advances in metabolite identification. Bioanalysis. 2011;3:1769–82. doi: 10.4155/bio.11.155. [DOI] [PubMed] [Google Scholar]
- Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Research. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Lin C, Wu H, Tjeerdema RS, Viant MR. Evaluation of metabolite extraction strategies from tissue samples using NMR metabolomics. Metabolomics. 2007;3:55–67. [Google Scholar]