Summary
The germinal center (GC) is a unique histological structure found in peripheral lymphoid organs. GCs provide an important source of humoral immunity by generating high affinity antibodies against a pathogen. The GC response is tightly regulated during clonal expansion, immunoglobulin modification, and affinity maturation, while its deregulation has a detrimental effect on immune function, leading to development of diseases such as lymphoma and autoimmunity. LRF (lymphoma/leukemia-related factor), encoded by the ZBTB7A gene, is a transcriptional repressor belonging to the POK (POZ and Krüppel)/ZBTB (zing finger and BTB) protein family. LRF was originally identified as a PLZF (promyelocytic leukemia zinc finger) homologue that physically interacts with BCL6 (B-cell lymphoma 6), whose expression is required for GC formation and associated with non-Hodgkin’s lymphoma. Recently, our group demonstrated that LRF plays critical roles in regulating lymphoid lineage commitment, mature B-cell development, and the GC response via distinct mechanisms. Here, we review POK/ZBTB protein function in lymphoid development, with particular emphasis on the role of LRF in GC B cells.
Keywords: germinal center, LRF, POKEMON, ZBTB, POZ, BTB
Introduction
Activation of naive B cells occurs during the T-cell-dependent (TD) antibody response to exogenous antigen in secondary lymphoid organs. They enter either into extrafollicular areas, where differentiating into antibody-secreting short-lived plasma cells or into B-cell follicles, in which germinal centers (GCs) are established. The GC is a distinct microanatomical site that generates memory B cells and plasma cells. Plasma cells produce high affinity antibodies against invading microorganisms. The GC reaches maximal size within roughly two weeks after exposure to the antigen and then disappears within several weeks. In the GC, B cells undergo two major modifications: somatic hypermutation (SHM) and class switch recombination (CSR) to enable efficient affinity maturation and effector function, respectively. SHM modifies the immunoglobulin variable region (IgV) of rearranged B-cell antibody genes during the immune response, which is associated with DNA strand breaks and introduces single nucleotide exchanges, small deletions, and/or duplication (1, 2). On the other hand, CSR mediates isotype switching (or class switching), which occurs within and outside of GCs in both T-cell-dependent and -independent responses (3). Dysregulation of this process often leads to malignant transformation and development of autoimmune diseases.
Little is known about transcriptional programs regulating GC B-cell proliferation and survival. Few transcription factors [such as BCL6, Pou2AF1 (also known as OBF1 and OCAB], Bach2, Spi-B, Mef2c and IRF8) are known to function in GC B-cells. Pou2AF1 is reportedly essential for the B-cell response to antigens and required for GC formation (4). Bach2 is critical for CSR and SHM of immunoglobulin genes: GC B-cells are not observed in Bach2-deficient mice (5). Mice lacking Spi-B, an Ets family transcription factor closely related to PU.1, exhibit severe abnormalities in B-cell function and selective TD humoral immune responses, such as dramatic defects in GC formation and maintenance (6). Mef2c loss promotes defects in B-cell proliferation and survival: Mef2c-deficient GC B-cells show an impaired GC response to TD antigens (7). IRF8 (interferon regulatory factor 8) is also reportedly implicated in GC formation (8).
Poxvirus and zinc finger (POZ) and Krüppel-type (POK) proteins, also known as ZBTB [zinc finger and BTB (Broad complex, Tramtrack, and Bric-à-brac]) proteins, are part of an emerging family of transcription factors with critical functions in development, differentiation, and oncogenesis (9–15). Here, we review recent findings relevant to POK/ZBTB proteins, particularly the transcription factor LRF, in lymphoid development and lymphomagenesis.
The BTB/POZ zinc finger proteins
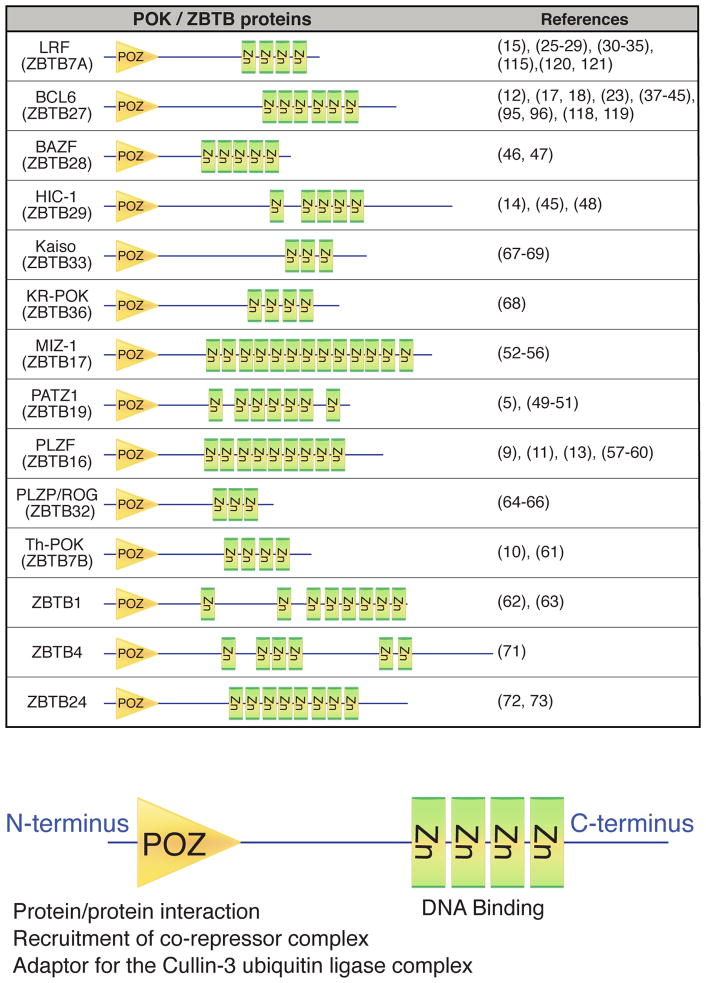
POK/ZBTB proteins exhibit C-terminal zinc fingers and an N-terminal BTB/POZ domain. The former recognize and bind specific DNA sequences, while the BTB/POZ domain mediates homodimerization and/or heterodimerization and interacts with other proteins (16–18). POK proteins generally function as transcriptional repressors via BTB/POZ-mediated recruitment of co-repressors such as N-CoR (nuclear co-repressor), SMRT (silencing mediator of retinoic acid and thyroid hormone receptor)(also known as N-CoR2), mSin3A, BCOR (BCL6 interacting corepressor), CtBP (for C-terminal binding protein), or HDACs (histone deacetylases) to promoter regions (19–24). In the human genome, 43 genes encode POK/ZBTB proteins (16), and several of these are implicated in lymphocyte development and/or lymphomagenesis (Fig. 1).
Fig. 1. Structure of POK family members.
The evolutionarily conserved POZ domain (yellow triangles) functions in homo- and heterodimerization and recruits corepressor complexes. Zinc fingers (green squares) mediate specific DNA sequence recognition and binding within gene-regulatory regions. The function of these family members as transcription factors is described in the indicated references.
LRF (for leukemia/lymphoma related factor), which is encoded by the ZBTB7A gene and formerly known as POKEMON (POK erythroid myeloid ontogenic factor), was originally identified as a PLZF (promyelocytic leukemia zinc finger) homologue interacting with BCL6 (B-cell lymphoma 6) (25). Its human homologue was cloned as a factor that binds to the inducer of short transcripts (IST), a bipartite human immunodeficiency virus (HIV) promoter element. The IST promotes synthesis of HIV short transcripts via formation of transcriptional complexes that cannot efficiently undergo elongation (26). Thus, Pessler et al. (27) named the gene FBI-1 (for factor binding to IST-1) and proposed that FBI-1 acts as a transcriptional termination or pausing factor that acts on transcriptionally-engaged polymerases. OCZF (for osteoclast-derived zinc finger), a rat homologue of LRF, was identified by immunoscreening with an osteoclast-specific monoclonal antibody (clone Kat6) using a phage cDNA library generated from rat osteoclasts (28). Kukita et al. (28) proposed that OCZF is required for terminal osteoclast differentiation. Recently, it was also reported that LRF/FBI-1/OCZF is induced by RANKL (receptor activator of NF-κB ligand), a key regulator of dendritic cell and osteoclast function, and highly expressed in multinucleated lacunar osteoclasts of rheumatoid arthritis (RA) patients (29).
LRF acts as a proto-oncogene via repressing the p19Arf (alternative reading frame) tumor suppressor, and LRF-deficient mouse embryonic fibroblasts (MEFs) show a replicative senescence phenotype due to de-repression of the p19Arf gene (15). In fact, growth defects in LRF-deficient MEFs are fully reversed by genetic loss of the p19Arf gene. Conversely, LRF overexpression combined with that of other oncogenes leads to oncogenic transformation of primary MEFs, and transgenic mice in which LRF is ectopically expressed in immature T and B cells (lckEμ-LRF/Pokemon mice) develop fatal T-lymphoblastic leukemia/lymphoma (15). Furthermore, LRF is expressed at high levels in 60–80% of diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL) cases (15). Recent reports also indicate that LRF may function as a proto-oncogene through ARF-independent mechanisms (30–32).
LRF is broadly expressed in hematopoietic lineage cells, but its expression is particularly high in erythroblasts and GC B cells (15, 33–35). Studies of LRF knockout mice demonstrate its essential role in terminal erythroid differentiation (34). LRF deficiency promotes apoptosis of terminally differentiating erythroblasts (such as polychromatic and orthochromatic erythroblasts) due to high levels of the pro-apoptotic factor BIM (Bcl-2 interacting mediator of cell death). Genetic loss of Bim partially rescues both the anemia phenotype and embryonic lethality seen in LRF knockout mice. GATA1, an erythroid specific transcription factor, directly binds the LRF promoter and transactivates LRF. LRF, in turn, transcriptionally represses BIM, suppressing erythroblast apoptosis at terminal stages of differentiation (33). Of note, Yu et al. (33, 36) recently reported that LRF binds to GATA1 occupancy sites of GATA1 direct target genes. LRF’s role in lymphoid development and function will be discussed in detail below.
BCL6 (also known as LAZ3 and ZBTB27) is a proto-oncogene originally cloned from the breakpoint of a chromosomal translocation in DLBCL. Gene rearrangements and/or somatic hypermutations of the BCL6 gene result in dysregulated BCL6 expression and subsequent lymphomagenesis (37). BCL6 is normally expressed in GCs and downregulated as GC B cells mature toward plasma cells (38). BCL6-deficient mice exhibit normal B-cell development but cannot form GCs, indicating its essential role in GC establishment and/or maintenance (12, 39). Several other BCL-6 functions have been reported: in GC responses, BCL-6 inhibits expression of genes such as CD69, signal transducer and activator of transcription 1 (STAT1), and the costimulatory receptor CD80 (40), which function in B-cell activation in TD immune responses; BCL-6 blocks plasma cell differentiation by repressing expression of the transcription factor Prdm1 [also known as B-lymphocyte-induced maturation protein 1 (BLIMP1)] (41); and BCL-6 represses memory cell differentiation (42). Since BCL6 is also required for development of T-follicular helper cells (43, 44), it presumably functions through both B-cell intrinsic and extrinsic mechanisms. Recently, an unexpected role for BCL6 in leukemia stem cells was reported (45): Duy et al. (45) reported that BCL6 functions as a key factor in a transcriptional network in BCR-ABL+ B-acute lymphoblastic leukemia (B-ALL), regulating apoptotic responses to imatinib treatment. The role of BCL6 in GC formation is described in greater detail elsewhere in this issue.
BAZF (for BCL6-associated zinc finger protein) (also known as BCL6b or ZBTB28) protein is most closely related to BCL6, to which it is 65% similar in the BTB/POZ domain and 94% in the zinc finger region (46). BAZF is a transcriptional repressor, but it requires BCL6 for its function. BAZF-deficient mice display defects in generation of CD8 T-cell memory (47), although the role of BAZF in GC B cells is not known.
HIC1 (for hypermethylated in cancer 1) (also known as ZBTB29) controls osteosarcoma and mammary gland carcinoma formation by cooperating with p53 (48). HIC1 heterozygous mice are predisposed to an age- and gender-dependent spectrum of malignant tumors (14). The HIC1 gene is frequently inactivated in DLBCL cases (45), and one third of HIC1 heterozygous female mice develop lymphoma (14).
PATZ1 (POZ and AT hook containing zinc finger 1)(also known as MAZR, ZFP278 and ZBTB19) negatively regulates the CD8 gene complex during the CD4/CD8 double negative (DN) to double positive (DP) transition by recruiting NCOR (49) and represses Th-POK expression during the CD4 versus CD8 cell fate decision (50). PATZ1 physically interacts with BACH2 (5, 51), a BTB transcription factor required for GC formation (5, 51). Based on data reported in these papers, it is tempting to speculate that PATZ1 and BACH2 function cooperatively in GC formation.
MIZ1 (Myc-interacting zinc-finger protein1) (also known as ZBTB17) functions as a transcriptional activator or repressor depending on its binding partners and forms a complex with the bHLH (basic helix-loop-helix) leucine zipper transcription factor Myc (52, 53). MIZ1 interacts with BCL6 to suppress the cyclin-dependent kinase inhibitor p21 (encoded by the CDKN1A gene) and induce cell cycle arrest in GC B cells (54). Binding of Myc to MIZ1 is required to antagonize cell cycle arrest and cellular senescence induced by transforming growth factor-β (TGFβ) in lymphocytes (55). Recently, Kosa et al. (56) reported that MIZ1 regulates interleukin-7 (IL-7) receptor signaling at early B-cell commitment. It is not yet clear whether MIZ1, BCL6, and Myc function coordinately in the context of GC B-cells.
PLZF (promyelocytic leukemia zinc finger) (also known as ZBTB16) was originally identified as the fusion partner of the retinoic acid receptor α (RAR) gene in acute promyelocytic leukemia harboring translocation t(11;17)(q23;q21) (13). PLZF plays critical roles in oncogenesis, development, and stem cell maintenance (9, 57, 58). Plzf-null mice display severe defects in development of invariant natural killer T-cells (iNKT), indicating a pleiotropic role for PLZF in iNKT cell development (9, 11, 59). PLZF is also expressed in a subset of mature γδ T-cells, and production of IFN-γ and IL-4 in response to T-cell receptor (TCR) stimuli is impaired in Plzf-deficient γδ T-cells. While interaction and colocalization of PLZF and BCL6 have been reported (60), the relevance of that interaction to GCs is not yet known.
Studies indicate that several other POK/ZBTB proteins are relevant to lymphoid development and/or lymphomagenesis. Th-POK (T-helper-inducing POZ/Krüppel-like factor) (also known as ZBTB7B) reportedly functions as a master regulator of T-cell lineage commitment specifically for differentiation of CD4+ T-helper (Th) cells undergoing CD4+ versus CD8+ lineage specification and commitment (10, 61). ZBTB1, which was originally identified as an efficient target of small ubiquitin-like modifiers (SUMO) (62), functions as a critical determinant of T-cell development and lymphopoiesis. Mice harboring a point mutation within the POZ domain of Zbtb1 showed a cell-intrinsic T-cell deficiency and competitive failure of lymphoid reconstitution in spite of normal development of hematopoietic progenitors in the bone marrow (BM) (63). PLZP/ROG (PLZF-like zinc finger protein) (also known as FAZF, TZFP, ROG and ZBTB32) heterodimerizes with PLZF (64), and PLZP-deficient mice show both cell cycle and cytokine production defects in T cells (65). PLZP overexpression in Th cells downregulates expression of cytokine genes, including Th1 and Th2 (66). Kaiso (also known as ZBTB33), which was first isolated through its interaction with the Armadillo-repeat protein catenin p120, functions in intestinal cancer development (67). Although Kaiso negatively regulates canonical Wnt signaling in the Xenopus model system (68), Kaiso-deficient mice display resistance to intestinal cancer (69). KR-POK (70) (kidney cancer-related POK; also known as ZBTB36) and ZBTB4(71) physically interact with MIZ1 and repress p21 expression. Finally, germ line deletion of the ZBTB24 gene was recently identified in some patients with immunodeficiency, centromere instability, and facial anomalies (ICF) syndrome, a rare autosomal recessive disease. Such patients usually exhibit fatal respiratory and gastrointestinal infections due to hypogammaglobulinemia, regardless of normal lymphocyte counts (72, 73), suggesting that ZBTB24 plays a pleiotropic role in the immune system.
Role of LRF in early B-cell development
Hematopoietic stem cells (HSCs) constantly generate a large number of specialized cell types and at the same time replenish the stem cell pool. Common lymphoid progenitors (CLPs), defined as lineage (Lin)−IL-7Rα+Flt3+Sca-1loc-Kitlo, are among the earliest lymphoid-restricted precursors (74). CLPs enter into the B-cell differentiation pathway upon expression of the B-cell marker B220. Immunoglobulin rearrangements and B-cell receptor (BCR) assembly that follow give rise to immature B cells, which leave the BM and enter the periphery where they further differentiate to mature B cells through several transitional stages. Of importance, expression of the pre-BCR provides a vital checkpoint for functionality in early B-cell development. Furthermore, BCR expression is required for B-cell development and survival in the periphery (75).
B-cell development in the BM occurs in sequential steps characterized by specific gene expression programs and combinations of surface molecules. For example, B-cell development is impaired in mice carrying a deletion in PU.1, a member of the Ets domain-containing transcription factor family (76). Ikaros knockout (KO) mice fail to generate B cells, T cells, NK cells, and dendritic cells (77), while the transition from pro- to pre-B-cells is impeded in mice expressing a hypomorphic form of Ikaros (78). E2A (77, 78) and early B-cell factor (EBF) (also known as OLF1) (79) are essential for the transition from prepro-B- to pro-B-cells, while paired box protein 5 (PAX5) is a key transcription factor that regulates pro-B- to pre-B differentiation (80). These transcription factors have stage-specific functions in B-cell development but also function cooperatively in transcriptional networks.
Although deletion of the Zbtb7a gene in mice results in embryonic lethality due to severe anemia likely caused by increased apoptosis of late-stage erythroblasts (33), examination of B lymphopoiesis at 14.5 d.p.c reveals a significantly reduced number of CD19+B220+ B cells (33, 34). Cre-lox mediated LRF inactivation at HSC/progenitor stages in adult mice (LRFFlox/Flox Mx1-Cre+) promotes development of double positive (DP) T cells in the BM at the expense of B lymphopoiesis (34). The number of pro-B, pre-B, and immature B cells is drastically reduced in LRFFlox/Flox Mx1-Cre+ mice, while prepro-B cells increase (34). Despite their B220 positivity, LRF-deficient ‘prepro-B’ cells express CD25, a marker of immature thymic T cells. mRNAs that encode pre-BCR components (such as Igα, Ig β, and VpreB1), terminal deoxynucleotidyl transferase (TdT), and Rag recombinases, are significantly reduced in LRF-deficient prepro-B-cells, while Notch1/3 mRNA and Notch target genes such as Hes-1 are upregulated. Furthermore, LRF-deficient prepro-B cells resemble normal thymic double negative stage 3 (DN3) T cells (CD25+CD44−) and can be readily differentiated into CD4/8 DP T cells when co-cultured with stromal cells overexpressing the Notch ligand Delta-like 1 (34).
Notch is essential for normal T-cell lineage commitment in the BM. In particular, Notch1 is necessary for the early stage of T-cell development. Notch1 inactivation in mouse HSCs results in marked reduction in thymic T-cells and ectopic B-cell development in the thymus, while B-cell development in the BM remains intact. LRF conditional knockout mouse phenotypes resemble those of mice overexpressing the intracellular domain of Notch1 (ICN1), in that ectopic CD4/CD8 DP cell development is induced at the expense of B-cell development in the BM (81, 82). Strikingly, aberrant DP-T cells in the BM of LRFFlox/Flox Mx1-Cre+ mice completely disappear and normal B-cell development resumes after treatment with a gamma secretase inhibitor (GSI), a potent inhibitor of Notch signaling (34). These data indicate that defects in early lymphoid fate determination seen in LRFFlox/Flox Mx1-Cre+ mice are caused by Notch-dependent mechanisms.
Despite its critical role in early lymphoid lineage commitment, LRF is dispensable to maintain BM B cells. B-cell-specific LRF conditional knockout mice (LRFFlox/FloxMb-1 Cre+ mice), in which Cre expression is restricted to B cells after the pro-B-cell stage, show normal B-cell development in the BM (35), consistent with the fact that GSI treatment restores normal B-cell development in LRFFlox/Flox Mx1-Cre+ mice. Taken together, LRF is essential for B-cell lineage commitment at HSC/progenitor stages via interfering with the Notch pathway, while it is dispensable for maintenance of ‘committed’ B-cell precursors in the BM.
Role of LRF in mature B-cell development
Mouse mature B-cells are categorized into follicular (FOB) or marginal zone (MZB) cells. Each population resides in specific anatomical locations and has distinct properties. About 10% of mature B cells are MZB cells, which reside in the outer white pulp of the spleen between the marginal sinus and the red pulp and respond vigorously to blood-borne pathogens (83, 84). B cells activated by antigen have cognate interaction with T cells in the T/B-cell interface of secondary lymphoid organs and then either become immunoglobulin-secreting plasma cells or enter primary follicles to initiate the GC reaction (86, 87).
In mice, BM-derived immature B-cells mature into FOB (B220+CD19+AA4.1−CD1d−CD23+IgMlo/−CD21lo) or MZB (B220+CD19+AA4.1−CD1d+CD23−IgMhiCD21hi) cells through transient transitional (T1 and T2) B-cell stages (88). Multiple signaling pathways, including Notch, BCR, and NF-κB, function to regulate MZB development (88). Notch2 expression levels increase with B-cell maturation and Notch signaling is required for MZB cell development during B-cell maturation in the spleen. Inactivation of Notch2 or components of the Notch pathway such as RBP-jk, Mastermind-like 1 (MAML1) or the Notch ligand DLL1 promotes defects in MZB cell development (89–92). DLL1 is expressed on the luminal face of venules found in red pulp of the spleen and in the marginal zone. Notch2 and DLL1 weakly interact, but that interaction is strengthened by the lunatic fringe and manic fringe glycosyltranserases (91). Mindbomb 1, an E3 ligase that regulates DLL1 endocytosis, is also required for marginal zone B-cell development (92). By contrast, MZB cell numbers increase in mice lacking the MSX2-interacting protein (MINT), a suppressor of Notch signaling (67).
In LRFFlox/FloxMb1 Cre+ mice, the proportion and absolute number of FOB cells in spleen decrease, while both proportion and number of MZB cells significantly increases (35). Skewed differentiation toward MZB cells in LRFFlox/FloxMb1 Cre+ mice is masked by either genetic loss of Notch2 (LRF/Notch2 double knockout mice) or treatment with anti-DLL1 antibody (35). These data do not provide direct evidence for functional interaction between LRF and Notch2. However, considering that the Notch target Hes1 is upregulated in LRF-deficient transitional B cells, LRF may antagonize Notch2-DLL1-mediated signaling at a transitional B-cell stage. Considering that DLL4, but not DLL1, inhibition enables normal lymphoid development in the BM in LRFFlox/Flox Mx-1 Cre+ mice (S.U. Lee and T. Maeda, unpublished data), LRF may regulate Notch pathways in a ligand-dependent and cell type-specific manner.
Role of LRF in GC formation and function
Generation of effector B cells starts when newly formed B cells migrate from the BM to secondary lymphoid organs, including spleen, lymph nodes, Peyer’s patches, and tonsils. GCs form after an orchestrated series of cellular interactions between antigen-presenting dendritic cells, CD4+ Th cells, and antigen-specific B cells at the border between B- and T-cell areas. Those GCs provide distinct histologic structures where rapidly proliferating B cells undergo genetic modification of immunoglobulin genes and are selected according to the ability to recognize high affinity antigen. Most cells with low affinity B-cell receptors (BCRs) or lacking BCR expression die due to apoptosis (93). At the final stage of the GC reaction, memory B cells and long-lived plasma cells develop. Plasma cells are effector cells producing a high level of antibodies, while memory B cells maintain a rapid immunological response upon later exposure to the same antigen (37). Surprisingly, GCs are not always required for affinity maturation, because lymphotoxin-α-deficient mice, which lack GCs, exhibit measurable affinity maturation (94). However, GCs are necessary for more efficient affinity maturation in the humoral immune response.
BCL6 is a key regulator of the GC reaction, and BCL6 knockout mice cannot produce high affinity antibodies due to lack of GCs (12, 39). BCL6 expression enables GC B cells to survive physiological stresses such as DNA double-strand breaks. BCL6 suppresses growth-arrest and apoptotic responses by transcriptionally repressing the p53 tumor suppressor gene and/or the CDKN1A gene in collaboration with MIZ1 (54, 95). BCL6 may also regulate GC B-cell survival by suppressing ATR (ataxia telangiectasia and Rad3 related), a critical sensor of DNA damage (96).
LRF and BCL6 interact in vitro (25), and their expression overlaps in GCs (Fig. 2), implying that LRF also functions in GCs. As expected, GC B cells are significantly reduced in LRFFlox/Flox Mb-1 Cre+ mice after immunization with TD antigen (35). Although BCL6 knockout mice reportedly show complete loss of GC formation (39), a few GC B cells were observed and overall GC structures, albeit small, remain intact in LRFFlox/Flox mb-1 Cre+ mice (35). Furthermore, reduced GC B-cell number is seen in LRF conditional knockout mice (LRFFlox/Flox Cγ1 Cre+), in which expression of Cre recombinase is efficiently induced in the majority of GC B cells generated in response to immunization with TD antigens (97), indicating that LRF is necessary for maintenance and function of GC B cells rather than commitment to GC B cells. As previously noted, LRFFlox/Flox Mb-1 Cre+ mice show reduced numbers of FOB cells, which are rescued by Notch2 loss. However, GC formation defects are also seen in LRF/Notch2 double knockout mice, suggesting that reduced FOB cell numbers are not the primary cause of defective GC formation in LRFFlox/Flox Mb-1 Cre+ mice.
Fig. 2. LRF protein is highly expressed in GC B-cells, as is Bcl6.
Immunohistochemical analysis for LRF in the spleen of an immunized WT mouse (left) and Bcl6 staining in an adjacent section (right) (picture adopted from Sakurai et al (35)). Bcl6 is exclusively expressed in GC B-cells [strong brown staining in the right panel (arrowheads)], while LRF is highly expressed in GC-B cells as well as in other cell types (e.g. erythroid cells).
Although competing views exist as evident in the other reviews in this volume, a classical view of the GCs is that they are divided into two anatomically distinct regions: a dark zone (DZ) consisting of large, mitotically active B cells known as centroblasts, and a light zone (LZ) exhibiting smaller, nondividing B cells known as centrocytes, as well as follicular dendritic cells (FDCs), antigen-specific follicular T helper cells, and macrophages (98). In the DZ where lymphocytes are closely packed, centroblasts undergo rapid proliferation and SHM of antibody variable region genes and then become smaller centrocytes that travel to the LZ. Centrocytes express a wide range of affinity surface antibodies for a given antigen, and selected centrocytes present antigen to helper T cells in the LZ and promote differentiation into antibody-secreting plasma cells or memory B cells. In this classical model of GC reaction, selection would require cell migration from the DZ to the LZ. However, findings from recent real time imaging experiments are not fully consistent with the classical model. B-cell trafficking between the DZ and LZ appears to be mediated by chemokines possibly produced by stromal cells (99). Although the DZ and LZ are difficult to precisely delineate based on cell surface marker expression, immunohiostochemical and genetic evidence supports the notion that centroblasts, which express higher levels of the CXC-chemokine receptor 4 (CXCR4) in the DZ, migrate towards a CXCL12/SDF1 gradient originating in the LZ (99). CXCL13 is more abundant in the LZ, and the CXCL13-CXCL5 chemokine and receptor pair is needed for GC B-cell accumulation in the LZ. Significantly, CXCR4 protein expression controls positioning of cells between the DZ and LZ. CXCR4hi+ GC B cells, which are more abundant in the DZ and in actively cycling cells, are significantly reduced in LRF-deficient mice compared with controls (15). LRF-deficient GCB cells proliferate less rapidly than wildtype cells, as revealed by microarray analysis as well as 5-ethynyl-2′-deoxyuridine (EdU) incorporation assays (15).
A very rapid proliferation rate, in which a complete cell cycle requires only 6 to 12 h, is the hallmark of centroblasts, from which few B cells bearing enhanced antigen-binding immunoglobulin are selected (100, 101). GC B cells undergo programmed cell death (or apoptosis) by default if they are not rescued by anti-apoptotic signals (102). GC B cells (B220+CD19+CD38dimFas+) are reportedly more prone to apoptosis than are non-GC B-cells (B220+CD19+CD38hi+Fas−) (36). Although they downregulate anti-apoptotic markers such as BCL-2, centroblasts express several pro-apoptotic molecules, which promote rapid elimination of B cells with non-functional or non-binding antibodies (100, 103). In addition to proliferation defects, LRF-deficient GC B cells undergo apoptosis at a high frequency. Levels of the active form of caspase-3 and the cleaved form of Poly (ADP-ribose) polymerase (PARP), whose formation is catalyzed by caspase-3, markedly increase in LRF-deficient mice (15).
The p53 pathway is critical to control the cell cycle and apoptosis in GC B cells, and p53 expression is normally absent in GC B cells showing hyper-proliferation. p19Arf is a negative regulator of the ubiquitin-protein isopeptide ligase Mdm2, which functions in p53 degradation in a ubiquitin-dependent manner. Thus, p19Arf repression stimulates cell proliferation by decreasing p53 activity. BCL6 antagonizes cell-cycle arrest and apoptosis in GC B-cells through p53-dependent and -independent mechanisms (54, 95). In contrast, LRF specifically binds to and represses the p19Arf promoter in MEFs, while maintaining expression of the p16Ink4a gene, which encodes a different tumor suppressor at the same Ink4a-Arf locus intact (15). In LRF-deficient GC B-cells, p19Arf mRNA levels are significantly increased, while p53 mRNA levels remain unchanged (35). p19Arf upregulation is also confirmed at protein levels by western blot and immunofluorescence. Of note, p21, which inhibits activities of cyclin-cdk2 complexes and is a p53 transcriptional target, is also upregulated in the absence of the LRF gene. Thus, derepression of the p19Arf gene accounts, at least in part, for impaired proliferation and increased apoptosis seen in LRF-deficient GC B-cells (35). Since LRF and BCL6 synergize to enhance MEF proliferation (15), both may act together in the context of GC B cells.
During GC responses, immunoglobulin genes in activated B-cells are modified by SHM and CSR to produce genetically diverse and high affinity antibodies. Both SHM and CSR require activity of activation-induced cytidine deaminase (AID) (104). AID deficiency leads to the complete absence of CSR before or after immunization with TD antigens and also abrogates SHM of the specific VH gene in response to NP (4-hydroxy-3-nitrophenylacetyl hapten)-conjugated TD antigen. AID initiates gene conversion of immunoglobulin genes by deoxycytidine deamination, followed by error-prone mismatch- or base-excision DNA repair. AID-mediated deamination promotes formation of DNA double-strand breaks. LRF-deficient GC B cells express Aid mRNA levels equivalent to those of controls, indicating that AID function per se is likely intact (35). However, among serum immunoglobulin titers tested, those of class-switched antibodies (IgG1, IgG2b, and IgG3) were perturbed in LRF-deficient mice. Furthermore, primary and secondary antibody responses to NP-specific TD-antigen are severely impaired in LRFFlox/FloxMb-1 Cre+ mice and in LRFFlox/Flox Cγ1 Cre+ mice (18). Most antibody-forming cells in a given immune reaction die in secondary lymphoid organs within a few days (105). Ten to twenty percent of plasma cells in the BM survive for more than 2 weeks (106). Such extended survival requires both specific signals from the BM and molecular competence to respond to them. LRF-deficient mice exhibit fewer BM long-lived plasma (B220−CD138+) cells (35). Along with the fact that LRF-deficient B cells proliferate less rapidly than controls, it is conceivable that impaired CSR seen in LRF knockout mice is due to lack of absolute GC B-cell numbers rather than defective Aid function. Although SHM is closely linked to cell division (107), mutation frequencies in rearranged V186.2 genes upon in response to NP-conjugated TD antigen, which represent the degree of SHM, were not grossly altered in LRF-deficient GC B-cells relative to controls (35). Remaining LRF-deficient GC B-cells, in which Aid expression is intact, may be sufficient to generate diverse mutations.
Taken together, defects in GC B-cell proliferation and survival lead to reduced GC B-cell numbers, which accounts for defective CSR seen in LRF knockout mice. LRF likely helps GC B-cells to proliferate and survive genotoxic stress, such as SHM and CSR, by regulating p53 pathways via p19Arf repression.
Aberrations in GC formation are common features of lymphomagenesis, particularly in B-cell non-Hodgkin lymphomas (B-NHLs). Most B-NHLs show SHM in immunoglobulin gene variable regions, indicating that tumor cells are derived from B-cells emerging from GCs (108). Reciprocal translocations between IgH genes and several oncogenes, including c-MYC (Burkitt’s lymphoma), BCL-2 (FL), and BCL-6 (DLBCL) are characteristic of NHL and likely arise from aberrant resolution of Ig CSR-associated DNA double-stranded breaks (109). In addition, aberrant SHM has been suggested to result in mutations in the proto-oncogenes PIM1, c-MYC, and PAX5, contributing to DLBCL development (110).
Among POK/ZBTB proteins, BCL6 and PLZF are implicated in leukemia/lymphoma pathogenesis as, respectively, a proto-oncogene in non-Hodgkin lymphoma and tumor suppressor in acute promyelocytic leukemia (111, 112). LRF could also function as a proto-oncogne via ARF repression in cooperation with other oncogenes (15). Transgenic mice overexpressing LRF driven by the lckEμ enhancer/promoter develop aggressive tumors, massive thymic enlargement, splenomegaly, hepatomegaly, and tumor infiltration into the BM (15). In human NHL (130 DLBCL and 290 FL cases), lymphomas positive for both LRF and BCL6 proteins show a higher proliferative index than do controls. Furthermore, LRF positivity is predictive of better overall survival in DLBCL. LRF and BCL6 double positivity predicts an even better clinical outcome for overall survival (15).
DLBCL can be divided into prognostically different subgroups exhibiting GC B-cell-like and activated B-cell-like (ABC) phenotypes (113). LRF is highly expressed in all 11 NHL cell lines examined regardless of subtype [5 GC-, 3 ABC (activated B cell-like)- and 3 Burkitt-type] (35). Short hairpin RNA (shRNA)-mediated LRF knockdown was toxic to the lymphoma lines OCI-Ly-1, OCI-Ly-3, and Ramos, an effect that was not rescued by p21 knockdown (35), suggesting a p53-independent LRF function. Mechanisms underlying toxic effects of LRF shRNA include Bim upregulation and/or p53-independent ARF function.
LRF dimerization in B-cell function
POK/ZBTB proteins form homo- or hetero-dimeric complexes through interaction of BTB/POZ domains, and dimerization is critical for cellular localization, DNA-binding, and transcriptional activity (16, 114). Interestingly, experimental evidence indicates that the BTB/POZ domain alone is sufficient to mediate self-association of POK proteins (21). Nonetheless, complex formation does not always occur between and/or among POK protein homologues despite high homology in the BTB/POZ domain. For example, BAZF and its close homolog BCL6 (65% identical in the BTB/POZ domain) form a heterodimer necessary for BAZF repressor activity, while LRF does not interact with its close homologue PLZF (25). On the other hand, LRF and BCL6 form a heterodimer requiring both their BTB/POZ domain and zinc-fingers (25). The crystal structure of LRF’s BTB domain has been solved and suggests that LRF interaction with BCL6 but not with PLZF may be due to differing electrostatic properties of their POZ domains (115, 116). The physiological relevance of POK protein dimerization has been intensively investigated in recent years. A complex containing both MIZ-1 and BCL6 suppresses transcription of cell cycle arrest genes in GC B cells, suggesting a functional significance of dimer formation in the GC reaction.
The LRF BTB/POZ domain forms an obligate homodimer and LRF interacts with DNA as a dimer (116, 117). Our group demonstrated that obligate dimerization of LRF plays a critical role in GC B cells. Among 37 interface residues that mediate LRF-POZ dimerization, 4 (I11, I20, L21, and Q27) have been shown via computational analysis to be critical for dimerization (35). The I20A-mutant in particular cannot repress p19Arf reporter activity or restore growth of LRF−/− MEFs, indicating that LRF dimerization is required for p19Arf repression. Furthermore, LRF-deficient B-cells transduced with wild-type rather than dimerization-deficient (I20A) LRF can form GCs when transferred to lethally-irradiated recipient mice (35). Therefore, LRF is essential for GC B formation in a B-cell-intrinsic manner and dimer formation is a prerequisite for its function.
LRF function in HSCs and T cells
In addition to supporting a role for LRF in B lymphopoiesis and lymphomagenesis, our unpublished work suggests an underlying function for LRF in HSC self-renewal and T-cell development in thymus. We previously showed that the number of LSKs (the classical definition of HSCs) and CLPs (defined as Lin−Sca1+c-Kit+IL7Rα+ without Flt3) was unchanged in LRFFlox/FloxMx1 Cre+ mice (34). However, those two fractions were found to consist in large part of T-cell precursors (S.U. Lee and T. Maeda, unpublished data). Our recent multicolor FACS analysis using additional surface markers (e.g. Flt3, CD25, Vcam-1, CD48, CD150, CD34) revealed that numbers of the most primitive LT-HSCs [CD150+CD48−Flt3−Vcam-1+/−IL7Rα −LSK (Lin−Sca-1+c-Kit+)] were significantly reduced and Flt3+ lymphoid progenitors, such as lymphoid-biased multi-potential progenitors (LMPPs)(CD150−CD48+Flt3+Vcam-1 +/−IL7Rα −LSK) and Flt3+CLPs (Lin−CD150−CD48+Flt3+Vcam-1−IL7Rα+), were barely detected in LRF conditional knockout mice (S.U. Lee and T. Maeda, unpublished data). Moreover, serial BM transplantation and 5-FU (5-fluorouracil) challenge experiments confirmed that LRF functions to maintain the HSC pool. Overall, LRF is indispensable for hematopoietic homeostasis by preventing HSCs from responding to Notch mediated T-instructive signals in the BM niche. Also, we observed that LRF overexpression blocks differentiation of CD8+ cytotoxic T cells in the thymus (S.U. Lee and T. Maeda, unpublished data), suggesting that LRF is involved in T-cell lineage commitment in thymus as does Th-POK (50).
Concluding remarks
POK/ZBTB family proteins function in multiple biological processes, some of which critically regulate development of specific lineages in the immune system, promote oncogenesis and maintain stem cells. LRF is unique among POK/ZBTB proteins in that it plays diverse, cell-type specific roles, such as p19Arf repression in MEFs and GC B-cells, Notch repression in HSC/progenitors, and Bim repression in erythroblasts (15, 33, 34). Using a series of stage/lineage-specific LRF conditional knockout mice, we have demonstrated that LRF is a novel factor functioning in regulation of mature B-cell lineage fate and GC B-cell maintenance via distinct mechanisms.
Although our findings imply a physiological function for BCL6 and LRF in GCs, it remains unknown whether LRF and BCL6 act synergistically during GC formation to maintain GC B-cell proliferation and survival. Most of all, the results showing that shRNA-mediated LRF knockdown is toxic to a subset of lymphoma lines provides important insight that LRF could be a therapeutic target for B-cell malignancies. Since LRF dimer formation is required for its function in GC B cells, the binding interfaces of the LRF dimer may be an attractive target for pharmacological blockage of LRF function. Alternatively, interaction between the LRF POZ/BTB domain and as yet uncharacterized corepressors could also be an attractive target for drug development, as successfully achieved in the case of BCL6 (118, 119).
Little is known regarding how LRF expression is regulated, except that its upregulation is mediated by GATA1 in erythroblasts (33, 36) and RANKL in osteoclasts (29). In GC B cells, LRF may be regulated via post-translational mechanisms, since, despite high LRF protein expression in GC B cells, LRF mRNA levels in GC B cells are lower than that in non-GC B cells (35). miRNA-mediated LRF regulation was also recently proposed: miR-20a overexpression reduced LRF expression and upregulated p19Arf levels in MEFs, leading to replicative senescence (120). Other miRNAs, such as miR-28 and miR-505, also control cellular proliferation and survival in an LRF-dependent manner in MEFs (121). These miRNAs may also be viable therapeutic targets and a powerful intervention tool in regulating LRF gene expression.
Here, we have discussed recent findings relevant to LRF, a POK/ZBTB protein, in lymphoid development and lymphomagenesis. Although in vivo animal studies have demonstrated key pleiotropic roles for LRF in hemato/lymphoid development and oncogenesis, further studies are warranted to determine how LRF functions in a cell-context specific manner.
Fig. 3. Regulation of LRF through two distinct mechanisms.
LRF regulates mature B cell (FOB versus MZB) lineage fate determination presumably by counteracting the Notch2/Dll1 pathway. LRF also regulates GC B-cell proliferation and survival by repressing ARF.
Acknowledgments
The authors thank current and past members of Maeda lab. We also thank Elise Lamar for critical reading of the manuscript. This publication was made possible in part by grants to T. Maeda from STOP Cancer, the V Foundation, Margaret E. Early Medical Research Trust, the Tim Nesvig Lymphoma Research Fund, the American Society of Hematology, and the NIH (1R01AI084905-01A1).
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Bross L, Fukita Y, McBlane F, Demolliere C, Rajewsky K, Jacobs H. DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity. 2000;13:589–597. doi: 10.1016/s1074-7613(00)00059-5. [DOI] [PubMed] [Google Scholar]
- 2.Goossens T, Klein U, Kuppers R. Frequent occurrence of deletions and duplications during somatic hypermutation: implications for oncogene translocations and heavy chain disease. Proc Natl Acad Sci U S A. 1998;95:2463–2468. doi: 10.1073/pnas.95.5.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toellner KM, Gulbranson-Judge A, Taylor DR, Sze DM, MacLennan IC. Immunoglobulin switch transcript production in vivo related to the site and time of antigen-specific B cell activation. J Exp Med. 1996;183:2303–2312. doi: 10.1084/jem.183.5.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schubart DB, Rolink A, Kosco-Vilbois MH, Botteri F, Matthias P. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 1996;383:538–542. doi: 10.1038/383538a0. [DOI] [PubMed] [Google Scholar]
- 5.Muto A, et al. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429:566–571. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- 6.Su GH, et al. Defective B cell receptor-mediated responses in mice lacking the Ets protein, Spi-B. EMBO J. 1997;16:7118–7129. doi: 10.1093/emboj/16.23.7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilker PR, et al. Transcription factor Mef2c is required for B cell proliferation and survival after antigen receptor stimulation. NatImmunol. 2008;9:603–612. doi: 10.1038/ni.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CH, et al. Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J Exp Med. 2006;203:63–72. doi: 10.1084/jem.20051450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barna M, Hawe N, Niswander L, Pandolfi PP. Plzf regulates limb and axial skeletal patterning. Nat Genet. 2000;25:166–172. doi: 10.1038/76014. [DOI] [PubMed] [Google Scholar]
- 10.He X, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 11.Costoya JA, et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- 12.Ye BH, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 13.Koken MH, et al. Leukemia-associated retinoic acid receptor alpha fusion partners, PML and PLZF, heterodimerize and colocalize to nuclear bodies. Proc Natl Acad Sci U S A. 1997;94:10255–10260. doi: 10.1073/pnas.94.19.10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen WY, et al. Heterozygous disruption of Hic1 predisposes mice to a gender-dependent spectrum of malignant tumors. Nat Genet. 2003;33:197–202. doi: 10.1038/ng1077. [DOI] [PubMed] [Google Scholar]
- 15.Maeda T, et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433:278–285. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- 16.Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang CC, Ye BH, Chaganti RS, Dalla-Favera R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci U S A. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhordain P, et al. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc Natl Acad Sci U S A. 1997;94:10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmad KF, et al. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell. 2003;12:1551–1564. doi: 10.1016/s1097-2765(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 20.Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA-and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- 21.Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 22.Melnick A, et al. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol Cell Biol. 2002;22:1804–1818. doi: 10.1128/MCB.22.6.1804-1818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghetu AF, Corcoran CM, Cerchietti L, Bardwell VJ, Melnick A, Prive GG. Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol Cell. 2008;29:384–391. doi: 10.1016/j.molcel.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q, et al. Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proc Natl Acad Sci U S A. 2007;104:829–833. doi: 10.1073/pnas.0610590104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Davies JM, et al. Novel BTB/POZ domain zinc-finger protein, LRF, is a potential target of the LAZ-3/BCL-6 oncogene. Oncogene. 1999;18:365–375. doi: 10.1038/sj.onc.1202332. [DOI] [PubMed] [Google Scholar]
- 26.Ratnasabapathy R, Sheldon M, Johal L, Hernandez N. The HIV-1 long terminal repeat contains an unusual element that induces the synthesis of short RNAs from various mRNA and snRNA promoters. Genes Dev. 1990;4:2061–2074. doi: 10.1101/gad.4.12a.2061. [DOI] [PubMed] [Google Scholar]
- 27.Pessler F, Pendergrast PS, Hernandez N. Purification and characterization of FBI-1, a cellular factor that binds to the human immunodeficiency virus type 1 inducer of short transcripts. Mol Cell Biol. 1997;17:3786–3798. doi: 10.1128/mcb.17.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kukita A, Kukita T, Ouchida M, Maeda H, Yatsuki H, Kohashi O. Osteoclast-derived zinc finger (OCZF) protein with POZ domain, a possible transcriptional repressor, is involved in osteoclastogenesis. Blood. 1999;94:1987–1997. [PubMed] [Google Scholar]
- 29.Kukita A, et al. The transcription factor FBI-1/OCZF/LRF is expressed in osteoclasts and regulates RANKL-induced osteoclast formation in vitro and in vivo. Arthritis Rheum. 2011;63:2744–2754. doi: 10.1002/art.30455. [DOI] [PubMed] [Google Scholar]
- 30.Lee DK, et al. FBI-1 enhances transcription of the nuclear factor-kappaB (NF-kappaB)-responsive E-selectin gene by nuclear localization of the p65 subunit of NF-kappaB. J Biol Chem. 2005;280:27783–27791. doi: 10.1074/jbc.M504909200. [DOI] [PubMed] [Google Scholar]
- 31.Jeon BN, Yoo JY, Choi WI, Lee CE, Yoon HG, Hur MW. Proto-oncogene FBI-1 (Pokemon/ZBTB7A) represses transcription of the tumor suppressor Rb gene via binding competition with Sp1 and recruitment of co-repressors. J Biol Chem. 2008;283:33199–33210. doi: 10.1074/jbc.M802935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi WI, et al. Proto-oncogene FBI-1 represses transcription of p21CIP1 by inhibition of transcription activation by p53 and Sp1. J Biol Chem. 2009;284:12633–12644. doi: 10.1074/jbc.M809794200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maeda T, et al. LRF is an essential downstream target of GATA1 in erythroid development and regulates BIM-dependent apoptosis. Dev Cell. 2009;17:527–540. doi: 10.1016/j.devcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeda T, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakurai N, et al. The LRF transcription factor regulates mature B cell development and the germinal center response in mice. J Clin Invest. 2011;121:2583–2598. doi: 10.1172/JCI45682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu M, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 38.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 39.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 40.Niu H, Cattoretti G, Dalla-Favera R. BCL6 controls the expression of the B7-1/CD80 costimulatory receptor in germinal center B cells. J Exp Med. 2003;198:211–221. doi: 10.1084/jem.20021395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173:1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- 42.Kuo TC, Shaffer AL, Haddad J, Jr, Choi YS, Staudt LM, Calame K. Repression of BCL-6 is required for the formation of human memory B cells in vitro. J Exp Med. 2007;204:819–830. doi: 10.1084/jem.20062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duy C, et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cellsto survive BCR-ABL1 kinase inhibition. Nature. 2011;473:384–388. doi: 10.1038/nature09883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okabe S, et al. BAZF, a novel Bcl6 homolog, functions as a transcriptional repressor. Mol Cell Biol. 1998;18:4235–4244. doi: 10.1128/mcb.18.7.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Broxmeyer HE, et al. Aberrant regulation of hematopoiesis by T cells in BAZF-deficient mice. Mol Cell Biol. 2007;27:5275–5285. doi: 10.1128/MCB.01967-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen W, et al. Epigenetic and genetic loss of Hic1 function accentuates the role of p53 in tumorigenesis. Cancer Cell. 2004;6:387–398. doi: 10.1016/j.ccr.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 49.Bilic I, et al. Negative regulation of CD8 expression via Cd8 enhancer-mediated recruitment of the zinc finger protein MAZR. Nat Immunol. 2006;7:392–400. doi: 10.1038/ni1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakaguchi S, et al. The zinc-finger protein MAZR is part of the transcription factor network that controls the CD4 versus CD8 lineage fate of double-positive thymocytes. Nat Immunol. 2010;11:442–448. doi: 10.1038/ni.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kobayashi A, et al. A combinatorial code for gene expression generated by transcription factor Bach2 and MAZR (MAZ-related factor) through the BTB/POZ domain. Mol Cell Biol. 2000;20:1733–1746. doi: 10.1128/mcb.20.5.1733-1746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider A, Peukert K, Eilers M, Hanel F. Association of Myc with the zinc-finger protein Miz-1 defines a novel pathway for gene regulation by Myc. Curr Top Microbiol Immunol. 1997;224:137–146. doi: 10.1007/978-3-642-60801-8_14. [DOI] [PubMed] [Google Scholar]
- 53.Staller P, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 54.Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6:1054–1060. doi: 10.1038/ni1245. [DOI] [PubMed] [Google Scholar]
- 55.van Riggelen J, et al. The interaction between Myc and Miz1 is required to antagonize TGFbeta-dependent autocrine signaling during lymphoma formation and maintenance. Genes Dev. 2010;24:1281–1294. doi: 10.1101/gad.585710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kosan C, et al. Transcription factor miz-1 is required to regulate interleukin-7 receptor signaling at early commitment stages of B cell differentiation. Immunity. 2010;33:917–928. doi: 10.1016/j.immuni.2010.11.028. [DOI] [PubMed] [Google Scholar]
- 57.He LZ, et al. Two critical hits for promyelocytic leukemia. Mol Cell. 2000;6:1131–1141. doi: 10.1016/s1097-2765(00)00111-8. [DOI] [PubMed] [Google Scholar]
- 58.Hobbs RM, Seandel M, Falciatori I, Rafii S, Pandolfi PP. Plzf regulates germline progenitor self-renewal by opposing mTORC1. Cell. 2010;142:468–479. doi: 10.1016/j.cell.2010.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dhordain P, et al. Colocalization and heteromerization between the two human oncogene POZ/zinc finger proteins, LAZ3 (BCL6) and PLZF. Oncogene. 2000;19:6240–6250. doi: 10.1038/sj.onc.1203976. [DOI] [PubMed] [Google Scholar]
- 61.Sun G, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 62.Matic I, et al. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol Cell. 2010;39:641–652. doi: 10.1016/j.molcel.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 63.Siggs OM, Li X, Xia Y, Beutler B. ZBTB1 is a determinant of lymphoid development. J Exp Med. 2011 doi: 10.1084/jem.20112084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoatlin ME, et al. A novel BTB/POZ transcriptional repressor protein interacts with the Fanconi anemia group C protein and PLZF. Blood. 1999;94:3737–3747. [PubMed] [Google Scholar]
- 65.Piazza F, Costoya JA, Merghoub T, Hobbs RM, Pandolfi PP. Disruption of PLZP in mice leads to increased T-lymphocyte proliferation, cytokine production, and altered hematopoietic stem cell homeostasis. Mol Cell Biol. 2004;24:10456–10469. doi: 10.1128/MCB.24.23.10456-10469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miaw SC, Choi A, Yu E, Kishikawa H, Ho IC. ROG, repressor of GATA, regulates the expression of cytokine genes. Immunity. 2000;12:323–333. doi: 10.1016/s1074-7613(00)80185-5. [DOI] [PubMed] [Google Scholar]
- 67.Daniel JM. Dancing in and out of the nucleus: p120(ctn) and the transcription factor Kaiso. Biochim Biophys Acta. 2007;1773:59–68. doi: 10.1016/j.bbamcr.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 68.Park JI, et al. Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev Cell. 2005;8:843–854. doi: 10.1016/j.devcel.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 69.Prokhortchouk A, et al. Kaiso-deficient mice show resistance to intestinal cancer. Mol Cell Biol. 2006;26:199–208. doi: 10.1128/MCB.26.1.199-208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee KM, et al. The proto-oncoprotein KR-POK represses transcriptional activation of CDKN1A by MIZ-1 through competitive binding. Oncogene. 2011 doi: 10.1038/onc.2011.331. [DOI] [PubMed] [Google Scholar]
- 71.Weber A, et al. Zbtb4 represses transcription of P21CIP1 and controls the cellular response to p53 activation. EMBO J. 2008;27:1563–1574. doi: 10.1038/emboj.2008.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chouery E, et al. A novel deletion in ZBTB24 in a Lebanese family with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Clin Genet. 2011 doi: 10.1111/j.1399-0004.2011.01783.x. [DOI] [PubMed] [Google Scholar]
- 73.de Greef JC, et al. Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Am J Hum Genet. 2011;88:796–804. doi: 10.1016/j.ajhg.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 75.Lam KP, Kuhn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 76.Polli M, Dakic A, Light A, Wu L, Tarlinton DM, Nutt SL. The development of functional B lymphocytes in conditional PU. 1 knock-out mice. Blood. 2005;106:2083–2090. doi: 10.1182/blood-2005-01-0283. [DOI] [PubMed] [Google Scholar]
- 77.Georgopoulos K, et al. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 78.Kirstetter P, Thomas M, Dierich A, Kastner P, Chan S. Ikaros is critical for B cell differentiation and function. Eur J Immunol. 2002;32:720–730. doi: 10.1002/1521-4141(200203)32:3<720::AID-IMMU720>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 79.Carsetti R, Kohler G, Lamers MC. Transitional B cells are the target of negative selection in the B cell compartment. J Exp Med. 1995;181:2129–2140. doi: 10.1084/jem.181.6.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rolink AG, Andersson J, Melchers F. Characterization of immature B cells by a novel monoclonal antibody, by turnover and by mitogen reactivity. Eur J Immunol. 1998;28:3738–3748. doi: 10.1002/(SICI)1521-4141(199811)28:11<3738::AID-IMMU3738>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 81.Pui JC, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 82.Yan XQ, et al. A novel Notch ligand, Dll4, induces T-cell leukemia/lymphoma when overexpressed in mice by retroviral-mediated gene transfer. Blood. 2001;98:3793–3799. doi: 10.1182/blood.v98.13.3793. [DOI] [PubMed] [Google Scholar]
- 83.Balazs M, Martin F, Zhou T, Kearney J. Blood dendritic cells interact with splenic marginal zone B cells to initiate T-independent immune responses. Immunity. 2002;17:341–352. doi: 10.1016/s1074-7613(02)00389-8. [DOI] [PubMed] [Google Scholar]
- 84.Pillai S, Cariappa A, Moran ST. Marginal zone B cells. Annu Rev Immunol. 2005;23:161–196. doi: 10.1146/annurev.immunol.23.021704.115728. [DOI] [PubMed] [Google Scholar]
- 85.Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature. 2008;455:764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-specific memory B cell development. Annu Rev Immunol. 2005;23:487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
- 87.Kerfoot SM, et al. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity. 2011;34:947–960. doi: 10.1016/j.immuni.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pillai S, Cariappa A. The follicular versus marginal zone B lymphocyte cell fate decision. Nat Rev Immunol. 2009;9:767–777. doi: 10.1038/nri2656. [DOI] [PubMed] [Google Scholar]
- 89.Saito T, et al. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 2003;18:675–685. doi: 10.1016/s1074-7613(03)00111-0. [DOI] [PubMed] [Google Scholar]
- 90.Gibb DR, et al. ADAM10 is essential for Notch2-dependent marginal zone B cell development and CD23 cleavage in vivo. J Exp Med. 2010;207:623–635. doi: 10.1084/jem.20091990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tan JB, et al. Lunatic and manic fringe cooperatively enhance marginal zone B cell precursor competition for delta-like 1 in splenic endothelial niches. Immunity. 2009;30:254–263. doi: 10.1016/j.immuni.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 92.Song R, et al. Mind bomb 1 in the lymphopoietic niches is essential for T and marginal zone B cell development. J Exp Med. 2008;205:2525–2536. doi: 10.1084/jem.20081344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 94.Matsumoto M, et al. Affinity maturation without germinal centres in lymphotoxin-alpha-deficient mice. Nature. 1996;382:462–466. doi: 10.1038/382462a0. [DOI] [PubMed] [Google Scholar]
- 95.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 96.Ranuncolo SM, et al. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8:705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- 97.Casola S, et al. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc Natl Acad Sci U S A. 2006;103:7396–7401. doi: 10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Camacho SA, Kosco-Vilbois MH, Berek C. The dynamic structure of the germinal center. Immunol Today. 1998;19:511–514. doi: 10.1016/s0167-5699(98)01327-9. [DOI] [PubMed] [Google Scholar]
- 99.Allen CD, et al. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat Immunol. 2004;5:943–952. doi: 10.1038/ni1100. [DOI] [PubMed] [Google Scholar]
- 100.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 101.De Wit L, et al. Motor and functional recovery after stroke: a comparison of 4 European rehabilitation centers. Stroke. 2007;38:2101–2107. doi: 10.1161/STROKEAHA.107.482869. [DOI] [PubMed] [Google Scholar]
- 102.Liu YJ, Joshua DE, Williams GT, Smith CA, Gordon J, MacLennan IC. Mechanism of antigen-driven selection in germinal centres. Nature. 1989;342:929–931. doi: 10.1038/342929a0. [DOI] [PubMed] [Google Scholar]
- 103.Klein U, et al. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci U S A. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 105.Smith KG, Hewitson TD, Nossal GJ, Tarlinton DM. The phenotype and fateof the antibody-forming cells of the splenic foci. Eur J Immunol. 1996;26:444–448. doi: 10.1002/eji.1830260226. [DOI] [PubMed] [Google Scholar]
- 106.Hofer T, et al. Adaptation of humoral memory. Immunol Rev. 2006;211:295–302. doi: 10.1111/j.0105-2896.2006.00380.x. [DOI] [PubMed] [Google Scholar]
- 107.French DL, Laskov R, Scharff MD. The role of somatic hypermutation in the generation of antibody diversity. Science. 1989;244:1152–1157. doi: 10.1126/science.2658060. [DOI] [PubMed] [Google Scholar]
- 108.Kuppers R, Klein U, Hansmann ML, Rajewsky K. Cellular origin of human B-cell lymphomas. N Engl J Med. 1999;341:1520–1529. doi: 10.1056/NEJM199911113412007. [DOI] [PubMed] [Google Scholar]
- 109.Ramiro AR, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 110.Pasqualucci L, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 111.Dalla-Favera R, et al. BCL-6 in diffuse large-cell lymphomas. Important Adv Oncol. 1996:139–148. [PubMed] [Google Scholar]
- 112.Kerckaert JP, Deweindt C, Tilly H, Quief S, Lecocq G, Bastard C. LAZ3, a novel zinc-finger encoding gene, is disrupted by recurring chromosome 3q27 translocations in human lymphomas. Nat Genet. 1993;5:66–70. doi: 10.1038/ng0993-66. [DOI] [PubMed] [Google Scholar]
- 113.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 114.Melnick A, et al. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol Cell Biol. 2000;20:6550–6567. doi: 10.1128/mcb.20.17.6550-6567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schubot FD, Tropea JE, Waugh DS. Structure of the POZ domain of human LRF, a master regulator of oncogenesis. Biochem Biophys Res Commun. 2006;351:1–6. doi: 10.1016/j.bbrc.2006.09.167. [DOI] [PubMed] [Google Scholar]
- 116.Stogios PJ, Chen L, Prive GG. Crystal structure of the BTB domain from the LRF/ZBTB7 transcriptional regulator. Protein Sci. 2007;16:336–342. doi: 10.1110/ps.062660907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pessler F, Hernandez N. Flexible DNA binding of the BTB/POZ-domain protein FBI-1. J Biol Chem. 2003;278:29327–29335. doi: 10.1074/jbc.M302980200. [DOI] [PubMed] [Google Scholar]
- 118.Polo JM, et al. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med. 2004;10:1329–1335. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- 119.Parekh S, Prive G, Melnick A. Therapeutic targeting of the BCL6 oncogene for diffuse large B-cell lymphomas. Leuk Lymphoma. 2008;49:874–882. doi: 10.1080/10428190801895345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Poliseno L, et al. The proto-oncogene LRF is under post-transcriptional control of MiR-20a: implications for senescence. PLoS One. 2008;3:e2542. doi: 10.1371/journal.pone.0002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Verduci L, et al. MicroRNA (miRNA)-mediated interaction between leukemia/lymphoma-related factor (LRF) and alternative splicing factor/splicing factor 2 (ASF/SF2) affects mouse embryonic fibroblast senescence and apoptosis. J Biol Chem. 2010;285:39551–39563. doi: 10.1074/jbc.M110.114736. [DOI] [PMC free article] [PubMed] [Google Scholar]