Abstract
Covalent labeling and mass spectrometry are seeing increased used together as a way to obtain insight into the 3-dimensional structure of proteins and protein complexes. Several amino acid specific (e.g. diethylpyrocarbonate) and non-specific (e.g. hydroxyl radicals) labeling reagents are available for this purpose. Diethylpyrocarbonate (DEPC) is a promising labeling reagent because it can potentially probe up to 30% of the residues in the average protein and gives only one reaction product, thereby facilitating mass spectrometric analysis. It was recently reported, though, that DEPC modifications are labile for some amino acids. Here, we show that label loss is more significant and widespread than previously thought, especially for Ser, Thr, Tyr, and His residues, when relatively long protein digestion times are used. Such label loss ultimately decreases the amount of protein structural information that is obtainable with this reagent. We find, however, that the number of DEPC modified residues, and thus protein structural information, can be significantly increased by decreasing the time between the covalent labeling reaction and the mass spectrometric analysis. This is most effectively accomplished using short (e.g. 2 h) proteolytic digestions with enzymes such as immobilized chymotrypsin or Glu-C rather than using methods (e.g. microwave or ultrasonic irradiation) that accelerate proteolysis in other ways. Using short digestion times, we show that the percentage of solvent accessible residues that can be modified by DEPC increases from 44% to 67% for cytochrome c, 35% to 81% for myoglobin, and 76% to 95% for β-2-microglobulin. In effect, these increased numbers of modified residues improve the protein structural resolution available from this covalent labeling method. As compared to typical overnight digestion conditions, the short digestion times decrease the average distance between modified residues from 11 Å to 7 Å for myoglobin, 13 Å to 10 Å for cytochrome c, and 9 Å to 8 Å for β-2-microglobulin.
Introduction
Mass spectrometry (MS) is playing an ever increasing role in the study of protein 3-dimensional (3D) structure because more commonly used methods, such as NMR and X-ray crystallography, have some limitations with regard to protein molecular size, conformational flexibility, aggregation propensity, and/or limited sensitivity. Mass spectrometry alone cannot provide direct information about protein 3D structure in solution, and therefore must be used with labeling techniques such as H/D exchange [1–6], cross-linking [7–14], covalent labeling [15–17], and non-covalent labeling [18–20]. Alternatively, ion mobility mass spectrometry can also be used to study protein structure in the gas phase [21–23]. Our group has been investigating covalent labeling because such methods are complementary to H/D exchange and cross-linking, and in some cases covalent labeling offers some advantages over these approaches. In comparison to H/D exchange, for example, covalent labeling techniques provide structural information about amino acid side chains, while having limited back-exchange and scrambling. They also offer more straightforward data analysis as compared to cross-linking methods.
Covalent labeling approaches use reagents that non-specifically or specifically modify solvent exposed amino acids, and these modified residues are identified after proteolytic digestion and tandem mass spectrometry (MS/MS). Differential reactivity of side chains can indicate changes in protein structure, especially upon binding to a metal, ligand, or another protein [15–17]. Indeed, covalent labeling methods have significant merit when it comes to studying the interfaces of protein-protein complexes because these methods probe the solvent accessibility of amino acid side chains, which are critically important in mediating protein-protein interactions.
Numerous amino acid specific (e.g. succinimides for Lys residues) and non-specific (e.g. hydroxyl radicals) reagents have been used for covalent labeling. We have recently demonstrated that diethylpyrocarbonate (DEPC) has great promise as a labeling reagent for studying protein structure [24–28]. Despite many studies using DEPC, even with MS-based detection [29–34], it had been generally thought that this reagent reacts almost exclusively with His residues; however, in recent studies we have shown that DEPC can react with Lys, Tyr, Cys, Ser, and Thr residues in addition to His [24–27]. In fact, DEPC can potentially probe up to 30% of the residues in the average protein. This degree of coverage could make DEPC a very general surface mapping reagent for proteins having typical numbers of His, Tyr, Ser, Thr, Lys, and Cys residues. Because reactions with DEPC also only produce one type of product, this reagent has nice advantages when compared to non-specific reagents (e.g. hydroxyl radicals) that produce numerous product types. In effect, sites modified by DEPC can be more readily and sensitively identified by MS because modified peptide signals are not diluted across many products.
Although DEPC has great promise as a labeling reagent, we have found that DEPC does not always efficiently label all solvent accessible His, Tyr, Ser, Thr, Lys, and Cys residues in a given protein. One possible reason for this is that the reaction of DEPC with some amino acids is reversible with half-lives depending on the modified amino acid. For example, N-carbethoxyimidazole, which is the modified form of the histidine side chain, has a reported half-life of approximately 55 h at pH 7, 2 h at pH 2, and 18 min at pH 10 [35]. Lys and Tyr modifications are thought to be irreversible and reversible, respectively [36], and modifications of Ser and Thr residues are reversible with half-lives that are probably under 20 h [24]. Reversal of the covalent modification can lead to the loss of structural information especially if traditional digestion procedures are used to generate peptide fragments for bottom-up sequencing. This problem is especially critical for Ser and Thr residues because of their short half-lives and the fact that Ser and Thr are the third and seventh most common amino acids in proteins [37], accounting for about 13% of the sequence of an average protein. Methods that can reduce this reversibility will be very helpful in obtaining more information from DEPC as a labeling reagent.
In the work described here, we have investigated the reversibility of several different DEPC-modified amino acids in peptides and proteins and find that DEPC label loss occurs more readily than previously thought. Using these data, we then show that the labeling (and thus structural) information obtained with DEPC as a reagent can be substantially increased if the time between the covalent labeling reaction and MS analysis is reduced. We find that short digestion times minimize label reversibility and consequently enable the detection of twice as many modified amino acids in some cases, thereby increasing protein structural resolution.
Experimental Methods
Materials
Chymotrypsin was purchased from Roche Diagnostics (Indianapolis, IN) and immobilized chymotrypsin and triethylamine acetate (pH 8.0) were from Princeton Separations (Adelphia, NJ). Human β-2-microglobulin (β2m) was purchased from Fitzgerald Industries International, Inc (Concord, MA). Diethylpyrocabonate (DEPC), imidazole, iodoacetamide, ammonium bicarbonate, sulfo-N-hydroxysuccinimide acetate (NHSA), tris(2-carboxyethyl)phosphine (TCEP), equine heart cytochrome c, equine skeletal muscle myoglobin and Glu-C were obtained from Sigma-Aldrich (St. Louis, MO). Ammonium acetate, methanol, acetonitrile, and acetic acid were obtained from Fisher Scientific (Fair Lawn, NJ). Tris(hydroxymethyl)-aminomethane (Tris) was purchased from EM Science (Gladstone, NJ). Centricon molecular weight cutoff (MWCO) filters were from Millipore (Burlington, MA). Deionized water was generated from a Millipore (Burlington, MA) Simplicity 185 water purification system. All the peptides used in this study, except GH, GGH, and GGM were obtained from American Peptide Company (Sunnyvale, CA). The peptides GH, GGH, and GGM were purchased from Sigma-Aldrich (St. Louis, MO).
Covalent Modification
Carbethoxylation with DEPC: Stock solutions of DEPC were prepared in acetonitrile. The DEPC reactions of peptides and proteins were performed for 1 min at 37 °C and were initiated by adding DEPC in a molar excess of 3 or 4. The total reaction volume for the experiments was 100 µL, and the total amount of acetonitrile added was 1%. The reactions were quenched after 1 min by adding 10 mM imidazole [24].
Acetylation with NHSA: Stock solutions of NHSA were prepared in water. The labeling of peptides with 100 molar excess of NHSA was carried out for 3 min at 37 °C. The reactions were quenched by adding 200 mM Tris.
Proteolytic Digestion
Before proteolytic digestion, the modified proteins were cleaned with a 10,000 MWCO filter and reconstituted in deionized water to a final concentration of 250 µM. Cleaned β2m samples were reacted with TCEP (protein:TCEP=1:40) to reduce the disulfide bond. Iodoacetamide was added simultaneously at room temperature for 30 min in the dark to block the reduced Cys residues. β2m and the other proteins were incubated with 10% (v/v) acetonitrile at 50 °C for 45 min prior to digestion. Chymotrypsin was then added in a triethylamine buffer at pH 8.0 to yield a final enzyme/substrate ratio of 1:20. For conventional overnight digestion, the samples were incubated at 37 °C for 16 h. For the microwave-assisted digestions, the protein/enzyme mixture was irradiated in an MARS 230/6 microwave oven (CEM Corp., Matthews, NC) for 8 min. For the ultrasound-assisted digestions, the protein/enzyme mixture was exposed to an ultrasonic field for 4 h at 37 °C using a Branson 3510R-MTH Ultrasonic Cleaner. The digestions were stopped by inactivating the enzymes via the addition of acetic acid. For the 2 h digestions, immobilized chymostrypsin was added to yield an enzyme/substrate ratio of 1:10, and the protein/enzyme mixture was incubated at 37 °C. After the digestion with the immobilized chymotrypsin, the reaction mixture was centrifuged for 2 min at around 9000 relative centrifugal force to separate the enzyme from the protein. The enzyme Glu-C was used with an enzyme to protein ratio of 1:20 in an ammonium bicarbonate buffer at pH 8.0. The enzyme/protein mixture was incubated at 25 °C for 4 to 6 h. Acetic acid was then added to inactivate the enzyme and stop the digestion. In all cases, the modified proteins were immediately analyzed after the digestions.
Instrumentation
All mass spectral measurements were carried out on a Bruker AmaZon (Billerica, MA) quadrupole ion trap mass spectrometer equipped with an electrospray ionization source. Typically, the needle voltage was kept at ~4 kV, and the capillary temperature was set to 250 °C. Either CID or ETD was used to generate tandem mass spectra. For the CID experiments, an activation time of 40 ms was used, and the amplitude of the resonance excitation voltage during the CID experiment was optimized to ensure efficient ion dissociation. In the ETD experiments, low m/z cutoff (LMCO) values ranging from 50 to 180 and reaction times ranging from 50 ms to 180 ms were used. When sequencing the peptides by CID or ETD, ions with abundances at least 10 times greater than the noise were considered as product ions.
HPLC-MS analyses were conducted using an HP1100 (Agilent, Wilmington, DE) with a Discovery C18 column (15 cm × 2.1 mm, 5 µm particle size, Supelco, St. Louis, MO). Peptide mixtures or peptide fragments from the proteolytic digests were eluted using a gradient of methanol containing 0.1% acetic acid that increased from 10 to 100% methanol for 30 min at a flow rate of 0.25 mL/min.
Results and Discussion
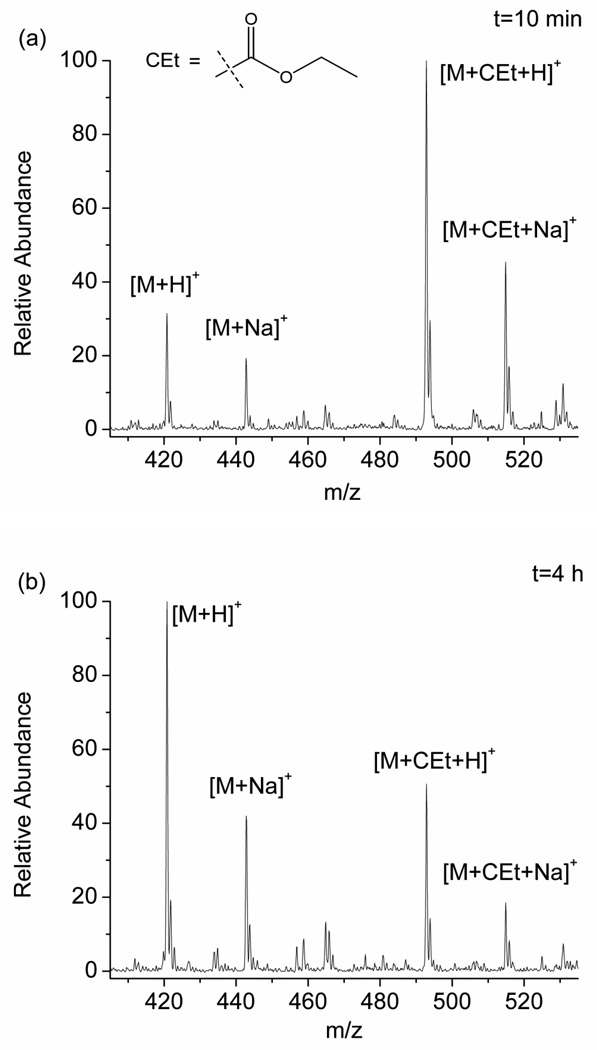
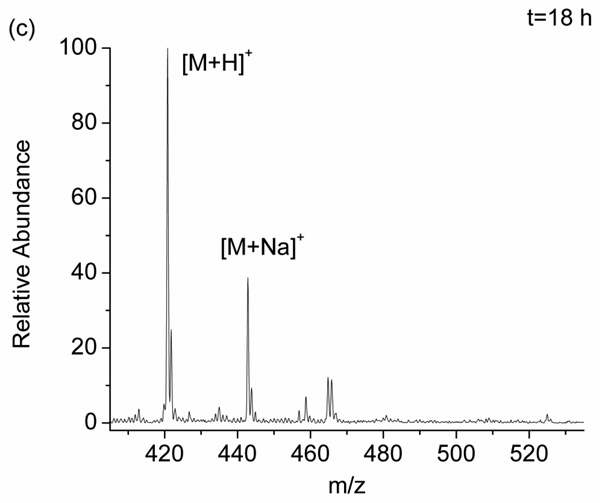
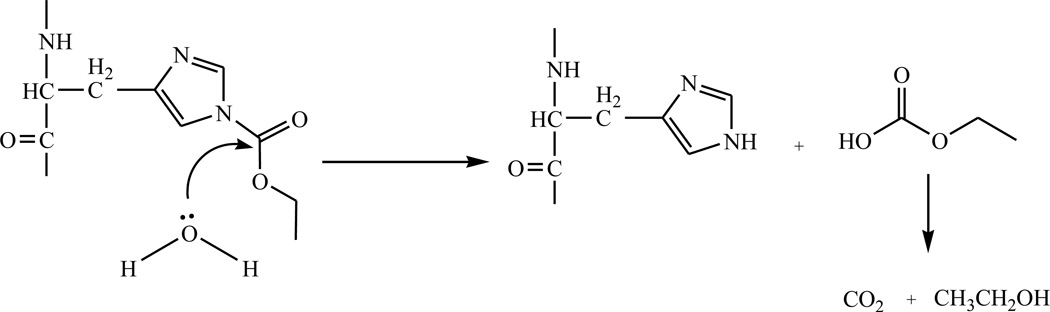
To better understand the scope of the DEPC hydrolysis reaction, we investigated a series of DEPC-modified peptides and proteins. Figure 1 shows an example of the modification reversibility for the peptide pGlu-HPG after the peptide was modified by DEPC and allowed to sit in solution for 10 min, 4 h, and 18 h at 37 °C. The mass spectra in Figure 1 show that the peaks corresponding to the modified peptide (m/z 493, [M+CEt+H]+ and m/z 515, [M+CEt+Na]+) decrease in abundance the longer the peptide sits in solution. This observation indicates that the modified His residue, which was confirmed by MS/MS, becomes unmodified over time. The percent modification, which is equal to the sum of the ion abundances of [M+CEt+H]+ and [M+CEt+Na]+ divided by the sum of the abundances of [M+CEt+H]+, [M+CEt+Na]+, [M+H]+, and [M+Na]+, decreases from 77 ± 2 % at 10 min to 33.6 ± 0.5 % after 4 h to 0 % after 18 h. Scheme 1 illustrates the hydrolysis reaction that labeled histidine undergoes [35]. It should be noted that the DEPC-related product of the hydrolysis reaction, ethyl hydrogen carbonate, is inherently unstable in water just like all the other alkyl hydrogen carbonates, and it readily decomposes into carbon dioxide and ethanol [37].
Figure 1.
Mass spectra of the DEPC-modified peptide pGlu-HPG after allowing the peptide to sit in solution for (a) 10 min, (b) 4 h, and (c) 18 h after quenching the reaction with imidazole.
Scheme 1.
Hydrolysis of carbethoxylated histidine
Similar experiments were performed with a series of other peptides containing modified His, Tyr, Lys, and N-termini (Table 1). For peptides with multiple modification sites, such as RPKPQQFFGLM-NH2 and MEHFRWGK, LC was used to separate the isomers prior to MS analysis. Two conclusions can be drawn from the data in Table 1. First, DEPC modifications to Lys residues and the N-termini are not reversible during an 18 h period. Second, His and Tyr residues more readily lose their label at rates that appear to depend on the surrounding amino acid residues. The behavior of His and Tyr is similar to that noted in our previous work on Ser- and Thr-containing peptides [24]. Interestingly, despite a reported half-life of 55 h for modified imidazoles at pH = 7 [35], we find that His residues in peptides can have much shorter half lives in some cases.
Table 1.
DEPC modification percentages for selected peptides after sitting in solution for various time periods after reaction with DEPC. All the experiments were repeated three times, and the means and standard deviations are reported.
| Peptide sequence | Modification | Modification percentages2 | ||
|---|---|---|---|---|
| sites1 | 10 min | 4 h | 18 h | |
| pGlu-HPG | His | 77 ± 2 | 33.6 ± 0.5 | 0.6 ± 0.93 |
| 4GHK | His | 68 ± 2 | 28 ± 6 | 1 ± 1 |
| 4GH | His | 72 ± 3 | 7 ± 1 | 0.5 ± 0.6 |
| 4KHG-NH2 | His | 53 ± 1 | 32 ± 3 | 11 ± 2 |
| 4GGH | His | 41 ± 1 | 18 ± 5 | 0.8 ± 0.2 |
| pGlu-LYENK | Tyr | 11.5 ± 0.8 | 4 ± 0.2 | 2.9 ± 0.2 |
| Ac-YVKD-CHO | Tyr | 22 ± 1 | 16 ± 2 | 16.4 ± 0.8 |
| 4FYGPV | Tyr | 10 ± 2 | 4 ± 2 | 1.6 ± 0.1 |
| RPKPQQFFGLM-NH2 | N-terminus | 71 ± 3 | 67 ± 4 | 72 ± 3 |
| Lys | 2.9 ± 0.3 | 2.8 ± 0.1 | 2.9 ± 0.2 | |
| MEHFRWGK | N-terminus | 22 ± 2 | 22 ± 1 | 23 ± 2 |
| His | 19 ± 1 | 13 ± 2 | 4 ± 2 | |
| Lys | 2 ± 1 | 1.8 ± 0.3 | 2.0 ± 0.3 | |
| GGM | N-terminus | 64 ± 4 | 65 ± 4 | 65 ± 2 |
Modification sites were confirmed by MS/MS.
Modification percentages are equal to the sum of the ion abundances of [M+CEt+H]+ and [M+CEt+Na]+ divided by the sum of the abundances of [M+CEt+H]+, [M+CEt+Na]+, [M+H]+, and [M+Na]+ (e.g. see Figure 1)
This modified peptide is not detected in every experiment.
Sulfo-N-hydroxysuccinimide acetate was used to block the N-termini and Lys side chains.
The modification reversibility of intact proteins was also investigated, and the results indicate that modification losses for whole proteins are not as rapid as with smaller peptides. The modification percentage for intact cytochrome c is 56 ± 3% when measured 10 minutes after the modification is quenched; while its modification percentage is 57 ± 2% when measured after 24 hours (see Figure S1 in the Supplemental Information). Similar experiments with myoglobin and β-2-microglobulin (β2m) indicate modification percentages of 83 ± 2% (10 min) and 73 ± 5% (24 h) and 68 ± 3% (10 min) and 49 ± 3% (24 h), respectively. One possible explanation for the lower modification loss from the intact proteins is that the protein buries the hydrophobic carbethoxy group in its interior after modification, thereby slowing the hydrolysis reaction that results in the cleavage of this group.
The effect of digestion conditions on labeling information
Because label loss can be substantial for some residues during a standard enzymatic digest time period (e.g. 18 h), we investigated different digestion procedures to identify the conditions that minimize label loss. Microwave [39, 40], ultrasonic irradiation [41–43] and immobilized enzymes [44] are convenient ways to accelerate protein digestions, so each of these methods was compared to conventional overnight digestion. For each digestion condition, the protein was first labeled with DEPC under conditions known to minimize structural changes to the protein. DEPC-protein reaction plots (Figure S2 in Supplemental Information) were used to ensure protein structural integrity during the labeling reactions just as described in previous work [24]. Results show that each protein maintains its structural integrity under the conditions used here. After enzymatically digesting the modified proteins, both ETD and CID were used to sequence the peptide fragments and identify the modification sites. Figure S3 gives two example tandem mass spectra to illustrate that modification sites can be determined with single amino acid resolution.
The DEPC modification sites and percentages obtained after digestion under different conditions indicate that the 2 h digestion with immobilized chymotrypsin provides the most labeling information (Table 2), both in terms of the number of modification sites found and the modification percentages. Using β2m as an example, we find that the 2 h digestion is extremely informative (Table 2). Given that residues with ≥ 30% solvent accessibility are usually considered to be solvent exposed [45], the percentage of the surface exposed residues that can be labeled (i.e. Ser, Thr, Lys, His, Cys, Tyr, and N-terminus, see Table S1 in Supplemental Information) is 95% with the 2 h digestion as compared to 76%, 38%, and 52% for the overnight, microwave-assisted, and ultrasound-assisted digestions, respectively. In addition, the measured modification levels with the 2 h digestion are higher in most cases. A similar trend is seen for cytochrome c (Table 2). The same modification sites are observed under both overnight and the 2 h digestion conditions, but fewer modification sites are observed after microwave and ultrasonic digestion conditions, even though all of the digestion conditions provide > 90% sequence coverage. About 45% of the modifiable residues are found labeled with the overnight and 2 h digestions, whereas only 33% of the modifiable surface exposed residues are measured after the microwave and ultrasound-assisted digestions. While the same number of modified sites is observed in cytochrome c after overnight and 2 h digestions, the measured modification levels for some amino acids are found to be higher in the 2 h digestion data (Table 2). The data for myoglobin indicates that the 2 h digestion results in a slightly higher percentage of measured modified residues (39%), whereas for the overnight (35%), microwave (29%), and ultrasound-assisted (29%) digestions lower percentages of modified residues are measured (Table 2). Again, the recovery of modifications is higher in almost all cases when the 2 h digestion is used.
Table 2.
A comparison of the modification percentage under several different digestion conditions for β-2-microglobulin, cytochrome c and myoglobin. All the experiments were repeated three times, and the means and standard deviations are reported.
| residue | overnight | 2 h | microwave | ultrasonic | 2+24 h | % SASA ratio1 |
|---|---|---|---|---|---|---|
| β-2-microglobulin2 | ||||||
| H13 | 10 ± 3 | 42.7 ± 0.7 | 14 ± 9 | 27.7 ± 0.3 | 10 ± 2 | 50.8 |
| H31 | 0.1 ± 0.13 | 7 ± 1 | N.D.4 | 2 ± 23 | N.D. | 32.7 |
| H51 | N.D. | 10 ± 1 | N.D. | N.D. | 0.7 ± 0.73 | 54.7 |
| S11 | 10 ± 5 | 8 ± 1 | 7 ± 3 | 5 ± 3 | 7 ± 3 | 11.4 |
| S20 | 1.1 ± 0.5 | 0.7 ± 0.1 | 0.8 ± 0.4 | 0.4 ± 0.3 | 0.7 ± 0.3 | 65.0 |
| S28 | 0.6 ± 0.2 | 0.31 ± 0.03 | N.D. | N.D. | N.D. | 19.7 |
| S33 | 2.4 ± 0.8 | 1.3 ± 0.3 | N.D. | 1 ± 13 | N.D. | 66.4 |
| S55 | 2 ± 23 | 3.4 ± 0.6 | N.D. | N.D. | N.D. | 55.9 |
| S57/K58 | 2 ± 23 | 3.3 ± 0.6 | N.D. | N.D. | N.D. | 35.2/96.3 |
| Y10 | N.D. | 0.065 ± 0.008 | N.D. | N.D. | N.D. | 41.9 |
| Y67/T68 | N.D. | 1.6 ± 0.8 | N.D. | N.D. | N.D. | 22.5/2.5 |
| Y78 | N.D. | 0.7 ± 0.3 | N.D. | N.D. | N.D. | 6.0 |
| N-terminus | 2.8 ± 0.4 | 2.4 ± 0.4 | 3 ± 2 | 2.3 ± 0.6 | 3.7 ± 0.7 | 85.0 |
| K6 | 0.20 ± 0.06 | 0.20 ± 0.07 | 0.2 ± 0.23 | 0.24 ± 0.05 | N.D. | 80.1 |
| K19 | 1.1 ± 0.3 | 4.1 ± 0.1 | 2 ± 1 | 3 ± 1 | 1.1 ± 0.2 | 69.0 |
| K41 | 2.3 ± 0.5 | 2.6 ± 0.7 | N.D. | N.D. | 1.9 ± 0.9 | 14.1 |
| K48 | 13 ± 2 | 8 ± 2 | N.D. | 13 ± 1 | 12 ± 2 | 81.8 |
| K75 | 0.6 ± 0.1 | 0.4 ± 0.2 | N.D. | N.D. | N.D. | 92.3 |
| K91 | 1.5 ± 0.3 | 1.4 ± 0.3 | 1 ± 13 | 1.0 ± 0.3 | 1.0 ± 0.2 | 61.5 |
| K94 | 2.3 ± 0.4 | 2.1 ± 0.5 | 2 ± 1 | 1.4 ± 0.4 | 1.5 ± 0.3 | 67.8 |
| % labeled5 | 76% | 95% | 38% | 52% | 48% | |
| cytochrome C2 | ||||||
| K25 | 30 ± 5 | 24 ± 2 | 13 ± 6 | 7 ± 4 | 23 ± 1 | 90.7 |
| K39 | 17 ± 2 | 20 ± 3 | 18 ± 4 | 19 ± 5 | 25 ± 3 | 72.9 |
| K53 | 2.6 ± 0.2 | 4.6 ± 0.5 | 3 ± 1 | 3.4 ± 0.9 | 1.1 ± 0.7 | 63.0 |
| K55 | 6 ± 1 | 9.1 ± 0.5 | 3 ± 33 | 7 ± 3 | 9.6 ± 0.7 | 36.5 |
| K60 | 1.6 ± 0.1 | 1.60 ± 0.08 | N.D. | N.D. | N.D. | 50.0 |
| K72 | 60 ± 3 | 56 ± 3 | N.D. | N.D. | 60 ± 10 | 75.3 |
| K73 | 18 ± 2 | 27 ± 1 | N.D. | N.D. | 17 ± 7 | 62.6 |
| K86 | 7 ± 1 | 10 ± 1 | 7 ± 2 | 8 ± 1 | 1 ± 13 | 41.6 |
| K87 | 9.3 ± 0.6 | 9.5 ± 0.9 | 8 ± 1 | 10.0 ± 0.3 | 8.1 ± 0.6 | 86.8 |
| K88 | 1.6 ± 0.1 | 1.7 ± 0.2 | 1.4 ± 0.2 | 1.77 ± 0.04 | 1.4 ± 0.1 | 94.0 |
| K99 | 45 ± 3 | 40 ± 10 | 30 ± 10 | 24 ± 2 | 14 ± 6 | 44.2 |
| K100 | 24 ± 2 | 22 ± 3 | 14 ± 6 | 14 ± 2 | 30 ± 10 | 50.4 |
| % labeled5 | 45% | 45% | 33% | 33% | 41% | |
| Myoglobin2 | ||||||
| H24 | N.D. | 3.1 ± 0.4 | N.D. | N.D. | N.D. | 2.7 |
| H36 | 1.9 ± 0.7 | 6.4 ± 0.5 | 2.9 ± 0.9 | 1.0 ± 0.1 | 3.2 ± 0.5 | 32.6 |
| H81 | 9 ± 5 | 38 ± 1 | 18 ± 2 | 9 ± 1 | 1 ± 13 | 89.1 |
| H116 | 11 ± 7 | 33.1 ± 0.7 | N.D. | N.D. | N.D. | 43.9 |
| S117 | 0.6 ± 0.4 | 1.74 ± 0.04 | N.D. | N.D. | N.D. | 51.3 |
| N-terminus | 11 ± 1 | 16 ± 3 | 19 ± 1 | 13 ± 2 | 39 ± 2 | 97.8 |
| K16 | 1.8 ± 0.7 | 1.3 ± 0.8 | 1 ± 23 | 2.3 ± 0.3 | 0.8 ± 0.83 | 22.1 |
| K42 | 0.24 ± 0.08 | 0.81 ± 0.06 | 0.4 ± 0.1 | 0.12 ± 0.02 | 0.40 ± 0.07 | 33.0 |
| K45 | 0.24 ± 0.08 | 0.81 ± 0.06 | 0.4 ± 0.1 | 0.12 ± 0.02 | 0.40 ± 0.07 | 54.9 |
| K77 | 19 ± 6 | 42 ± 8 | 45 ± 8 | 19 ± 3 | 3 ± 53 | 50.5 |
| K78 | 36 ± 6 | 37 ± 4 | 44 ± 6 | 27 ± 8 | 2 ± 23 | 46.2 |
| K79 | 6 ± 3 | 13 ± 2 | 14 ± 3 | 6 ± 3 | 1 ± 13 | 53.6 |
| % labeled5 | 35% | 39% | 29% | 29% | 29% | |
SASA was calculated using GETAREA 1.1 [45]. 1.4 Å was used as the probe radius, and the calculated SASA percentage is the ratio of the SASA of the side chain in the protein to the SASA of the side chain (X) in the unstructured Gly-X-Gly tripeptide. Residues with %SASA values that exceed 50% are typically considered to be solvent exposed, and residues with ratios less than 20% are typically considered to be buried. We chose 30% as the cutoff for determining if a residue is solvent exposed.
The PDB IDs for β-2-microglobulin, cytochrome c and myoglobin that were used to determine SASA were 1JNJ, 1AKK and 1DWR, respectively. For β-2-microglobulin, 1JNJ consists of 20 NMR structures, and so the reported SASA values are the average from these 20 structures.
This modified peptide is not detected in every experiment.
N.D. indicates that modified peptide is not detected in any experiment under these conditions.
% labeled corresponds to the percentage of the surface exposed modifiable (i.e. His, Lys, Cys, Ser, Thr, and Tyr) residues that are found to be labeled. A complete list of modifiable residues in β-2-microglobulin, cytochrome c and myoglobin and their calculated SASA values can be found in Table S1 in the Supplemental Information.
The relatively low recovery of modified amino acids after microwave and ultrasound-assisted digestions is interesting. The results suggest that in addition to accelerating the digestion reactions, microwaves and ultrasound also accelerate hydrolysis of the DEPC-modified residues, even to greater extents than they accelerate proteolysis, thereby increasing the modification losses. Several papers report that microwave irradiation can accelerate common organic transformations such as hydrolysis reactions [46, 47]. Moreover, microwaves also accelerate the hydrolysis of peptide bonds in the absence of enzymes [48, 49]. Ultrasonic irradiation is also known to accelerate hydrolysis reactions [50]. Therefore, it appears that microwave- and ultrasound-assisted digestion methods are not good choices when trying to identify residues modified by DEPC.
To confirm that the shortened digestion times enable more modifications to be retained before MS analysis, labeled proteins were digested for 2 h with immobilized chymotrypsin, and then, after removing the immobilized chymotrypsin, the resulting peptide fragments were incubated at 37 °C for 24 h to monitor if further label loss occurred (Table 2, 6th column). The data clearly indicate that label loss occurs as a result of the peptide fragments remaining in solution for an additional 24 h. These observations confirm that label loss occurs over time and that shorter digestion times help minimize label loss, so that a greater number and percentage of modified residues can be retained and measured. The extents of label loss follow the trends observed in Table 1 and in our previous work [24]. The modification percentages for the N-termini and Lys residues do not substantially decrease after 24 h, indicating that they maintain their labels. In contrast, His, Tyr, Ser, and Thr residues more readily lose their labels after sitting in solution for 24 h. These experiments also help us understand the label loss data for the undigested proteins described above. Cytochrome c did not lose any DEPC modifications (56 ± 3% to 57 ± 2%) after sitting in solution for 24 h, while β2m lost more labels (68 ± 3% to 49 ± 3%) after sitting in solution for 24 h. Almost all of the residues labeled in cytochrome c are Lys residues, which undergo little label loss. For β2m, more than half of the modification sites are His, Tyr, Ser, or Thr residues, which are the amino acids that most easily lose the modification.
It is important to note, however, that the amount of label loss observed for the intact β2m after 24 h is not as great as expected based on the 2+24 h results in Table 2. Less label loss in the case of the undigested protein suggests that intact modified proteins might partially bury the DEPC modified side chains, thereby slowing hydrolysis. If this is the case, then we might be able to increase modification recoveries by generating larger proteolytic fragments that partially protect the modified residues from hydrolysis.
Glu-C and middle-down sequencing
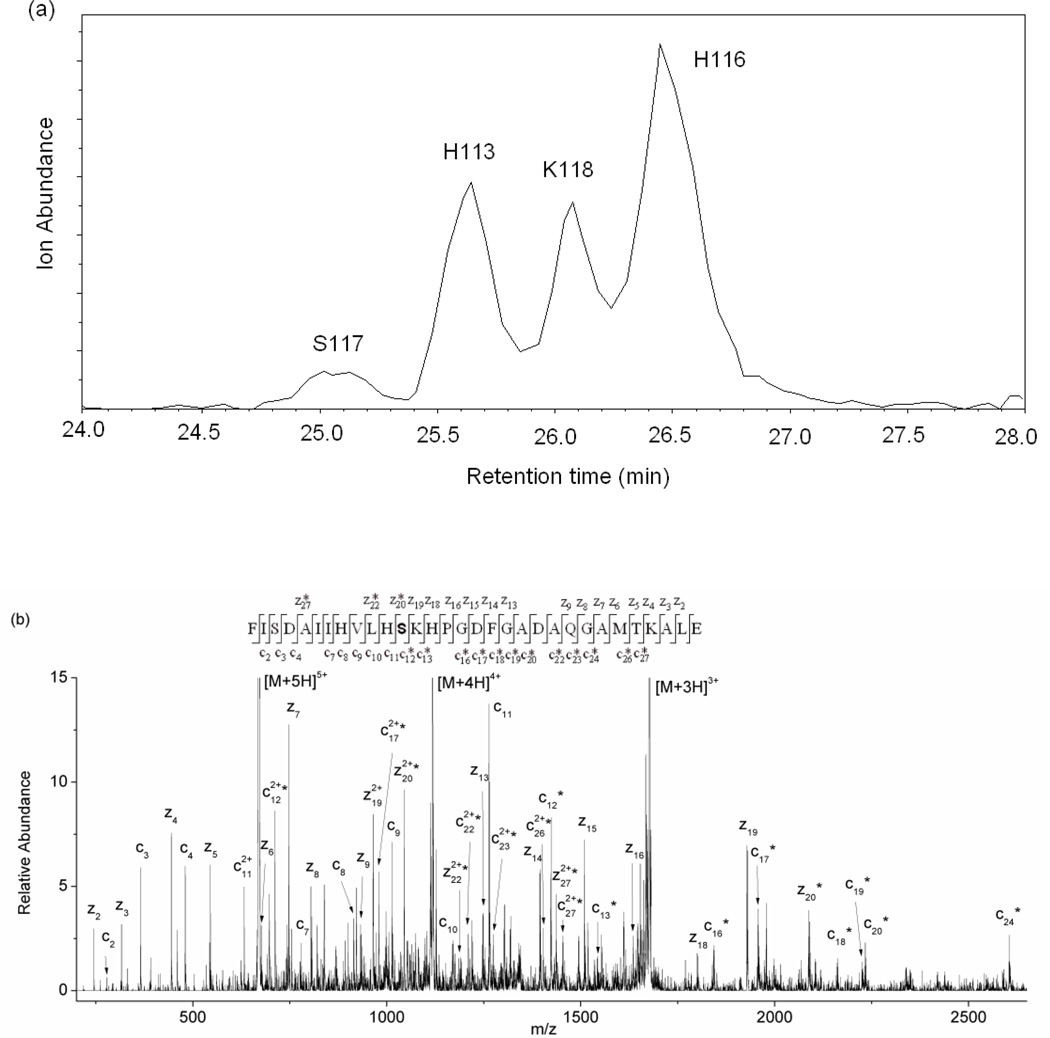
To test the idea that longer peptide fragments may protect the modified residues from hydrolysis, we used the enzyme Glu-C to generate longer proteolytic fragments, while still maintaining a short overall digestion time. Glu-C cleaves peptide bonds on the C-terminal side of Glu residues. The fragments that are typically produced are large enough that this could be considered a form of middle-down sequencing [51–55]. The larger peptide fragments produced by Glu-C lead to more peptide fragments with multiple modification sites. In many cases, LC is able to separate these peptide isomers, so that MS/MS can readily confirm the modification sites. For example, the peptide fragment, 106FISDAIIHVLHSKHPGDFGADAQGAMTKALE136, from myoglobin was found to be modified at H113, H116, S117, and K118. Each of these modified forms of the peptide can be separated by LC, and the modification sites can be determined by ETD (Figure 2). In other cases, modified peptide isomers elute at the same time, but ETD of these isomers almost always enables the multiple modification sites to be unambiguously identified. For example, the peptide fragment, 137LFRNDIAAKYKE148, from myoglobin was found to be modified at K145 and K147 even though these modified forms elute together (Figure S4). Unfortunately, the inability to completely separate all modified peptide forms makes it difficult to accurately determine the modification percentages for all the modified sites.
Figure 2.
(a) Extracted ion chromatogram (EIC) of m/z 671, which is the +5 ion of the DEPC-modified peptide 106FISDAIIHVLHSKHPGDFGADAQGAMTKALE136 from myoglobin. (b) An example ETD tandem mass spectrum of the first chromatographic peak of the modified Phe106-Glu136 fragment, illustrating how ETD can identify the modified amino acid. A series of unmodified z ions from z2 to z19, modified z20, z22, and z27 ions, a series of unmodified c ions from c2 to c11, and a series of modified c ions from c12 to c27 confirm that Ser117 is the site of modification. The product ions with an asterisk are the product ions that contain the DEPC modification.
Because only 45% and 39% of the modifiable surface exposed residues of cytochrome c and myoglobin are labeled using the 2 h digestion procedure, we only digested these proteins with Glu-C. 95% of the modifiable residues in β2m are already labeled, so middle-down sequencing is not expected to provide significantly more information for this protein. In addition, the relatively high number of Glu residues in β2m yields peptide fragments that are similar in size to those generated by chymotrypsin, thereby providing no real advantage. In contrast, Glu-C generates peptide fragments for cytochrome c and myoglobin that have, on average, almost twice as many residues as those produced by chymotrypsin.
When Glu-C is used to digest DEPC-modified cytochrome c and myoglobin, we found six new modification sites for cytochrome c and 17 new sites for myoglobin as compared to the 2 h digestion with chymotrypsin. The six new sites that are modified in cytochrome c are all Lys residues. Overall, the measured modification sites in this protein upon Glu-C digestion are: Lys5, Lys7, Lys8, Lys22, Lys25, Lys27, Lys39, Lys53, Lys55, Lys60, Lys72, Lys73, Lys79, Lys86, Lys87, Lys88, Lys99, and Lys100 (Table S1 in Supplemental Information). The presence of newly found modified Lys residues suggests that some Lys residues can undergo hydrolysis at faster rates in some circumstances even though the data in Tables 1 and 2 indicate the label loss reaction is slow for Lys. The inability to find additional modified His, Thr, Ser, or Tyr residues might be because there are only 7 of these residues that have SASA percentages above 30% and most of them have SASA percentages close to 30%. Overall, a total of 18 residues were found to be modified, which corresponds to 67% of the modifiable residues in cytochrome c (see Table S1 for a complete list of modifiable residues and their modification status). This is a clear improvement over the 45% obtained with the 2 h digestion with chymotrypsin.
For myoglobin, a total of 27 modification sites were found. These sites are: the N-terminus, His36, His48, His64, His81, His97, His113, His116, Tyr103, Tyr146, Ser92, Ser117, Thr95, Lys42, Lys45, Lys50, Lys62, Lys63, Lys77, Lys78, Lys79, Lys87, Lys96, Lys98, Lys118, Lys145, and Lys147 (Table S1). As indicated before, 17 of these sites are new. Eight of these new sites are His, Tyr, Ser, or Thr residues. Two of these newly identified sites, Tyr146 and Ser92, have low SASA percentages (Table S1). Even though the modification percentages for these residues are very low (i.e. < 1 %), observing these sites modified might suggest that a small fraction of the proteins are structurally perturbed, thereby exposing these residues. Our previous DEPC labeling experiments with myoglobin gave no evidence for any perturbations to the protein structure under identical reaction conditions [24], but the ability to find more modified sites with the current conditions could be revealing that some fraction of the proteins do undergo slight structural perturbations. This possibility highlights the advantage of being able to identify more labeled sites. The remaining nine newly discovered sites in myoglobin are Lys residues. Considering these data as a whole, we find that 81% of the solvent exposed modifiable residues are labeled when the protein is analyzed using Glu-C and middle-down sequencing (see Table S1 for a complete list of modifiable residues and their modification status). This percentage of measured modification sites is a significant improvement over the 39% that was obtained with the 2 h digestion using chymotrypsin.
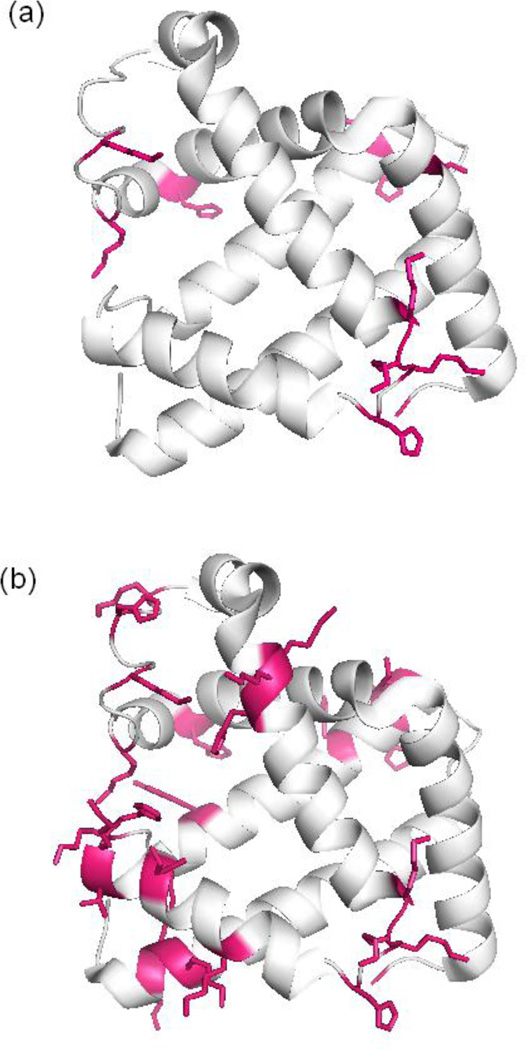
Increasing the number of measured modification sites is advantageous for protein structure determination. In effect, measuring more modified residues provides greater protein structural resolution. To quantify the improved resolution obtained with the shorter digestion conditions, we calculated the average distance between adjacent modification sites to arrive at an effective resolution for each digestion condition. To do this, we mapped the measured modification sites on the crystal or NMR structure of each protein (e.g. Figure 3), and then the distance between a given modified residue and the next closest modified residue was determined using Pymol [56]. The overall average of these distances was then calculated to provide an effective resolution. Figure 3 illustrates the measured modification sites mapped on the crystal structure of myoglobin using the data from two different digestion conditions. This figure shows that a greater effective resolution is obtained upon digesting the protein with Glu-C (Figure 3b) as compared to a conventional overnight digestion (Figure 3a). The protein structural resolution that is calculated upon the Glu-C digestion is 7 ± 2 Å, while the resolution from the overnight digestion data is 11 ± 3 Å. Conducting a similar calculation for all three proteins under the different digestion conditions indicates that in all cases the structural resolution is greater when a shorter digestion time is used (Table 3).
Figure 3.
Mapping of the measured modification sites on myoglobin. (a) Modification sites after conventional overnight digestion and (b) modification sites after digestion with Glu-C. The DEPC modified residues are indicated in red.
Table 3.
Calculated protein structural resolution from DEPC-based labeling after digesting the proteins under different conditions.
| myoglobin | cytochrome c | β-2-microglobulin | |
|---|---|---|---|
| Glu-C | 7 ± 21 | 10 ± 3 | -- |
| 2 h | 10 ± 3 | 13 ± 5 | 8 ± 3 |
| overnight | 11 ± 3 | 13 ± 5 | 9 ± 3 |
| microwave | 11 ± 3 | 14 ± 5 | 11 ± 5 |
| ultrasonic | 11 ± 3 | 14 ± 5 | 9 ± 3 |
| 2+24 h | 11 ± 3 | 13 ± 5 | 11 ± 5 |
The resolution was calculated by considering the average distance between adjacent modification sites after mapping the measured modification sites on the crystal or NMR structure of each protein.
Conclusions
By studying several peptides and proteins, we find that DEPC-modified amino acids are more labile than previously thought. This lability can lead to losses in protein structural information. Label loss is most significant for Ser, Thr, and Tyr residues but also occurs readily for His residues. By shortening the time between the modification reaction and MS analysis, though, the number of modification sites that are identified after labeling with DEPC can be increased, and this greater number of labeled sites translates into increased structural information. As expected, more extensive levels of DEPC modification are primarily due to limiting the hydrolysis of Ser, Thr, Tyr, and His residues. The decreased time between the modification reaction and protein sequencing analysis by MS can be achieved using shorter digestion times. Microwave and ultrasonic-irradiation are two ways to accelerate protein digestion, but these methods also accelerate hydrolysis to a greater extent, resulting in lower measured levels of modification. In contrast, a simple two-hour digestion with immobilized chymotrypsin was found to be superior for obtaining more structural information for the proteins studied here. Surprisingly, protein digestion using Glu-C coupled with middle-down sequencing provides the most extensive labeling information. Indeed, the number of labeled sites increases by 1.5-fold for cytochrome c and 2-fold for myoglobin, resulting in notable improvements in the structural resolution of this covalent labeling technique. This phenomenon is likely due to the production of larger peptide fragments that are able to partially bury the relatively hydrophobic carbethoxy group, thereby shielding the modified residues from hydrolysis. Overall, the amount of structural information that is accessible with DEPC indicates that it might be useful as a standalone labeling reagent when followed by short digestion times and middle-down sequencing, although alternate short-time digestion conditions would need to be optimized for more intractable proteins. Because this reagent also only produces a single reaction product, thereby simplifying the MS analysis, it may serve as a viable alternative to hydroxyl radical labeling.
Supplementary Material
Acknowledgements
This work was supported by a grant from the National Institutes of Health (R01 GM075092). We thank Prof. Julian Tyson for allowing us to use the microwave oven for the microwave-assisted digestions.
References
- 1.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 2.Engen JR. Analysis of Protein Conformation and Dynamics by Hydrogen/Deuterium Exchange MS. Anal. Chem. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoofnagle AN, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annu. Rev. Bioph. Biom. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 4.Marcsisin SR, Engen JR. Hydrogen exchange mass spectrometry: what is it and what can it tell us? Anal. Bioanal. Chem. 2010;397:967–972. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsutsui Y, Wintrode PL. Hydrogen/deuterium exchange-mass spectrometry: a powerful tool for probing protein structure, dynamics and interactions. Curr. Med. Chem. 2007;14:2344–2358. doi: 10.2174/092986707781745596. [DOI] [PubMed] [Google Scholar]
- 6.Englander SW. Hydrogen Exchange and Mass Spectrometry: A Historical Perspective. J. Am. Soc. Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrotchenko Evgeniy V, Borchers Christoph H. Crosslinking combined with mass spectrometry for structural proteomics. Mass Spectrom. Rev. 2010;29:862–876. doi: 10.1002/mas.20293. [DOI] [PubMed] [Google Scholar]
- 8.Sinz A. Investigation of protein-protein interactions in living cells by chemical crosslinking and mass spectrometry. Anal.Bioanal. Chem. 2010;397:3433–3440. doi: 10.1007/s00216-009-3405-5. [DOI] [PubMed] [Google Scholar]
- 9.Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 10.Steen H, Jensen ON. Analysis of protein-nucleic acid interactions by photochemical cross-linking and mass spectrometry. Mass Spectrom. Rev. 2002;21:163–182. doi: 10.1002/mas.10024. [DOI] [PubMed] [Google Scholar]
- 11.Singh P, Panchaud A, Goodlett DR. Chemical Cross-Linking and Mass Spectrometry As a Low-Resolution Protein Structure Determination Technique. Anal. Chem. 2010;82:2636–2642. doi: 10.1021/ac1000724. [DOI] [PubMed] [Google Scholar]
- 12.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Probing native protein structures by chemical cross-linking, mass spectrometry and bioinformatics. Mol. Cell. Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakravarti B, Lewis SJ, Chakravarti DN, Raval A. Three dimensional structures of proteins and protein complexes from chemical cross-linking and mass spectrometry a biochemical and computational overview. Curr. Proteomics. 2006;3:1–21. [Google Scholar]
- 14.Back JW, De Jong L, Muijsers AO, De Koster CG. Chemical cross-linking and mass spectrometry for protein structural modeling. J. Mol. Biol. 2003;331:303–313. doi: 10.1016/s0022-2836(03)00721-6. [DOI] [PubMed] [Google Scholar]
- 15.Mendoza VL, Vachet RW. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom. Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konermann L, Stocks BB, Pan Y, Tong X. Mass spectrometry combined with oxidative labeling for exploring protein structure and folding. Mass Spectrom. Rev. 2010;29:651–667. doi: 10.1002/mas.20256. [DOI] [PubMed] [Google Scholar]
- 17.Roeser J, Bischoff R, Bruins AP, Permentier HP. Oxidative protein labeling in mass-spectrometry-based proteomics. Anal. Bioanal. Chem. 2010;397:3441–3455. doi: 10.1007/s00216-010-3471-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu ZJ, Cheng SJ, Gailie DR, Julian RR. Exploring the Mechanism of Selective Noncovalent Adduct Protein Probing Mass Spectrometry Utilizing Site-Directed Mutagenesis To Examine Ubiquitin. Anal. Chem. 2008;80:3846–3852. doi: 10.1021/ac800176u. [DOI] [PubMed] [Google Scholar]
- 19.Ly T, Julian RR. Protein–Metal Interactions of Calmodulin and α-Synuclein Monitored by Selective Noncovalent Adduct Protein Probing Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2008;19:1663–1672. doi: 10.1016/j.jasms.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Ly T, Julian RR. Using ESI-MS to Probe Protein Structure by Site-Specific Noncovalent Attachment of 18-Crown-6. J. Am. Soc. Mass Spectrom. 2006;17:1209–1215. doi: 10.1016/j.jasms.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 21.Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH., Jr Ion mobility-mass spectrometry. J. Mass Spectrom. 2008;43:1–22. doi: 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- 22.Ruotolo BT, Hyung S-j, Robinson PM, Giles K, Bateman RH, Robinson CV. Ion mobility-mass spectrometry reveals long-lived, unfolded intermediates in the dissociation of protein complexes. Angew. Chem. Int. Ed. 2007;46:8001–8004. doi: 10.1002/anie.200702161. [DOI] [PubMed] [Google Scholar]
- 23.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung S-J, Robinson CV. Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 24.Mendoza VL, Vachet RW. Protein Surface Mapping Using Diethylpyrocarbonate with Mass Spectrometric Detection. Anal. Chem. 2008;80:2895–2904. doi: 10.1021/ac701999b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendoza VL, Baron-Rodriguez MA, Blanco C, Vachet RW. Structural Insights into the Pre-Amyloid Tetramer of β-2-Microglobulin from Covalent Labeling and Mass Spectrometry. Biochemistry. 2011;50:6711–6722. doi: 10.1021/bi2004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendoza VL, Antwi K, Baron-Rodriguez MA, Blanco C, Vachet RW. Structure of the Preamyloid Dimer of β-2-Microglobulin from Covalent Labeling and Mass Spectrometry. Biochemistry. 2010;49:1522–1532. doi: 10.1021/bi901748h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srikanth R, Mendoza VL, Bridgewater JD, Zhang G, Vachet RW. Copper Binding to β-2-Microglobulin and Its Pre-Amyloid Oligomers. Biochemistry. 2009;48:9871–9881. doi: 10.1021/bi901172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srikanth R, Wilson J, Burns CS, Vachet RW. Identification of the Copper(II) Coordinating Residues in the Prion Protein by Metal-Catalyzed Oxidation Mass Spectrometry: Evidence for Multiple Isomers at Low Copper(II) Loadings. Biochemistry. 2008;47:9258–9268. doi: 10.1021/bi800970m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glocker MO, Kalkum M, Yamamoto R, Schreurs J. Selective Biochemical Modification of Functional Residues in Recombinant Human Macrophage Colony-Stimulating Factor β (rhM-CSF β): Identification by Mass Spectrometry. Biochemistry. 1996;35:14625–14633. doi: 10.1021/bi961199o. [DOI] [PubMed] [Google Scholar]
- 30.Dage JL, Sun H, Halsall HB. Determination of diethylpyrocarbonate-modified amino acid residues in α1-acid glycoprotein by high performance liquid chromatography electrospray ionization-mass spectrometry and matrix-assisted laser desorption/ionization time-of-flight-mass spectrometry. Anal. Biochem. 1998;257:176–185. doi: 10.1006/abio.1997.2552. [DOI] [PubMed] [Google Scholar]
- 31.Kalkum M, Przybylski M, Glocker MO. Structure Characterization of Functional Histidine Residues and Carbethoxylated Derivatives in Peptides and Proteins by Mass Spectrometry. Bioconjugate Chem. 1998;9:226–235. doi: 10.1021/bc970162t. [DOI] [PubMed] [Google Scholar]
- 32.Tsubaki M, Kobayashi K, Ichise T, Takeuchi F, Tagawa S. Diethyl Pyrocarbonate Modification Abolishes Fast Electron Accepting Ability of Cytochrome b561 from Ascorbate but Does Not Influence Electron Donation to Monodehydroascorbate Radical: Identification of the Modification Sites by Mass Spectrometric Analysis. Biochemistry. 2000;39:3276–3284. doi: 10.1021/bi991883d. [DOI] [PubMed] [Google Scholar]
- 33.Jin XR, Abe Y, Li CY, Hamasaki N. Histidine-834 of Human Erythrocyte Band 3 Has an Essential Role in the Conformational Changes That Occur during the Band 3-Mediated Anion Exchange. Biochemistry. 2003;42:12927–12932. doi: 10.1021/bi0350809. [DOI] [PubMed] [Google Scholar]
- 34.Qin K, Yang Y, Mastrangelo P, Westaway D. Mapping Cu(II) binding sites in prion proteins by diethyl pyrocarbonate modification and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometric footprinting. J. Biol. Chem. 2002;277:1981–1990. doi: 10.1074/jbc.M108744200. [DOI] [PubMed] [Google Scholar]
- 35.Melchior WB, Jr, Fahrney D. Ethoxyformylation of proteins. Reaction of ethoxyformic anhydride with α-chymotrypsin, pepsin, and pancreatic ribonuclease at pH 4. Biochemistry. 1970;9:251–258. doi: 10.1021/bi00804a010. [DOI] [PubMed] [Google Scholar]
- 36.Miles EW. Modification of histidyl residues in proteins by diethylpyrocarbonate. Method. Enzymol. 1977;47:431–442. doi: 10.1016/0076-6879(77)47043-5. [DOI] [PubMed] [Google Scholar]
- 37.Trinquier G, Sanejouand Y-H. Which effective property of amino acids is best preserved by the genetic code? Protein Eng. 1998;11:153–169. doi: 10.1093/protein/11.3.153. [DOI] [PubMed] [Google Scholar]
- 38.Solomons TWG, Fryhle C. Organic Chemistry. 9th edn. New York: John wiley & Sons; 1998. [Google Scholar]
- 39.Lopez-Ferrer D, Canas B, Vazquez J, Lodeiro C, Rial-Otero R, Moura I, Capelo JL. Sample treatment for protein identification by mass spectrometry-based techniques. TrAC-Trend. Anal. Chem. 2006;25:996–1005. [Google Scholar]
- 40.Pramanik BN, Mirza UA, Ing YH, Liu Y-H, Bartner PL, Weber PC, Bose AK. Microwave-enhanced enzyme reaction for protein mapping by mass spectrometry: a new approach to protein digestion in minutes. Protein Sci. 2002;11:2676–2687. doi: 10.1110/ps.0213702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez-Ferrer D, Capelo JL, Vazquez J. Ultra Fast Trypsin Digestion of Proteins by High Intensity Focused Ultrasound. J. Proteome Res. 2005;4:1569–1574. doi: 10.1021/pr050112v. [DOI] [PubMed] [Google Scholar]
- 42.Carreira RJ, Cordeiro FM, Moro AJ, Rivas MG, Rial-Otero R, Gaspar EM, Moura I, Capelo JL. New findings for in-gel digestion accelerated by high-intensity focused ultrasound for protein identification by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J. Chromatogr. A. 2007;1153:291–299. doi: 10.1016/j.chroma.2006.09.078. [DOI] [PubMed] [Google Scholar]
- 43.Rial-Otero R, Carreira RJ, Cordeiro FM, Moro AJ, Fernandes L, Moura I, Capelo JL. Sonoreactor-Based Technology for Fast High-Throughput Proteolytic Digestion of Proteins. J. Proteome Res. 2007;6:909–912. doi: 10.1021/pr060508m. [DOI] [PubMed] [Google Scholar]
- 44.Duan J, Sun L, Liang Z, Zhang J, Wang H, Zhang L, Zhang W, Zhang Y. Rapid protein digestion and identification using monolithic enzymatic microreactor coupled with nano-liquid chromatography-electrospray ionization mass spectrometry. J. Chromatogr. A. 2006;1106:165–174. doi: 10.1016/j.chroma.2005.11.102. [DOI] [PubMed] [Google Scholar]
- 45.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 1998;19:319–333. [Google Scholar]
- 46.Roberts BA, Strauss CR. Toward Rapid, "Green", Predictable Microwave-Assisted Synthesis. Accounts Chem. Res. 2005;38:653–661. doi: 10.1021/ar040278m. [DOI] [PubMed] [Google Scholar]
- 47.Gedye RN, Smith FE, Westaway KC. The rapid synthesis of organic compounds in microwave ovens. Can. J. Chemistry. 1988;66:17–26. [Google Scholar]
- 48.Lill JR, Ingle ES, Liu PS, Pham V, Sandoval WN. Microwave-assisted proteomics. Mass Spectrom. Rev. 2007;26:657–671. doi: 10.1002/mas.20140. [DOI] [PubMed] [Google Scholar]
- 49.Yu HM, Chen ST, Chiou SH, Wang KT. Determination of amino acids on Merrifield resin by microwave hydrolysis. J. Chromatogr. 1988;456:357–362. doi: 10.1016/0021-9673(86)80032-2. [DOI] [PubMed] [Google Scholar]
- 50.Luque de Castro MD, Priego-Capote F. Ultrasound-assisted preparation of liquid samples. Talanta. 2007;72:321–334. doi: 10.1016/j.talanta.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 51.Ge Y, Rybakova IN, Xu Q, Moss RL. Top-down high-resolution mass spectrometry of cardiac myosin binding protein C revealed that truncation alters protein phosphorylation state. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12658–12663. S12658/1–S12658/12. doi: 10.1073/pnas.0813369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warren MRE, Parker CE, Mocanu V, Klapper D, Borchers CH. Electrospray ionization tandem mass spectrometry of model peptides reveals diagnostic fragment ions for protein ubiquitination. Rapid Commun. Mass Spectrom. 2005;19:429–437. doi: 10.1002/rcm.1798. [DOI] [PubMed] [Google Scholar]
- 53.Garcia BA, Siuti N, Thomas CE, Mizzen CA, Kelleher NL. Characterization of neurohistone variants and post-translational modifications by electron capture dissociation mass spectrometry. Int. J. Mass Spectrom. 2007;259:184–196. [Google Scholar]
- 54.Young NL, Di Maggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. High throughput characterization of combinatorial histone codes. Mol. Cell. Proteomics. 2009;8:2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garcia BA, Thomas CE, Kelleher NL, Mizzen CA. Tissue-Specific Expression and Post-Translational Modification of Histone H3 Variants. J. Proteome Res. 2008;7:4225–4236. doi: 10.1021/pr800044q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeLano WL. San Carlos, CA, USA: DeLano Scientific; 2002. The PyMol Molecular Graphics System. httP://www.pymol.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.