Abstract
Background
The protein targets for general anesthetics remain unclear. A tool to predict anesthetic binding for potential binding targets is needed. In this study, we explore whether a computational method, AutoDock, could serve as such a tool.
Methods
High-resolution crystal data of water soluble proteins (cytochrome C, apoferritin and human serum albumin), and a membrane protein (a pentameric ligand-gated ion channel from Gloeobacter violaceus, GLIC) were used. Isothermal titration calorimetry (ITC) experiments were performed to determine anesthetic affinity in solution conditions for apoferritin. Docking calculations were performed using DockingServer with the Lamarckian genetic algorithm and the Solis and Wets local search method (https://www.dockingserver.com/web). Twenty general anesthetics were docked into apoferritin. The predicted binding constants are compared with those obtained from ITC experiments for potential correlations. In the case of apoferritin, details of the binding site and their interactions were compared with recent co-crystallization data. Docking calculations for six general anesthetics currently used in clinical settings (isoflurane, sevoflurane, desflurane, halothane, propofol, and etomidate) with known EC50 were also performed in all tested proteins. The binding constants derived from docking experiments were compared with known EC50s and octanol/water partition coefficients for the six general anesthetics.
Results
All 20 general anesthetics docked unambiguously into the anesthetic binding site identified in the crystal structure of apoferritin. The binding constants for 20 anesthetics obtained from the docking calculations correlate significantly with those obtained from ITC experiments (p=0.04). In the case of GLIC, the identified anesthetic binding sites in the crystal structure are among the docking predicted binding sites, but not the top ranked site. Docking calculations suggest a most probable binding site located in the extracellular domain of GLIC. The predicted affinities correlated significantly with the known EC50s for the six commonly used anesthetics in GLIC for the site identified in the experimental crystal data (p=0.006). However, predicted affinities in apoferritin, human serum albumin, and cytochrome C did not correlate with these six anesthetics’ known experimental EC50s. A weak correlation between the predicted affinities and the octanol/water partition coefficients was observed for the sites in GLIC.
Conclusion
We demonstrated that anesthetic binding sites and relative affinities can be predicted using docking calculations in an automatic docking server (Autodock) for both water soluble and membrane proteins. Correlation of predicted affinity and EC50 for six commonly used general anesthetics was only observed in GLIC, a member of a protein family relevant to anesthetic mechanism.
Background
The introduction of general anesthetics into clinical practice for surgical operations and dental extractions in 1842 is one of the most important steps in the development of modern medicine. General anesthetics are widely used daily across the world for most of the surgical cases, interventional examination and therapy, and sedation. However, the mechanism of general anesthetics remains unclear. Many hypotheses have been proposed and some suggest that proteins in the central nervous system might be the target of the general anesthetic action.1–8 We and others have demonstrated that inhaled and IV anesthetics share identical sites in multiple proteins, which suggests that they may also share similar protein targets, most likely membrane protein, in the central nervous system.2,9–11 However, protein targets that are specifically responsible for states of anesthesia have not been well identified in the central nervous system mainly because of the scarce amount of the individual protein in the central nervous system.
Although membrane proteins are considered to be the most probable target of general anesthetics, only a few proteins have been identified to have a specific interaction with general anesthetics. Many techniques have been developed and used to identify and explore direct anesthetic interactions with proteins; these include hydrogen exchange,10 isothermal titration calorimetry (ITC),9 fluorescence spectroscopy,10,12 photo-affinity labeling,8 magnetic resonance imaging,13 and crystallographic studies.2,9,11 Among these techniques, the structural approaches provide atomistic information about the different interactions within the protein, especially interactions in the binding site. Despite the increase in the number of structures deposited in the PDB (Protein Data Bank; http://www.rcsb.org/pdb), few structures are relevant to protein-anesthetic interactions. Due to the difficulties in obtaining such anesthetic-protein structures, it would be desirable to have a tool able to predict anesthetic binding interactions using available high-resolution protein structures.
Ligand-protein docking is a molecular modeling technique that can predict binding interactions. Pharmacological research uses docking techniques for a variety of purposes, most notably the virtual screening of large chemical databases in order to select potential drug candidates when the protein target(s) have been revealed. In the case of inhaled anesthetics, not only the identities but also the locations of their protein target(s) are unclear. It is possible to select a particular anesthetic and screen all proteins in the PDB bank to predict possible specific interactions using computational docking algorithms. However, it is important to evaluate the reliability of such docking tools.
AutoDock is an automated docking program that predicts the interaction between proteins and different ligands.14,15 AutoDock has been successfully used in designing new drugs16 and revealing medication mechanisms.17 It is one of the most frequently used applications for ligand and protein interaction predictions.18 In this study, we performed such predictions for the case of general anesthetics in combination with recently available crystallization data for a water soluble proteins (cytochrome C, apoferritin and human serum albumin, HSA) and for a membrane protein (a pentameric ligand-gated ion channel from Gloeobacter violaceus; GLIC).11,19 The binding constants for apoferritin derived from docking experiments for 20 anesthetics were compared with those obtained from ITC; the binding constants for all the tested proteins for six clinical general anesthetics were correlated with their known anesthetic EC50s.20
Methods
Waiver from the IRB at the University of Pennsylvania is assumed because there is no human subject or animal involved in this study. Horse serum apoferritin was purchased from Sigma-Aldrich (St. Louis, MO, USA). All other chemicals were purchased from Fisher Scientific as A.C.S. grade or higher, and were used without further purification unless otherwise specified.
Docking
Docking calculations were performed using DockingServer (http://www.dockingserver.com).21 Semiempirical charges calculated by MOPAC2009 were added to the ligand atoms (http://openmopac.net/MOPAC2009.html).22 Nonpolar hydrogen atoms were merged, and rotatable bonds were defined. Docking calculations were performed on multiple protein structures including, apoferritin, HSA, cytochrome C and GLIC. The high-resolution structure of apoferritin was solved in our lab (PDB code: 3F33).11 The high-resolution structures of GLIC (PDB code:3P4W),19 HSA (PDB code:1E7C)2 and cytochrome C (PDB code: 3NBS)23 were obtained from PDB. Cytochrome C was used as a negative control for anesthetic binding site evaluation because we have demonstrated that it has no specific interaction with anesthetics using various direct binding techniques.24,25 Both proteins, HSA and apoferritin have been used as models for anesthetic binding interactions.2,9–11,26 Using AutoDock tools, essential hydrogen atoms, Kollman united atom type charges, and solvation parameters were added to the different proteins. Affinity (grid) maps of 50×50×80 Å grid points for apoferritin, cytochrome C, and HSA; and 25×25×50 Å grid points for GLIC with 0.375 Å spacing in both cases, were generated using the Autogrid program.27,28
Anesthetic binding search were performed using the Solis and Wets local search method with a Lamarckian genetic algorithm.29 Initial position, orientation, and torsions of the anesthetic molecules were set randomly. During the search, a translational step of 0.2 Å and torsion steps of 5° were applied. Docking calculations are the result of 100 independent runs with each run terminating after a maximum of 2500000 energy evaluations. Two indices that may be used for ranking of the docking results include the free energy of binding and the frequency of the most probable binding site(s). Frequency represents the percent of these runs that resulted in similar geometry (within a root mean square deviation tolerance of 2.0 Å). In this study, we chose the predicted site with a dominant frequency and a dominant energy (1 kcal lower than the rest) in order to unambiguously identify the best prediction. If there was discrepancy of the predicted dominant frequency and dominant energy, the site with above 10% frequency and with the lowest free energy of binding was chosen as the most probable binding site. The estimated binding constant (Kd) is derived from ΔG=−RTlnK equation, where ΔG is directly calculated during docking runs using the Autodock scoring function.
Twenty general anesthetics were selected for docking studies with apoferritin (Table 1). Also, six general anesthetics (Table 2) currently used in clinical settings were selected for docking calculations with GLIC, apoferritin, HSA, and cytochrome C because the EC50s for these six general anesthetics are available20. The coordinates of the tested ligands were obtained from the PubChem database (http://pubchem.ncbi.nlm.nih.gov/). PyMOL was used to generate the graphical renderings (Version 1.3, Schrödinger, LLC.; http://www.pymol.org/).
Table 1.
Anesthetics Tested for Docking and Isothermal Titration Calorimetry
| Propofol (1) |
| 2-isopropyl-6-propylphenol (2) |
| Thiopental (3) |
| 2,6-di-sec-butylphenol (4) |
| 2,6-diethylphenol (5) |
| Enflurane (6) |
| Methoxyflurane (7) |
| 2-ethyl-6-methylphenol (8) |
| 2-isopropylphenol (9) |
| Isoflurane (10) |
| Halothane (11) |
| Sevoflurane (12) |
| 1-chloro-1,2,2-trifluorocyclobutane (13) |
| Aminoanthracene (14) |
| Trichloroethylene (15) |
| 2,6-dimethyl phenol (16) |
| Chloroform (17) |
| Fluroxene (18) |
| Benzene (19) |
| Phenol (20) |
The numbers in parentheses are used as labels for the anesthetics in Figure 4.
Table 2.
Anesthetics tested for docking to Gloeobacter violaceus
| Halothane |
| Isoflurane |
| Sevoflurane |
| Desflurane |
| Propofol |
| Etomidate |
Isothermal Titration calorimetry
ITC is a technique that can measure the full thermodynamic profile of two molecules binding directly in solution conditions. The method consists of ultrasensitive measurements of the heat changes that occur when ligand and protein are mixed. By fitting the heat signal of multiple systematic injections of ligand into protein solutions, thermodynamic profiles, including the association constant (Ka) and stoichiometry (n), can be determined. We and others have demonstrated good agreement of ITC results with data derived from other techniques.9,26,30,31 In this study, titrations were performed at 20 °C using a Microcal, Inc. VP ITC (Northampton, MA). The sample vessel contained 25 µM apoferritin in 130 mM NaCl, 20 mM NaHPO4, pH 7.0, and the reference vessel contained water. Ligand (1.5 mM) was titrated into the protein sample cell, using volumes of 20 µL for each titration, with 5-min intervals between titrations. The signals of ligand into buffer, buffer into protein, and buffer into buffer were subtracted after separate titrations. Origin 5.0 (Microcal Software, Inc., Northampton, MA) was used to fit thermodynamic parameters to the heat profiles. The following formula was used for data fitting:
where K is the binding constant, n is the number of sites, V0 is the active volume involved in interaction, Mt is the total concentration of protein in V0, Xt is the total concentration of ligand, H is the molar heat of ligand binding, and Q is the total heat content of the solution contained in V0 (determined relative to zero for the unliganded species) at fractional saturation.
Correlation Analysis
To further evaluate the accuracy of the docking algorithm to correctly predict the interaction of proteins and anesthetics, the predicted affinity obtained from docking experiments for the tested anesthetics with apoferritin was compared with the experimental affinity obtained from ITC experiments using linear regression. In a similar fashion, the predicted affinity from docking calculations for the tested anesthetics with apoferritin, HSA, cytochrome C, and GLIC was compared with the known EC50 for these anesthetics available in the literature.20 Linear regression calculation was performed using a GraphPad software (Prism version 5.02; La Jolla, CA; http://www.graphpad.com/). A p value less than 0.05 was considered statistically significant for a linear correlation.
Results
Predicted Location of Anesthetic Binding Sites in 3 Proteins with Co-crystal Structures
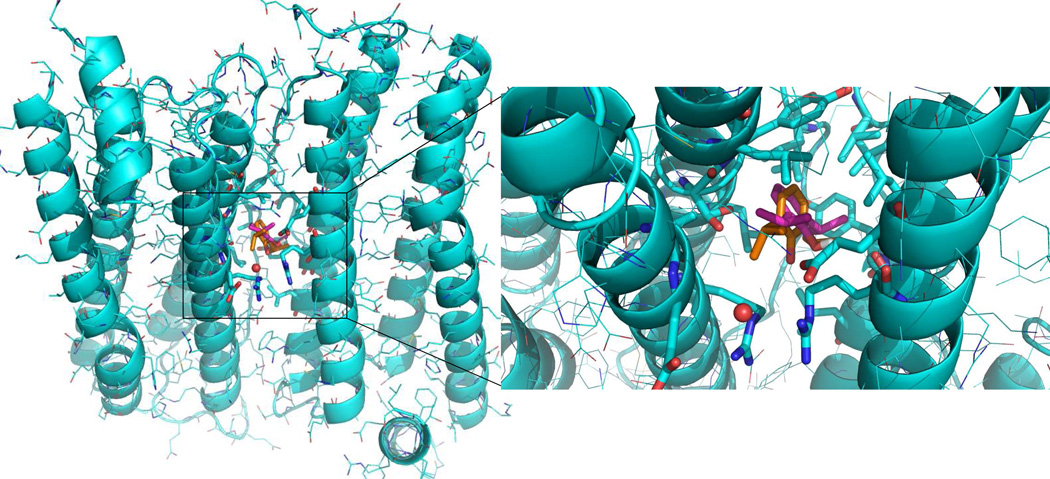
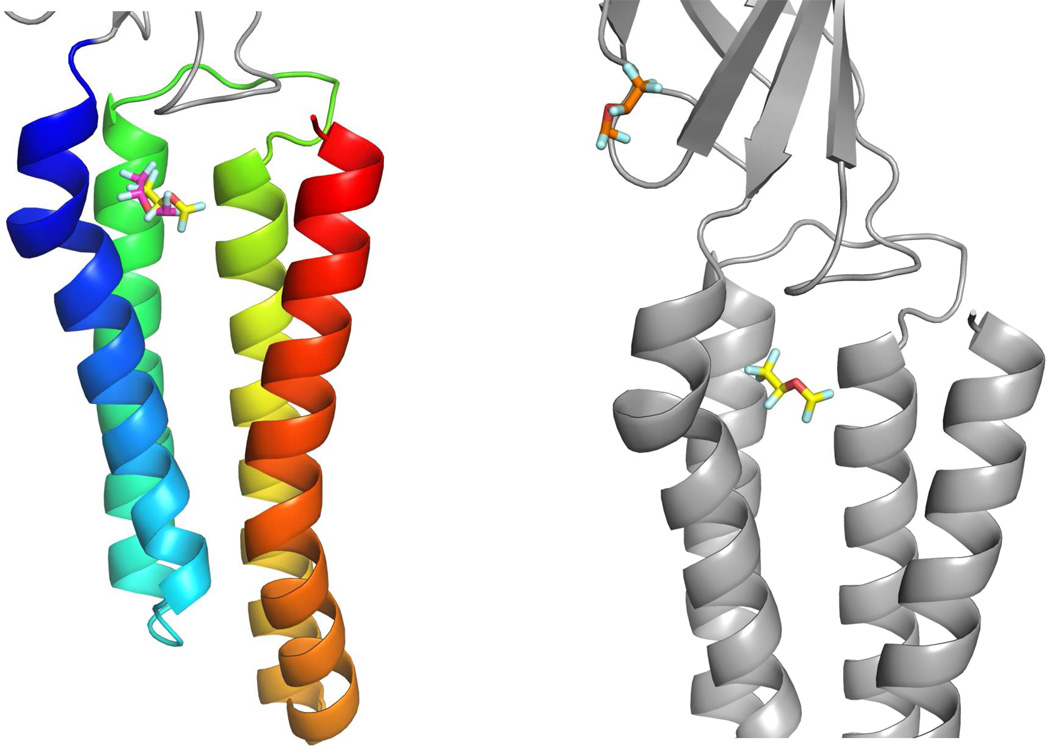
Top ranked sites from the docking calculations for all 20 general anesthetics are consistent with the anesthetic binding site identified in the crystallographic structures for apoferritin. In the case of propofol for example, the predicted orientation of the ligand overlaps very closely to the orientation in the crystallographic data as shown in Figure 1. In the case of GLIC, the anesthetic binding site observed in the crystal structure is among the top three ranked potential binding sites for the six tested anesthetics in docking prediction. As indicated in Figure 2a, one of the predicted desflurane geometries overlaps well with the orientation of the ligand in the crystal structure of GLIC. While the most probable (top ranked) site for propofol and etomidate is consistent with the crystallographic binding site in GLIC, the predicted most probable site identified for desflurane, using the same criteria from the server (http://www.dockingserver.com),21 is not consistent with the one identified in the crystal structure.19 As indicated in Figure 2b, an additional binding site not identified from the crystallographic data for GLIC is predicted for desflurane. Propofol was also docked into this extracellular site with less probability. The interacting residues for the top ranked site for the inhaled anesthetics tested with GLIC are listed in Table 3.The top ranked binding sites identified in HSA for anesthetics (propofol and halothane) are consistent with those of the crystal data (data not shown). Finally, there are no available co-crystal data for cytochrome C with anesthetics for comparison purposes and no common sites for anesthetics were identified from docking calculations.
Figure 1.
Propofol molecule binds to the same site in apoferritin both from experimental data (orange sticks) and docking prediction (magenta). Water molecules (red spheres) and residues in the binding site proximity are highlighted.
Figure 2.
Transmembrane and extracellular domains of the pentameric ligand-gated ion channel from Gloeobacter violaceus, GLIC co-crystallized with desflurane (PDB: 3P4W). Figure 2a depicts the transmembrane helices in a rainbow range color from blue in α1 to red in α4. The desflurane molecule from the X-ray structure is yellow while the predicted structure from docking is magenta. Figure 2b displays the most probable binding site predicted by docking (orange) and located in the extracellular domain of GLIC. The predicted structure (yellow) docked in the crystallographic binding site is also shown.
Table 3.
Interacting residues for the most probable binding site for inhaled anesthetics for Gloeobacter violaceus
| Sevoflurane | Desflurane | Isoflurane | Halothane |
|---|---|---|---|
| GLU82 | GLU82 | ASN83 | ASN83 |
| ASN83 | ASN83 | ALA84 | ALA84 |
| ALA84 | ALA84 | ASP 86 | SER107 |
| ARG 85 | ASP 86 | SER107 | ALA108 |
| ASP 86 | SER107 | ALA108 | ARG109 |
| SER107 | ALA108 | ARG109 | |
| ALA108 | ARG109 | ||
| ARG109 |
Predicted and Actual Affinities of Anesthetics for Apoferritin
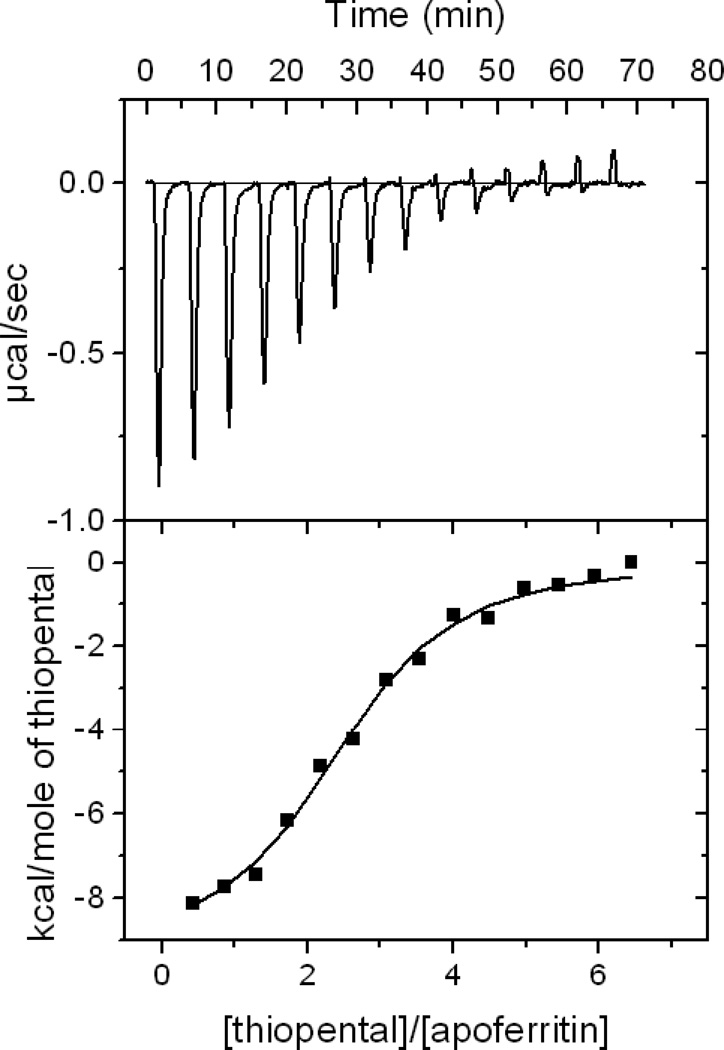
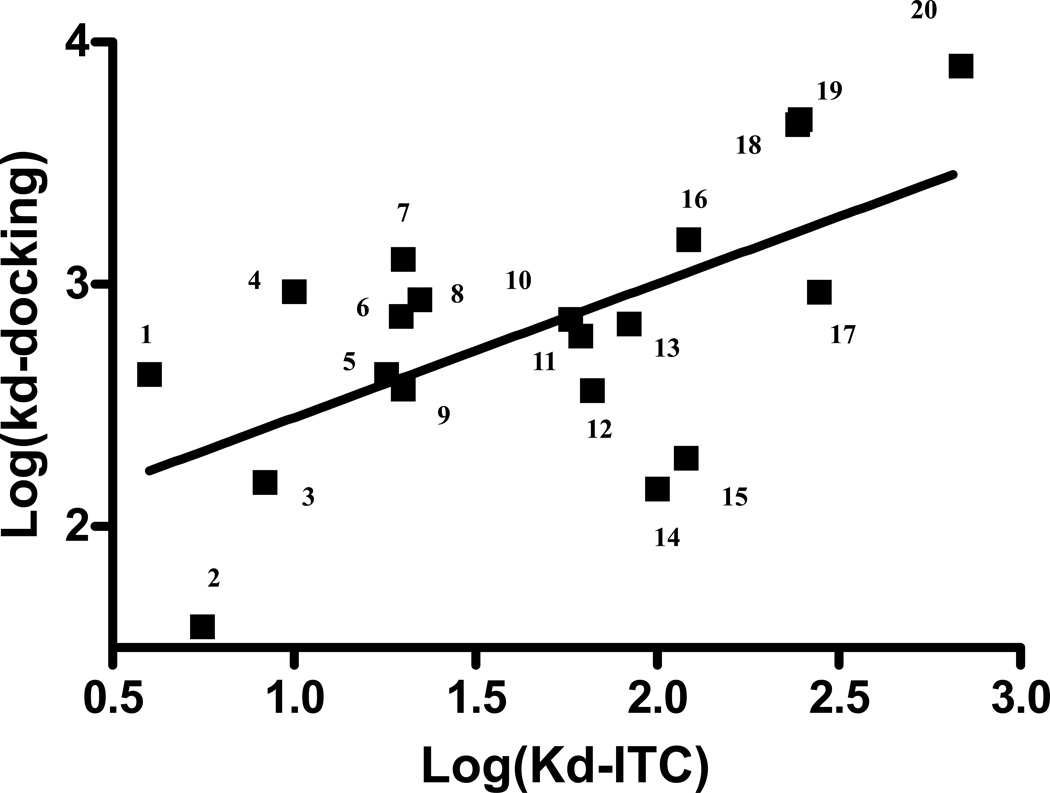
As reported previously,9,31 the interaction between all the tested anesthetics to protein exhibited a heat release process and the interaction is saturable with the increase of the concentration of each anesthetic. As shown in Figure 3, the heat release diminished to almost zero when enough thiopental are titrated into apoferritin. The binding constants (Kd-docking) for the 20 anesthetics obtained from the docking calculations correlate significantly with the dissociation constant (Kd-ITC) obtained from ITC experiments as shown in Figure 4 (p = 0.04). Docking predictions underestimate the affinities, but the differences are only at most 7mM when compared with the values from ITC.
Figure 3.
Isotherm of thiopental titration (0.25 mM) into apoferritin (0.01 mM) indicating a heat release process and saturable site from an isothermal titration calorimetry experiment. Please refer to the curve fitting equation in the text. Buffer condition is 130 NaCl, 20 mM NaHPO4 and pH 7.0.
Figure 4.
Significant correlations between predicted affinity for the top ranked binding site (Kd-docking) obtained from docking and dissociation constant from the isothermal titration calorimetry (Kd -ITC) experiment are observed (p = 0.004). Twenty general anesthetics (ethers, phenol analogues and barbiturate) are included (numbers are compounds listed in Table 1). These data indicate that the predicted affinity from docking could predict relative strength of binding between anesthetics and their protein targets.
Predicted Anesthetic Affinities and known Experimental Anesthetic EC50s
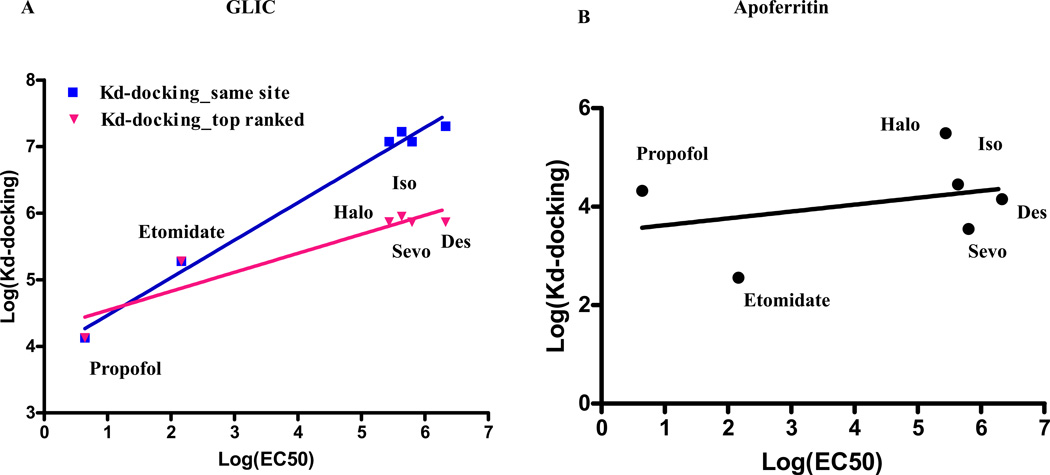
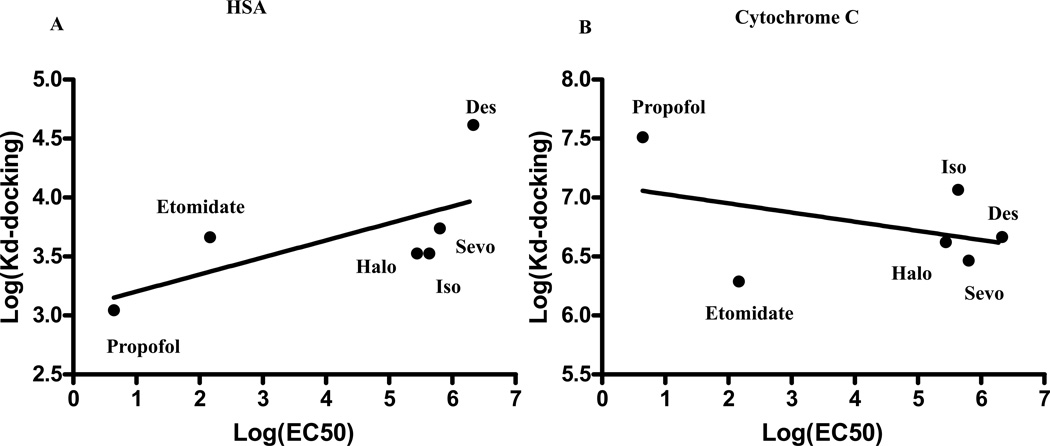
The predicted affinity of the tested general anesthetics for GLIC shows significant correlation with the available EC50 data as shown in Figure 5A (p = 0.018). Interestingly, there is a correlation whether the affinity of the most probable site predicted by docking is used or the affinity of the site identified by both docking and crystal data is used. It seems that the affinity for the site in the transmembrane helix correlates better (r2= 0.98, p<0.0001) than the affinity for the most probable site ranked by docking and located in the extracellular domain (r2= 0.89, p=0.006). Figure 5 also demonstrates that the predicted affinity from docking correlates with anesthetic potency for a protein (GLIC) that is a member of a protein family having a potential relationship to anesthetic mechanism, but not for a water soluble protein (apoferritin) that has no potential role in pharmacological action. There are no significant correlations between known EC50s for these commonly used anesthetics and the predicted affinity from docking in soluble protein that does not bind anesthetics (cytochrome C) or has no potential role in pharmacological action for general anesthetics (p = 0.18 for Figure 6B, p=0.52 for Figure 5B, and p=0.16 Figure 6A for cytochrome C, apoferritin, and HSA respectively).
Figure 5.
Significant correlations between predicted affinity for both the top ranked binding site (Kd-docking_top ranked) and the site consistent with the one in crystal structure from docking (Kd-docking_same site) and known EC50s for six clinically used anesthetics (Table 2) in pentameric ligand-gated ion channel (GLIC) are shown in 5A. No significant correlation was observed in the case of apoferritin as shown in 5B. These data demonstrates that the predicted affinity from docking correlates with anesthetic potency in a protein (channel protein, GLIC) that has a potential relationship with anesthetic mechanism, but not in a protein (apoferritin as water soluble protein) that has no potential role in pharmacological action. Halo: halothane; Sevo: sevoflurane; Iso: isoflurane; Des: desflurane.
Figure 6.
No significant correlation between the predicted affinity of the top ranked binding site (Kd-docking) and EC50s of six clinically used anesthetics were observed with either human serum albumin (HSA, panel A) or cytochrome C as shown panel B, respectively. These data along with data presented in Figure 5B demonstrate that no significant correlation between known EC50s for these commonly used anesthetics (Table 2) and the predicted affinities for the top ranked binding sites from docking in soluble proteins that has no binding with anesthetics or has no potential role in pharmacological action for general anesthetics. Halo: halothane; Sevo: sevoflurane; Iso: isoflurane; Des: desflurane.
Predicted Anesthetic Affinities and known octanol/water partition coefficients:32
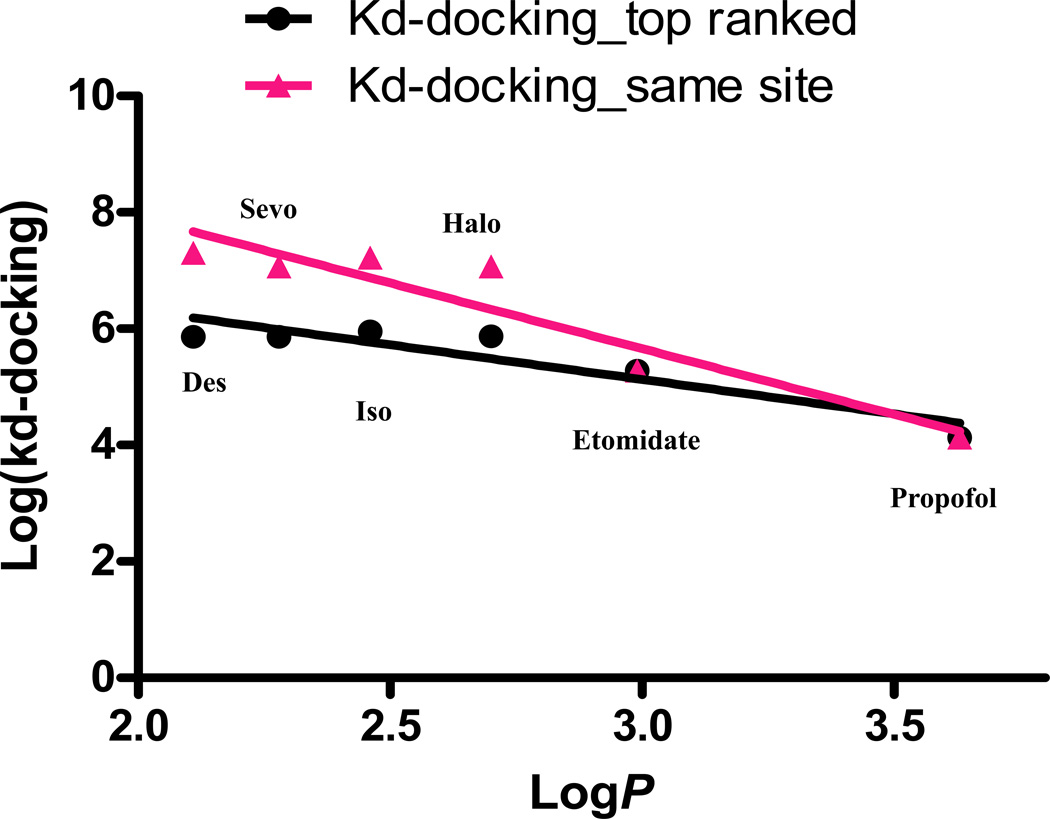
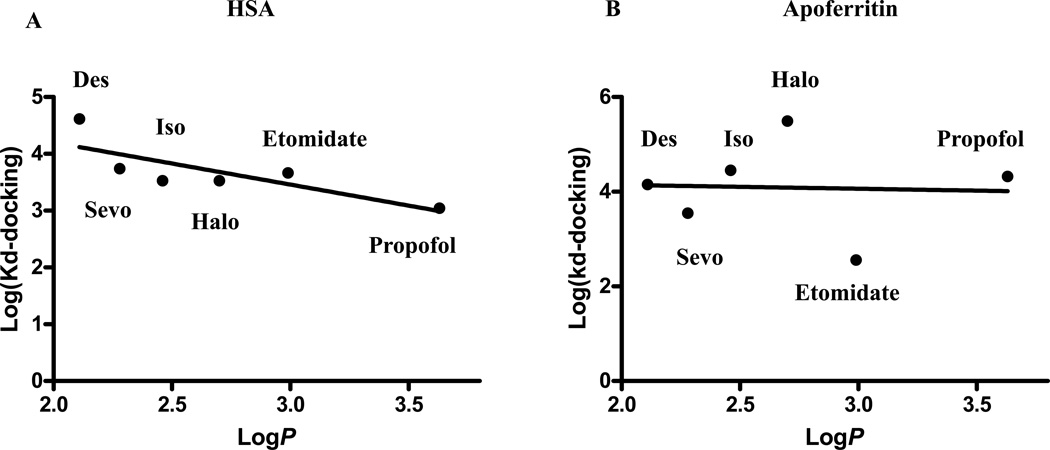
A weak but significant correlation between the predicted affinities and the known octanol/water partition coefficients of the tested general anesthetics in GLIC (Figure 7). There is no significant correlation between the predicted affinities and the known octanol/water partition coefficients in HSA (Figure 8A) and apoferritin (Figure 8B).
Figure 7.
Significant correlations between predicted affinity for both the top ranked binding site (Kd-docking_top ranked; r2= 0.85, p=0.0087) and the site consistent with the one in the crystal structure from docking (Kd-docking_same site; r2= 0.89, p=0.0051) for six clinically used anesthetics with known logP (Table 2) in pentameric ligand-gated ion channel (GLIC) are shown. Halo: halothane; Sevo: sevoflurane; Iso: isoflurane; Des: desflurane.
Figure 8.
No significant correlations between predicted affinity and known logP for six clinically used anesthetics (Table 2) with HSA (panel A; r2= 0.63, p=0.059) and apoferritin (panel B; r2= 0.002, p=0.93) are shown. Halo: halothane; Sevo: sevoflurane; Iso: isoflurane; Des: desflurane. HSA: human serum albumin.
Discussion
In this study, we demonstrated that binding sites for general anesthetics for both water soluble proteins (apoferritin and HSA) and a membrane protein (GLIC, a ligand-gated ion channel protein) can be predicted reasonably well using docking algorithms. The calculated affinity correlates well with Kds obtained from ITC experiments for apoferritin and with known EC50s for GLIC.
Docking as an alternative tool for anesthetic binding site identification
Molecular docking is an essential method in structure-based drug design when a specific target protein structure is known via high-resolution techniques including nuclear magnetic resonance, X-ray crystallography and reliable computational modeling.14,18,27,33,34 Multiple molecular docking programs have been developed; many of them with good interactive interfaces and tools that help to set up the calculations easily. In this study, a web-based docking server was tested. The avoidance of acquiring powerful computational infrastructure for running the programs is one of the most important advantages to using such a server.
In this study, the docking algorithm identified the binding sites consistent with crystallographic data in both water soluble protein and membrane protein. These results suggest that a docking algorithm is a useful alternative tool to identify probable anesthetic binding sites. Possible applications include (i) the ability to choose representative ligands to screen all available high-resolution protein structures for possible interactions with anesthetics; (ii) the ability to select a model protein structure that might represent an anesthetic target to screen available chemical compounds for the discovery of new anesthetics; (iii) the ability to provide atomistic details about the orientation of the ligands inside the binding pocket (also referred to as the binding mode or pose) which could be used to design more potent and selective anesthetics for safer clinical applications.35 The advantage of using computational docking algorithms is that no actual protein is needed for large scale predicting experiments and also no complicated settings and preparations are needed. In contrast, many other techniques require a large amount of protein, something that is often a significant challenge, especially in the case of membrane proteins. Binding mode and atomic interactions between ligand and protein can also be predicted and analyzed in detail. Another advantage is that no solubility issue is involved for both membrane proteins and hydrophobic ligands.
Docking is currently in a relatively mature stage of development, but it is still far from perfect.18,36 Many docking programs predict known protein bound poses with averaged accuracies of about 1.5–2 Å with reported success rates in the range of 70–82%.35,37 False positive results are the major disadvantages to the computational aspects of a docking algorithm. The docking output usually gives multiple possibilities of interactions either represented in different binding sites or in different orientations in the same binding sites. It is difficult to know the reliability of predictions using only the docking method. Thus, it is important to verify these findings using binding studies, functional assays and structural determinations.
In this study, the predicted anesthetic binding sites in apoferritin and HSA are consistent with crystallographic data. The most probable binding site for inhaled anesthetics in GLIC is in the extracellular domain rather than the one identified by crystallographic study in the transmembrane domain. It is unclear from this study whether this is a false positive of docking or a binding site not resolved in the crystal structure. In the experimental crystal data, detergent is used to solubilize the protein, and it is possible that the detergent may have excluded this anesthetic binding site. Interestingly, a recent study indicated that the proton activation site(s) of GLIC is located in the extracellular domain, as observed electrophysiologically using Xenopus oocytes that over-express a chimera in which the extracellular domain of GLIC is fused to the transmembrane domain of the human α1 glycine receptor.38 We included the interacting residues in Table 3 for all the tested anesthetics for potential mutagenesis studies to verify this potential anesthetic binding site in the extracellular domain of GLIC.
The role of ITC
ITC has been successfully used to investigate the interaction between anesthetics and proteins.9,26,30,31 It is a unique tool to characterize molecular interaction by monitoring heat changes when specific interactions occur between molecules. The advantage of using ITC is that no specific modification is needed for target molecules in solution conditions. A typical ITC experiment takes only a few hours, and the protein can be recycled after anesthetic titration. Therefore, ITC may be a good method to screen and/or confirm anesthetic-protein binding quickly. If the experiment is designed properly, the binding constant and the number of binding sites as well as enthalpic and entropic components of binding affinity can be obtained. We have used ITC as a tool to identify new binding proteins for general anesthetics successfully and as a tool to confirm anesthetic binding predicted or observed using other techniques.9,26,30,31
In the present study, ITC was used as a tool to obtain binding constants for all the tested anesthetics to apoferritin. Although ITC measures the global affinity of macromolecules, it is predictable that the overall affinities of anesthetics to apoferritin obtained from ITC correlate well with those obtained from docking calculations because there is only one binding site in apoferritin, as demonstrated previously via co-crystallization.9 Thus, a model with one binding site was used to fit the isotherm data. The measured affinity correlates well with that obtained from docking calculations. The tendency to underestimate the affinity by docking may be caused by the absence of solvent effect and direct participation of water molecules in protein–ligand interactions. Additionally, the lack of protein flexibility upon ligand binding could also play an important role. The disadvantage of using ITC for direct binding studies is that purified macromolecules and ligands are needed. False negative results may be caused by a low concentration of target molecules; thus, solubility is a very important limiting factor.
Predicted affinity, known EC50 and octanol/water partition coefficients
Although only six general anesthetics used in the clinical setting with known EC50s were included, only the docking-predicted affinity for GLIC correlates well with their known EC50 values. There is no significant correlation of the predicted affinity for three soluble proteins (cytochrome C, apoferritin and HSA) with the known EC50s for these six general anesthetics. There is no doubt that these soluble proteins have no apparent relationship with anesthetic mechanism(s). This indicates the possibility of this automatic docking algorithm and correlation calculation for predicting anesthetic binding site(s) and confirms its potential use as a tool for searching anesthetic binding targets related to anesthetic mechanism(s). Further analysis of the relationship of the predicted affinities with octanol/water partition coefficients demonstrates a weak correlation in GLIC, but not in apoferritin or HSA. In addition to revealing the potential binding site, these findings indicate that docking is revealing something more interesting than mere hydrophobicity.
Precautions in data interpretation
Although binding entropy and the solvation/desolvation of ligands are considered in Autodock, the "apparent" calculated free energy change is approximate and derived from the static interaction between ligand and the rigid protein. Significant deviation from true affinity should be expected. While the Kd derived from ITC may reflect the true affinity, significant error may occur if the number of binding sites and the character of the binding sites are unknown because the total heat profile is dictated by both number of binding sites and affinity. In the case of apoferritin, both the number and character of binding sites are known. The EC50 reflects the potency of each compound, and in the case of general anesthetics, stronger affinity to the target protein could be reflected as stronger potency. However, the correlation between potency and affinity of a specific protein does not necessarily indicate that the protein is related to the mechanism of action.
In conclusion, we demonstrated that anesthetic binding sites and relative affinities can be predicted using docking calculations in an automatic docking server (Autodock) for both water soluble and membrane proteins. Correlation of predicted affinity and EC50 for six commonly used general anesthetics was only observed in GLIC, a membrane protein with potential relevance for understanding anesthetic mechanisms.
Acknowledgement
Funding: This research was supported by departmental funding from the department of Anesthesiology and Critical Care at the University of Pennsylvania, funding from Foundation for Anesthesia Education and Research (Rochester, MN; PI:RL), and support from NIH K08-GM-093115-01(PI:RL). Jeffery G. Saven acknowledges support from NIH (P01GM055876) and NSF (NSEC DMR08-32802).
Dr. Renyu Liu appreciates the great mentoring from Dr. Roderic G. Eckenhoff. The authors thank the technical support from Jin Xi and Jingyuan Ma.
This manuscript was handled by: Steven L. Shafer, MD
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
This report was previously presented, in part, at the ASA 2009.
DISCLOSURES:
Name: Renyu Liu, MD, PhD
Contribution: This author helped design the study, conduct the study, analyze the data, and write the manuscript
Attestation: Renyu Liu has seen the original study data, reviewed the analysis of the data, approved the final manuscript, and is the author responsible for archiving the study files
Name: Jose Manuel Perez-Aguilar, BA, PhD
Contribution: This author helped conduct the study, analyze the data, and write the manuscript
Attestation: Jose Manuel Perez-Aguilar has seen the original study data, reviewed the analysis of the data, and approved the final manuscript
Name: David Liang, BA
Contribution: This author helped conduct the study, analyze the data, and write the manuscript
Attestation: David Liang has seen the original study data, reviewed the analysis of the data, and approved the final manuscript
Name: Jeffery G. Saven, PhD
Contribution: This author helped design the study, analyze the data, and write the manuscript
Attestation: Jeffery G. Saven has seen the original study data, reviewed the analysis of the data, and approved the final manuscript
Contributor Information
Renyu Liu, Department of Anesthesiology and Critical Care, Hospital of University of Pennsylvania, Philadelphia, Pennsylvania.
Jose Manuel Perez-Aguilar, Department of Chemistry, University of Pennsylvania, Philadelphia, Pennsylvania.
David Liang, Department of Anesthesiology and Critical Care, Hospital of University of Pennsylvania, Philadelphia, Pennsylvania.
Jeffery G. Saven, Department of Chemistry, University of Pennsylvania, Philadelphia, Pennsylvania.
References
- 1.Bertaccini EJ, Trudell JR, Franks NP. The common chemical motifs within anesthetic binding sites. Anesth Analg. 2007;104:318–324. doi: 10.1213/01.ane.0000253029.67331.8d. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharya AA, Curry S, Franks NP. Binding of the general anesthetics propofol and halothane to human serum albumin. High resolution crystal structures. J Biol Chem. 2000;275:38731–38738. doi: 10.1074/jbc.M005460200. [DOI] [PubMed] [Google Scholar]
- 3.Eckenhoff RG. Do specific or nonspecific interactions with proteins underlie inhalational anesthetic action? Mol Pharmacol. 1998;54:610–615. [PubMed] [Google Scholar]
- 4.Eckenhoff RG. A noble approach to mechanisms. Anesth Analg. 1998;87:239–241. doi: 10.1097/00000539-199808000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Eckenhoff RG, Johansson JS. Molecular interactions between inhaled anesthetics and proteins. Pharmacol Rev. 1997;49:343–367. [PubMed] [Google Scholar]
- 6.Franks NP, Lieb WR. Do general anaesthetics act by competitive binding to specific receptors? Nature. 1984;310:599–601. doi: 10.1038/310599a0. [DOI] [PubMed] [Google Scholar]
- 7.Franks NP, Lieb WR. Seeing the light: protein theories of general anesthesia. Anesthesiology. 2004;101:235–237. doi: 10.1097/00000542-200407000-00034. [DOI] [PubMed] [Google Scholar]
- 8.Xi J, Liu R, Asbury GR, Eckenhoff MF, Eckenhoff RG. Inhalational anesthetic-binding proteins in rat neuronal membranes. J Biol Chem. 2004;279:19628–19633. doi: 10.1074/jbc.M313864200. [DOI] [PubMed] [Google Scholar]
- 9.Liu R, Loll PJ, Eckenhoff RG. Structural basis for high-affinity volatile anesthetic binding in a natural 4-helix bundle protein. Faseb J. 2005;19:567–576. doi: 10.1096/fj.04-3171com. [DOI] [PubMed] [Google Scholar]
- 10.Liu R, Yang J, Ha CE, Bhagavan NV, Eckenhoff RG. Truncated human serum albumin retains general anaesthetic binding activity. Biochem J. 2005;388:39–45. doi: 10.1042/BJ20041224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vedula LS, Brannigan G, Economou NJ, Xi J, Hall MA, Liu R, Rossi MJ, Dailey WP, Grasty KC, Klein ML, Eckenhoff RG, Loll PJ. A unitary anesthetic binding site at high resolution. J Biol Chem. 2009;284:24176–24184. doi: 10.1074/jbc.M109.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Strzalka J, Tronin A, Johansson JS, Blasie JK. Mechanism of interaction between the general anesthetic halothane and a model ion channel protein, II: Fluorescence and vibrational spectroscopy using a cyanophenylalanine probe. Biophys J. 2009;96:4176–4187. doi: 10.1016/j.bpj.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma D, Tillman TS, Tang P, Meirovitch E, Eckenhoff R, Carnini A, Xu Y. NMR studies of a channel protein without membranes: structure and dynamics of water-solubilized KcsA. Proc Natl Acad Sci U S A. 2008;105:16537–16542. doi: 10.1073/pnas.0805501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodsell DS. Computational docking of biomolecular complexes with AutoDock. Cold Spring Harb Protoc. 2009;5 doi: 10.1101/pdb.prot5200. pdb prot5200. [DOI] [PubMed] [Google Scholar]
- 15.Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 16.Kamel MM, Ali HI, Anwar MM, Mohamed NA, Soliman AM. Synthesis, antitumor activity and molecular docking study of novel Sulfonamide-Schiff's bases, thiazolidinones, benzothiazinones and their C-nucleoside derivatives. Eur J Med Chem. 2010;45:572–580. doi: 10.1016/j.ejmech.2009.10.044. [DOI] [PubMed] [Google Scholar]
- 17.Mao WJ, Lv PC, Shi L, Li HQ, Zhu HL. Synthesis, molecular docking and biological evaluation of metronidazole derivatives as potent Helicobacter pylori urease inhibitors. Bioorg Med Chem. 2009;17:7531–7536. doi: 10.1016/j.bmc.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 18.Sousa SF, Fernandes PA, Ramos MJ. Protein-ligand docking: current status and future challenges. Proteins. 2006;65:15–26. doi: 10.1002/prot.21082. [DOI] [PubMed] [Google Scholar]
- 19.Nury H, Van Renterghem C, Weng Y, Tran A, Baaden M, Dufresne V, Changeux JP, Sonner JM, Delarue M, Corringer PJ. X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature. 2011;469:428–431. doi: 10.1038/nature09647. [DOI] [PubMed] [Google Scholar]
- 20.Weng Y, Yang L, Corringer PJ, Sonner JM. Anesthetic sensitivity of the Gloeobacter violaceus proton-gated ion channel. Anesth Analg. 2010;110:59–63. doi: 10.1213/ANE.0b013e3181c4bc69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bikadi Z, Hazai E. Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J Cheminform. 2009;1:15. doi: 10.1186/1758-2946-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart JJ. MOPAC: a semiempirical molecular orbital program. J Comput Aided Mol Des. 1990;4:1–105. doi: 10.1007/BF00128336. [DOI] [PubMed] [Google Scholar]
- 23.Hirota S, Hattori Y, Nagao S, Taketa M, Komori H, Kamikubo H, Wang Z, Takahashi I, Negi S, Sugiura Y, Kataoka M, Higuchi Y. Cytochrome c polymerization by successive domain swapping at the C-terminal helix. Proc Natl Acad Sci U S A. 2010;107:12854–12859. doi: 10.1073/pnas.1001839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan K, Meng QC, Johansson JS, Eckenhoff RG. Low-affinity analytical chromatography for measuring inhaled anesthetic binding to isolated proteins. Anal Biochem. 2002;301:308–313. doi: 10.1006/abio.2001.5506. [DOI] [PubMed] [Google Scholar]
- 25.Eckenhoff RG, Pidikiti R, Reddy KS. Anesthetic stabilization of protein intermediates: myoglobin and halothane. Biochemistry. 2001;40:10819–10824. doi: 10.1021/bi010691r. [DOI] [PubMed] [Google Scholar]
- 26.Liu R, Eckenhoff RG. Weak polar interactions confer albumin binding site selectivity for haloether anesthetics. Anesthesiology. 2005;102:799–805. doi: 10.1097/00000542-200504000-00016. [DOI] [PubMed] [Google Scholar]
- 27.Morris GM, Goodsell DS, Huey R, Olson AJ. Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J Comput Aided Mol Des. 1996;10:293–304. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
- 28.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solis FJ, Wets RJB. Minimization by Random Search Techniques. Mathematics of Operations Research. 1981;6:19–30. [Google Scholar]
- 30.Liu R, Meng Q, Xi J, Yang J, Ha CE, Bhagavan NV, Eckenhoff RG. Comparative binding character of two general anaesthetics for sites on human serum albumin. Biochem J. 2004;380:147–152. doi: 10.1042/BJ20031652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang T, Johansson JS. An isothermal titration calorimetry study on the binding of four volatile general anesthetics to the hydrophobic core of a four-alpha-helix bundle protein. Biophys J. 2003;85:3279–3285. doi: 10.1016/S0006-3495(03)74746-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urban BW, Bleckwenn M, Barann M. Interactions of anesthetics with their targets: non-specific, specific or both? Pharmacol Ther. 2006;111:729–770. doi: 10.1016/j.pharmthera.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Osterberg F, Morris GM, Sanner MF, Olson AJ, Goodsell DS. Automated docking to multiple target structures: incorporation of protein mobility and structural water heterogeneity in AutoDock. Proteins. 2002;46:34–40. doi: 10.1002/prot.10028. [DOI] [PubMed] [Google Scholar]
- 34.Rosenfeld RJ, Goodsell DS, Musah RA, Morris GM, Goodin DB, Olson AJ. Automated docking of ligands to an artificial active site: augmenting crystallographic analysis with computer modeling. J Comput Aided Mol Des. 2003;17:525–536. doi: 10.1023/b:jcam.0000004604.87558.02. [DOI] [PubMed] [Google Scholar]
- 35.Thomsen R, Christensen MH. MolDock. a new technique for high-accuracy molecular docking. J Med Chem. 2006;49:3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 36.Halperin I, Ma B, Wolfson H, Nussinov R. Principles of docking: An overview of search algorithms and a guide to scoring functions. Proteins. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 37.Kellenberger E, Rodrigo J, Muller P, Rognan D. Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins. 2004;57:225–242. doi: 10.1002/prot.20149. [DOI] [PubMed] [Google Scholar]
- 38.Duret G, Van Renterghem C, Weng Y, Prevost M, Moraga-Cid G, Huon C, Sonner JM, Corringer PJ. Functional prokaryotic-eukaryotic chimera from the pentameric ligand-gated ion channel family. Proc Natl Acad Sci U S A. 2011;108:12143–12148. doi: 10.1073/pnas.1104494108. [DOI] [PMC free article] [PubMed] [Google Scholar]