Abstract
DNA replication of the mitochondrial genome is unique in that replication is not primed by RNA derived from dedicated primases, but instead by extension of processed RNA transcripts laid down by the mitochondrial RNA polymerase. Thus, the RNA polymerase serves not only to generate the transcripts but also the primers needed for mitochondrial DNA replication. The interface between this transcription and DNA replication is not well understood but must be highly regulated and coordinated to carry out both mitochondrial DNA replication and transcription. This review focuses on the extension of RNA primers for DNA replication by the replication machinery and summarizes the current models of DNA replication in mitochondria as well as the proteins involved in mitochondrial DNA replication, namely, the DNA polymerase γ and its accessory subunit, the mitochondrial DNA helicase, the single-stranded DNA binding protein, toposiomerase I and IIIα and RNaseH1.
1. Introduction
The mitochondrial (mt) genome is a multicopy closed circular genome of 16,569 bp that codes for 13 proteins involved in the electron transport chain, 22 transfer RNA genes, and 2 ribosomal RNAs required for mitochondrial protein synthesis of the 13 polypeptides. Cells contain several thousand copies of mtDNA spread out over hundreds of mitochondria. The mtDNA is located in discrete nucleoids in the inner mitochondrial matrix of the mitochondrion that contain between 1–2 copies of mtDNA [1]. MtDNA is replicated by an assembly of proteins in a replisome consisting of DNA polymerase γ (pol γ), the mitochondrial single-stranded DNA binding protein (mtSSB), mitochondrial DNA helicase, topoisomerases and RNaseH activities (Table I and Fig. 1).
Table 1.
Human mitochondrial DNA replication proteins
| Enzyme | Size | Human Chromosome |
|---|---|---|
| DNA polymerase γ | ||
| POLG | 140 kDa | 15q25 |
| POLG2 | 55 kDa | 17q23-24 |
| Single-Stranded DNA binding protein (mtSSB) | 15 kDa | 7q34 |
| Helicase | ||
| C10orf2 (Twinkle) | 77 kDa | 10q24 |
| Topoisomerases | ||
| Topo Imt | 67 kDa | 8q24.3 |
| Topo IIIα | 112 kDa | 17p12-11.2 |
| RNase H1 | 32 kDa | 19p13.2 |
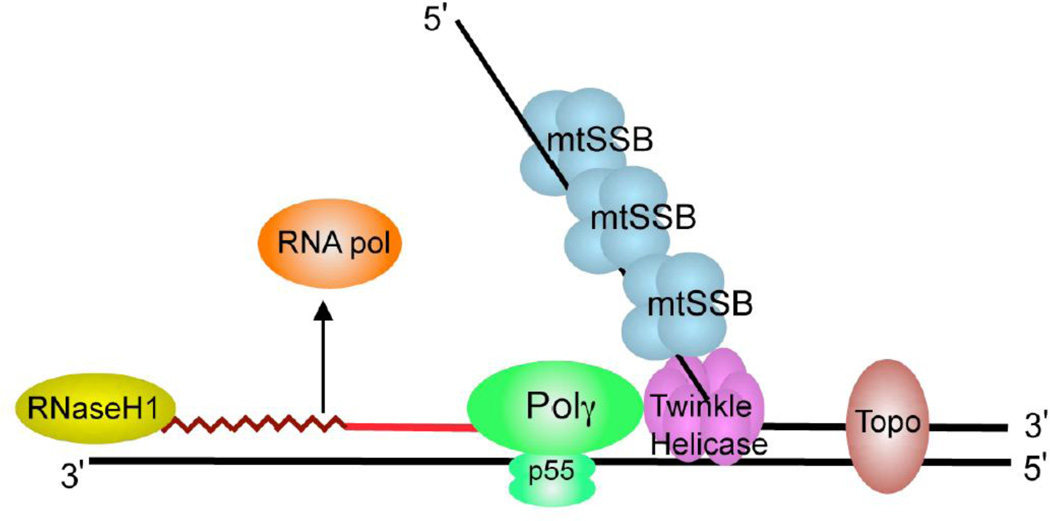
Figure 1. Schematic diagram of a mitochondrial DNA replication fork showing the critical proteins required for DNA replication.
The nascent DNA synthesized by pol γ (green) is shown as a solid red line, while the RNA primer (jagged red line) created by the mitochondrial RNA polymerase (orange) is being degraded by RNase H1 (yellow). The mitochondrial DNA helicase (purple) unwinds the downstream DNA forming a single-stranded loop which is coated with mtSSB (light blue). Topoisomerases (brown) work to relieve torsional tension in the DNA created by unwinding.
1.1 Current models of mitochondrial DNA replication
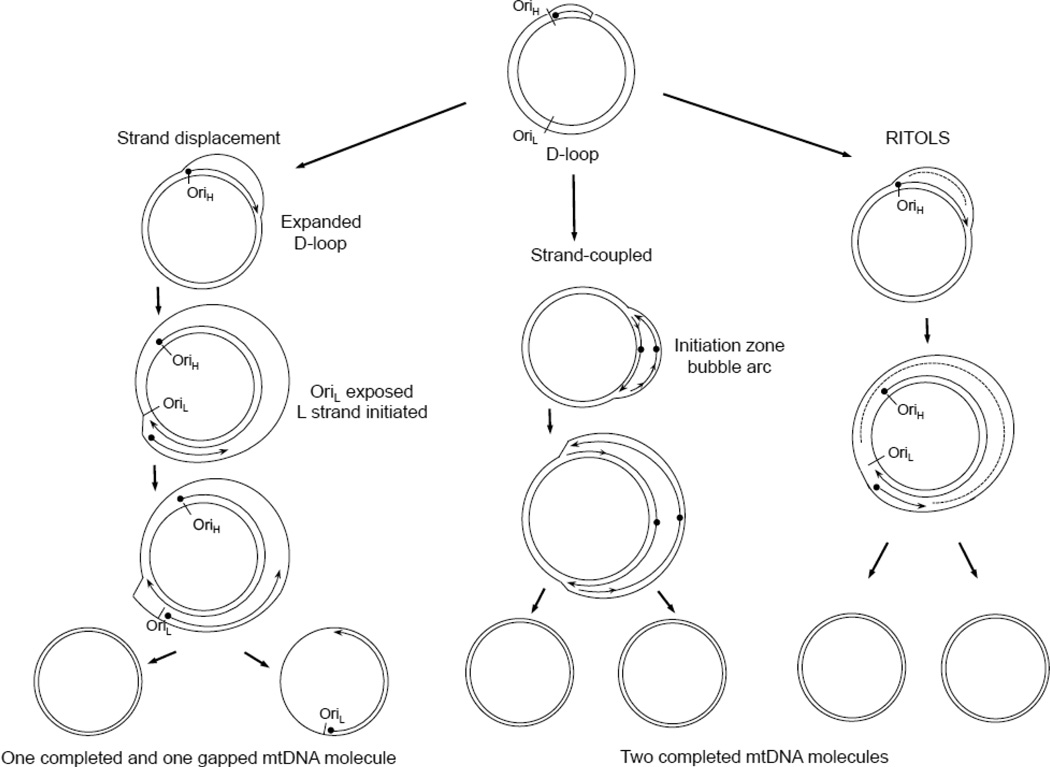
Two modes of DNA replication have been proposed to copy the mitochondrial genome, an asynchronous strand displacement model and a strand-coupled bidirectional replication model [2–5]. In the asynchronous strand displacement model, mtDNA is replicated in an asymmetric fashion where DNA synthesis is primed by transcription through the H-strand origin within the D-loop [6]. After two-thirds of the nascent H-strand is replicated, the L-strand origin is exposed, allowing initiation of nascent L-strand synthesis. In the strand-coupled model, bidirectional replication is initiated from a zone near OriH followed by progression of the two forks around the mtDNA circle [7]. Because subsequent research has shown that the strand-coupled replication intermediates contain RNaseH sensitive sites, an alternative mechanism of the strand-coupled model includes the idea that the lagging strand is initially laid down by RNA before being converted to DNA (Fig. 2), and has been termed RITOLS (RNA Incorporated Throughout Lagging Strand). [8] In all of these models, initiation is primed by an RNA primer and the DNA polymerization reaction is performed by the pol γ holoenzyme.
Figure 2. Models of mtDNA replication.
Left panel. The asymmetric or strand displacement model. Replication of the H-strand is initiated at OriH with accompanying displacement of the H-strand thus forming a D-loop. This synthesis proceeds until OriL is exposed where synthesis of the L-stand is initiated in the opposite direction. Middle panel. The strand-coupled model. Bidirectional replication is initiated from a zone near OriH followed by progression of the two forks around the mtDNA circle. Right panel. The RITOLS model. Replication of the leading strand initiates similar to the strand-displacment model but the lagging strand is initially transcribed as RNA (dashed line) before being converted to DNA.
In the asynchronous strand displacement model, transcription initiates replication of mtDNA at OriH within the D-loop at the light-strand promoter (LSP) [9]. The primer for initiation of mtDNA replication at OriH is generated by processing the transcript starting at LSP [6, 10]. Pol γ initiates H-strand synthesis by extending the RNA primer [6, 11, 12]. When nascent H-strand synthesis is ~70% complete, the replication fork exposes the major origin for L-strand synthesis (OriL), allowing initiation of L-strand synthesis on the displaced H-strand to proceed in the opposite direction [13–15]. L-strand replication is initiated near the WANCY tRNA coding region that in a single-strand form is postulated to assume a stable stem-loop structure, and DNA synthesis proceeds along the entire length of the mtDNA strand and terminates after H-strand replication is completed [16]. In support of the strand-displacement model, L-strand replication has also been shown to initiated in vitro by the mitochondrial RNA polymerase [17].
The strand-coupled model of mtDNA replication is based on the ribonucleotide substitution pattern in mtDNA and analysis of replication intermediates by 2D-gel electrophoresis [7, 8, 18]. 2D-gel electrophoresis revealed two types of replication intermediates [18], one of which is resistant to nucleases that digest single-stranded DNA consistent with conventional duplex replication intermediates from symmetric, semi-discontinuous DNA replication with coupled leading and lagging strand DNA synthesis. A second class of replication intermediates, presumably derived from the strand-asynchronous mechanism of mtDNA replication, is sensitive to single-strand nuclease and is most abundant in cultured cells not treated with ethidium bromide. Although this initial report suggested coexistence of both the asynchronous and strand-coupled modes of mtDNA replication [18], later findings by the same authors indicate that mammalian mtDNA replication proceeds mainly, if not exclusively, by a strand-coupled mechanism [7, 8]. Subsequently, replication intermediates from highly purified mitochondria were demonstrated to be essentially duplex throughout their length, although they contain RNA/DNA hybrid regions, which result from the infrequent incorporation of ribonucleotides [8]. The authors suggested that in vitro RNaseH treatment or the process of extracting mtDNA from crude mitochondria leads to degradation of these ribonucleotide-rich regions and produces the partially single-stranded molecules previously assumed to arise by the asynchronous mechanism [18]. Analysis of mitochondrial DNA from rats, mice and humans revealed that mtDNA replication initiates at multiple origins that are distributed across a four kilobase fragment downstream from the 3’-end of the displacement loop, and that DNA replication is restricted to one direction after fork arrest near OriH [7]. Further mapping of prominent free 5’ ends identified two regions of start sites, one corresponding to OriH for the strand-asynchronous model, and the other several hundred nucleotides toward the non-coding D-loop region corresponding to a possible bidirectional replication origin [19].
Analysis of two-dimensional agarose gel electrophoresis also shows that mtDNA contains ribonucleotide incorporation throughout the lagging strand, RITOLS, which is depicted in Figure 2 alongside the other models of mtDNA replication. Yasukawa et al. has demonstrated that these RITOLS map to the major non-coding region of birds and mammals and occur unidirectionally. One of the more prominent RITOLS that is utilized for DNA replication starts in the region of OriL [20]. Transmission EM and antibodies specific for RNA/DNA hybrid molecules shows that duplex DNA intermediates can be found throughout mtDNA and contain extensive RNA tracts on one strand, indicative of a strand-coupled model [21]. However, Brown et al. observe that many mitochondrial transcripts also form stable R-loops, similar to the well-documented R-loop at the leading strand replication origin (OriH) [22]. These stable but nonreplicative, partially hybridized RNA/DNA duplexes raise questions about the function of RITOLS. There are certain elements in both models that are well supported by experimentation, but it is clear that further studies are needed to illuminate whether both models predominate in nature or are products of experimental artifacts.
Regardless of the mode of replication, both models require extension of an RNA primer and it is accepted that this is mediated by the transcription machinery to generate a nascent RNA that is cleaved, processed, and extended by DNA pol γ. DNA replication is carried out by pol γ, the mtDNA helicase Twinkle, mtSSB, topoisomerases, RNaseH1 and a host of transcription initiation and termination factors. RNA polymerase is recruited to the LSP by TFAM and TFB2 to initiate transcription and generate near-genomic length transcripts [23–25]. These transcripts from the LSP can be cleaved or prematurely terminated to generate primers for DNA replication [11, 12]. While the RNA polymerase is highly processive on double-stranded DNA, in vitro, the mitochondrial RNA polymerase will synthesize short RNA primers on single-stranded DNA of 25–75 nucleotides that can be extended by DNA pol γ [26]. Further evidence has suggested that the RNA polymerase synthesizes a 25 nt RNA primer at OriL, after it becomes single-stranded and adopts a stem-loop structure [17]. This RNA primer is then extended by pol γ to complete the initiation of lagging strand DNA replication [17]. Thus, RNA polymerase initiates RNA primers for DNA pol γ at OriH and OriL.
2. Human mitochondrial DNA polymerase, catalytic subunit and accessory subunit
Of the 16 DNA polymerases present in eukaryotes, pol γ is the only DNA polymerase found in the mammalian mitochondria and hence bears the burden of replicating and repairing the entire 16.5 kb circular mitochondrial genome [27, 28]. The human pol γ holoenzyme consists of a catalytic subunit (encoded by POLG at chromosomal locus 15q25) and a dimeric form of its accessory subunit (encoded by POLG2 at chromosomal locus 17q24.1). The catalytic subunit is a 140 kDa enzyme (p140) that has DNA polymerase, 3'→5' exonuclease, and 5′ - deoxyribose phosphate (dRP) lyase activities [29]. The accessory subunit is a 55 kDa protein (p55) required for tight DNA binding and processive DNA synthesis [30].
Mutations in the POLG gene were first discovered in 2001 as the cause of progressive external ophthalmoplegia (PEO) [31] and have been found to be a frequent cause of mitochondrial disease resulting from either mtDNA depletion disorders, including Alpers and myocerebrohepatopathy spectrum disorder, or mtDNA deletion diseases such as the aforementioned PEO, ataxia-neuropathy syndromes, myoclonus epilepsy myopathy sensory ataxia, and related disorders [32–34]. Although not as frequent as POLG disorders, mutation in POLG2 can cause PEO and PEO-like symptoms [35, 36].
2.1 Biochemical properties
DNA pol γ displays robust activity in vitro with diverse primer-template substrates including natural DNA substrates and homopolymeric templates like poly(dA)·oligo(dT) and poly(dC)·oligo(dG). The activity of pol γ is a magnitude of order higher for homopolymeric primer-template substrates that contain high primer density compared to natural DNA substrates [37]. Pol γ can also perform efficient catalysis on ribohomopolymeric primer-templates like poly(rA)·oligo(dT12–18) in the presence of Mn2+ as the metal co-activator, which accelerates the catalysis of DNA polymerases at the expense of fidelity [38]. However, in vivo, DNA polymerases use Mg2+ as the co-activator, which promotes high fidelity polymerization. DNA pol γ also performs reverse transcription with a higher catalytic efficiency than HIV-1 reverse transcriptase, but the reverse transcription activity of pol γ is far less efficient than its replication of natural DNA sequences [39]. While intriguing, the physiological significance of DNA pol γ reverse transcription still needs to be elucidated. Related to this novel pol γ function, recent studies using steady state kinetic analysis have shown that the polymerase does not discriminate between deoxyribonucleotide incorporation opposite to a single deoxyribonucleotide or ribonucleotide in the template, suggesting that the single nucleotide reverse transcription activity of pol γ is very efficient [40]. Since pol γ is the sole DNA polymerase in human mitochondria, its ability to utilize a wide variety of substrates allows its involvement in all DNA transactions of the mitochondrial genome unlike the 15 nuclear DNA polymerases, which have specialized roles in replication and repair processes.
Recombinant human DNA pol γ overproduced in the baculovirus system and purified from insect (Sf9) cells is active over a broad pH range from 7.5 to 9.5, and requires divalent cations like Mg2+ or Mn2+, depending on the substrates used in polymerase reactions. In addition, the enzyme has a very high affinity for dNTP binding as expected from a replicative polymerase, but is strongly inhibited by dideoxynucleotides. The protein is resistant to inhibition by aphidicolin, and the sulfhydryl blocking agent N-ethylmaleimide (NEM) inhibits the polymerase activity of the catalytic subunit > 90% at 0.5 mM NEM [37]. However, the holoenzyme displayed nearly complete resistance to NEM up to 1 mM [30], suggesting that the p55 accessory subunit protects the catalytic subunit more than 100-fold from inhibition by NEM. This property of the accessory subunit has been biochemically exploited to determine physical interactions between the subunits of the holoenzyme [41]. The optimal salt concentration for pol γ activity also depends on the presence of p55 and on the substrate used. On homopolymeric poly(rA)·oligo(dT12–18) substrates, pol γ is active at 75 mM NaCl while the optimal salt is 25 mM for the poly(dA)·oligo(dT12–18) substrate. However, the activity of pol γ is significantly inhibited at >100 mM NaCl on activated salmon sperm DNA substrates [37]. Reconstitution of the heterotrimeric holoenzyme abolishes the salt sensitivity and the complex is active over a broader range of salt concentration (75 mM to 175 mM). In addition, the processivity of the catalytic subunit is enhanced as much as 50-fold upon interaction with the accessory subunit [30]. The interaction of the accessory subunit with the catalytic subunit also enhances the affinity of the complex for DNA, which increases the processivity of the holoenzyme.
The N-terminal region of the pol γ catalytic subunit comprises an exonuclease domain responsible for degrading newly incorporated nucleotides in the nascent DNA strand. This robust activity depends on the conserved aspartic acid and glutamic acid residues [37]. The 3’→5’ exonuclease has a broad pH spectrum, requires a divalent metal cation for its activity, and is stimulated by moderate to high concentrations of NaCl. The exonuclease can efficiently degrade single-stranded DNA and shows mild preference for 3’-terminal mismatches in double-stranded DNA [42, 43]. Mutations of the catalytic aspartates in yeast mitochondrial DNA polymerase, Mip1, showed a 1,440-fold increase in the enzyme’s mutation frequency in addition to incorrect nucleotide incorporation and proficient mismatch extension [44]. Use of the human exonuclease-proficient pol γ [37] demonstrated that pol γ has high DNA synthesis fidelity due to high nucleotide selectivity and efficient proofreading [45]. The exonuclease proofreading function offered a >20-fold enhancement of misincorporation fidelity [45]. Surprisingly, the exonuclease activity of the catalytic subunit is compromised upon interaction with its accessory subunit [45–47]. This could be due to the bias displayed by the accessory subunit upon interaction with the catalytic core to position the primer-terminus region towards the polymerase domain of the holoenzyme [48]. Alternatively, the tight binding to DNA offered by associating with the p55 subunit promotes highly processive DNA synthesis at the cost of slightly reduced fidelity [30].
2.2 Crystal structure
In 2009, Lee et al. described the three dimensional (3D) structure of the pol γ holoenzyme at 3.2 Å resolution using the exonuclease-deficient version of the catalytic subunit and the homodimeric accessory subunit lacking a four α-helix bundle at the dimer interface [48]. The p140 catalytic subunit contains an N-terminal exonuclease and a C-terminal polymerase domain connected by a long linker or spacer domain (~400 residues). The polymerase domain includes three subdomains, termed the “palm,” “finger,” and “thumb”. The crystal structure also revealed that a small region near the N-terminus of the spacer domain made up a portion of the thumb subdomain. The spacer region contains two subdomains, the intrinsic processivity (IP) subdomain and an accessory-interacting determinant (AID). These subdomains are essential for the interaction of p140 with the p55 dimer, which results in a highly processive holoenzyme. Sequence alignment of the catalytic subunit of pol γ revealed that the AID region in the spacer domain is conserved only in mammals and not in Xenopus, Drosophila and other lower eukaryotic organisms such as Saccharomyces cerevisiae or Schizosaccharomyces pombe [49]. Interestingly, the organisms that lack this AID region are also devoid of the accessory subunit.
The heterotrimeric 3D structure also contained a dimer of the accessory subunit. While the accessory subunit used for these structural studies lacked a helix bundle, the crystal structure revealed that this region of p55 is not involved in interaction with the catalytic subunit. The structure of the holoenzyme was also very unique in that the catalytic subunit mainly interacted with only one monomer of the dimeric accessory subunit [48]. Each monomer of the p55 dimer contributes differently to the overall processivity of the holoenzyme; the monomer proximal to p140 increases the DNA binding affinity of the complex whereas the distal monomer enhances the rate of polymerization. While the distal subunit of p55 contacts p140 mainly through only two of its residues (Glu-394 forms a salt bridge with Arg-232 of p140, and Arg-122 weakly contacts Gln-540 of p140 through van der Waals interactions), the proximal subunit of p55 makes extensive contact with the catalytic core of the holoenzyme predominantly via hydrophilic interactions with the N-terminal region of the p140 thumb subdomain (Glu-454 to Asp-469 and Arg-579) and positively charged residues in the palm subdomain of p140. Furthermore, the C-terminal region of the proximal p55 monomer forms hydrophobic contacts with the L-helix (Val-543 to Leu-558) of the AID of p140 [48].
2.3 Ribonucleotide incorporation
During DNA replication, DNA pol γ must preferentially incorporate deoxyribonucleotides over ribonucleotides, and it must discriminate between these two in an environment in which ribonucleotides are more abundant [50, 51]. In 1973, Grossman et al documented that mature mouse and human mtDNA contain ribonucleotides at a frequency of ~10 ribonucleotides per genome [52]. Even though pol γ selectively incorporates deoxyribonucleotides during mtDNA synthesis, mammalian mtDNA contains at least 10–30 individual ribonucleotides spaced at ~500-bp intervals across the entire mitochondrial genome [8, 52]. Ribonucleotides in mtDNA may arise from incomplete digestion of randomly spaced RNA primers, as suggested in the RITOLS model, or incorrectly incorporated during DNA synthesis. Given that pol γ is highly processive and is therefore unlikely to fall off of DNA during mtDNA replication, pol γ is the likely source of ribonucleotide incorporation into mtDNA. Preliminary analysis revealed that pol γ can incorporate a ribonucleotide into DNA [39], and a recent study identified that human pol γ was able to differentially discriminate against each of the four ribonucleoside triphosphates [40]. The identity of the base played a role in discrimination, as rGTP was discriminated against dGTP only 1,100-fold; the discrimination for rCTP versus dCTP was 6,600-fold, and for rATP versus dATP, discrimination was 9,300-fold. Interestingly, pol γ exhibited a 77,000-fold discrimination against rUTP compared to dTTP. Prior work suggests that the stringent discrimination against rUTP is not due to the absence of the C5-methyl group compared to dTTP, because porcine liver DNA pol γ binds dUTP with only a 3-fold lower efficiency compared to dTTP [53]. Hence, the majority of discrimination against rUTP is primarily due to the 2’-OH group.
A recent study by Wheeler et al. revealed that in rat tissue mitochondria, the rNTP/dNTP levels varied depending on the identity of the nucleotide and tissue, but in general ATP, UTP, CTP, and GTP levels were ~1000-fold, 9 to 73-fold, 6 to 12-fold and 2 to 26-fold higher compared to their respective deoxyribonucleotide counterparts [54]. In the study of the discrimination by the human pol γ against ribonucleotides, the nucleotide binding affinity (Km) appeared to be the greater determinant in the kinetic analysis [40]. Thus, the higher ribonucleotide concentration in mitochondria likely contributes to the high degree of ribonucleotide incorporation observed in mammalian mtDNA. However, since one source of mitochondrial dNTP pools appears to involve the reduction of ribonucleotides by ribonucleotide reductases [32, 55], the up-regulation of the reductase enzyme may alter this balance by reducing rNTP pools and increasing dNTP pools. Nevertheless, neither the biological origin nor the consequences of the presence of ribonucleotides in mtDNA is well understood.
Several studies suggest that the mitochondrial transcription machinery primes the initiation of mtDNA replication [6, 11, 12, 17, 24, 26]. Recent studies using single-nucleotide incorporation and steady-state kinetic analysis revealed that the mtDNA polymerase could extend primers with a terminal ribonucleotide, albeit with a 3–14-fold lower efficiency compared to a deoxyribonucleotide-terminated primer [40]. This difference appears to be mainly due to reduced nucleotide binding affinity and catalysis seen with the pol γ/DNA complex containing a terminal ribonucleotide since the enzyme displayed similar affinity for DNA/DNA and DNA/RNA substrates [39].
Both models of mtDNA replication, the asynchronous strand displacement model and the coupled replication model, agree that the 16.5 kb human mitochondrial genome is replicated by the pol γ holoenzyme [2]. As discussed earlier in section 1.1, in the asynchronous model, pol γ extends the H-strand DNA from an RNA primer, suggesting that it can initiate DNA synthesis from a ribonucleotide [6]. However, in the RITOLS model of mtDNA replication, the replication intermediates contain regions of RNA/DNA heteroduplexes and extensive RNA-rich sequences in the lagging strand, indicating a role for pol γ in incorporating these ribonucleotides during nascent strand synthesis [7, 8, 19, 20]. RNase H enzymes can remove the RNA incorporated into DNA, where RNase H1 processively cleaves long stretches of RNA/DNA hybrids and RNase H2 removes singly incorporated ribonucleoside residues in DNA. While both H1 and H2 enzymes are found in the nucleus, only RNase H1 has been implicated in the mitochondria [56]. Thus, the lack of a known RNase H2 activity in mitochondria helps explain why the single ribonucleotide residues persist in the mitochondrial genome, regardless of their source. Since pol γ possesses reverse transcriptase activity, these single ribonucleotides present in the template DNA strand could be bypassed during replication.
Studies investigating the physiological consequence of reverse transcriptase activity by human pol γ opposite a single ribonucleotide found that pol γ stalls after incorporating a single deoxyribonucleotide [57]. In addition, recent results reveal that the efficiency of bypassing a single ribonucleotide in the template DNA was 51% compared to a control template containing no ribonucleotides [40]. Templates with longer stretches of RNA reduced the bypass efficiency because pol γ stalled while bypassing long stretches of ribonucleotides in DNA. Pol γ was able to bypass 4 and 8 consecutive ribonucleotides in the template with only 29% and 14% efficiency, respectively, compared to the control reactions. The cause of this decline in catalytic activity is possibly due to the A-form conformation of a RNA-DNA heteroduplex, which is not well tolerated by most replicative DNA polymerases [40, 58].
2.4 Base excision repair in mitochondria
Pol γ is also implicated in base excision repair (BER) of mtDNA. BER in mitochondria can occur either by single-nucleotide BER (SN-BER) or long-patch BER (LP-BER) pathways [59]. In both cases, a specific DNA glycosylase removes the damaged DNA base and an AP endonuclease cleaves the DNA strand 5′ to the abasic site leaving a single nucleotide gap containing a 5′ -dRP group. In SN-BER, this 5′ -dRP moiety is then removed and the resulting single nucleotide gap is filled by pol γ making the substrate suitable for ligation by ligase III [60]. The rate of the dRP lyase reaction is considerably slow for pol γ compared to pol β, which is involved in nuclear BER. Since the removal of the dRP moiety by pol γ proceeds via a β-elimination reaction mechanism involving the formation of a Schiff base intermediate between the dRP-containing DNA substrate and the active site lysine residue (the Schiff base nucleophile) of the enzyme, the enzyme-dRP intermediate can be trapped as a covalent complex using sodium borohydride. Recently, LP-BER activity has been reported in mitochondrial extracts, which involves the concerted efforts of several proteins like the flap endonuclease FEN-1, which removes the 5’- flap DNA displaced by pol γ, and DNA2, a DNA helicase with endonuclease activity that processes the expanding 5’-flap structure [61–64]. In addition, ligase III is involved in sealing the gaps in DNA. However, the location of the dRP lyase active site in pol γ has remained elusive. In an attempt to delineate the dRP lyase region of pol γ, partial proteolysis of a trapped enzyme/DNA complex identified a 58 kDa fragment covalently crosslinked with the DNA substrate containing the dRP moiety. Amino-terminal sequence analysis of this 58 kDa protease-resistant fragment revealed that the polypeptide was a C-terminal fragment of the full-length enzyme that begins with the Gly723 residue. This fragment comprises the polymerase domain of pol γ and a small region of the C-terminal portion of the spacer domain of unknown function (Kasiviswanathan and Copeland, unpublished data). Mapping the dRP lyase active site in a family A DNA polymerase will be significant in elucidating the mitochondrial BER pathway.
3. The mitochondrial DNA helicase, C10orf2 gene product or Twinkle helicase
The C10orf2 gene encodes a 5’ → 3’ helicase [65] with significant sequence homology to the C-terminal end of T7 gene product 4 (gp4) helicase-primase [65, 66]. The C10orf2 gene product contains five helicase sequence motifs in the C-terminal half of the protein, but it lacks the primase-associated sequences found in T7 gp4. However, similar to T7 gp4 [67], the mitochondrial helicase has been observed as both a hexamer and a heptamer depending on salt and cofactors [68, 69]. This helicase co-localizes with mtDNA in mitochondrial nucleoids and forms punctate mitochondrial fluorescence reminiscent of twinkling stars from which it derives its name, Twinkle [65]. Transgenic mouse lines overexpressing wild-type Twinkle demonstrated up to a 3-fold increase in mtDNA copy number in muscle and heart, and reduction of Twinkle expression in cultured human cells by RNAi dramatically decreased mtDNA copy number in cultured human cells [70]. These observations suggest that in addition to playing a role in mtDNA maintenance, the mtDNA helicase Twinkle is essential for regulation of mtDNA copy number in mammals [70].
To help determine the specific function of Twinkle in mtDNA transactions, several lines of work have focused on the biochemical characterization of the enzyme. The helicase activity of Twinkle is specifically stimulated by human mtSSB [71], and the enzyme alone was found to only unwind short oligonucleotide substrates. With the help of mtSSB, Twinkle can promote rolling-circle DNA synthesis by the pol γ holoenzyme in vitro by unwinding dsDNA at the replication fork [72]. To further characterize the activity of the mitochondrial helicase, deletions and amino acid substitutions in the C-terminus of the Drosophila enzyme demonstrate its essential role in helicase function [73]. In addition, alanine substitution mutagenesis of conserved residues in a putative N-terminal primase domain of Drosophila mtDNA helicase suggests a lack of primase activity [74], consistent with the overall lack of homology to T7 gp4 in this area of the protein [65]. Despite the lack of evidence for primase activity in mitochondrial helicases, these enzymes may still play a role in elongating the RNA-primers required to initiate mitochondrial DNA replication. Recent evidence suggests that the mitochondrial DNA helicase Twinkle can load onto closed circular DNA without a specialized helicase loader [75]. Furthermore, in concert with pol γ, Twinkle could support replication of a closed circular double-stranded DNA template containing a D-loop reminiscent of that found in vivo. This loading onto a preformed substrate and supporting efficient extension of a primer by pol γ has direct implications for understanding initiation of mtDNA replication.
Disease mutations in the C10orf2 gene have been evaluated in vivo and by study of the recombinant helicase harboring these mutations as a homogeneous population. Overexpression of mutations in C10orf2 associated with adPEO in cultured human or Schneider cells results in stalled mtDNA replication or depletion of mtDNA [76–78]. Analysis of the recombinant helicase purified from baculoviral infected insect cells has demonstrated defects in the helicase due to disease mutations [79, 80]. Disease mutations in the linker region were shown to disrupt protein hexamerization and abolish DNA helicase activity [79]. Four mutations in the N-terminal domain demonstrated a dramatic decrease in ATPase activity [80]. A comprehensive study of recombinant disease variants overproduced and purified from E. coli has revealed that all of the disease variants display some level of activity, but many have partial accompanying defects. In over 20 different variants tested, mild to moderate defects were seen in helicase activity, ATP hydrolysis, and stability [81]. All of the variants displayed efficient DNA binding. This study emphasizes the need to optimize in vitro conditions for biochemical analysis of disease variants [81]. The moderate defects demonstrated in vitro is consistent with the delayed onset of the autosomal dominant PEO associated with mutation of C10orf2.
4. The mitochondrial single-stranded DNA binding protein
Mitochondrial single-stranded DNA binding protein was discovered in an analysis of protein-mtDNA complexes derived from rat liver mitochondria that had been lysed with SDS, which revealed nucleoprotein fibrils within the single-stranded portions of both stable and expanding D-loops in replicative intermediates of rat liver mtDNA [82]. The asynchronous model of mtDNA replication predicts the existence of large regions of single-stranded DNA, and the abundant presence of mtSSB in these nucleoprotein fibrils strongly suggests that the mtSSB protein is an essential component of the mtDNA replication machinery. Mitochondrial SSBs have been isolated and cloned from yeast and various animal sources and are between 13 and 16 kDa [83–90]. The native form of the mammalian mtSSB is a tetramer, with a molecular weight of 56 kDa [90, 91]. The gene for the human mtSSB has been cloned [88], and the crystal structure has been determined [92]. The DNA is proposed to wrap around the tetrameric mtSSB through electropositive patches guided by flexible loops [92]. The mtSSB tetramer has high affinity for DNA, and its DNA binding site encompasses 8 to 17 nucleotides [84, 85, 93].
Although no direct interaction of mtSSB with the transcription machinery has been demonstrated, numerous interactions with the replication proteins and other factors have been documented. In vitro experiments with mtSSB added to purified pol γ demonstrated significant stimulation of the polymerase activity on various primer-template substrates [94, 95]. In Drosophila, mtSSB increases mitochondrial DNA synthesis almost 40-fold, and fruit flies with a mutated mtSSB gene display significant mtDNA depletion and dysfunction of the respiratory chain [96, 97]. In humans, an 8-fold stimulation of human pol γ was noted with human mtSSB [95]. This functional interaction with the human enzyme is modest but is negatively modulated by the terminal regions of the mtSSB, as C- and N- terminal deletion variants of human mtSSB stimulate more DNA synthesis than full length human mtSSB [98]. In a minimal in vitro replication system, human mtSSB stimulates DNA synthesis by pol γ and Twinkle to form products of 16 kb in length compared to 2 kb generated by pol γ and Twinkle alone [72]. Stimulation of human pol γ by human mtSSB is specific as no stimulation of bacterial and nuclear DNA polymerase is observed by mtSSB [95]. In addition to pol γ, mtSSB also specifically stimulates the mitochondrial DNA helicase Twinkle. Whereas E. coli SSB could not enhance the activity of the enzyme, mtSSB could promote unwinding of DNA by Twinkle [71, 98]. Recent work with human mtSSB variants demonstrated strikingly different effects on pol γ and Twinkle [99]. Human mtSSB variants that displayed defects in stimulating pol γ had unaltered effect on Twinkle. Conversely, mtSSB variant that showed reduced stimulation of helicase activity stimulated pol γ like wild type mtSSB. These experiments demonstrate distinct structural elements in the mtSSB for the functional interaction with pol γ and helicase [99].
5. Topoisomerases
DNA topoisomerases (topos) are ubiquitous enzymes that alter the topology of DNA by reversibly breaking the phosphodiester DNA backbone and allowing a second DNA strand to pass through this break (for review, [100]). While all topos induce DNA breaks via an active site tyrosine residue, they are classified as either type I (A or B) or type II (A or B) based on their mechanism of action. Type I topos break one strand of DNA at a time, while type II topos induce a concerted double-stranded break in the DNA. Topoisomerase-induced reversible strand breaks enable DNA transactions to occur as if there were no constraints. As a result, topos play essential roles in DNA transactions such as relieving supercoiling stress during replication and transcription. To date, six topoisomerases have been identified in humans. Two of the six are type II enzymes: topoisomerases IIα and IIβ. The remainder are type I enzymes: topoisomerases IIIα and IIIβ, topoisomerase I (top1), and mitochondrial topoisomerase I (top1mt). Of these six, two have been convincingly demonstrated to function in the mitochondria, and they are described below. While it is likely that other topos play a role in mitochondrial DNA transactions, minimal data is available to conclusively describe and define their identity and role(s).
5.1 Mitochondrial topoisomerase I
Until about a decade ago, it was thought that humans only had a single gene encoding for topoisomerase I. Work by Zhang et al. altered this perception when the group discovered a gene duplication of nuclear topo I that encoded for a polypeptide with an N-terminal mitochondrial targeting sequence, top1mt [101]. Top1mt is a type IB topoisomerase that requires a divalent metal and alkaline pH for optimal activity, and studies have shown that this enzyme has been adapted to optimally function specifically in the mitochondria [102]. Top1mt localizes to the mitochondria, and it is highly expressed in tissues rich in mitochondria such as the heart and brain [101]. Furthermore, top1mt can be trapped to form covalent complexes with mtDNA when treated with camptothecin, a known topoisomerase poison. Top1mt cleavage sites were mapped to an asymmetric cluster near the regulatory D-loop region of mtDNA. Treatment of mitochondria with camptothecin caused a reduction in the level of 7S DNA formation, suggesting that blocking top1mt activity reduces the level of D-loop formation in the mitochondria [103]. These results imply that top1mt may play a novel role in regulating mitochondrial replication and/or transcription.
To further address the role of top1mt in mitochondrial transcription versus replication, work by Dalla Rosa et al. suggests that top1mt primarily affects mitochondrial transcription [102]. In an elegantly designed experiment, nuclear top1 was targeted to the mitochondria and overexpressed, but the nuclear enzyme could not function in the mitochondria. Overexpression of nuclear top1 in the mitochondria instantaneously inhibited mitochondrial transcription, and replication was inhibited after a delay. Because the kinetics of transcription inhibition versus replication inhibition are distinct, the authors hypothesized that these effects were due to inhibition of RNA-primer synthesis that is required to initiate mtDNA replication. If this were the case, one would expect that overexpression of nuclear top1 in the mitochondria would preferentially inhibit replication initiation, but replication that has already begun could still proceed. Indeed, 2D agarose gel electrophoresis of mtDNA replication intermediates showed that intermediates indicative of replication initiation were significantly reduced whereas intermediates of ongoing replication were only marginally reduced [102]. This result supports a role for top1mt in mtDNA transcription initiation, and also suggests that an alternate topoisomerase can relieve supercoiling stress during mtDNA replication. Furthermore, unpublished work shows that TOP1mt knockout mice are viable, suggesting that yet another topoisomerase may complement top1mt activity in transcription [104]. Whether TOP1mt knockout mice exhibit a pathological phenotype indicative of a mitochondrial disease remains to be determined, but work by Wang et al. identified human TOP1mt as a mitochondrial disease candidate, which points to an important role for top1mt in mitochondrial function [105].
5.2 Topoisomerase IIIα
While top1mt arose from a gene duplication that resulted in a dedicated mitochondrial enzyme, topo IIIα serves dual roles in the mitochondria and nucleus. Topo IIIα is a type IA topoisomerase that was previously thought to only function in the nucleus until 2002, when work by Wang et al. demonstrated the existence of two start codons (M1 and M26) for the topo IIIα polypeptide [100]. In HeLa cells, translation from the first codon produces an enzyme with a mitochondrial targeting sequence that directs the enzyme to the mitochondria. Translation from the second codon results in an enzyme that is primarily localized to the nucleus. This study was the first to conclusively demonstrate the existence of a type IA topoisomerase in the human mitochondria [100].
While progress has been made on the function of topo IIIα in the nucleus, little is known about its function in the mitochondria. In the nucleus, topo IIIα helps resolve recombination intermediates, but recombination of human mitochondrial DNA is rare. Clues from other organisms point to an essential role for topo IIIα in mitochondrial DNA maintenance. Wu et al. showed that topo IIIα is required to maintain the mtDNA genome in Drosophila [106]. Flies without the mitochondrial form of topo IIIα can survive to adulthood, but with accompanying fertility defects. The most dramatic effect occurs in females, which are completely sterile because the eggs they lay fail to hatch in part because of dramatically reduced mtDNA content [106]. Complementing the studies in Drosophila, additional clues about topo IIIα’s function come from trypanosomes. Trypanosomes are protozoans that possess a topologically intricate mtDNA (kinetoplast DNA) network in the form of thousands of interlocked circles. This network contains several dozen maxicircles interlocked with thousands of minicircles. In trypanosomes, replication of these mtDNA minicircles was found to require a type IA topoisomerase, named topo IAmt. Scocca and Shapiro demonstrated that without this essential type IA topoisomerase, θ-type intermediates accumulate because the final mtDNA minicircle intertwines were not resolved at the end of replication [107]. Additional studies in bacteria show that topo III, a type IA topo, cooperates with RecQ and SSB to resolve converging replication forks at the end stage of replication [108]. Taken together, these results suggest that topo IIIα may serve an essential role in mtDNA maintenance by removing the last few intertwines of the parental mtDNA strands at the end of replication. More studies are needed to define the specific function of topo IIIα in the mitochondria, and to establish whether it is a candidate disease locus for patients suffering from mitochondrial disease.
5.3 Other mitochondrial topoisomerases
To our knowledge, top1mt and topo IIIα are the only two topoisomerases that have been convincingly shown to localize to the mitochondria in humans. Furthermore, additional evidence suggests that these two enzymes are functionally important for mitochondrial transcription and/or replication. While these two type I topoisomerases play important roles in the mitochondria, additional studies suggest that type II topoisomerases may function in the mitochondria as well. Type II topoisomerases are ubiquitous throughout nature and it would come as no surprise if they were found to be involved in mitochondrial DNA transactions, especially given the circular nature of mtDNA. A truncated form of topo IIβ, a type II topoisomerase, was putatively identified in bovine mitochondria based on MALDI-TOF analysis of an isolated polypeptide found to exhibit topoisomerase activity [109]. Other studies suggest the presence of a bacterial-like type II topoisomerase in mammalian mitochondria based on the fact that the type II topo-targeting antibacterial drug ciprofloxacin induces the formation of protein-linked mtDNA breaks in cells (for review, [110]). These studies provide evidence that other topoisomerases are present in mammalian mitochondria, but a considerable amount of work is needed to confirm these findings and define the function of these enzymes in mitochondrial transcription and replication.
6. RNase H activity in mitochondria
As mentioned above in sections 1.1 and 2.3, RNase H enzymes are necessary for the removal of RNA that is found in DNA. Two forms of RNase H exist, H1 and H2, where RNase H1 functions to processively cleave long stretches of RNA/DNA hybrids and RNase H2 removes singly incorporated ribonucleoside 5’-monophosphate (rNMP) residues in DNA. Only RNase H1 has been implicated in the mitochondria as determined by deletion of the mouse RNaseHI gene [56]. Mouse embryos carrying the RNaseH1 gene knockout are embryonic lethal due to a significant decrease in mtDNA content [56]. While most RNase H1 predominantly localizes to the nucleus, a fraction of the protein is found in mitochondria. This dual localization is reminiscent of topo IIIα as discussed in section 5.2, and it results from differential translation of two potential in-frame start codons. Translation from the first codon produces an RNase H1 enzyme with a mitochondrial targeting sequence whose expression is tightly controlled since too little or too much RNase H1 in the mitochondria can lead to cell death [111]. Possible roles for RNase H1 may be removal of RNA primers at origins of replication on the leading (OH) and lagging (OL) DNA strand, or during processing of Okazaki fragments proposed in the coupled replication model of mtDNA replication.
7. Conclusion
The proteins and enzymes mentioned in this review must work in a coordinated fashion to initiate and carry out mtDNA replication. Figure 1 depicts a schematic cartoon of how these proteins must function in a mtDNA replication fork. While many experiments have demonstrated functional interactions between these proteins, it is unknown whether physical interactions occur between the transcription and replication proteins. Given the promiscuity of pol γ to utilize a variety of DNA and RNA substrates, a definitive interaction between the RNA polymerase and pol γ may not be needed. Future experiments are needed to address this interaction.
Highlights.
Mitochondrial DNA replication is primed by transcription
An asynchronous and coupled model of mtDNA replication have been proposed
MtDNA is replicated by pol γ, helicase, SSB, topo and RNaseH1
Acknowledgements
We thank Drs. Matthew Longley and Matthew Young for critically reading this manuscript. This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (ES 065078 and ES 065080).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain
References
- 1.Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown TA, Cecconi C, Tkachuk AN, Bustamante C, Clayton DA. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes & Development. 2005;19:2466–2476. doi: 10.1101/gad.1352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogenhagen DF, Clayton DA. Concluding remarks: The mitochondrial DNA replication bubble has not burst. Trends Biochem Sci. 2003;28:404–405. doi: 10.1016/S0968-0004(03)00165-8. [DOI] [PubMed] [Google Scholar]
- 4.Bogenhagen DF, Clayton DA. The mitochondrial DNA replication bubble has not burst. Trends Biochem Sci. 2003;28:357–360. doi: 10.1016/S0968-0004(03)00132-4. [DOI] [PubMed] [Google Scholar]
- 5.Holt IJ, Jacobs HT. Response: The mitochondrial DNA replication bubble has not burst. Trends Biochem Sci. 2003;28:355–356. doi: 10.1016/S0968-0004(03)00133-6. [DOI] [PubMed] [Google Scholar]
- 6.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 7.Bowmaker M, Yang MY, Yasukawa T, Reyes A, Jacobs HT, Huberman JA, Holt IJ. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J Biol Chem. 2003;278:50961–50969. doi: 10.1074/jbc.M308028200. [DOI] [PubMed] [Google Scholar]
- 8.Yang MY, Bowmaker M, Reyes A, Vergani L, Angeli P, Gringeri E, Jacobs HT, Holt IJ. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 2002;111:495–505. doi: 10.1016/s0092-8674(02)01075-9. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Exp Physiol. 2003;88:41–56. doi: 10.1113/eph8802514. [DOI] [PubMed] [Google Scholar]
- 10.Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee DY, Clayton DA. Properties of a primer RNA-DNA hybrid at the mouse mitochondrial DNA leading-strand origin of replication. J Biol Chem. 1996;271:24262–24269. doi: 10.1074/jbc.271.39.24262. [DOI] [PubMed] [Google Scholar]
- 12.Xu B, Clayton DA. RNA-DNA hybrid formation at the human mitochondrial heavy-strand origin ceases at replication start sites: an implication for RNA-DNA hybrids serving as primers. Embo J. 1996;15:3135–3143. [PMC free article] [PubMed] [Google Scholar]
- 13.Robberson DL, Clayton DA. Replication of mitochondrial DNA in mouse L cells and their thymidine kinase - derivatives: displacement replication on a covalently-closed circular template. Proceedings of the National Academy of Sciences of the United States of America. 1972;69:3810–3814. doi: 10.1073/pnas.69.12.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tapper DP, Clayton DA. Mechanism of replication of human mitochondrial DNA. Localization of the 5' ends of nascent daughter strands. J Biol Chem. 1981;256:5109–5115. [PubMed] [Google Scholar]
- 15.Kang D, Miyako K, Kai Y, Irie T, Takeshige K. In vivo determination of replication origins of human mitochondrial DNA by ligation-mediated polymerase chain reaction. J Biol Chem. 1997;272:15275–15279. doi: 10.1074/jbc.272.24.15275. [DOI] [PubMed] [Google Scholar]
- 16.Wong TW, Clayton DA. In vitro replication of human mitochondrial DNA: accurate initiation at the origin of light-strand synthesis. Cell. 1985;42:951–958. doi: 10.1016/0092-8674(85)90291-0. [DOI] [PubMed] [Google Scholar]
- 17.Fuste JM, Wanrooij S, Jemt E, Granycome CE, Cluett TJ, Shi Y, Atanassova N, Holt IJ, Gustafsson CM, Falkenberg M. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol Cell. 2010;37:67–78. doi: 10.1016/j.molcel.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 18.Holt IJ, Lorimer HE, Jacobs HT. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100:515–524. doi: 10.1016/s0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- 19.Yasukawa T, Yang MY, Jacobs HT, Holt IJ. A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol Cell. 2005;18:651–662. doi: 10.1016/j.molcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Yasukawa T, Reyes A, Cluett TJ, Yang MY, Bowmaker M, Jacobs HT, Holt IJ. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25:5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pohjoismaki JL, Holmes JB, Wood SR, Yang MY, Yasukawa T, Reyes A, Bailey LJ, Cluett TJ, Goffart S, Willcox S, Rigby RE, Jackson AP, Spelbrink JN, Griffith JD, Crouch RJ, Jacobs HT, Holt IJ. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J Mol Biol. 2010;397:1144–1155. doi: 10.1016/j.jmb.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown TA, Tkachuk AN, Clayton DA. Native R-loops persist throughout the mouse mitochondrial DNA genome. J Biol Chem. 2008;283:36743–36751. doi: 10.1074/jbc.M806174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clayton DA. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- 24.Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 25.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 26.Wanrooij S, Fuste JM, Farge G, Shi Y, Gustafsson CM, Falkenberg M. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11122–11127. doi: 10.1073/pnas.0805399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burgers PM, Koonin EV, Bruford E, Blanco L, Burtis KC, Christman MF, Copeland WC, Friedberg EC, Hanaoka F, Hinkle DC, Lawrence CW, Nakanishi M, Ohmori H, Prakash L, Prakash S, Reynaud CA, Sugino A, Todo T, Wang Z, Weill JC, Woodgate R. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J Biol Chem. 2001;28:28. doi: 10.1074/jbc.R100056200. [DOI] [PubMed] [Google Scholar]
- 28.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 29.Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase gamma in Mitochondrial DNA Replication and Repair. Chemical Reviews. 2006;106:383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- 30.Lim SE, Longley MJ, Copeland WC. The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J Biol Chem. 1999;274:38197–38203. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- 31.Van Goethem G, Dermaut B, Lofgren A, Martin JJ, Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nature genetics. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- 32.Copeland WC. Inherited mitochondrial diseases of DNA replication. Annu Rev Med. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong LJ, Naviaux RK, Brunetti-Pierri N, Zhang Q, Schmitt ES, Truong C, Milone M, Cohen BH, Wical B, Ganesh J, Basinger AA, Burton BK, Swoboda K, Gilbert DL, Vanderver A, Saneto RP, Maranda B, Arnold G, Abdenur JE, Waters PJ, Copeland WC. Molecular and clinical genetics of mitochondrial diseases due to POLG mutations. Hum Mutat. 2008;29:E150–E172. doi: 10.1002/humu.20824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saneto RP, Naviaux RK. Polymerase gamma disease through the ages. Dev Disabil Res Rev. 2010;16:163–174. doi: 10.1002/ddrr.105. [DOI] [PubMed] [Google Scholar]
- 35.Longley MJ, Clark S, Yu Wai Man C, Hudson G, Durham SE, Taylor RW, Nightingale S, Turnbull DM, Copeland WC, Chinnery PF. Mutant POLG2 Disrupts DNA Polymerase gamma Subunits and Causes Progressive External Ophthalmoplegia. Am J Hum Genet. 2006;78:1026–1034. doi: 10.1086/504303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young MJ, Longley MJ, Li FY, Kasiviswanathan R, Wong LJ, Copeland WC. Biochemical analysis of human POLG2 variants associated with mitochondrial disease. Hum Mol Genet. 2011;20:3052–3066. doi: 10.1093/hmg/ddr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Longley MJ, Ropp PA, Lim SE, Copeland WC. Characterization of the native and recombinant catalytic subunit of human DNA polymerase gamma: identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry. 1998;37:10529–10539. doi: 10.1021/bi980772w. [DOI] [PubMed] [Google Scholar]
- 38.Copeland WC, Lam NK, Wang TS. Fidelity studies of the human DNA polymerase alpha. The most conserved region among alpha-like DNA polymerases is responsible for metal- induced infidelity in DNA synthesis. J Biol Chem. 1993;268:11041–11049. [PubMed] [Google Scholar]
- 39.Murakami E, Feng JY, Lee H, Hanes J, Johnson KA, Anderson KS. Characterization of novel reverse transcriptase and other RNA-associated catalytic activities by human DNA polymerase gamma: importance in mitochondrial DNA replication. J Biol Chem. 2003;278:36403–36409. doi: 10.1074/jbc.M306236200. [DOI] [PubMed] [Google Scholar]
- 40.Kasiviswanathan R, Copeland WC. Ribonucleotide discrimination and reverse transcription by the human mitochondrial DNA polymerase. J Biol Chem. 2011 doi: 10.1074/jbc.M111.252460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan SSL, Longley MJ, Copeland WC. The common A467T mutation in the human mitochondrial DNA polymerase (POLG) compromises catalytic efficiency and interaction with the accessory subunit. J Biol Chem. 2005;280:31341–31346. doi: 10.1074/jbc.M506762200. [DOI] [PubMed] [Google Scholar]
- 42.Kaguni LS, Olson MW. Mismatch-specific 3'----5' exonuclease associated with the mitochondrial DNA polymerase from Drosophila embryos. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:6469–6473. doi: 10.1073/pnas.86.17.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olson MW, Kaguni LS. 3'-->5' exonuclease in Drosophila mitochondrial DNA polymerase. Substrate specificity and functional coordination of nucleotide polymerization and mispair hydrolysis. J Biol Chem. 1992;267:23136–23142. [PubMed] [Google Scholar]
- 44.Foury F, Vanderstraeten S. Yeast mitochondrial DNA mutators with deficient proofreading exonucleolytic activity. Embo J. 1992;11:2717–2726. doi: 10.1002/j.1460-2075.1992.tb05337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Longley MJ, Nguyen D, Kunkel TA, Copeland WC. The Fidelity of Human DNA Polymerase gamma with and without Exonucleolytic Proofreading and the p55 Accessory Subunit. J Biol Chem. 2001;276:38555–38562. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- 46.Pinz K, Bogenhagen D. The influence of the DNA polymerase γ accessory subunit on base excision repair by the catalytic subunit. DNA Repair (Amst) 2006;5:121–128. doi: 10.1016/j.dnarep.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 47.Johnson AA, Johnson KA. Exonuclease proofreading by human mitochondrial dna polymerase. J Biol Chem. 2001;276:38097–38107. doi: 10.1074/jbc.M106046200. [DOI] [PubMed] [Google Scholar]
- 48.Lee YS, Kennedy WD, Yin YW. Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations. Cell. 2009;139:312–324. doi: 10.1016/j.cell.2009.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kasiviswanathan R, Copeland WC. Biochemical analysis of the G517V POLG variant reveals wild-type like activity. Mitochondrion. 2011 doi: 10.1016/j.mito.2011.08.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao G, Orlova M, Georgiadis MM, Hendrickson WA, Goff SP. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grossman LI, Watson R, Vinograd J. The presence of ribonucleotides in mature closed-circular mitochondrial DNA. Proceedings of the National Academy of Sciences of the United States of America. 1973;70:3339–3343. doi: 10.1073/pnas.70.12.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mosbaugh DW. Purification and characterization of porcine liver DNA polymerase gamma: utilization of dUTP and dTTP during in vitro DNA synthesis. Nucleic Acids Res. 1988;16:5645–5659. doi: 10.1093/nar/16.12.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wheeler LJ, Mathews CK. Nucleoside triphosphate pool asymmetry in mammalian mitochondria. J Biol Chem. 2011;286:16992–16996. doi: 10.1074/jbc.M111.236968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, de Lonlay P, Paquis-Flucklinger V, Arakawa H, Nakamura Y, Munnich A, Rotig A. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nature genetics. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 56.Cerritelli SM, Frolova EG, Feng C, Grinberg A, Love PE, Crouch RJ. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol Cell. 2003;11:807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 57.Lee HR, Johnson KA. Fidelity and processivity of reverse transcription by the human mitochondrial DNA polymerase. J Biol Chem. 2007;282:31982–31989. doi: 10.1074/jbc.M705392200. [DOI] [PubMed] [Google Scholar]
- 58.Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Copeland WC, Longley MJ. DNA2 resolves expanding flap in mitochondrial base excision repair. Mol Cell. 2008;32:457–458. doi: 10.1016/j.molcel.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Identification of 5'-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Akbari M, Visnes T, Krokan HE, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst) 2008;7:605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 62.Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, 3rd, Shen B, Demple B. Removal of Oxidative DNA Damage via FEN1-Dependent Long-Patch Base Excision Repair in Human Cell Mitochondria. Mol Cell Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, Shen B. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem. 2008;283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, Santoro L, Toscano A, Fabrizi GM, Somer H, Croxen R, Beeson D, Poulton J, Suomalainen A, Jacobs HT, Zeviani M, Larsson C. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nature genetics. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 66.Garrido N, Griparic L, Jokitalo E, Wartiovaara J, Van Der Bliek AM, Spelbrink JN. Composition and dynamics of human mitochondrial nucleoids. Mol Biol Cell. 2003;14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toth EA, Li Y, Sawaya MR, Cheng Y, Ellenberger T. The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol Cell. 2003;12:1113–1123. doi: 10.1016/s1097-2765(03)00442-8. [DOI] [PubMed] [Google Scholar]
- 68.Ziebarth TD, Farr CL, Kaguni LS. Modular architecture of the hexameric human mitochondrial DNA helicase. J Mol Biol. 2007;367:1382–1391. doi: 10.1016/j.jmb.2007.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ziebarth TD, Gonzalez-Soltero R, Makowska-Grzyska MM, Nunez-Ramirez R, Carazo JM, Kaguni LS. Dynamic effects of cofactors and dna on the oligomeric state of human mitochondrial DNA helicase. J Biol Chem. 2010;285:14639–14647. doi: 10.1074/jbc.M109.099663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tyynismaa H, Sembongi H, Bokori-Brown M, Granycome C, Ashley N, Poulton J, Jalanko A, Spelbrink JN, Holt IJ, Suomalainen A. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum Mol Genet. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 71.Korhonen JA, Gaspari M, Falkenberg M. TWINKLE Has 5' -> 3' DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J Biol Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 72.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. Embo J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matsushima Y, Kaguni LS. Functional importance of the conserved N-terminal domain of the mitochondrial replicative DNA helicase. Biochim Biophys Acta. 2009;1787:290–295. doi: 10.1016/j.bbabio.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsushima Y, Farr CL, Fan L, Kaguni LS. Physiological and biochemical defects in carboxyl-terminal mutants of mitochondrial DNA helicase. J Biol Chem. 2008;283:23964–23971. doi: 10.1074/jbc.M803674200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jemt E, Farge G, Backstrom S, Holmlund T, Gustafsson CM, Falkenberg M. The mitochondrial DNA helicase TWINKLE can assemble on a closed circular template and support initiation of DNA synthesis. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wanrooij S, Goffart S, Pohjoismaki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsushima Y, Kaguni LS. Differential phenotypes of active site and human adPEO mutations in drosophila mitochondrial DNA helicase expressed in schneider cells. J Biol Chem. 2007;282:9436–9444. doi: 10.1074/jbc.M610550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goffart S, Cooper HM, Tyynismaa H, Wanrooij S, Suomalainen A, Spelbrink JN. Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum Mol Genet. 2009;18:328–340. doi: 10.1093/hmg/ddn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Korhonen JA, Pande V, Holmlund T, Farge G, Pham XH, Nilsson L, Falkenberg M. Structure-function defects of the TWINKLE linker region in progressive external ophthalmoplegia. J Mol Biol. 2008;377:691–705. doi: 10.1016/j.jmb.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 80.Holmlund T, Farge G, Pande V, Korhonen J, Nilsson L, Falkenberg M. Structure-function defects of the twinkle amino-terminal region in progressive external ophthalmoplegia. Biochim Biophys Acta. 2009;1792:132–139. doi: 10.1016/j.bbadis.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 81.Longley MJ, Humble MM, Sharief FS, Copeland WC. Disease variants of the human mitochondrial DNA helicase encoded by C10orf2 differentially alter protein stability, nucleotide hydrolysis and helicase activity. J Biol Chem. 2010;285:29690–29702. doi: 10.1074/jbc.M110.151795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Van Tuyle GC, Pavco PA. The rat liver mitochondrial DNA-protein complex: displaced single strands of replicative intermediates are protein coated. J Cell Biol. 1985;100:251–257. doi: 10.1083/jcb.100.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Van Dyck E, Foury F, Stillman B, Brill SJ. A single-stranded DNA binding protein required for mitochondrial DNA replication in S. cerevisiae is homologous to E. coli SSB. Embo J. 1992;11:3421–3430. doi: 10.1002/j.1460-2075.1992.tb05421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mignotte B, Barat M, Mounolou JC. Characterization of a mitochondrial protein binding to single-stranded DNA. Nucleic Acids Res. 1985;13:1703–1716. doi: 10.1093/nar/13.5.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoke GD, Pavco PA, Ledwith BJ, Van Tuyle GC. Structural and functional studies of the rat mitochondrial single strand DNA binding protein P16. Arch Biochem Biophys. 1990;282:116–124. doi: 10.1016/0003-9861(90)90094-f. [DOI] [PubMed] [Google Scholar]
- 86.Tiranti V, Barat-Gueride B, Bijl J, DiDonato S, Zeviani M. A full-length cDNA encoding a mitochondrial DNA-specific single-stranded DNA binding protein from Xenopus laevis. Nucleic Acids Res. 1991;19:4291. doi: 10.1093/nar/19.15.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ghrir R, Lecaer JP, Dufresne C, Gueride M. Primary structure of the two variants of Xenopus laevis mtSSB, a mitochondrial DNA binding protein. Arch Biochem Biophys. 1991;291:395–400. doi: 10.1016/0003-9861(91)90152-9. [DOI] [PubMed] [Google Scholar]
- 88.Tiranti V, Rocchi M, DiDonato S, Zeviani M. Cloning of human and rat cDNAs encoding the mitochondrial single- stranded DNA-binding protein (SSB) Gene. 1993;126:219–225. doi: 10.1016/0378-1119(93)90370-i. [DOI] [PubMed] [Google Scholar]
- 89.Stroumbakis ND, Li Z, Tolias PP. RNA- and single-stranded DNA-binding (SSB) proteins expressed during Drosophila melanogaster oogenesis: a homolog of bacterial and eukaryotic mitochondrial SSBs. Gene. 1994;143:171–177. doi: 10.1016/0378-1119(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 90.Li K, Williams RS. Tetramerization and single-stranded DNA binding properties of native and mutated forms of murine mitochondrial single-stranded DNA-binding proteins. J Biol Chem. 1997;272:8686–8694. doi: 10.1074/jbc.272.13.8686. [DOI] [PubMed] [Google Scholar]
- 91.Longley MJ, Smith LA, Copeland WC. Preparation of Human Mitochondrial Single-Stranded DNA-Binding Protein. Methods Mol Biol. 2009;554:73–85. doi: 10.1007/978-1-59745-521-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang C, Curth U, Urbanke C, Kang C. Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4 A resolution. Nat Struct Biol. 1997;4:153–157. doi: 10.1038/nsb0297-153. [DOI] [PubMed] [Google Scholar]
- 93.Thommes P, Farr CL, Marton RF, Kaguni LS, Cotterill S. Mitochondrial single-stranded DNA-binding protein from Drosophila embryos. Physical and biochemical characterization. J Biol Chem. 1995;270:21137–21143. doi: 10.1074/jbc.270.36.21137. [DOI] [PubMed] [Google Scholar]
- 94.Mignotte B, Marsault J, Barat GM. Effects of the Xenopus laevis mitochondrial single-stranded DNA-binding protein on the activity of DNA polymerase gamma. Eur J Biochem. 1988;174:479–484. doi: 10.1111/j.1432-1033.1988.tb14123.x. [DOI] [PubMed] [Google Scholar]
- 95.Genuario R, Wong TW. Stimulation of DNA polymerase gamma by a mitochondrial single-strand DNA binding protein. Cell Mol Biol Res. 1993;39:625–634. [PubMed] [Google Scholar]
- 96.Maier D, Farr CL, Poeck B, Alahari A, Vogel M, Fischer S, Kaguni LS, Schneuwly S. Mitochondrial single-stranded DNA-binding protein is required for mitochondrial DNA replication and development in Drosophila melanogaster. Mol Biol Cell. 2001;12:821–830. doi: 10.1091/mbc.12.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Farr CL, Matsushima Y, Lagina AT, 3rd, Luo N, Kaguni LS. Physiological and biochemical defects in functional interactions of mitochondrial DNA polymerase and DNA-binding mutants of single-stranded DNA-binding protein. J Biol Chem. 2004;279:17047–17053. doi: 10.1074/jbc.M400283200. [DOI] [PubMed] [Google Scholar]
- 98.Oliveira MT, Kaguni LS. Functional roles of the N- and C-terminal regions of the human mitochondrial single-stranded DNA-binding protein. PLoS One. 2010;5:e15379. doi: 10.1371/journal.pone.0015379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Oliveira MT, Kaguni LS. Reduced stimulation of recombinant Pol {gamma} and mtDNA helicase by variants of mitochondrial single-stranded DNA-binding protein (mtSSB) correlates with defects in mtDNA Replication in animal cells. J Biol Chem. 2011 doi: 10.1074/jbc.M111.289983. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 101.Zhang H, Barcelo JM, Lee B, Kohlhagen G, Zimonjic DB, Popescu NC, Pommier Y. Human mitochondrial topoisomerase I. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10608–10613. doi: 10.1073/pnas.191321998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dalla Rosa I, Goffart S, Wurm M, Wiek C, Essmann F, Sobek S, Schroeder P, Zhang H, Krutmann J, Hanenberg H, Schulze-Osthoff K, Mielke C, Pommier Y, Boege F, Christensen MO. Adaptation of topoisomerase I paralogs to nuclear and mitochondrial DNA. Nucleic Acids Res. 2009;37:6414–6428. doi: 10.1093/nar/gkp708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang H, Pommier Y. Mitochondrial topoisomerase I sites in the regulatory D-loop region of mitochondrial DNA. Biochemistry. 2008;47:11196–11203. doi: 10.1021/bi800774b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang H, Meng LH, Pommier Y. Mitochondrial topoisomerases and alternative splicing of the human TOP1mt gene. Biochimie. 2007;89:474–481. doi: 10.1016/j.biochi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 105.Wang W, Shen P, Thiyagarajan S, Lin S, Palm C, Horvath R, Klopstock T, Cutler D, Pique L, Schrijver I, Davis RW, Mindrinos M, Speed TP, Scharfe C. Identification of rare DNA variants in mitochondrial disorders with improved array-based sequencing. Nucleic Acids Res. 2011;39:44–58. doi: 10.1093/nar/gkq750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu J, Feng L, Hsieh TS. Drosophila topo IIIalpha is required for the maintenance of mitochondrial genome and male germ-line stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6228–6233. doi: 10.1073/pnas.1001855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Scocca JR, Shapiro TA. A mitochondrial topoisomerase IA essential for late theta structure resolution in African trypanosomes. Mol Microbiol. 2008;67:820–829. doi: 10.1111/j.1365-2958.2007.06087.x. [DOI] [PubMed] [Google Scholar]
- 108.Suski C, Marians KJ. Resolution of converging replication forks by RecQ and topoisomerase III. Mol Cell. 2008;30:779–789. doi: 10.1016/j.molcel.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Low RL, Orton S, Friedman DB. A truncated form of DNA topoisomerase IIbeta associates with the mtDNA genome in mammalian mitochondria. Eur J Biochem. 2003;270:4173–4186. doi: 10.1046/j.1432-1033.2003.03814.x. [DOI] [PubMed] [Google Scholar]
- 110.Rowe TC, Weissig V, Lawrence JW. Mitochondrial DNA metabolism targeting drugs. Adv Drug Deliv Rev. 2001;49:175–187. doi: 10.1016/s0169-409x(01)00133-8. [DOI] [PubMed] [Google Scholar]
- 111.Suzuki Y, Holmes JB, Cerritelli SM, Sakhuja K, Minczuk M, Holt IJ, Crouch RJ. An upstream open reading frame and the context of the two AUG codons affect the abundance of mitochondrial and nuclear RNase H1. Mol Cell Biol. 2010;30:5123–5134. doi: 10.1128/MCB.00619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]