Abstract
Actin polymerization is required for Chlamydia trachomatis entry into nonphagocytic host cells. Host and chlamydial actin nucleators are essential for internalization of chlamydiae by eukaryotic cells. The host cell Arp2/3 complex and the chlamydial translocated actin recruiting phosphoprotein (Tarp) are both required for entry. Tarp and the Arp2/3 complex exhibit unique actin polymerization kinetics individually, but the molecular details of how these two actin nucleators cooperate to promote bacterial entry is not understood. In this study we provide biochemical evidence that the two actin nucleators act synergistically by co-opting the unique attributes of each to enhance the dynamics of actin filament formation. This process is independent of Tarp phosphorylation. We further demonstrate that Tarp colocalization with actin filaments is independent of the Tarp phosphorylation domain. The results are consistent with a model in which chlamydial and host cell actin nucleators cooperate to increase the rate of actin filament formation.
Keywords: Chlamydia trachomatis, Tarp, Arp2/3 complex, Actin
INTRODUCTION
Chlamydia trachomatis is the most prevalent sexually transmitted bacterium in the United States [1]. Chlamydia can infect the genital tract of both men and women. A majority of infections in women are asymptomatic, which if not detected, can lead to pelvic inflammatory disease, ectopic pregnancy and infertility [2]. Worldwide, C. trachomatis infection of the eye (trachoma) is the leading cause of preventable blindness with an estimated 40 million active trachoma infections in 2009 [3].
Chlamydiae display a unique developmental cycle in which the extracellular infectious elementary bodies (EBs) invade human epithelial cells. Once internalized the EB differentiates into a reticulate body (RB) within a membrane bound vacuole called an inclusion and undergoes several rounds of replication before differentiating back to EBs, which are released from the infected cell and can initiate a new infection [4; 5].
C. trachomatis entry into a human cell is dependent on host cytoskeletal rearrangements triggered by bacterial attachment to the host cell surface. [6]. Host cells treated with drugs such as cytochalasin D that disrupt actin polymerization inhibit chlamydial entry [6]. In addition, chlamydial entry is inhibited by disruption of the host actin related protein 2 and 3 nucleating (Arp2/3) complex [7; 8]. A chlamydial type III secreted effector protein termed Tarp, for translocated actin recruiting phosphoprotein, has also been implicated in the bacterial induced cytoskeletal changes that permit EB entry [9]. Tarp harbors distinct actin binding and oligomerization domains which allow for the clustering of multiple actin monomers to nucleate a new actin filament [10]. Antibodies with specificity to the Tarp actin binding domain, when microinjected into host cells prior to C. trachomatis infection, inhibited bacterial entry [11]. Taken together, these findings implicate both host and bacterial actin nucleation complexes in driving cytoskeletal changes required for C. trachomatis entry. The molecular details of Tarp and Arp2/3 complex interactions are unknown.
Each of these two independent actin nucleators employs unique strategies for the generation of new actin filaments. The host cell Arp2/3 complex is comprised of seven proteins including the actin related protein 2 and 3 polypeptides which share homology with actin [12; 13; 14]. The Arp2/3 complex is regulated by host cell machinery such as those members of the Wiskott-Aldrich syndrome family proteins (WASP) whose members are themselves regulated by signal transduction cascades[15; 16]. The Arp2/3 complex associates with existing actin filaments to nucleate a new actin filament forming a branch at an angle of approx. 70 degrees [12]. Conversely, Tarp is a single chlamydial polypeptide of 1005 amino acids, which harbors distinct phosphorylation, oligomerization and actin binding domains [9; 10; 17]. Tarp is able to associate with globular actin to nucleate the formation of linear actin filaments without activation from a nucleation promoting factor [10].
C. trachomatis Tarp is rapidly phosphorylated by host tyrosine kinases such as Abl, Syk and Src family members following translocation into the host cell[18; 19]. Phosphorylated Tarp has been shown to associate with phosphoinositide 3-kinase (PI3K) and Src homology 2 (SH2) domain containing transforming protein 1 (SCH-1) via their respective SH2 domains [20; 21]. Additionally, phosphorylated Tarp had been suggested to be implicated in the GTPase mediated activation of the host cell Arp2/3 complex [20]. Consequently, a role for Tarp phosphorylation in bacterial entry is controversial as chemical inhibitors which prevent Tarp phosphorylation do not inhibit chlamydial entry [18]. Furthermore, Tarp orthologs from other chlamydial species are not phosphorylated suggesting they have evolved mechanisms to circumvent any requirement for Tarp phosphorylation [11; 17; 22].
We demonstrate here that Tarp and the Arp2/3 complex cooperate to increase the rate of actin polymerization and were accompanied by a concomitant increase in branched actin filaments observed by scanning electron microscopy. We show that Tarp phosphorylation does not alter the cooperation between Tarp and the Arp2/3 complex. Furthermore, Tarp colocalization with actin filaments is independent of the Tarp phosphorylation domain. Taken together our results indicate that bacterial and host cell derived actin nucleators work cooperatively to increase the rate of actin filament formation and promote chlamydial entry.
MATERIALS & METHODS
SDS-PAGE and immunoblotting
Proteins were separated on SDS-10% polyacrylamide gels and stained with coomassie R-250 (Pierce) or transferred to 0.45µm pure nitrocellulose transfer and immobilization membrane (Schleicher & Schuell, Keene, NH). Immunoblotting employed peroxidase conjugated secondary antibodies (Chemicon International, Temecula, CA) and Supersignal West Pico chemiluminescent substrate (Pierce, Rockford, IL). The anti-phosphotyrosine 4G10 monoclonal antibody was purchased from Upstate (Millipore). Polyclonal rabbit antibodies directed towards C. trachomatis L2 LGV 434 Tarp was developed at Rocky Mountain Laboratories as previously described [9].
Cloning, protein expression and purification
An in frame glutathione-S-transferase (GST) and polyhistidine C. trachomatis L2 LGV 434 Tarp fusion protein was generated by PCR amplifying the corresponding coding regions from C. trachomatis genomic DNA (QIAGEN genomic purification kit, Valencia CA) using custom synthesized oligonucleotide primers (Integrated DNA Technologies, Coralville, IA) engineered with SalI, SacI or NotI linkers. PCR products were purified (QIAGEN), digested with restriction enzymes (New England Biolabs, Beverly, MA) and subcloned into linearized pGEX-6P-1 to generate translational fusions with GST at the N-terminus and polyhistidine at the C-terminus. The PCR fragments described above harboring tarP were also cloned into pEGFP-C3 (BD Biosciences Clontech) to allow for the ectopic expression of eGFP-Tarp in HeLa cells.
pGEX-6P-1 plasmids were transformed into BL21 strain of E. coli (Novagen, Madison WI). Protein expression and purification were performed according to the procedures outlined for Ni Sepharose 6 Fast Flow and Glutathione sepharose 4B in the Bulk GST Purification Module (GE health sciences, Piscataway, NY).
Pyrene assay
The rate of actin polymerization in the presence of Tarp and the Arp2/3 complex was monitored according to the methods outlined in the Actin Polymerization Biochem Kit BK003 (Cytoskeleton, Denver CO). Briefly, monomeric pyrene labeled actin was prepared by diluting 500 µg of lyophilized pyrene actin into 5 mls of 5 mM Tris (pH 8.0), 0.2 mM CaCl2 and 0.2 mM ATP (G-buffer) and incubating for 1 hour at room temperature followed by an additional hour of incubation at 4°C. Monomeric pyrene actin was obtained by collecting the supernatant following a 2 hour, 100,000 rcf, 4°C spin in a Beckman Optima TLX Ultracentrifuge using a TLA 100.3 rotor (Beckman Coulter Inc., Fullerton, CA). Approximately 40 µg of pyrene labeled actin was gently mixed with 2–5 µg of test proteins in a volume of 500 µl for 5 minutes prior to the addition of 1/20th volume of polymerization buffer (500 mM KCl, 20mM MgCl2 and 10mM ATP). The reaction was monitored over one hour with an LS 55 Luminescence spectrophotometer equipped with a biokinetics accessory and directed by FL WinLab software version 4.0 (Perkin Elmer, Beaconsfield, BUCKS, UK) with 2.5 nm bandwidth at 365 nm excitation wavelength and 2.5 nm bandwidth at 407 nm emission wavelength.
Scanning electron microscopy
Actin filaments were added to an SEM type 3 mount and sputter coated with 8–10nm chromium. Coated filaments were examined in a Hitachi S5200 scanning electron microscope at 30 kV accelerating voltage.
Transfection of HeLa cells and indirect immunofluorescence microscopy
Hela cells (2 × 105) were seeded in 6 well plates with coverslips and grown for 24 hours in DMEM containing 10% FBS. Cells were then transfected with transfection mixture containing 8 µl of Fugene HD (promega) and 2.5 mg of respective plasmid. Following 24 hours, cells were fixed by adding 4 % paraformaldehyde and incubating at 4° C for 15 minutes. Cells were then treated with ice cold 0.4 % Triton-X for 10 minutes, followed by blocking with 5% BSA for 45 minutes. To visualize tyrosine phosphorylated protein, cells were first incubated with anti-phosphotyrosine primary antibody (upstate) at 1:1000 dilutions in 0.5 % BSA at RT for 45 minutes followed by incubation with anti-mouse secondary antibody conjugated to Alexa 350 (invitrogen). To simultaneously visualize actin, phalloidin conjugated to Alexa 568 (invitrogen) was added to the above mixture containing secondary antibodies. To stain for Arp2/3 complex, cells were first incubated with anti-Arp3 primary antibody (upstate) at 1:100 dilutions in 0.5 % BSA at RT for 45 minutes followed by incubation with anti- rabbit secondary antibody conjugated to Alexa 594 (invitrogen). Coverslips were rinsed and mounted in Prolong Gold antifade reagent (invitrogen). Cells were examined with a Zeiss Axio Observer A1 microscope equipped with phase contrast and epifluorescence optics. Images were obtained using an AxioCam MRm camera controlled by AxioVision 4.8.2 and further processed using Adobe Photoshop CS2.
RESULTS
Tarp and activated Arp2/3 cooperate to polymerize actin
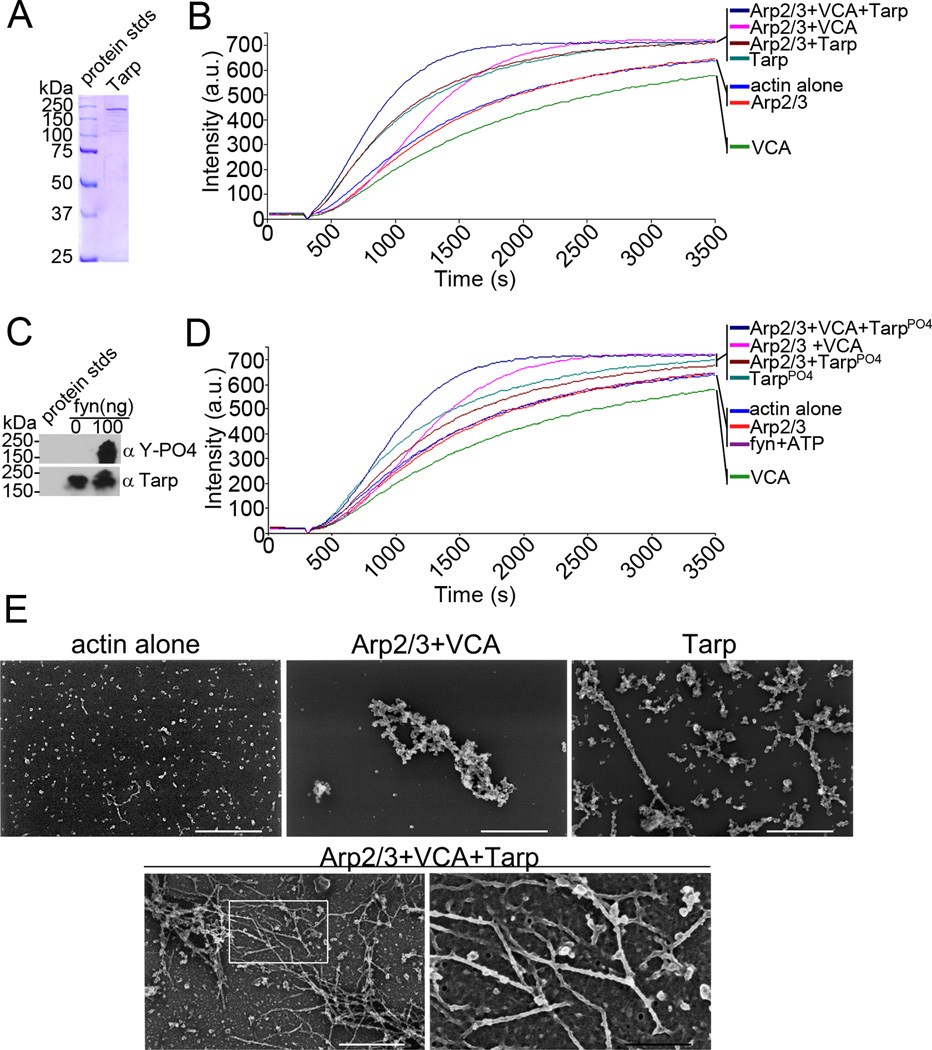
We have previously shown that the actin filaments produced by chlamydial Tarp are linear [10], whereas actin filaments produced via activated Arp2/3 complex from existing filaments result in branched actin filaments [13]. To test whether the chlamydial and host cell actin nucleators cooperate, we examined the rate of actin polymerization in a pyrene actin polymerization assay following the addition of each purified actin nucleator singularly and together (Fig. 1). Chlamydial Tarp and activated Arp2/3 complex (Arp2/3 complex and the nucleation promoting factor verprolin, cofilin, acidic (VCA) domain of the Wiskott-Aldrich Syndrome protein (WASp)) were able to polymerize actin at a greater rate than actin alone controls (Fig. 1B) [10; 16]. Arp2/3 mediated actin nucleation, similar to actin alone controls, exhibits a lag phase in which actin monomers assemble to form a nucleus from which linear actin filament formation proceeds. As the concentration of small linear actin filaments increases, the activated Arp2/3 complex is able to associate to the linear filaments and initiate the formation of a new filament (branched actin) from the existing filament [23; 24; 25]. Conversely, a lag phase is not observed when chlamydial Tarp is added to the pyrene assay indicating that there is no requirement for existing actin filaments to promote Tarp mediated actin nucleation. The rate of actin filament formation increased when Tarp and the activated Arp2/3 complex were added in the same actin polymerization reaction indicating the two actin nucleators cooperate (Fig. 1B). To test whether phosphorylation of Tarp altered its cooperation with the Arp2/3 complex to polymerize actin, recombinant Tarp was phosphorylated with purified Fyn kinase (Fig. 1C) and examined in the pyrene actin polymerization assay (Fig. 1D). Phosphorylated Tarp did not alter the kinetics of actin polymerization as compared to that of nonphosphorylated Tarp, nor did it alter cooperation with the Arp2/3 complex. To examine whether Tarp might activate the Arp2/3 complex directly by serving as a nucleation-promoting factor (NPF) in vitro, pyrene actin polymerization assays were performed in the presence of Tarp or phosphorylated Tarp (Tarp-PO4) and the Arp2/3 complex in the absence of the VCA peptide, which activates the Arp2/3 complex. The rate of actin polymerization did not change relative to Tarp alone (with or without phosphorylation) suggesting that Tarp does not activate the Arp2/3 complex directly (Fig. 1B, D). Actin filaments removed from a pyrene actin polymerization assay at 10 minutes post-delivery of the actin polymerization buffer were analyzed by scanning electron microscopy (Fig 1E). Electron micrography revealed an increase in the amount of branched actin filaments observed in the presence of both the Arp2/3 complex and Tarp compared to activated Arp2/3 complex alone suggesting that the two actin nucleators cooperate to increase the local concentration of branched actin.
Fig. 1.
Tarp and Arp2/3 complex cooperate to polymerize actin. (A) Purified recombinant Tarp employed for these studies was resolved by SDS-PAGE and visualized by Coomassie blue staining. (B) Pyrene actin polymerization in the presence of Tarp and activated Arp2/3 complex (Arp2/3+VCA+Tarp) was compare to actin polymerization kinetics initiated by Tarp alone (Tarp), activated Arp2/3 complex alone (Arp2/3+VCA) and Tarp with an inactive Arp2/3 complex (Arp2/3+Tarp). Actin alone (actin), inactive Arp2/3 complex (Arp2/3) and the Verprolin, Cofilin, Acidic (VCA) domain of N-WASP served as additional controls. Bacterial and host cell nucleators were incubated with pyrene conjugated actin and actin polymerization was measured by an increase in fluorescence of pyrenyl-actin incorporated into actin filaments (Intensity (a.u.)) over approximately 1 hour (Time (s)) following the addition of polymerization buffer at 300 seconds. (C) Purified recombinant Tarp was phosphorylated by the Src family kinase member fyn. Tarp proteins were incubated with 100ng of purified fyn kinase and ATP and following a short incubation were suspended in protein sample buffer and resolved by SDS-PAGE followed by transfer to nitrocellulose. Immunoblots were performed with Tarp (α Tarp) and phosphotyrosine (α Y-PO4) specific antisera. (D) Pyrene actin polymerization assays as B but with phosphorylated Tarp. Additional controls include actin polymerization assays performed in the presence of fyn kinase and phosphorylation buffer containing ATP (fyn+ATP)(E) Electron micrographs of actin filaments nucleated by Tarp and the Arp2/3 complex. Protein samples were removed from actin polymerization experiments similar to those shown in B at 900 seconds (10 minutes after the addition of polymerization buffer) and actin filaments were visualized by scanning electron microscopy. White scale bar represents 400 nm and black scale bar represents 150 nm.
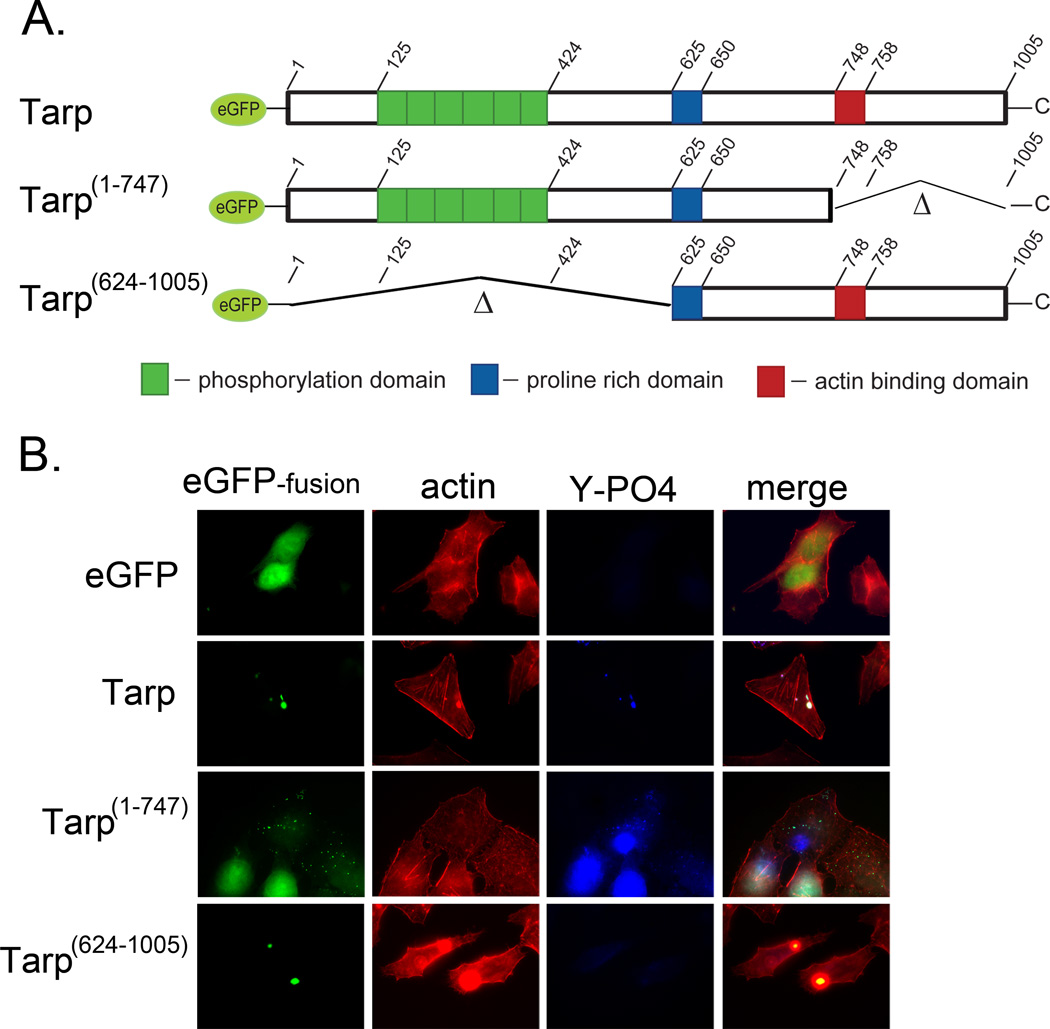
Ectopically expressed eGFP-Tarp colocalizes with actin filaments but not the Arp2/3 complex
Previous studies have examined actin colocalization with ectopically expressed eGFP-Tarp fusion proteins [9; 17]. Tarp fusion proteins aggregate in the host cell cytosol and colocalize with actin filaments. The actin colocalization is clearly observed with fusion proteins containing the entire Tarp protein sequence as well as the C-terminal half of Tarp [17]. Subsequent studies have specifically identified the actin binding alpha helix present between amino acids 748–758 as responsible for Tarp mediated actin binding [10]. Conversely, the ectopically expressed N-terminus and repeat region (phosphorylation domain) of Tarp do not aggregate in the cytosol and appear to localize to the host cell nucleus [17]. A new eGFP-Tarp fusion protein was engineered which harbors the phosphorylation domain and the proline rich domain, responsible for Tarp aggregation, but lacks the actin binding domain (Fig. 2A). Host cells expressing this eGFP fusion protein (Tarp1–747) displayed small discrete aggregates in the host cell cytosol that were phosphorylated and did not localize with actin filaments (Fig.2B), whereas eGFP-Tarp lacking the phosphorylation domain but harboring the actin binding domain (Tarp624–1005) did localize with actin filaments. Interestingly neither the ectopically expressed full length nor mutant eGFP-Tarp proteins colocalized with the Arp2/3 complex which is consistent with previous reports in which the Arp2/3 complex was not detected in Tarp binding studies (Supplemental Fig. 2)[10]. Taken together, these data demonstrate that the Tarp phosphorylation domain is not required for colocalization with actin and suggests the phosphorylation domain alone is not sufficient to drive the cytoskeletal changes observed surrounding the ectopically expressed full length Tarp but that the proline-rich oligomerization domain may play a prominent role in aggregation of ectopically expressed Tarp.
Fig. 2.
Actin filaments co-localize with ectopically expressed Tarp in a phosphorylation domain independent manner. Tarp proteins ectopically expressed as enhanced green fluorescence protein fusions were examined for their ability to localize with actin filaments. (A) Schematic of the eGFP-Tarp fusion proteins indicating the location of the actin binding domain (red box), the proline rich domain (blue box) and the tyrosine rich phosphorylation domain (green boxes). Δ indicates amino acids deleted in the mutant eGFP –Tarp fusion proteins and numbers indicate amino acid positions encoded within the C. trachomatis tarP gene (B) Host cells expressing eGFP (eGFP) alone or eGFP fusions (eGFP-fusion) of full length Tarp (Tarp), or deletion mutants lacking the phosphorylation domain (Tarp 624–1005) or actin binding domain (Tarp 1–747) but harboring the proline rich domain (PRD) required for protein aggregation were fixed and stained with Alexa fluor 568 conjugated phalloidin (actin), phosphotyrosine specific antisera (Y-PO4) and goat anti-mouse conjugated to Alexa 350.
DISCUSSION
We have previously hypothesized that chlamydial entry relies upon the coordinated efforts of Tarp and the Arp2/3 complex acting cooperatively to alter the host cell cytoskeleton and promote chlamydial entry [10]. The Arp2/3 complex is highly regulated by host cell proteins called nucleation promoting factors (NPFs) such as those members of the Wiskott-Aldrich syndrome family proteins (WASP) whose members are themselves regulated by Rho family of GTPases [16; 26]. Nucleation promoting factors are not restricted to eukaryotic origins and in some cases are derived from pathogens such as Listeria monocytogenes (ActA) and Rickettsia conorii (RickA) [27; 28]. Although Tarp alone is capable of directly nucleating actin filaments, a role for Tarp as a NPF for the Arp2/3 complex had not been investigated. The addition of Tarp (with or without phosphorylation) to the Arp2/3 complex in the absence of known NPFs produced no changes to the rate of actin polymerization detected by pyrene actin polymerization assays compared to that of Tarp alone. Conversely, when Tarp and activated Arp2/3 were added together the rate of actin polymerization increased relative to Tarp alone (Fig. 1B). Our results suggest that Tarp’s ability to rapidly initiate the formation of linear actin filaments provided substrate for the Arp2/3 complex to form branched actin. These results were supported by visualization of the actin filaments by scanning electron microscopy. Furthermore, the Tarp and Arp2/3 complex cooperation was not dependent on Tarp phosphorylation as Tarp proteins phosphorylated by Fyn kinase were also able to cooperate with the Arp2/3 complex.
A comparison of Tarp orthologs from different species and serovars of Chlamydia has shed light on the conserved nature of the characterized Tarp protein domains [11; 17; 22]. Interestingly, it appears that different Tarp orthologs can employ different mechanisms to nucleate actin depending on their respective number of actin binding domains [11]. Conversely, the phosphorylation domain is restricted to serovars of C. trachomatis, suggesting Tarp phosphorylation may initiate signal transduction cascades not utilized by other chlamydial species. Phosphorylated Tarp has been shown to associate with the host cell adapter protein SHC-1 and this association is implicated in altering expression of host cell genes pertinent to cell growth and host cell survival [21]. Phosphorylated Tarp peptides have also been shown to immunoprecipitate a complex of proteins containing Sos1 and Vav2, two Rac guanine nucleotide exchange factors thought to participate in WAVE2 and Arp2/3 complex recruitment [20]. To examine whether Tarp was capable of recruiting actin in the absence of an actin binding domain, via the Tarp phosphorylation domain, eGFP-Tarp fusion proteins were expressed in HeLa cells that were missing the actin binding domain, but still harbored the proline rich domain required for Tarp aggregation. eGFP-Tarp proteins were phosphorylated and did not colocalize with filamentous actin consistent with the belief that Tarp/actin colocalization is a function of the C-terminal actin binding domain and does not require the phosphorylation domain.
Collectively, the data demonstrate that the Arp2/3 complex and Tarp actin nucleators cooperate to increase the rate of actin filament formation, however, Tarp does not directly activate the Arp2/3 complex. While an indirect role for phosphorylated Tarp mediated Arp2/3 complex activation is possible, the eGFP-Tarp mutant proteins harboring the phosphorylation domain alone were unable to recruit actin filaments in vivo. Our data advocates that Tarp signaling alone is insufficient to activate the Arp2/3 complex. Arp2/3 complex activation during chlamydial entry is likely to be multifactorial as multiple signals converge on the host actin nucleating complex. Arp2/3 complex activation might be accomplished in part via EB binding to host cell receptors. Fibroblast Growth Factor Receptors (FGFRs) were recently implicated in EB entry and are known to activate the Arp2/3 complex by association with Ras GTPase activating-like protein 1 (IQGAP1) [30; 31]. Presently, a role for IQGAP1 in chlamydial entry has not been examined and requires further investigation. Although Arp2/3 complex activation during EB entry remains unclear, our results indicate that bacterial and host actin nucleators cooperate biochemically to increase the local concentration of actin filaments required for invasion.
Highlights.
Arp2/3 and Tarp cooperate to increase the rate of actin polymerization.
Tarp does not directly activate the Arp2/3 complex.
Tarp aggregation with actin filaments is independent of Tarp phosphorylation.
Arp2/3 chemical inhibitors disrupt chlamydial entry
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to recognize members of Mollie W. Jewett laboratory for helpful discussions as well as acknowledge the technical assistance of Talia Chavez and Brenda Nguyen. This work was supported by the NIAID, NIH K award 5K22AI81729-2 and the University of Central Florida to T.J.J. and the intramural research program of the NIAID, NIH to T.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.CDC Grand Rounds: Chlamydia prevention: challenges and strategies for reducing disease burden and sequelae. MMWR Morb Mortal Wkly Rep. 60:370–373. [PubMed] [Google Scholar]
- 2.Schachter J. Infection and disease epidemiology. In: Stephens RS, editor. Chlamydia; Intracellular biology, pathogenesis, and immunity. Washington, D.C.: ASM Press; 1999. pp. 139–169. [Google Scholar]
- 3.Mariotti SP, Pascolini D, Rose-Nussbaumer J. Trachoma: global magnitude of a preventable cause of blindness. Br J Ophthalmol. 2009;93:563–568. doi: 10.1136/bjo.2008.148494. [DOI] [PubMed] [Google Scholar]
- 4.Hybiske K, Stephens RS. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc Natl Acad Sci U S A. 2007;104:11430–11435. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moulder JW. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carabeo RA, Grieshaber SS, Fischer E, Hackstadt T. Chlamydia trachomatis induces remodeling of the actin cytoskeleton during attachment and entry into HeLa cells. Infect Immun. 2002:3793–3803. doi: 10.1128/IAI.70.7.3793-3803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carabeo RA, Dooley CA, Grieshaber SS, Hackstadt T. Rac interacts with Abi-1 and WAVE2 to promote an Arp2/3-dependent actin recruitment during chlamydial invasion. Cell Microbiol. 2007;9:2278–2288. doi: 10.1111/j.1462-5822.2007.00958.x. [DOI] [PubMed] [Google Scholar]
- 8.Hybiske K, Stephens RS. Mechanisms of Chlamydia trachomatis entry into nonphagocytic cells. Infect Immun. 2007;75:3925–3934. doi: 10.1128/IAI.00106-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A. 2004;101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jewett TJ, Fischer ER, Mead DJ, Hackstadt T. Chlamydial TARP is a bacterial nucleator of actin. Proc Natl Acad Sci U S A. 2006;103:15599–15604. doi: 10.1073/pnas.0603044103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jewett TJ, Miller NJ, Dooley CA, Hackstadt T. The conserved Tarp actin binding domain is important for chlamydial invasion. PLoS Pathog. 2010;6:e1000997. doi: 10.1371/journal.ppat.1000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mullins RD, Stafford WF, Pollard TD. Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. J Cell Biol. 1997;136:331–343. doi: 10.1083/jcb.136.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol. 1997;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgs HN, Pollard TD. Regulation of actin polymerization by Arp2/3 complex and WASp/Scar proteins. J Biol Chem. 1999;274:32531–32534. doi: 10.1074/jbc.274.46.32531. [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi H, Miki H, Suetsugu S, Ma L, Kirschner MW, Takenawa T. Two tandem verprolin homology domains are necessary for a strong activation of Arp2/3 complex-induced actin polymerization and induction of microspike formation by N-WASP. Proc Natl Acad Sci U S A. 2000;97:12631–12636. doi: 10.1073/pnas.190351397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clifton DR, Dooley CA, Grieshaber SS, Carabeo RA, Fields KA, Hackstadt T. Tyrosine phosphorylation of the chlamydial effector protein Tarp is species specific and not required for recruitment of actin. Infect Immun. 2005;73:3860–3868. doi: 10.1128/IAI.73.7.3860-3868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jewett TJ, Dooley CA, Mead DJ, Hackstadt T. Chlamydia trachomatis tarp is phosphorylated by src family tyrosine kinases. Biochem Biophys Res Commun. 2008;371:339–344. doi: 10.1016/j.bbrc.2008.04.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehlitz A, Banhart S, Hess S, Selbach M, Meyer TF. Complex kinase requirements for Chlamydia trachomatis Tarp phosphorylation. FEMS Microbiol Lett. 2008;289:233–240. doi: 10.1111/j.1574-6968.2008.01390.x. Engel. [DOI] [PubMed] [Google Scholar]
- 20.Lane BJ, Mutchler C, Al Khodor S, Grieshaber SS, Carabeo RA. Chlamydial entry involves TARP binding of guanine nucleotide exchange factors. PLoS Pathog. 2008;4:e1000014. doi: 10.1371/journal.ppat.1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehlitz A, Banhart S, Maurer AP, Kaushansky A, Gordus AG, Zielecki J, Macbeath G, Meyer TF. Tarp regulates early Chlamydia-induced host cell survival through interactions with the human adaptor protein SHC1. J Cell Biol. 2010;190:143–157. doi: 10.1083/jcb.200909095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lutter EI, Bonner C, Holland MJ, Suchland RJ, Stamm WE, Jewett TJ, McClarty G, Hackstadt T. Phylogenetic analysis of Chlamydia trachomatis Tarp and correlation with clinical phenotype. Infect Immun. 2010;78:3678–3688. doi: 10.1128/IAI.00515-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amann KJ, Pollard TD. Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc Natl Acad Sci U S A. 2001:15009–15013. doi: 10.1073/pnas.211556398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amann KJ, Pollard TD. The Arp2/3 complex nucleates actin filament branches from the sides of pre-existing filaments. Nat Cell Biol. 2001;3:306–310. doi: 10.1038/35060104. [DOI] [PubMed] [Google Scholar]
- 25.Volkmann N, Amann KJ, Stoilova-McPhie S, Egile C, Winter DC, Hazelwood L, Heuser JE, Li R, Pollard TD, Hanein D. Structure of Arp2/3 complex in its activated state and in actin filament branch junctions. Science. 2001;293:2456–2459. doi: 10.1126/science.1063025. [DOI] [PubMed] [Google Scholar]
- 26.Miki H, Miura K, Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- 27.Gouin E, Egile C, Dehoux P, Villiers V, Adams J, Gertler F, Li R, Cossart P. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature. 2004;427:457–461. doi: 10.1038/nature02318. [DOI] [PubMed] [Google Scholar]
- 28.Welch MD, Rosenblatt J, Skoble J, Portnoy DA, Mitchison TJ. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- 29.Vingadassalom D, Campellone KG, Brady MJ, Skehan B, Battle SE, Robbins D, Kapoor A, Hecht G, Snapper SB, Leong JM. Enterohemorrhagic E. coli requires N-WASP for efficient type III translocation but not for EspFU-mediated actin pedestal formation. PLoS Pathog. 2010;6:e1001056. doi: 10.1371/journal.ppat.1001056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bensenor LB, Kan HM, Wang N, Wallrabe H, Davidson LA, Cai Y, Schafer DA, Bloom GS. IQGAP1 regulates cell motility by linking growth factor signaling to actin assembly. J Cell Sci. 2007;120:658–669. doi: 10.1242/jcs.03376. [DOI] [PubMed] [Google Scholar]
- 31.Kim JH, Jiang S, Elwell CA, Engel JN. Chlamydia trachomatis Co-opts the FGF2 signaling pathway to enhance infection. PLoS Pathog. 7:e1002285. doi: 10.1371/journal.ppat.1002285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.