Abstract
Daphnia pulex is the first crustacean to have its genome sequenced. The genome project provides new insight and data into how an aquatic crustacean may respond to environmental stressors, including toxicants. We cloned Daphnia pulex HR96 (DappuHR96), a nuclear receptor orthologous to the CAR/PXR/VDR group of nuclear receptors. In Drosophila melanogaster, (hormone receptor 96) HR96 responds to phenobarbital exposure and has been hypothesized as a toxicant receptor. Therefore, we set up a transactivation assay to test whether DappuHR96 is a promiscuous receptor activated by xenobiotics and endobiotics similar to the constitutive androstane receptor (CAR) and the pregnane X-receptor (PXR). Transactivation assays performed with a GAL4-HR96 chimera demonstrate that HR96 is a promiscuous toxicant receptor activated by a diverse set of chemicals such as pesticides, hormones, and fatty acids. Several environmental toxicants activate HR96 including estradiol, pyriproxyfen, chlorpyrifos, atrazine, and methane arsonate. We also observed repression of HR96 activity by chemicals such as triclosan, androstanol, and fluoxetine. Nearly 50% of the chemicals tested activated or inhibited HR96. Interestingly, unsaturated fatty acids were common activators or inhibitors of HR96 activity, indicating a link between diet and toxicant response. The omega-6 and omega-9 unsaturated fatty acids linoleic and oleic acid activated HR96, but the omega-3 unsaturated fatty acids alpha-linolenic acid and docosahexaenoic acid inhibited HR96, suggesting that these two distinct sets of lipids perform opposing roles in Daphnia physiology. This also provides a putative mechanism by which the ratio of dietary unsaturated fats may affect the ability of an organism to respond to a toxic insult. In summary, HR96 is a promiscuous nuclear receptor activated by numerous endo- and xenobiotics.
Keywords: HR96, Daphnia, unsaturated fatty acids, nuclear receptor, transactivation
1. Introduction
Daphnia sp. (water fleas) are small planktonic crustaceans in the order Cladocera, a diverse group of branchiopods, found all over the world. They are important components of aquatic ecosystems that feed on algae and are a food source for fish (Carpenter et al., 1987). Daphnia have short, typically parthenogenic, life-cycles and in turn are easy to maintain under laboratory conditions. Daphnia are widely used in ecotoxicology testing, and their sensitivity to toxicants has been correlated to other animal species in acute and chronic toxicity tests (LeBlanc, 1984; Maki, 1979). Daphnia pulex is the first crustacean and first aquatic arthropod whose genome has been sequenced (Colbourne et al., 2011). Therefore, the Daphnia genome may provide significant information about an individual’s ability to adapt and acclimate to environmental stressors within an aquatic environment, especially because there is a wealth of knowledge on daphnid ecology. Therefore, given the database of information on the effects of toxicants on Daphnia, and the completion of the Daphnia genome, the tools are available to better estimate how organisms adapt or acclimate to environmental stressors including xenobiotics (Baldwin et al., 2009).
Twenty-five nuclear receptors were identified from Daphnia pulex, including an ortholog to Drosophila melanogaster HR96 (DHR96) (Thomson et al., 2009). HR96 is a putative toxicant receptor in the NR1J group of nuclear receptors that are most closely related to the NR1I (VDR/CAR/PXR) group of nuclear receptors found in vertebrates (Bertrand et al., 2004). PXR and CAR are adopted orphan nuclear receptors activated by a variety of ligands, including bile acids, steroids, and several xenobiotics (Kretschmer and Baldwin, 2005), and in turn induce the expression of phase I–III detoxication enzymes (Hernandez et al., 2009; Qatanani and Moore, 2005; Swales and Negishi, 2004). HR96 has been found in most of the arthropod species tested including fruit fly [Drosophila melanagaster] (Fisk and Thummel, 1995; King-Jones et al., 2006), beetles [Tribolium castaneum] (Xu et al., 2010), honey bee [Apis mellifera] (Valverde et al., 2006), and Daphnia pulex (Thomson et al., 2009), but is missing in the pea aphid [Acyrthosiphon pisum] genome (Christiaens et al., 2010). Orthologs of HR96 such as DAF12, NHR8, and NHR48 have also been sequenced from Caenorhabditis elegans (Antebi et al., 2000; Maglich et al., 2001).
In Drosophila, DHR96 controls metabolic and stress-response genes, implicating it in coordinating toxicant response pathways, and it is selectively expressed in tissues involved in the metabolism and depuration of xenobiotics (King-Jones et al., 2006). For example, DHR96 modulates phenobarbital-mediated induction of detoxication genes, including Cyp6d1 (King-Jones et al., 2006; Lin et al., 2011). Drosophila lacking DHR96 do not induce detoxification genes in response to phenobarbital treatment (King-Jones et al., 2006). DAF-12, NHR-8, and NHR-48 from C. elegans are also orthologs of the HR96 receptors, and NHR-8 has been shown to be a toxicant receptor involved in regulation of detoxication enzymes and the protection of C. elegans from chemicals (Lindblom et al., 2001). Therefore, we hypothesize that Daphnia pulex HR96 (DappuHR96) is a promiscuous toxicant receptor that is activated by a relatively large set of diverse chemicals.
In addition, studies demonstrate a key role for DHR96 in dietary fat utilization and metabolism in Drosophila, indicating that the regulation of lipid metabolism is an ancestral function of the VDR/PXR/CAR/DHR96 nuclear receptor groups (Horner et al., 2009; Sieber and Thummel, 2009). DHR96 acts as a key regulator of the Niemann-Pick type C gene family, as well as of other genes involved in cholesterol uptake, metabolism, and transport (Bujold et al., 2010). DHR96 also plays an essential role in triglyceride homeostasis. DHR96 mutants are sensitive to starvation, have reduced levels of triglycerides in the fat body and midgut, and are resistant to diet-induced obesity, while DHR96 overexpression leads to starvation resistance and increased triglyceride levels (Sieber and Thummel, 2009).
Daphnia species have been shown to sequester specific lipids from their diet. Typically, the fatty acid composition of Daphnia reflects their diet; however, during starvation or when food quality is poor the Daphnia preferentially retain the omega-3 (n-3) fatty acids, eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA), and the omega-6 (n-6) fatty acid, arachidonic acid (ARA) (Brett et al., 2006). Temperature has also been shown to regulate lipid levels (Filho et al., 2011) with specific retention of EPA under low temperature conditions (Schlechtriem et al., 2006). In addition, EPA and ARA have been shown to enhance juvenile growth and EPA is a crucial fatty acid constituent of daphnid eggs (Becker and Boersma, 2005). It is generally considered that diet effects toxicity and several toxicants have been shown to alter cellular energy allocation especially lipid levels (De Coen and Janssen, 1997), but specific fatty acids have not been associated with toxicity nor has any mechanism by which lipids alter toxicity in Daphnia been proposed to our knowledge.
Therefore, phylogenetic analysis was performed to demonstrate DappuHR96 is an NR1J member, and transactivation assays were performed to determine if DappuHR96 is activated by environmental chemicals, steroid hormones, bile acids, and fatty acids. Our results indicate that DappuHR96 is as a promiscuous xenobiotic and endobiotic receptor. We propose that opposing interactions by xenobiotics or dietary endobiotics may perturb the ability of an organism to respond to environmental toxicants.
2. Materials and Methods
2.1. Phylogenetics
Phylogenetic analyses of DappuHR96 and the related NR1I group of receptors was performed using methods described previously (Hannas et al., 2010; Thomson et al., 2009) with some modifications. DappuHR96 was compared to NR1I and NR1J members of other species available in GenBank, including Drosophila melanogaster (fruitfly), Homo sapiens (human), Mus musculus (mouse), Camponotus floridanus (Florida carpenter ant), Nasonia vitripennis (parasitic wasp), Ciona intestinalis (vase tunicate; sea squirt), C. elegans (roundworm), Apis mellifera (honeybee), and Ixodes scapularis (deer tick). NCBI accession numbers for the receptors used are available in Supplementary Data File 1. The DNA binding domain (DBD) and the ligand binding domain (LBD) of each receptor were identified using the conserved domain database (CDD) (Marchler-Bauer et al., 2007). Zf-C4 (pfam00105) was used to identify the DBD and Hormone Recep (pfam00104) was used to identify the LBD. Phylogenetic analysis was performed following ClustalX alignment using default parameters (Thompson et al., 1997). The sequences used to perform the phylogenetic analysis are provided as Supplementary Data File 2.
Trees were constructed using Bayesian Inference with MrBayes software version 3.1.2 (Ronquist and Huelsenbeck, 2003) on the Computational Biology Service Unit of Cornell University (http://cbsuapps.tc.cornell.edu/mrbayes.aspx). Phylogenetic trees were constructed using the “mixed-model” approach in which the Markov chain Monte Carlo sampler explores nine different fixed-rate amino acid substitution models implemented in MrBayes. We used 4 chains with runs of 5 million generations, chains sampled every 100 generations, a burnin of 10,000 trees with the WAG model. The Drosophila melanogaster ecdysone receptor was used as the outgroup.
Maximum parsimony and distance parameters were used to provide additional support for the phylogenetic relationships observed. Distance parameters were measured using PAUP 4.0b10 with default characteristics (mean character difference and among site rate variation), and full heuristic searches. Branch support was measured by bootstrap analysis with 1000 replicates. Parsimony was constructed using PAUP version 4.0b10 with heuristic searches, tree-bisection-reconnection, topological constraints not enforced, and multiple tree option in effect with an initial maximum tree setting at 100,000. Branch support was measured by bootstrapping with 10,000 replicates. Trees were visualized with FigTree (http://tree.bio.ed.ac.uk/software).
2.2. Daphnia culture and RNA extraction
Daphnia pulex “the chosen one” (Colbourne et al., 2011) were cultured in deionized, distilled water reconstituted with salts (192 mg/L CaS04·2H20, 192 mg/L NaHCO3, 120 mg/L MgS04, and 8 mg/L KCl, pH 8.2 ± 0.1) to produce moderately hard water (Baldwin and LeBlanc, 1994). Daphnids were housed in an environmental chamber at 21 ± 1°C with a 16:8 light-dark cycle, and fed Selenastrum capricornutum supplemented with fish food as described previously (Baldwin et al., 1995). RNA was extracted from fresh female Daphnia pulex with Tri-Zol (BioRad, Hercules, CA) according to the manufacturer’s directions. Homogenization was performed using a mini-bead beater (Bartlesville, OK). DNAse digestion was performed (Promega, Madison, WI), and D. pulex cDNA was synthesized with 2μg RNA using 200 units Moloney Murine Leukemia Virus-Reverse Transcriptase (MMLV-RT), a 10mM dNTP mixture, and 0.05 mg random hexamers (Promega Corporation, Madison, WI).
2.3. Cloning of DappuHR96
DappuHR96 was amplified using primers DappuHR96-F1 (5′-ATG GAA GAG TAC GTA ACG CTA ACC TCT CC-3′) and DappuHR96-R1 (5′-TCA TCT GGA CTT CAA GTC GAA GAT TTC AAT AAG C-3′). The PCR product was ligated into the pCR 2.1 vector (Invitrogen, Carlsbad, CA), and sequenced by MacrogenUSA (Rockville, MD).
The GAL4-HR96DEF chimeric plasmid was constructed using the Clontech In-Fusion Dry-Down PCR Cloning Kit (Clontech Laboratories, Mountain View, CA). The D, E and F domains of DappuHR96 were isolated by PCR using primer pair F-clontech-HR96-BamHI (5′-GAA TTC CCG GGG ATC GAA TTT ATC ATG TCT GAA GAA GAA CGA ACA GTA AAG AGA-3′) and R-clontech-HR96-XbaI (5′-CTG CGG CCG CTC TAG ATC ATC TGG ACT TCA AGT CGA AGA TTT CAA TAA GC-3′). The PCR product was then fused into the pBIND vector (Promega CheckMate Mammalian Two-hybrid system) in frame after the GAL4 DNA binding domain to construct a plasmid that expresses a chimeric protein consisting of the GAL4 DNA binding domain and DappuHR96 hinge, ligand binding and F domains (DEF) for use in transactivation assays.
2.4. Transactivation Assays
HepG2 cells (ATCC, Rockville MD) were cultured in phenol red-free Dulbecco’s Modified Eagle’s Medium (DMEM) (Mediatech, Manassas, VA), and supplemented with 10% fetal bovine serum (HyClone, Logan, UT), 1% L-glutamine (Invitrogen, Carlsbad, CA), and 1% Penicillin/Streptomycin (Invitrogen, Carlsbad, CA) at 37°C under 5% CO2.
Cells were plated in 12-well plates at 200,000 cells per well and then transfected the next day with GAL4-96DEF chimeric plasmid and 0.1 μg the pG5luc reporter plasmid (Promega CheckMate Mammalian Two-hybrid system) using Effectene Transfection Reagent (Qiagen, Valencia, CA). On the third day, cells were either treated with 0.1% DMSO or specific chemicals of interest dissolved in 0.1% DMSO as described previously (Hernandez et al., 2007; Mota et al., 2010). Luciferase activity was measured 24 hours later with the Steady-Glo Luciferase Assay System (Promega, Madison, WI). Transactivation assays were also performed in the presence of an inverse agonist (Triclosan, DHA) to test whether these chemicals could inhibit DappuHR96 activation (Baldwin and Roling, 2009). Chemicals used in the transactivation assays include the anthropogenic chemicals nonylphenol, lindane, chlorpyrifos, pyriproxyfen, monosodium methyl arsenate, atrazine, 2,4-D, cypermethrin, methoxychlor, parathion, bisphenol A, Phenobarbital, Triclosan, and fluoxetine; the dietary lipids cholesterol and corn oil, the unsaturated fats α-linolenic acid, linoleic acid, docosahexaenoic acid, eicosapentaenoic acid, arachidonic acid, and oleic acid, and the saturated fats palmitic acid, stearic acid, lithocholic acid, chenodeoxycholic acid, docosanoic acid, and decanoic acid; the hormones cortisol, corticosterone, androstenol, estradiol, 20-hydroxyecdysone, and methyl farnesoate. These chemicals were provided by a variety of sources, and Supplementary Data File 3 provides a list of the chemicals, chemical suppliers, CAS numbers, and concentrations used in the initial transactivation assays.
2.5. Statistics
Data are presented as the mean of triplicate assays ± standard error. Statistical significance was determined by ANOVA followed by Dunnett’s test as the post hoc test using the GraphPad Prizm 4.0 statistical and graphing package (La Jolla, CA). Dose-response curves were derived for several chemicals of interest. Chemical concentrations were log transformed and luminescent activity was normalized as described previously as a percent of maximal activation with 100% being the greatest measured activity of a chemical and 0% the lowest activity or the control (Baldwin and Roling, 2009; Mota et al., 2010). The EC50 value and 95% Confidence Interval were determined from the sigmoidal dose-response curves generated by GrapPad Prizm 4.0.
3. RESULTS
3.1. Phylogenetic Analysis
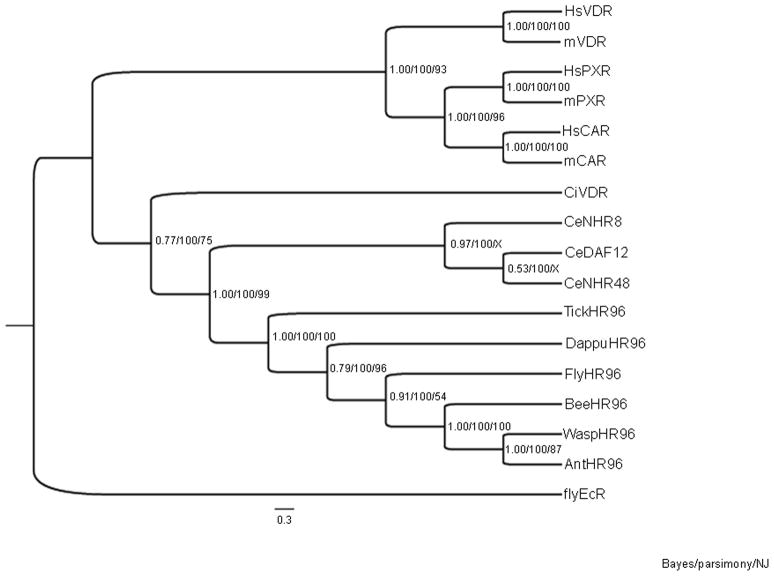
Primers developed based on the D. pulex genome (Thomson et al., 2009) were used to clone and sequence DappuHR96 (GenBank accession JQ026113). DappuHR96 is an ortholog to Drosophila melanogaster HR96 (DHR96 or flyHR96) and other insect HR96 receptors based on the phylogenetic analysis (Fig. 1). The NR1I (VDR/CAR/PXR) and NR1J (HR96) groups are related (Bertrand et al., 2004; Lin et al., 2011). Therefore, NR1I and NR1J receptors were compared phylogenetically to DappuHR96. The phylogenetic tree separates into two distinct clades; one containing the NR1I group and one containing the NR1J group (Fig. 1).
Fig. 1. Phylogenetic analysis of DappuHR96.
DappuHR96 was phylogenetically compared to nuclear receptors within the HR96 (NR1J) and VDR/CAR/PXR (NR1I) groups from several different species including Homo sapiens (Hs), Mus musculus (mouse), Drosophila melanogaster (fly), C. elegans (Ce), Ciona intestinalis (Ci), Ixodes scapularis (tick), Nasonia vitripennis (wasp), Apis mellifera (bee), and Camponotus floridanus (ant). Bayesian Inference, Maximum Parsimony, and Neighbor-Joining were used to confirm the phylogenetic relationship of HR96. The Bayesian tree is shown. Posterior probabilities from the Bayesian tree, and bootstrap support values from the Maximum Parsimony and Neighbor-Joining trees are separated by forward slashes in order from left to right. An X indicates an area of disagreement from the Bayesian tree. Drosophila melanogaster (fly) EcR was chosen as the outgroup. Accession numbers of the analyzed nuclear receptors are provided in Supplementary Data File 1, and the DBD and LBD sequences are provided in Supplementary Data File 2.
Interestingly, the sea squirt’s (Ciona intestinalis), a putative NR1I member, tentatively called CiVDR shows a greater relatedness to the other invertebrate receptors than to the other deuterostomes. DappuHR96 fits within the HR96 group; above the insect members and below the arachnid, Ixodes scapularis, which is consistent with the pancrustacea hypothesis of arthropod evolution (Regier et al., 2010). Direct comparisons of the DBD and LBD of DappuHR96 to the DBD and LBD of the other NR1I and NR1J members by Clustalw, demonstrates similarity among the receptors (Table 1). DappuHR96 and magnaHR96 show striking identity suggesting that the D. magna and D. pulex HR96 receptors have similar function. Overall, phylogenetic analysis demonstrates that DappuHR96 is a NR1J member.
Table 1.
Percent identities of the DNA binding domain (DBD) and ligand binding domain of Daphnia pulex HR96 (DappuHR96) to the DBD and LBD of other specie’s HR96.
| Nuclear Receptor | DBD | LBD |
|---|---|---|
| DappuHR96 | 100 | 100 |
| magnaHR96 | 98 | 96 |
| IsHR96 | 73 | 61 |
| flyHR96 | 77 | 60 |
| waspHR96 | 76 | 69 |
| CeNHR8 | 57 | 29 |
| HsVDR | 51 | 27 |
| HsCAR | 51 | 25 |
Magna = Daphnia magna, Is = Ixodes scapularis, fly = Drosophila melanogaster, wasp = Nasonia vitripennis, Ce = Caenorhabditis elegans, Hs = Homo sapiens
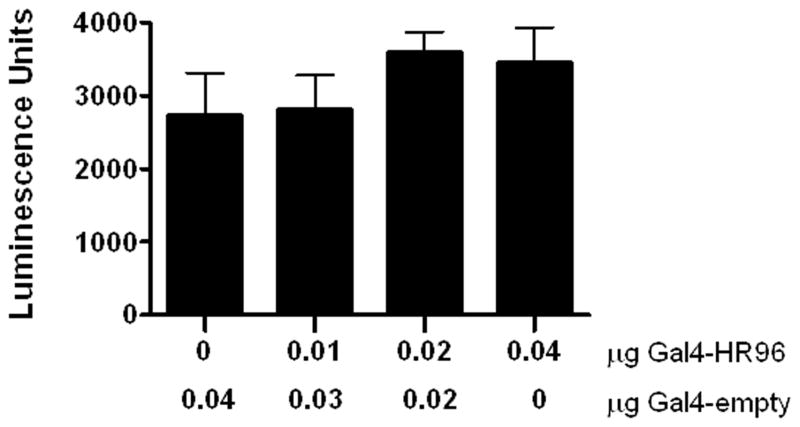
3.2. DappuHR96 does not repress luciferase activity without exogenous ligand
Some nuclear receptors, such as PXR, have been found to repress gene expression in the absence of ligands because of their recruitment of co-repressors (Ourlin et al., 2003; Takeshita et al., 2002). Therefore, cells were transfected with increasing doses of the GAL4-HR96DEF chimeric expression plasmid and the pG5luc reporter gene to determine if HR96 is a ligand-independent repressor of gene expression in HepG2 cells. To ensure that each well received equal amounts of plasmid, the empty pBIND plasmid was co-transfected in each well so that the plasmid concentration was 0.04 μg in all of the wells. HR96 had no effect on the constitutive transcription of the luciferase reporter in the absence of a chemical activator (Fig. 2).
Fig. 2. DappuHR96 does not repress luciferase activity.
HepG2 cells were transfected with a total of 0.04 mg of pBIND-GAL4-HR96-DEF and/or an empty pBIND-Gal4 plasmid and the pG5luc plasmid. Luciferase activity was measured with the Steady-Glo Luciferase Assay System. The presence of HR96 does not basally repress transactivation. Statistical analysis was performed by ANOVA followed by Dunnett’s post-hoc test using GraphPad Prism software (n = 4).
3.3. DappuHR96 is a promiscuous xenobiotic and endobiotic sensor
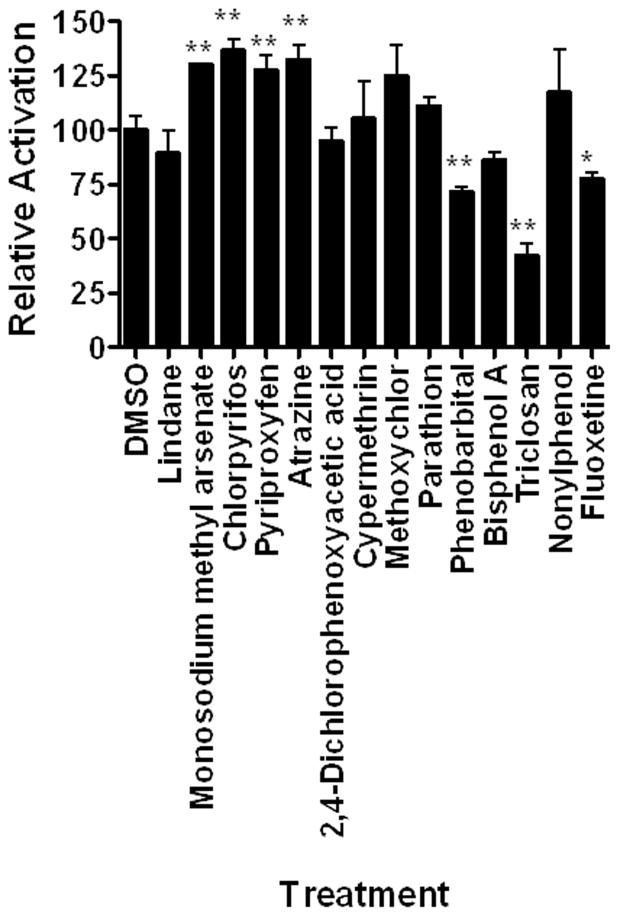
Transactivation assays were performed with numerous chemicals, including multiple xenobiotics, including several types of insecticides and herbicides, plasticizers, pharmaceuticals, and the antibiotic triclosan that is used in numerous consumer products (Fig. 3). Of the fourteen xenobiotics tested, four activated DappuHR96 activity and three reduced DappuHR96 activity. The inhibitors of DappuHR96 activity are considered inverse agonists or inverse activators because they reduce activity in the absence of a known ligand (Forman et al., 1998). None of the plasticizers, organochlorine insecticides, or cypermethrin activated HR96. DappuHR96 is activated by atrazine, monosodium methyl arsenate, chlorpyrifos, and pyriproxyfen; all various types of pesticides. DappuHR96 was inhibited weakly by Phenobarbital and Fluoxetine and strongly by Triclosan (Fig. 3). All three inhibitors have pharmaceutical activity.
Fig. 3. DappuHR96 activity is altered by several xenobiotics.
HepG2 cells were transfected with pBIND-GAL4-HR96-DEF and the pG5luc plasmid. Luciferase activity was measured following exposure to several xenobiotics. Statistical analysis was performed by ANOVA followed by Dunnett’s post-hoc test using GraphPad Prism software. An asterisk indicates p < 0.05 and two asterisks indicate a p < 0.01 (n = 4).
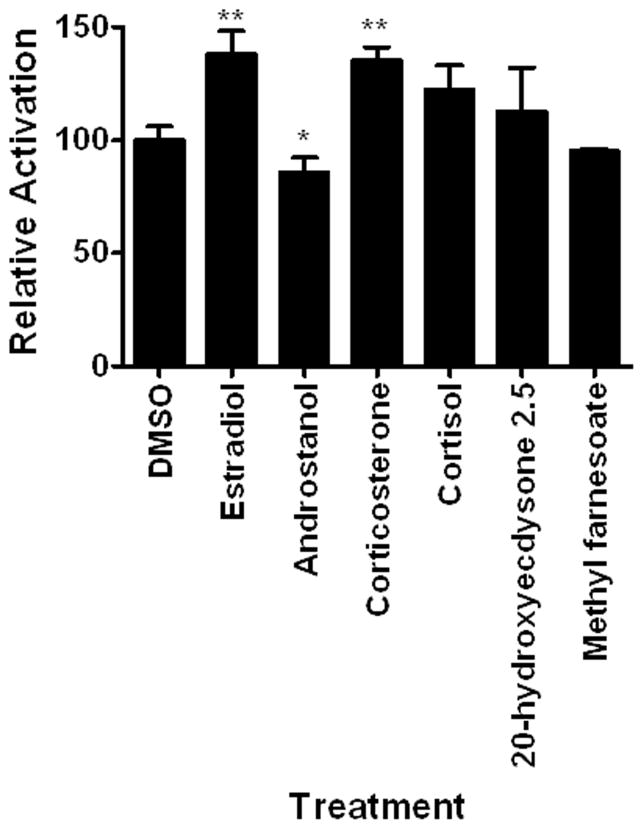
Six different hormones were investigated as potential DappuHR96 activators. Five are steroid hormones and methyl farnesoate is the unepoxidated juvenoid hormone found in crustaceans (Laufer and Biggers, 2001). Three of the hormones altered HR96 activity. Estradiol and corticosterone activated DappuHR96. Androstanol, a CAR inverse activator (Baldwin and Roling, 2009; Forman et al., 1998), is also a weak DappuHR96 inverse activator (Fig. 4).
Fig. 4. DappuHR96 is altered by select lipophilic hormones.
HepG2 cells were transfected with pBIND-GAL4-HR96-DEF and the pG5luc plasmid. Luciferase activity was measured following exposure to the hormones as described in the Materials and Methods. Statistical analysis was performed by ANOVA followed by Dunnett’s post-hoc test using GraphPad Prism software. An asterisk indicates p < 0.05 and two asterisks indicate a p < 0.01 (n = 4).
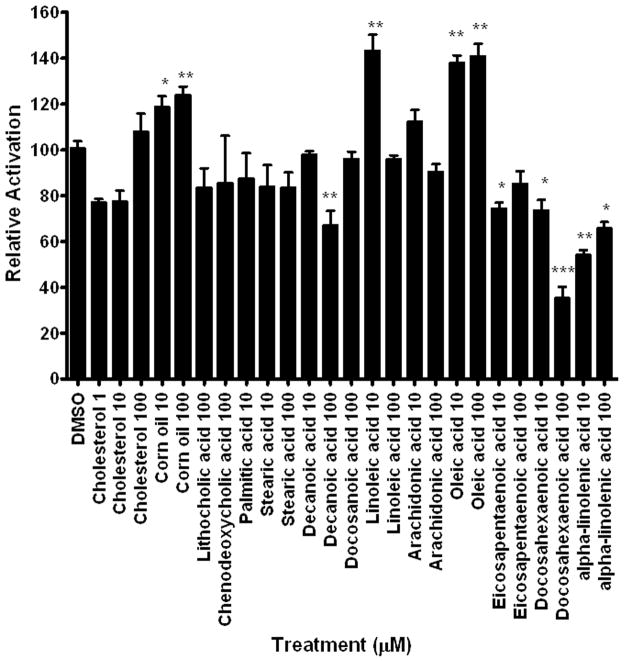
Fourteen lipids; primarily saturated fatty acids, bile acids, cholesterol, and unsaturated fatty acids were investigated as DappuHR96 activators (Fig. 5). None of the bile acids activated DappuHR96, one of the saturated fatty acids was a weak inverse activator, and cholesterol did not significantly alter DappuHR96 activity. Corn oil, which contains nearly 30% oleic acid and 53% linoleic acid (US Department of Agriculture, 2007) is an activator of DappuHR96. Both oleic acid and linoleic acid are DappuHR96 activators. All three omega-3 fatty acids tested, eicosapentaenoic acid, docosahexaenoic acid, and α-linolenic acid are DappuHR96 inverse activators. Overall, five of the six unsaturated fatty acids tested are either activators or inverse activators of DappuHR96 with the exception of arachidonic acid (Fig. 5).
Fig. 5. DappuHR96 activity is altered by multiple fatty acids, including several types of unsaturated fatty acids.
HepG2 cells were transfected with pBIND-GAL4-HR96-DEF and the pG5luc plasmid. Luciferase activity was measured following exposure to several fatty acids and bile acids. Statistical analysis was performed by ANOVA followed by Dunnett’s post-hoc test using GraphPad Prism software. An asterisk indicates p < 0.05, two asterisks indicate a p < 0.01, and three asterisks indicate a p < 0.001 (n = 4).
Overall, 34 chemicals were tested in transactivation assays. Nine chemicals activated DappuHR96, and eight chemicals inhibited DappuHR96 activity. Therefore, 50% of the chemicals (17/34) tested altered HR96 activity indicating that DappuHR96 is a promiscuous nuclear receptor similar to CAR and PXR (Hernandez et al., 2009).
3.4. Dose-response activation of DappuHR96
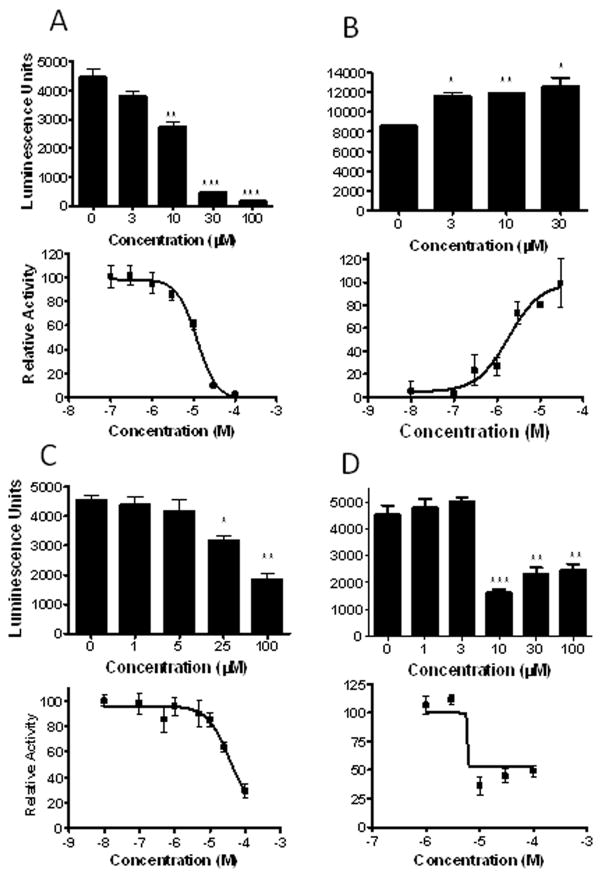
To verify transactivation and determine potency of some of the most robust activators and inactivators of DappuHR96, we performed dose-response transactivation assays. Two anthropogenic chemicals and two unsaturated fatty acids were examined (Fig. 6). Three of the four chemicals tested demonstrated a dose-dependent response; however, α-linolenic acid’s most potent response occurred at 10 μM (Fig. 6d). A similar response was observed in the previous transactivation assays (Fig. 5) where α-linolenic acid demonstrated a stronger response at 10 μM than 100 μM, and this may have adversely affected our ability to compute a 95% Confidence Interval (CI) (Table 2). We suspect that solubility may be an issue. Three of the four chemicals tested are inverse activators and have EC50’s ranging from 5.84 – 35.6 μM. Atrazine, which is an activator, has an EC50 of 1.85 μM (Table 2).
Fig. 6. Dose-response assays confirm DappuHR96 is altered by (A) triclosan, (B) atrazine, (C) docosahexaenoic acid, and (D) α-linolenic acid.
HepG2 cells were transfected with pBIND-GAL4-HR96-DEF and the pG5luc plasmid, and 24 hours later treated with chemicals at multiple concentrations (n = 4). Statistical analysis was performed by ANOVA followed by Dunnett’s post-hoc test using GraphPad Prism software. An asterisk indicates p < 0.05, two asterisks indicate a p < 0.01, and three asterisks indicate a p < 0.001. Dose-response curves were performed with concentrations from 0.01 to 100 μM with GraphPad Prism 4.0 software to determine EC50 values.
Table 2.
EC50 values and 95% Confidence Interval (CI) of some chemical activators and inverse agonists of Daphnia HR96.
| Chemical | EC50 (μM) | 95% CI (μM) | Direction |
|---|---|---|---|
| α-linolenic acid | 5.84 | DNC | inverse activator |
| Triclosan | 12.0 | 8.46 – 17.23 | inverse activator |
| Atrazine | 1.85 | 0.66 – 5.16 | activator |
| Docosahexaenoic acid (DHA) | 35.6 | 6.08 – 208.2 | inverse activator |
DNC: (Did not compute) GraphPad could not compute the 95% CI.
EC50: Concentration of chemicals that increased or decreased DappuHR96 activity to one-half of maximum (activators) or minimum (inverse activators) activity.
3.5. Antagonism of DappuHR96 activation by inverse activators
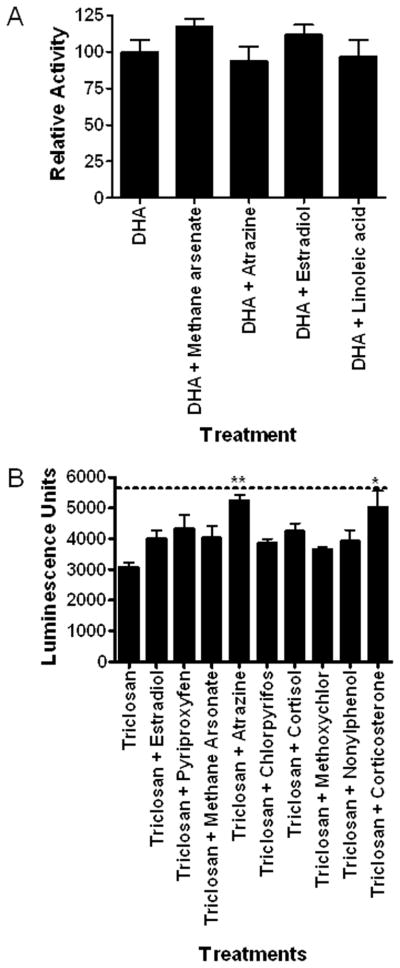
Organisms are not exposed to only one chemical at time. Therefore it is possible that DappuHR96 activity could be suppressed by dietary factors or other anthropogenic chemicals that inhibit DappuHR96 activity. Several chemicals that are DappuHR96 activators were tested in the presence of 100 μM DHA, an efficacious inverse activator (Fig. 7a). None of the chemicals tested activated DappuHR96 in the presence of DHA indicating that DHA is an efficacious antagonist of DappuHR96 activity. In addition, several DappuHR96 activators and a few chemicals that are not activators in the first set of assays were tested in the presence of 10 μM triclosan to determine if triclosan would antagonize HR96 activity or if it may increase the sensitivity of the assay (Fig. 7b) similar to androstanol’s role in CAR transactivation assays (Baldwin and Roling, 2009). Triclosan did not appear to increase the sensitivity of the assay as no new activators were found. Instead, similar to DHA, triclosan also antagonized DappuHR96 activity as some activators lost their ability to activate DappuHR96 in the presence of triclosan, including estradiol, pyriproxyfen, and chlorpyrifos (Fig. 7b). Furthermore, increasing concentrations of triclosan (10–30 μM) were able to completely antagonize atrazine-mediated transactivation of DappuHR96 (Fig. 8). Thus, the data demonstrates that inverse activators are able to antagonize the effects of DappuHR96 activators.
Fig. 7. Inverse activators can antagonize DappuHR96 activation.
Transactivation assays performed in the presence of the inverse activators (A) DHA and (B) triclosan demonstrate that DappuHR96 activation can be inhibited. The activation of most chemicals was inhibited by these inverse agonists. The dotted line indicates control luciferase activity without the presence of DappuHR96 activators or inverse activators. Statistical analysis was performed by ANOVA followed by Dunnett’s post-hoc test using GraphPad Prism software. An asterisk indicates p < 0.05, and two asterisks indicate a p < 0.01 (n = 4).
Fig. 8. Increasing concentrations of an inverse agonist can antagonize DappuHR96 activation.
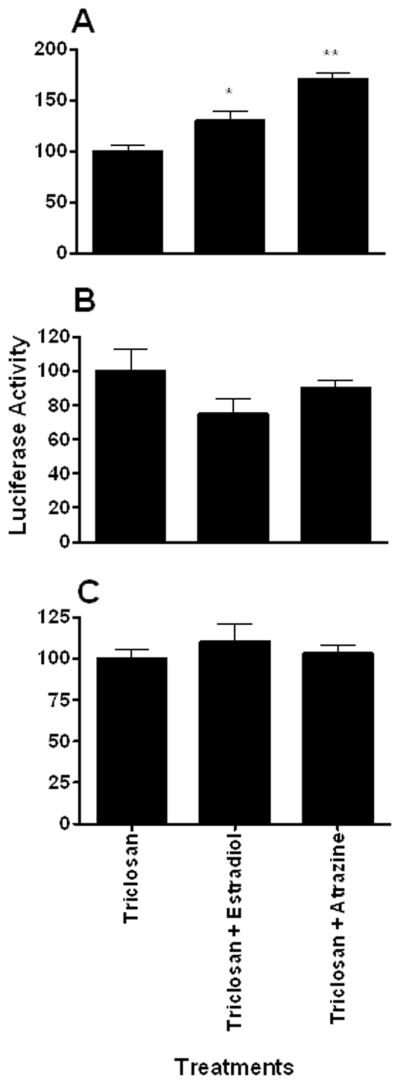
Transfected HepG2 cells were treated with estradiol or atrazine in the presence of (A) 10 μM, (B) 20 μM, or (C) 30 μM triclosan. Luciferase activity was measured 24 hours after treatment. Statistical analysis was performed by ANOVA followed by Dunnett’s post-hoc test using GraphPad Prism software. An asterisk indicates p < 0.05, and two asterisks indicate a p < 0.01 (n = 4).
4. DISCUSSION
To our knowledge this is the first study that demonstrates HR96 is a promiscuous receptor. Previously, DHR96 has been shown to induce Cyp6d1 as well as many other detoxification genes in response to Phenobarbital (King-Jones et al., 2006; Lin et al., 2011). DHR96 mutants are unable to mount this adaptive response (King-Jones et al., 2006) further indicating that DHR96 is a detoxification receptor. However, to our knowledge no studies demonstrated the promiscuity of DHR96. This study demonstrates that DappuHR96 is a promiscuous receptor similar to its relatives CAR and PXR (Table 3) (Baldwin and Roling, 2009; Blumberg et al., 1998; Hernandez et al., 2009).
Table 3.
Chemical activators of DappuHR96 compared to mammalian CAR, PXR, and VDR.
| Chemical | HR96 | CAR# | PXR# | VDR# |
|---|---|---|---|---|
| Methyl farnesoate | ↔ | nd | nd | nd |
| Cortisol | ↔ | ↔ | ↑ | nd |
| Corticosterone | ↑ | ↑ | ↑ | nd |
| Androstanol | ↓ | ↓ | ↑ | nd |
| Estradiol | ↑ | ↑ | ↑ | nd |
| 20-hydroxyecdysone | ↔ | nd | nd | nd |
| Pyriproxyfen (JHA) | ↑ | nd | nd | nd |
| Triclosan | ↓ | ↑ | ↑ | nd |
| Atrazine | ↑ | nd | ↔ | nd |
| Chlorpyrifos | ↑ | ↑ | ↑ | nd |
| Parathion | ↔ | ↑ | nd** | nd |
| Bisphenol A | ↔ | ↑ | ↑ | nd |
| Lindane | ↔ | nd | ↑ | nd |
| M. Methyl Arsonate | ↑ | ↑ | nd | nd |
| Phenobarbital | ↓ | ↑ | ↑ | nd |
| 2,4-D | ↔ | nd | nd | nd |
| Fluoxetine | ↓ | nd | ↑ | nd |
| Methoxychlor | ↔ | ↑ | ↑ | nd |
| Nonylphenol | ↔ | ↑ | ↑ | nd |
| Cypermethrin | ↔ | ↑ | ↑ | nd |
| Cholesterol | ↔ | ↔ | ↔ | ↔ |
| Lithocholic acid | ↔ | ↑ | ↑ | ↑ |
| Chenodeoxycholic acid | ↔ | ↑ | ↑ | ↔ |
| Palmitic acid | ↔ | nd | nd | nd |
| Steric acid | ↔ | nd | nd | nd |
| Decanoic acid | ↓ | nd | nd | nd |
| Docosanoic acid | ↔ | nd | nd | nd |
| Linoleic acid (n-6) | ↑ | ↑ | ↔ | ↕** |
| Oleic acid (n-9) | ↑ | nd | nd | nd |
| Arachidonic acid (n-6) | ↔ | ? | ? | ↕** |
| Eicosapentaenoic acid (n-3) | ↓ | ? | ? | ↕** |
| Docasahexaenoic acid (n-3) | ↓ | ↓ | ↔ | ↕** |
| α-Linolenic acid (n-3) | ↓ | nd | nd | nd |
| Corn oil (n-6) | ↑ | nd$ | nd | nd |
Data compiled from the following references (Adachi et al., 2005; Antolino-Lobo et al., 2011; Baldwin and Roling, 2009; Haussler et al., 2008; Hernandez et al., 2009; Ishizawa et al., 2008; Kliewer et al., 1998; Kretschmer and Baldwin, 2005; Lemaire et al., 2004; Lemaire et al., 2006; Staudinger et al., 2001; Yu et al., 2011; Zhang et al., 2004)
nd = not determined or not determined based on our literature searches
↔ = no effect
↓ = inverse agonist, antagonist, or inhibitor of activity
↑ = activator
Data obtained from binding assays so the directional effects (agonists, inverse agonists, antagonists) of these unsaturated fatty acids on VDR activity are unknown (Haussler et al., 2008).
? Whether these polyunsaturated fatty acids activate or inactivate CAR and PXR is unknown; however, there is evidence that they modulate activity and expression (Kuan et al., 2011)
nd** Methyl parathion is not an activator of PXR (Lemaire et al., 2004)
nd$ Linoleic acid, the major PUFA in corn oil is an activator of CAR (Finn et al., 2009)
DappuHR96 is clearly an HR96 ortholog to the insect HR96 genes (Fig. 1). In turn this receptor is evolutionarily related to the NR1I group (VDR/CAR/PXR) of nuclear receptors. Overall, there are a number of chemicals that interact with DappuHR96 and one of the NR1I group members (Table 3). Eight of the chemicals tested are DappuHR96 and CAR activators, seven chemicals are DappuHR96 and PXR activators, six chemicals activate DappuHR96, CAR, and PXR, and five chemicals have been shown to activate DappuHR96 but either do not activate CAR or PXR, or have not been tested. The chemicals steroids and toxicants demonstrate the promiscuous nature of DappuHR96 (Table 3). In addition, three chemicals are DappuHR96 and VDR activators. All three of these chemicals are unsaturated fatty acids (Haussler et al., 2008) suggesting an evolutionarily ancient role for the NR1I and NR1J receptors in recognition of unsaturated fatty acids. Interestingly, CAR is also activated by linoleic acid (Finn et al., 2009), which provides additional support that NR1I/J receptors may have been sensors for unsaturated fatty acids or similar molecules.
DappuHR96 was activated or inactivated by five of the six unsaturated fatty acids tested (Fig. 5). Interestingly, with the exception of arachidonic acid, an omega-6 fatty acid, all of the unsaturated fatty acids tested altered DappuHR96 activity. The omega-6 and omega-9 unsaturated fatty acids, linoleic and oleic acid, respectively, are activators of DappuHR96. All three omega-3 fatty acids tested (eicosapentaenoic acid, docosahexaenoic acid, and α-linolenic acid) repress DappuHR96 activity. This indicates a potentially unique interaction between the omega-3 and omega-6 fatty acids in which the omega-3 fatty acids inhibit DappuHR96 activation by omega-6 fatty acids or xenobiotics. In fact, 100 μM docosahexaenoic acid (DHA) repressed the activation of DappuHR96 by toxicants, estradiol, and the omega-6 fatty acid, linoleic acid (Fig. 7a). Therefore, DappuHR96 may be a mechanism that recognizes the ratio of omega-3 fatty acids to other fatty acids, or uses omega-3 fatty acids to antagonize the action of other unsaturated fatty acids.
Triclosan also represses DappuHR96 activation (Table 2; Fig. 6), and does so in a dose-dependent manner (Fig. 8). We hypothesized that an inverse agonist such as triclosan or DHA could increase the sensitivity of the assay similar to dihydroandrosterone (Baldwin and Roling, 2009) and androstanol (Forman et al., 1998; Tzameli et al., 2000) can for CAR. Transactivation of DappuHR96 by endo- and xenobiotics is not efficacious and rarely reaches 50%; however, it is consistent. The lack of strong activation may be due to low DappuHR96 activity in the HepG2 system, lack of specific coactivators, or high constitutive activity similar to CAR. Disappointingly, neither Triclosan nor DHA increased the efficacy and sensitivity of the transactivation assay at the concentrations tested, and instead they decreased the efficacy and sensitivity of the assay. Fewer chemicals were activators in their presence (Fig. 7). This may indicate a propensity for repressive activity by DappuHR96. It also indicates the complexity of interactions that could occur on this receptor to perturb typical toxicant or dietary acclimation responses.
Phenobarbital has been shown to activate DHR96 in vivo (King-Jones et al., 2006). Interestingly, Phenobarbital also acts like an inverse agonist of hCAR activity in vitro and as an hCAR activator in vivo. Whether other nonligand activators of hCAR or DappuHR96 may act similar is unknown. Phenobarbital has been shown to down-regulate CYP (testosterone hydroxylase) activity in Daphnia in vivo (Baldwin and LeBlanc, 1994). This could be due to Phenobarbital-mediated increases in testosterone conjugase activity in vivo, but a simpler answer would be that Phenobarbital inhibits HR96 in vitro and in vivo in Daphnia. Currently, the activity of the classic CAR activator on DappuHR96 in vivo is not known.
Work with Drosophiloa DHR96 supports the hypothesis that this receptor is both an inducer of drug metabolism enzymes and an endobiotic lipid sensor (Bujold et al., 2010; King-Jones et al., 2006; Lin et al., 2011; Sieber and Thummel, 2009). In Drosophila, DHR96 has been shown to bind cholesterol (Horner et al., 2009), and cholesterol is hypothesized to inhibit DHR96 activity (Bujold et al., 2010). DHR96 mutants are also unable to survive starvation conditions because of the inability to digest and store triglycerides, and Drosophila that overexpress DHR96 have higher lipid stores and are more likely to survive during starvation (Sieber and Thummel, 2009).
Similarly, VDR, CAR, and PXR all have roles in lipid and triglyceride metabolism. VDR binds several unsaturated fatty acids (Haussler et al., 2008), VDR-null mice are lean (Wong et al., 2009), and mice that overexpress VDR in adipocytes are obese due to decreased energy expenditure (Wong et al., 2011). PXR also increases hepatic steatosis as it down-regulates the expression of proteins involved in β-oxidation and increases the uptake of fatty acids (Gao and Xie, 2010; Nakamura et al., 2007). Conversely, CAR decreases hepatic steatosis and TCPOBOP-mediated CAR activation decreases fatty liver and obesity in high-fat diet induced obese and ob/ob mice. This does not occur in CAR-null mice (Dong et al., 2009; Gao et al., 2009). Taken together, the NR1I receptors are involved in an organism’s responses to xenobiotic and fatty acid metabolism. Our data indicates that DappuHR96 is also involved in both xenobiotic and fatty acid metabolism.
However, our assays did not demonstrate significant cholesterol repression of DappuHR96 activity (Fig. 5). Cholesterol is poorly soluble in water based media and its insolubility may increase the likelihood of precipitation. The activity of other poorly soluble lipids such as the unsaturated fatty acids could be measured though. Cholesterol is also already present in the HepG2 cells and therefore may be acting at its near maximal inhibitory capacity of DappuHR96 activity. At this time we can only speculate as to whether cholesterol interacts with DappuHR96; however, based on its similarity to DHR96 we hypothesize that cholesterol is found within the DappuHR96 LBD.
In summary, DappuHR96 is a promiscuous nuclear receptor that is activated or repressed by a number of xenobiotic and endobiotic compounds. Overall, 50% of the chemicals tested perturbed DappuHR96 activity, including 5 of the 6 unsaturated fatty acids tested, several xenobiotics, and a two steroids. The deactivation and activation of DappuHR96 by omega-3 and omega-6 fatty acids may indicate a mechanism by which diet and especially dietary fat perturb responses to specific environmental toxicants.
Supplementary Material
Highlights.
Daphnia HR96 (DappuHR96) is related to Drosophila HR96 and CAR, PXR, and VDR
DappuHR96 is a promiscuous xenobiotic and endobiotic receptor
The omega-3 unsaturated fatty acids tested inhibit DappuHR96 activity, and most of the omega-6/9 unsaturated fatty acids tested activate DappuHR96 activity.
The effects of unsaturated fatty acids on DappuHR96 indicate opposing roles of n-3 and n-6 fatty acids.
Overall, this data provides a putative mechanism of how dietary fat intake could alter an organism’s ability to respond to toxicants
Acknowledgments
The authors would like to thank Claudy Gay, Kiandra Scott, and LaToya Butler, who helped clone DappuHR96 and were supported by a Creative Inquiry project. The authors would also like to thank Travis Garriott, who was involved in the annotation of Daphnia pulex nuclear receptors. Funds for this project were provided by Turner Biosystems for a luminometer.org grant that provided the 20/20n luminometer, Clemson University start-up funds, Clemson University Creative Inquiry funds, a Public Service and Agriculture Next Generation Graduate Fellowships for Yangchun Li, a Fulbright Foreign Student Fellowship (15102663) for Elina Karimullina, and National Institutes of Health grant R15-ES017321 to William Baldwin.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 2000;14:1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Baldwin WS, LeBlanc GA. Identification of multiple steroid hydroxylases in Daphnia magna and their modulation by xenobiotics. Environ Toxicol Chem. 1994;13:1013–1021. [Google Scholar]
- Baldwin WS, Marko PB, Nelson DR. The Cytochrome P450 (CYP) gene superfamily in Daphnia pulex. BMC Genomics. 2009;10:169. doi: 10.1186/1471-2164-10-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin WS, Milam DL, LeBlanc GA. Physiological and biochemical perturbations in Daphnia magna following exposure to the model environmental estrogen diethylstilbestrol. Environ Toxicol Chem. 1995;14:945–952. [Google Scholar]
- Baldwin WS, Roling JA. A concentration addition model for the activation of the constitutive androstane receptor by xenobiotic mixtures. Toxicol Sci. 2009;107:93–105. doi: 10.1093/toxsci/kfn206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker C, Boersma M. Differential effects of phosphorus and fatty acids on Daphnia magna growth and reproduction. Limnol Oceanogr. 2005;50:388–397. [Google Scholar]
- Bertrand S, Brunet F, Escriva H, Parmentier G, Laudet V, Robinson-Rechavi M. Evolutionary genomics of nuclear receptors: from twenty-five ancestral genes to derived endocrine systems. Mol Biol Evol. 2004;21:1923–1937. doi: 10.1093/molbev/msh200. [DOI] [PubMed] [Google Scholar]
- Blumberg B, Sabbagh WJ, Juguilon H, Bolado JJ, van Meter CM, Ong ES, Evans RM. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 1998;12:3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett MT, Muller-Navarra DC, Ballantyne AP, Ravet ML, Goldman CR. Daphnia fatty acid composition reflects that of their diet. Limnol Oceanogr. 2006;51:2428–2437. [Google Scholar]
- Bujold M, Gopalakrishnan A, Nally E, King-Jones K. Nuclear receptor DHR96 acts as a sentinel for low cholesterol concentrations in Drosophila melanogaster. Mol Cell Biol. 2010;30:793–805. doi: 10.1128/MCB.01327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter SR, Kitchell JF, Hodgson JR, Cochran PA, Elser JJ, Elser MM, Lodge DM, Kretchmer D, He X, von Ende CN. Regulation of lake primary productivity by food web structure. Ecology. 1987;68:1863–1876. doi: 10.2307/1939878. [DOI] [PubMed] [Google Scholar]
- Christiaens O, Iga M, Velarde RA, Rougé P, Smagghe G. Halloween genes and nuclear receptors in ecdysteroid biosynthesis and signalling in the pea aphid. Insect Mol Biol. 2010;19:187–200. doi: 10.1111/j.1365-2583.2009.00957.x. [DOI] [PubMed] [Google Scholar]
- Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK, Bauer DJ, Caceres CE, Carmel L, Casola C, Choi JH, Detter JC, Dong Q, Dusheyko S, Eads BD, Frohlich T, Geiler-Samerotte KA, Gerlach D, Hatcher P, Jogdeo S, Krijgsveld J, Kriventseva EV, Kultz D, Laforsch C, Lindquist E, Lopez J, Manak R, Muller J, Pangilinan J, Patwardhan RP, Pitluck S, Pritham EJ, Rechtsteiner A, Rho M, Rogozin IB, Sakarya O, Salamov A, Schaack S, Shapiro H, Shiga Y, Skalitzky C, Smith Z, Souvorov A, Sung W, Tang Z, Tsuchiya D, Tu H, Vos H, Wang M, Wolf YI, Yamagata H, Yamada T, Ye Y, Shaw JR, Andrews J, Crease TJ, Tang H, Lucas SM, Robertson HM, Bork P, Zdobnov EM, Grigoriev IV, Lynch M, Boore JL. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coen WM, Janssen CR. The use of biomarkers in Daphnia magna toxicity test. IV. Cellular energy allocation: a new methodology to assess the energy budget of toxicant-stressed Daphnia populations. J Aquat Eco Stress Recov. 1997;6:43–55. [Google Scholar]
- Dong B, Saha PK, Huang W, Chen W, Abu-Elheiga LA, Wakil SJ, Stevens RD, Ilkayeva O, Newgard CB, Chan L, Moore DD. Activation of nuclear receptor CAR ameliorates diabetes and fatty liver disease. Proc Natl Acad Sci U S A. 2009;106:18831–18836. doi: 10.1073/pnas.0909731106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filho TUB, Soares AMVM, Loureiro S. Energy budget in Daphnia magna exposed to natural stressors. Envion Sci Pollut Res. 2011;18:655–662. doi: 10.1007/s11356-010-0413-0. [DOI] [PubMed] [Google Scholar]
- Finn RD, Henderson CJ, Scott CL, Wolf CR. Unsaturated fatty acid regulation of cytochrome P450 expression via a CAR-dependent pathway. Biochem J. 2009;417:43–54. doi: 10.1042/BJ20080740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk GJ, Thummel CS. Isolation, regulation, and DNA-binding properties of three Drosophila nuclear hormone receptor superfamily members. Proc Natl Acad Sci U S A. 1995;92:10604–10608. doi: 10.1073/pnas.92.23.10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Tzameli I, Choi HS, Chen J, Simha D, Seol W, Evans RM, Moore DD. Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature. 1998;395:612–615. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- Gao J, He J, Zhal Y, Wada T, Xie W. The constitutive androstane receptor is an anti-obesity nuclear receptor that improves insulin sensitivity. J Biol Chem. 2009;284:25984–25992. doi: 10.1074/jbc.M109.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Xie W. Pregnane X receptor and constitutive androstane receptor at the crossroads of drug metabolism and energy metabolism. Drug Metab Dispos. 2010;38:2091–2095. doi: 10.1124/dmd.110.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannas BR, Wang YH, Baldwin WS, Li Y, Wallace AD, LeBlanc GA. Interactions of the crustacean nuclear receptors HR3 and E75 in the regulation of gene transcription. Gen Comp Endocrinol. 2010;167:268–278. doi: 10.1016/j.ygcen.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussler MR, Haussler CA, Bartik L, Whitfield GK, Hsieh JC, Slater S, Jurutka PW. Vitamin D receptor: molecular signaling and actions of nutritional ligands in disease prevention. Nutr Rev. 2008;66(Suppl 2):S98–S112. doi: 10.1111/j.1753-4887.2008.00093.x. [DOI] [PubMed] [Google Scholar]
- Hernandez JP, Huang W, Chapman LM, Chua S, Moore DD, Baldwin WS. The environmental estrogen, nonylphenol, activates the constitutive androstane receptor (CAR) Toxicol Sci. 2007;98:416–426. doi: 10.1093/toxsci/kfm107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JP, Mota LC, Baldwin WS. Activation of CAR and PXR by dietary, environmental and occupational chemicals alters drug metabolism, intermediary metabolism, and cell proliferation. Curr Pharmacog Personal Med. 2009;7:81–105. doi: 10.2174/187569209788654005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner MA, Pardee K, Liu S, King-Jones K, Lajoie G, Edwards A, Krause HM, Thummel CS. The Drosophila DHR96 nuclear receptor binds cholesterol and regulates cholesterol homeostasis. Genes Dev. 2009;23:2711–2716. doi: 10.1101/gad.1833609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Jones K, Horner MA, Lam G, Thummel CS. The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metab. 2006;4:37–48. doi: 10.1016/j.cmet.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Kretschmer XC, Baldwin WS. CAR and PXR: Xenosensors of Endocrine Disrupters? Chem-Biol Interac. 2005;155:111–128. doi: 10.1016/j.cbi.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Laufer H, Biggers WJ. Unifying concepts learned from methyl farnesoate for invertebrate reproduction and post-embryonic development. Amer Zool. 2001;41:442–457. [Google Scholar]
- LeBlanc GA. Interspecies relationships in acute toxicity of chemicals to aquatic organisms. Environ Toxicol Chem. 1984;3:47–60. [Google Scholar]
- Lin GG, Kozaki T, Scott JG. Hormone receptor-like in 96 and Broad-Complex modulate phenobarbital induced transcription of cytochrome P450 CYP6D1 in Drosophila S2 cells. Insect Mol Biol. 2011;20:87–95. doi: 10.1111/j.1365-2583.2010.01047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblom TH, Pierce GJ, Sluder AE. A C. elegans orphan nuclear receptor contributes to xenobiotic resistance. Curr Biol. 2001;11:864–868. doi: 10.1016/s0960-9822(01)00236-6. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Sluder A, Guan X, Shi Y, McKee DD, Carrick K, Kamdar K, Willson TM, Moore JT. Comparison of complete nuclear receptor sets from the human Caenorhabditis elegans and Drosophila genomes. Genome Biol. 2001;2:research0029.0021–0029.0027. doi: 10.1186/gb-2001-2-8-research0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki AW. Correlation between Daphnia magna and fathead minnow (Pimephales promelas) chronic toxicity values for several classes of test substances. J Fish Res Board Can. 1979;36:411–421. [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:237–240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota LC, Hernandez JP, Baldwin WS. CAR-null mice are sensitive to the toxic effects of parathion: Association with reduced CYP-mediated parathion metabolism. Drug Metab Dispos. 2010;38:1582–1588. doi: 10.1124/dmd.110.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Moore R, Negishi M, Sueyoshi T. Nuclear pregnane X receptor crosstalk with FOXA2 to mediate the drug-induced regulation of lipid metabolism in fasting mouse liver. J Biol Chem. 2007;282:9768–9776. doi: 10.1074/jbc.M610072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ourlin JC, Lasserre F, Pineau T, Fabre JM, Sa-Cunha A, Maurel P, Vilarem MJ, Pascussi JM. The small heterodimer partner interacts with the pregnane X receptor and represses its transcriptional activity. Mol Endocrinol. 2003;17:1693–1703. doi: 10.1210/me.2002-0383. [DOI] [PubMed] [Google Scholar]
- Qatanani M, Moore DD. CAR, the continuously advancing receptor, in drug metabolism and disease. Curr Drug Metab. 2005;6:329–339. doi: 10.2174/1389200054633899. [DOI] [PubMed] [Google Scholar]
- Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, Wetzer R, Martin JW, Cunningham CW. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature. 2010;463:1079–1083. doi: 10.1038/nature08742. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schlechtriem C, Arts MT, Zellmer ID. Effect of temperature on the fatty acid composition and temporal trajectories of fatty acids in fasting Daphnia pulex (Crustacea, Cladocera) Lipids. 2006;41:397–400. doi: 10.1007/s11745-006-5111-9. [DOI] [PubMed] [Google Scholar]
- Sieber MH, Thummel CS. The DHR96 nuclear receptor controls triacylglycerol homeostasis in Drosophila. Cell Metab. 2009;10:481–490. doi: 10.1016/j.cmet.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swales K, Negishi M. Car, Driving into the Future. Mol Endocrinol. 2004;18:1589–1598. doi: 10.1210/me.2003-0397. [DOI] [PubMed] [Google Scholar]
- Takeshita A, Taguchi M, Koibuchi N, Ozawa Y. Putative role of the orphan nuclear receptor SXR (Steroid and Xenobiotics Receptor) in the Mechanism of CYP3A4 Inhibition by Xenobiotics. J Biol Chem. 2002;277:32453–32458. doi: 10.1074/jbc.M111245200. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The clustalx windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson SA, Baldwin WS, Wang YH, Kwon G, LeBlanc GA. Annotation, phylogenetics, and expression of the nuclear receptors in Daphnia pulex. BMC Genomics. 2009;10:500. doi: 10.1186/1471-2164-10-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzameli I, Pissios P, Schuetz EG, Moore DD. The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol Cell Biol. 2000;20:2951–2958. doi: 10.1128/mcb.20.9.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Department of Agriculture, Agricultural Research Service. USDA Nutrient Database for Standard Reference, Release 20. 2007 Retrieved November 15, 2011 from the Nutrient Data Laboratory Home Page: http://www.ars.usda.gov/ba/bhnrc/ndl.
- Valverde RA, Robinson GE, Fahrback SE. Nuclear receptors in the honey bee: annotation and expression in the adult brain. Insect Mol Biol. 2006;15:583–595. doi: 10.1111/j.1365-2583.2006.00679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KE, Kong J, Zhang W, Szeto FL, Ye H, Deb DK, Brady MJ, Li YC. Targeted expression of human vitamin D receptor in adipocytes decreases energy expenditure and induces obesity in mice. J Biol Chem. 2011;286:33804–33810. doi: 10.1074/jbc.M111.257568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KE, ZSzeto FL, Zhang W, Ye H, Kong J, Zhang Z, Sun XJ, Li YC. Involvement of the vitamin D receptor in energy metabolism: Regulation of uncoupling proteins. Am J Physiol Endocrinol Metab. 2009;296:E820–E828. doi: 10.1152/ajpendo.90763.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Tan A, Palli SR. The function of nuclear receptors in regulation of female reproduction and embryogenesis in the red flour beetle, Tribolium castaneum. J Insect Physiol. 2010;56:1471–1480. doi: 10.1016/j.jinsphys.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.