Abstract
BACKGROUND
Cardiac sodium channel β-subunit mutations have been associated with several inherited cardiac arrhythmia syndromes.
OBJECTIVE
To identify and characterize variations in SCN1Bb associated with Brugada (BrS) and sudden infant death syndromes (SIDS).
METHODS AND RESULTS
Patient 1 was a 44-y/o male with an ajmaline-induced Type-1 ST-segment elevation in V1 and V2 supporting the diagnosis of BrS. Patient 2 was a 62-y/o female displaying a coved-type BrS ECG who developed cardiac arrest during fever. Patient 3 was a 4-m/o female SIDS case. All known exons and intron borders of BrS and SIDS susceptibility genes were amplified and sequenced in both directions. A R214Q variant was detected in exon 3A of SCN1Bb (Navβ1B) in all three probands, but not in any other gene previously associated with BrS or SIDS. R214Q was identified in 4 of 807 ethnically-matched healthy controls (0.50%). Wild type (WT) and mutant genes were expressed in TSA201 cells and studied using whole-cell patch-clamp and co-immunoprecipitation techniques. Co-expression of SCN5A/WT+SCN1Bb/R214Q resulted in peak sodium channel current (INa) 56.5% smaller compared to SCN5A/WT+SCN1Bb/WT ( n=11–12, p<0.05 ). Co-expression of KCND3/WT+SCN1Bb/R214Q induced a Kv4.3 current (Ito) 70.6% greater compared with KCND3/WT+SCN1Bb/WT(n=10–11, p<0.01). Co-immunoprecipitation indicated structural association between Navβ1B and Nav1.5 and Kv4.3.
CONCLUSION
Our results suggest that R214Q variation in SCN1Bb is a functional polymorphism that may serve as a modifier of the substrate responsible for Brugada or SIDS phenotypes via a combined loss of function of INa and gain of function of Ito.
Keywords: Brugada Syndrome, Sudden Infant Death Syndrome, Arrhythmias, SCN1Bb, Sodium, Potassium
INTRODUCTION
Brugada syndrome (BrS) is an inherited cardiac arrhythmia disease characterized by an ST-segment elevation in the right precordial electrocardiogram (ECG) leads and a high incidence of sudden cardiac death (SCD). Reported in both children and the elderly, it is more common in males than females and typically first presents in the third or fourth decade of life.1 BrS has been associated with genetic variants in 11 different genes (SCN5A, GPD1L, CACNA1C, CACNB2b, SCN1B, KCNE3, SCN3B, KCNJ8, CACNA2D1, KCND3 and MOG1).2–5 Approximately 60% of BrS probands remain genotype-negative.
Sudden infant death syndrome (SIDS) is characterized by the sudden death of an infant (<1 y/o) that remains unexplained despite thorough examination. It is a major contributor to post-neonatal infant death, and is the third leading cause of infant mortality in USA.6 Over the past decade, postmortem genetic analysis or molecular autopsies of SIDS cases have identified a number of cardiac ion channel mutations associated with arrhythmia syndromes, including BrS, long QT syndrome (LQTS), short QT syndrome (SQTS), and catecholaminergic polymorphic ventricular tachycardia (CPVT).7 Mutations have been uncovered in genes encoding subunits of cardiac sodium, potassium and calcium channels, as well as in genes involved in trafficking or regulation of these channels. Approximately half of the cardiac channel mutation positive SIDS cases involve SCN5A.8
Nav channels are multi-subunit protein complexes comprised of pore-forming α subunits and multiple protein partners including regulatory β subunits. Four Navβ subunits have been identified in the human heart. Since 1995, mutations in Nav1.5 have been associated with multiple inherited cardiac diseases, including LQT3, BrS, cardiac conduction defect (CCD), atrial fibrillation (AF), sick sinus syndrome (SSS), atrial standstill, and SIDS.9 Mutations in sodium channel β-subunits have been directly associated with human cardiac disease only in the last 4 years.10–17 Here, we report a novel rare variant in SCN1Bb associated with BrS and SIDS, examine the structural association of the gene with both sodium channel current (INa) and transient outward potassium current (Ito) channels, and explore the functional effect of the variant on these two channels as a possible novel mechanism contributing to the phenotype. Our study is the first to demonstrate that the Navβ1B subunit has a structural and functional association with both Nav1.5 and Kv4.3 and that a genetic variation in this subunit can further modulate INa and Ito so as to modulate the arrhythmogenic substrate responsible for BrS and SIDS.
METHODS
Subjects
The study was conducted in conformance with the guidelines established by the local Human Review Board; written informed consent was obtained from all participants. Genomic DNA from the index cases was extracted from peripheral blood lymphocytes using a commercial kit (Gentra System, Puregene, Valencia, Calif). DNA from 476 patients clinically diagnosed with BrS or SIDS who were genotype-negative for variations in BrS1-4-susceptibility genes (317 males/159 females; 376 Caucasian; age at diagnosis, 6 hours to 72 years) were included in the study and screened for SCN1B variants. Control DNA collected from 807 healthy Caucasian subjects was screened for the SCN1Bb-R214Q mutations.
Genetic Analysis
Comprehensive open reading frame/splice site genetic analysis was performed using polymerase chain reaction (PCR), denaturing high performance liquid chromatography (DHPLC), and direct DNA sequencing. (See online supplement, including Online Table 1, for details)
Site-Directed Mutagenesis, Transfection and Electrophysiology Study
The ion channel variants were cloned by site-directed mutagenesis, expressed in TSA201 cells and studied using whole cell patch clamp techniques as detailed in the online supplement and reference.13
Co-immunoprecipitation and Western Blot
To identify the protein interaction of Navβ1b with Nav1.5 or Kv4.3, we used co-immunoprecipitation and western blot assay in plasma membrane of TSA201 cells. (See online supplement)
RESULTS
Clinical History
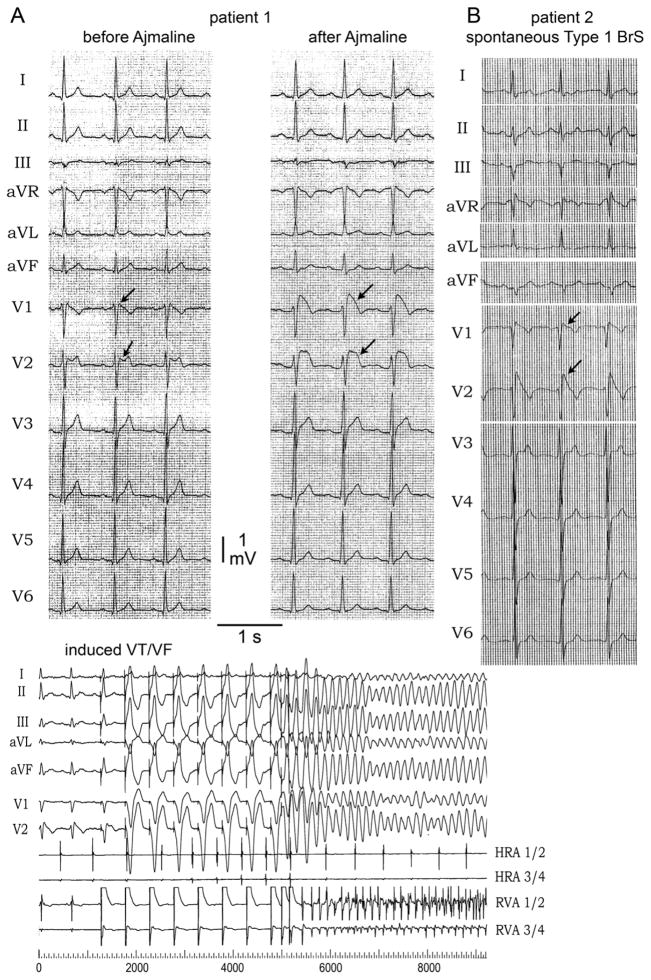
Patient 1 was a 44 year old male of European descent presenting because of a suspected Brugada ECG at baseline (Figure 1A). His medical history was unremarkable. The patient denied syncope or palpitations. There was no family history of SCD or syncope. Prominent ST segment elevation was induced in leads V1 and V2 following administration of 40 mg of ajmaline confirming the diagnosis of BrS (Figure 1A, upper panel; Online Table 2). The coved-type ST segment elevation was also documented to occur spontaneously during repetitive ECG recordings obtained during follow-up. Electrophysiological study revealed normal AV conduction (AH interval 104 ms and HV 44 ms). Wenckebach cycle length was achieved at a cycle length of 320 ms and sinus node recovery time was also normal (920 ms). Dual AV node physiology was seen without induction of atrioventricular nodal reentrant tachycardia (AVNRT). Ventricular fibrillation (VT) was inducible with two extrastimuli applied to the right ventricular outflow tract (RVOT) (Figure 1A, lower panel). An implantable cardioverter-defibrillator (ICD) was implanted for primary prevention; no shocks have been reported during follow-up.
Figure 1. ECG of the patient 1 and patient 2 with Brugada syndrome (BrS) sign.
A: (Upper panel) ECG at rest and 10 minutes after 40 mg of Ajmaline in patient 1. After sodium channel block challenge, ECG shows accentuation of R′ and development of a type 1 ST segment elevation in V1 and V2 (arrows). (Lower panel) Clinical electrophysiology study on patient 1. Ventricular tachycardia/ventricular fibrillation (VT/VF) was inducible with two extrastimulis. B: ECG of patient 2. It shows spontaneous huge accentuation of R′ and type 1 ST segment elevation in V1, and V2 (arrows).
Patient 2 was a 62 y/o female, who had cardiac arrest after developing diarrhea and fever. Echocardiography was normal. Two months after the cardiac arrest, the ECG exhibited a typical coved-type ST segment elevation in leads V1-V2 consistent with BrS. Atrioventricular (AV) conduction was within the normal range (Figure 1B, Online Table 2). She had significant neurological sequelae and, therefore, was not implanted with an ICD. Her maternal aunt died from SCD at the age of 19 y/o; and her son is asymptomatic with normal ECG. Unfortunately DNA was not available for the aunt who died suddenly and the son refused genetic testing.
Patient 3 was a Caucasian female from Mayo Clinic SIDS database who died at 4 months of age. An ECG was not available.
Molecular Genetics
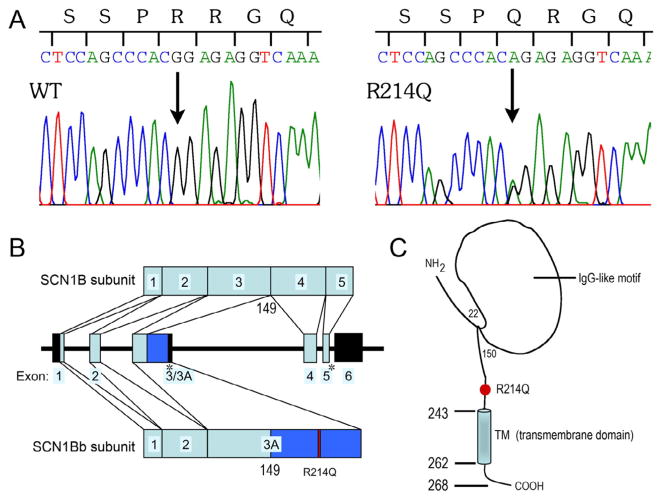
Of the genotype-negative probands, 184 cases diagnosed with BrS and 292 cases diagnosed with SIDS were screened for SCN1B variations, three were identified with the same variation in exon 3A of SCN1Bb. PCR-based sequencing analysis revealed a double peak in the sequence of SCN1Bb (Figure 2A) showing a G-to-A transversion at nucleotide 641, predicting a substitution of Glutamine (Q) for Arginine (R) at residue 214 (designated R214Q or p.Arg214Gln). This nucleotide change was observed in 4 of 807 controls (frequency, 0.50%); a frequency of 0.20% is reported in the 1000 genome project database (rs 66876876). The N-terminal region of the human SCN1Bb subunit is encoded by exons 1–3, whereas the novel C-terminal region is encoded via the extension of exon 3 to intron 3 (or partial intron 3 retention) with an in-frame stop codon. As the site of divergence between the SCN1B and SCN1Bb subunit cDNAs was located precisely at the exon 3/intron 3 boundary of the SCN1B gene, the human SCN1Bb is considered to be a splice variant of the SCN1B. R214 is located in exon 3A, and in the extra cellular domain (ECD) of SCN1Bb (Figure 2B and 2C). The 3 probands positive for SCN1Bb/R214Q were negative for the other candidate genes. All R214Q-positive cases also had the following non-synonymous heterozygous variants: L210P, S248Rand R250T, which had frequencies of 44%, 22% and 22%, respectively.
Figure 2. Genetic analysis of SCB1Bb/R214Q.
A: polymerase chain reaction -based sequence of SCN1Bb exon 3A showing wild-type (WT) and heterozygous G to A transversion at nucleotide 641 (arrow). It predicts a substitution of glutamine (CAG) for arginine (CGG) at position 214 (R214Q). B: Genomic structure of human Navβ1 gene. On chromosome 19, the SCN1B spans around 9 kb across six exons. SCN1Bb shares an identical N-terminal half (residues 1–149) with SCN1B, but contains a novel C-terminal half of less than 17% sequence identity with SCN1B. Exon 3A is an extended exon 3 (retention of part of intron 3) via alternative splicing. The dark blue region indicates the unique sequence of exon 3A compared with exon 3. Exons 1–5 (light blue boxes) encode the Navβ1 subunit, while exons 1, 2 and 3A (light and dark blue boxes) encode the Navβ1B subunit. The stop codon is indicated by an asterisk, the mutation is indicated as red box, and the untranslated regions are indicated using the black boxes. C: Predicted topology of Navβ1B. Red circle indicates the location of the mutant.
Electrophysiological characteristics of SCN5A co-expressed with SCN1Bb/wild-type (WT) and SCN1Bb/R214Q
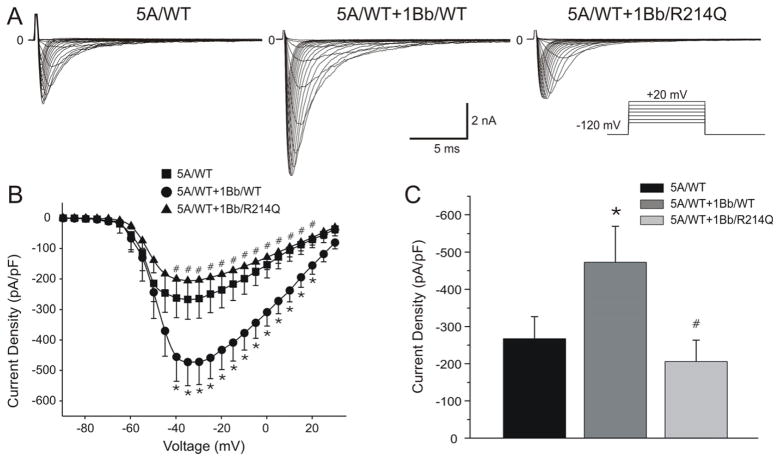
SCN5A/WT, SCN5A/WT+SCN1Bb/WT, and SCN5A/WT+SCN1Bb/R214Q were expressed in TSA201 cells to assess the effects of the rare variant on INa function. Figure 3 shows macroscopic currents recorded from these channels, together with the current-voltage (I-V) relationships. No shift was observed in the maximum peak inward current voltage for any of the channel types. Co-expression of SCN1Bb/WT with SCN5A/WT increased peak INa density from −267.33±64.59 pA/pF to −472.60±77.09 pA/pF (n=9 and 11, respectively; P<0.05). Co-expression of SCN1Bb/R214Q resulted in a peak INa current density of −205.70+-57.74 pA/pF (n=12), 56.5% smaller than SCN5A/WT+SCN1Bb/WT and 33.05% smaller than SCN5A/WT (P<0.05; Figure 3B and 3C).
Figure 3. Effect of SCN1Bb/R214Q on INa expressed in TSA201 cells.
A: Representative INa traces in cells expressing SCN5A/wild-type (WT) alone or co-transfected with SCN1Bb/WT or SCN1Bb/R214Q. Co-expression of SCN1Bb/R214Q produced loss of function. The inset shows the voltage-clamp protocol employed. B: Current-Voltage relationship for SCN5A/WT (n=9), SCN5A/WT + SCN1Bb/WT (n=11) and SCN5A/WT + SCN1Bb/R214Q (n=12). C: Bar graph of peak current density indicated significantly reduced for SCN5A/WT and SCN5A/WT + SCN1Bb/R214Q when compared to SCN5A/WT + SCN1Bb/WT (*P<0.05, compared with SCN5A/WT; #P <0.05, compared with SCN5A/WT+SCN1Bb/WT).
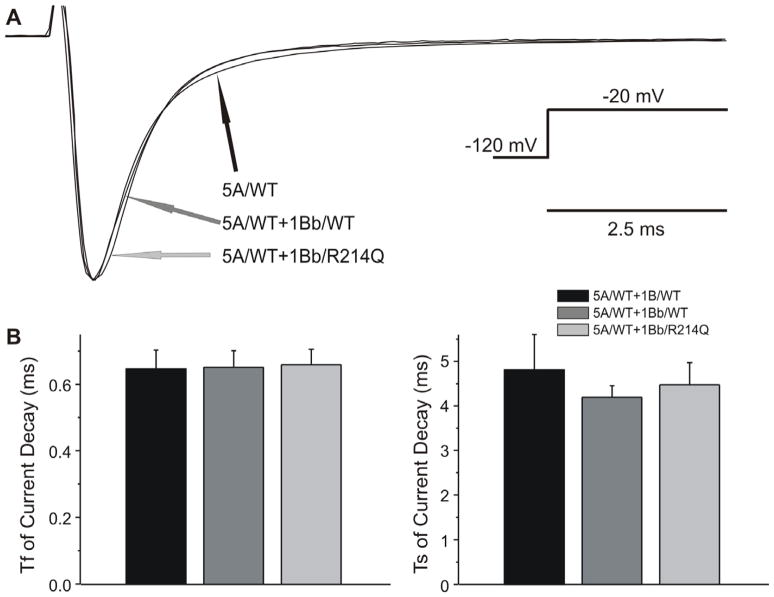
We fitted the decay of the current to the sum of 2 exponentials. At a potential of −20 mV, the time constants of the fast and slow component (τs and τf) of the current decay were similar among the 3 groups (Figure 4).
Figure 4. The effect of SCN1Bb subunit on current decay of INa.
A: Overlapping sodium channel current (INa) traces from cells expressing NaV1.5, in the absence and presence of SCN1Bb wild-type (WT) and variant. INa was evoked by +20mV depolarizing pulses from a holding potential of −120m V. Traces are shown normalized to their individual peak value. B and C: Current decay (τ fand τs) for all 3 groups fitted with double exponential function.
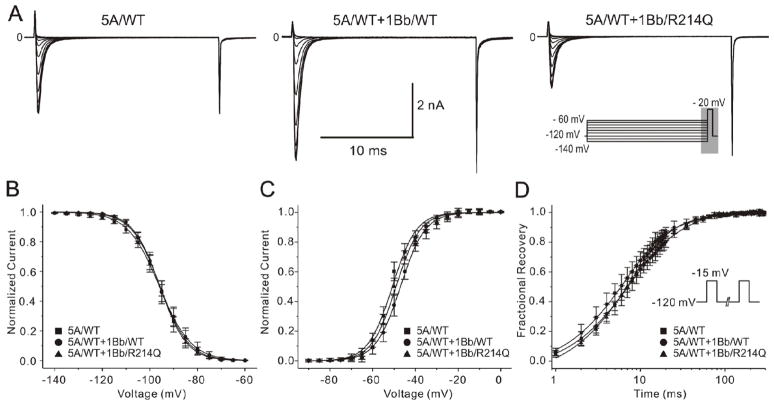
Figure 5 shows the results of steady-state activation and inactivation. No difference was observed in half-inactivation voltage (V1/2) or slop factor (k) between mutant INa channels and the 2 WT groups (Figure 5A and 5B). Steady-state activation was also similar among the 3 groups (Figure 5C). The results of recovery from inactivation, measured using a standard double paired-pulse protocol, are shown in Figure 5D. SCN5A/WT+SCN1Bb/WT had similar τf, but faster τs (P<0.05) compared with SCN5A/WT. Both τf and τs were slower in SCN5A/WT+SCN1Bb/R214Q compared with SCN5A/WT+SCN1Bb/WT (P<0.05 respectively; Figure 5D). R214Q caused no significant shift in the voltage–dependence of steady-state inactivation and activation, but decelerated recovery from inactivation, thus serving to further reduced sodium channel availability (see details in Online Table 3).
Figure 5. Functional characterization of SCN1Bb/R214Q on INa.
A: Representative traces recorded from wild-type (WT) and mutant channels in response to the voltage clamp protocol depicted on right middle inset designed to assess steady-state inactivation. B and C: Voltage dependence of inactivation and activation of SCN5A/WT and co-transfection of either SCN1Bb/WT or SCN1Bb/R214Q. Averaged values and the number of cells used are represented in Online Table 3. D: Recovery from fast inactivation of 3 groups determined using the two-pulse protocol shown in the inset. Fitting to a double-exponential function yielded the time constants demonstrated in Online Table 3. τf and τs in SCN5A/WT + SCN1Bb/R214Q were significantly slower as compared with SCN5A/WT + SCN1Bb/WT.
Electrophysiological characteristics of KCND3 co-expressed with WT and mutant SCN1Bb
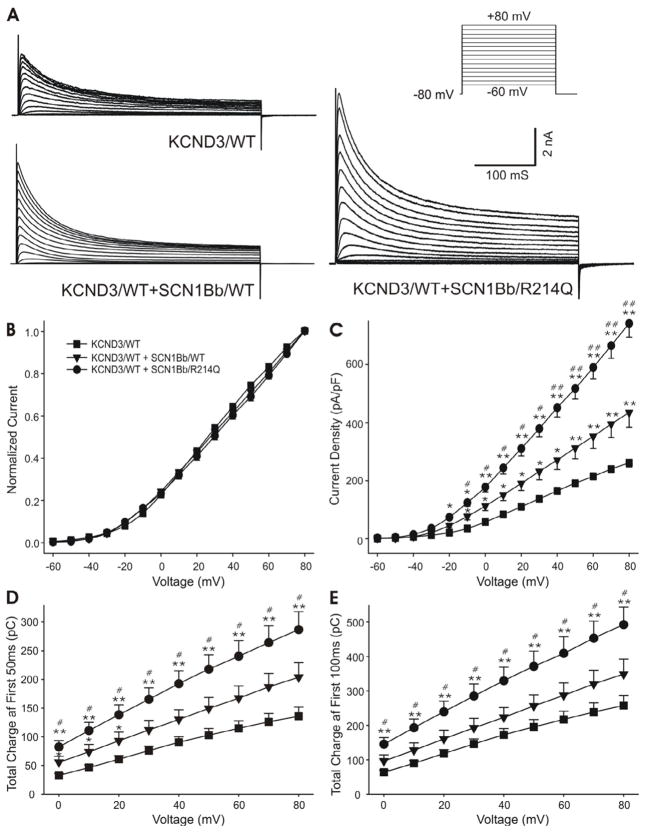
In light of the demonstration by Deschenes et al.18 of an effect of SCN1B to modulate Kv4.3 current, we sought to examine the hypothesis that SCN1Bb has a similar effect and that this action of the auxiliary subunit may be amplified by the rare variant. At +40 mV and +80 mV, co-expression of SCN1Bb/WT with KCND3/WT increased Ito density by approximately 64.59% and 66.17% (P<0.05 and P<0.01) over KCND3/WT; co-expression of SCN1Bb/R214Q with KCND3/WT enhanced the Ito density further (increasing by 173.51% and 183.55% over KCND3/WT current, P<0.01 respectively; and by 66.17% and 70.64% over KCND3/WT+SCN1Bb/WT current, P<0.01 respectively; Figure 6A and 6C). SCN1Bb, both WT and rare variant, did not alter the I-V relationship compared with KCND3/WT (Figure 6B), steady-state inactivation (Figure 7C and Online Table 4) or time to peak (Figure 7D), but significantly affected the other kinetics and voltage dependence of channel gating compared to KCND3/WT alone.
Figure 6. Effect of SCN1Bb/R214Q on Ito.
A: Representative Ito traces recorded from cells expressing KCND3/wild-type (WT) alone or co-transfected with SCN1Bb/WT or SCN1Bb/R214Q. Co-expression of SCN1Bb/R214Q produced gain of function. The inset shows the voltage-clamp protocol employed. B: Normalized current-voltage relationship for KCND3/WT, KCND3/WT + SCN1Bb/WT and KCND3/WT + SCN1Bb/R214Q. There is no significant change in the presence of SCB1Bb (WT/R214Q) compared with both WT groups. C: Raw current-voltage relationship for peak Ito current density. D and E: Total charge of Ito current during the first 50 ms and 100 ms as a function of voltage. For panel B–E: n=31, 10, 11 for KCND3/WT, KCND3/WT + SCN1Bb/WT, KCND3/WT + SCN1Bb/R214Q; *P<0.05, **P<0.01 compared with KCND3/WT; #P <0.05, # #P <0.01 compared with KCND3/WT+SCN1Bb/WT.
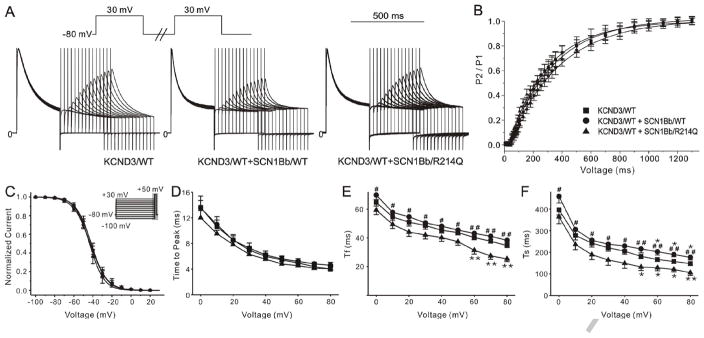
Figure 7. Functional characterization of SCN1Bb/R214Q on Ito.
A: Representative traces recorded from wild-type (WT) and mutant channels in response to the voltage clamp protocol depicted on the top left inset designed to assess recovery. P1 was normalized to the same amplitude. B: Recovery from inactivation of 3 groups. C: Voltage dependence of inactivation of KCND3/WT and co-transfection of either SCN1Bb/WT or SCN1Bb/R214Q. The protocol was displayed in the top inset of Figure 7C. Averaged values and the number of cells used are represented in Online Table 4. D: Time to peak current as a function of voltage. E and F: Kinetics of current decay (τf and τs) for all 3 groups fitted with double exponential function. *P<0.05, **P<0.01 compared with KCND3/WT; #P <0.05, # #P <0.01 compared with KCND3/WT+SCN1Bb/WT.
The effect of SCN1Bb/WT and SCN1Bb/R214Q on recovery of Ito from inactivation is shown in Figure 7A. τf with SCN1Bb/WT was slightly slower than KCND3/WT alone; τf and τs with co-expression of the SCN1Bb/R214Q were significantly slower as compared with two WT groups (Figure 7B and Online Table 4). A double-exponential function was fitted to the current decay of traces elicited by pulses from 0 mV to +80 mV. Figure 7E and 7F show inactivation time constants (τf and τs) for Ito at various potentials. KCND3/WT channel inactivation kinetics were faster at the more positive potentials (τf and τs decreased from 64.76 ± 2.55 ms and 396.53 ± 23.37 ms at 0 mV to 45.35 ± 2.05 ms and 204.85 ± 14.99 ms at +40 mV; n = 33). Co-expression of SCN1Bb/WT produced a moderate acceleration of inactivation as reflected by the slightly smaller time constant at all test potentials (P<0.05 for τf between +60 mV to +80 mV; P<0.05 for τs between +50 mV to +80 mV). With co-expression of SCN1Bb/R214Q, inactivation kinetics were slower compared to KCND3/WT alone (P<0.05 for τs between +60 mV to +80 mV) and KCND3/WT+SCN1Bb/WT (P<0.05 for τf and τs between 0 mV to +80 mV). Because co-expression of SCN1Bb/R214Q produces both an increase in peak current and a deceleration of current decay, we calculated the change in total charge contributing to the early phases of the action potential (AP). Figures 6D and 6E show that, compare with KCND3/WT and KCND3/WT+SCN1Bb/WT, there is a statistically significant increase in total charge during the first 50 and 100 ms of KCND3/WT+ SCN1Bb/R214Q current.
Co-immunoprecipitation Study
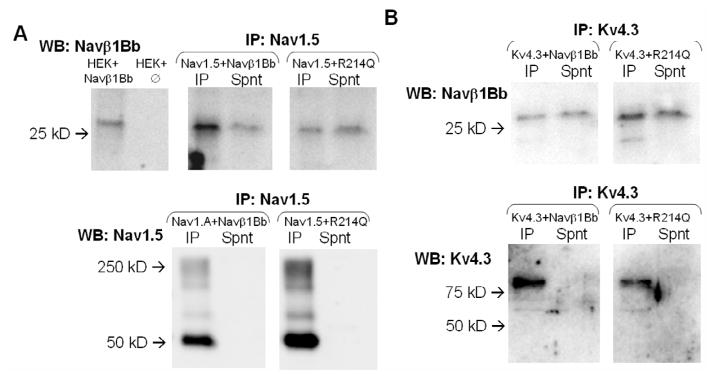
To determine whether SCN1Bb proteins associate with KCND3, we prepared extracts of TSA201 cells transfected with SCN1Bb and SCN5A, or SCN1Bb and KCND3 (n = 4). Figure 8A and 8B display western blots of the immunoprecipitated proteins showing a ~30 kDa SCN1Bb band using the anti-SCN1Bb antibody. These results provide evidence in support of an interaction of subsidiary SCN1Bb subunits (both WT and rare variant) with SCN5A and KCND3 subunits in the transfected cells.
Figure 8. Coimmunoprecipitation of Nav1.5/Kv4.3 and Navβ1b subunit.
Proteins were immunoprecipitated with (A) anti-Nav1.5 or (B) anti-Kv4.3 and immunoblotted with anti-Navβ1b (WT or R214Q variant) as indicated. Parallel western blot analysis of transfected and untransfected TSA201 cells was performed to confirm identity of labelled bands. Lower panels show that Nav1.5 and Kv4.3 are indeed fully precipitated. In both cases, the Navβ1b subunit appears in the pellet (IP) and in the supernatant (Spnt) of the immunoprecipitate. Each blot is representative of 4 experiments conducted under the same conditions.
DISCUSSION
Mutations in SCN1B have recently been associated with AF and CCD and two stop-codon mutations in SCN1Bb have been associated with a BrS/CCD (Online DISCUSSION and Online Table 5). In the present study, we identified a novel genetic variant in SCN1Bb in with BrS and SIDS, which is the first missense rare variant in SCN1Bb gene related to cardiac disease. In patient 1, VF was induced with programmed stimulation applied to the RVOT. In the case of patient 2, aborted SCD occurred in conjunction with diarrhea and fever, previously reported triggers,19, 20 and generally associated with SCN5A-mediated BrS.21 No mutation in SCN5A was found in this patient. The identification of the variant in patient 3 expands the spectrum of channelopathies associated with SIDS and provides further support for our hypothesis that, infants having rapid ventricular tachycardia, conduction abnormalities, (aborted) sudden death in the absence of structural or metabolic abnormalities are likely to have disease-causing genetic variants in cardiac depolarizing channels, including sodium and calcium channels.22
The SCN1Bb gene, which encodes a splice variant of Navβ1 subunit (termed Navβ1B), is located on chromosome 19 (19q13.1-q13.2). A 268 residue protein, with a calculated molecular mass of 30.4 kDa, is encoded by 807 bp open reading frame of the human Navβ1B subunit.23 It is formed through retention of intron 3, containing a stop codon. The predicted amino acid sequence of the retained intronic region exhibits very low homology between species, and the C-terminal region of the human Navβ1B is significantly different from that of the Navβ1 and Navβ1A subunits. The rare variant of c.641G>A in exon 3A results in substitution of a polar glutamine for a basic arginine at position 214. It is located at the ECD, which interacts with Nav1.5, and has been shown to be both necessary and sufficient for the modulation of Na+ channel characteristics.24
Navβ subunits have been proposed to exert multiple influences on function of the human Nav1.5, including altered activation, inactivation and recovery from inactivation; altered channel expression at the plasma membrane; as well as modulation of other related proteins in the cardiac sodium complex. Moreover, β-subunits modify not only the biophysical, but also the pharmacological properties of the cardiac sodium channel complex.25 Compared with those of other subunits, Navβ1-mediated effects on Nav1.5 are the most investigated.26 Expression of Nav1.5 in oocytes produces channels that inactivate rapidly in the absence of Navβ subunits.27 According to some reports, Navβ1 has no observable effect on SCN5A function.28 Most other groups have reported a Navβ1-dependent increase in INa amplitude. No detectable effects on channel kinetics or voltage-dependence are observed by several groups.29 A number of studies have reported a Navβ1-mediated 5–15mV depolarizing shift of steady-state inactivation,26, 30 or significant changes in the rate of recovery from inactivation.30 Makielski and co-workers reported modulation of channel sensitivity to lidocaine block with subtle changes in channel kinetics and gating properties in response to Navβ1 expression.31 As the majority of these studies focusing on Navβ1, similar studies have not been carried out for Navβ1B. In the present study, we demonstrate that Navβ1B/WT increases NaV1.5 density, consistent with previous studies conducted using CHO cells11 or Oocytes.23 The R214Q rare variant in Navβ1B blunted or inhibited the gain of function produced by Navβ1B/WT on INa density.
Ito, encoded by Kv4 family of potassium channels, is responsible for phase 1 of the AP. Recent studies indicate that co-expression of the Navβ1 subunit modulates the gating properties of Kv4.3 to closely recapitulate native Ito,18 and plays a key role in the structural association between subunits that comprise the Ito and INa channels.32 Coimmunoprecipitation studies revealed association of Kv4.2 or Kv4.3 with Navβ1 in ventricular myocardium. Inhibition of Navβ1 transcription in ventricular myocytes results in the reduction of mRNA and/or protein levels of Nav1.5, Kv4.2, Kv4.3, and KChIP2, and the marked decrease in INa and Ito. Together, these intriguing observations suggest that cardiac Nav1.5 may physically associate with Ito channels via β1 to form a macromolecular complex. Here, we first describe that Navβ1B subunit has a similar structural and functional association with hKv4.3, including co-localization with hKv4.3 protein and a gain of function of Ito. The increase in Kv4.3 current effected by co-expression of SCN1Bb/WT appears less significant than that of SCN1B/WT reported by Deschênes et al.18 Our study is the first to demonstrate that a rare genetic variant in Navβ subunit can produce a gain of function in hKv4.3 channel current. The R214Q variation in Navβ1B caused a prominent further augmentation of total charge due to an increase in Ito density and a slowing of the rate of inactivation. The remarkable gain in function of Ito likely contributes importantly to development of both the BrS and SIDS phenotypes. The combination of an increase in Ito and decrease in INa is expected to produce a prominent outward shift in net current active during the early phases of the epicardial AP, particularly in right ventricular epicardium where Ito is generally more prominent. This in turn would lead to accentuation of the right ventricular epicardial AP dome, heterogeneous loss of the AP dome, giving rise to phase 2 reentry and polymorphic VT.
The intracellular domain of β1 is critical for Nav1-ankyrin (G and B) interaction and channel modulation in vitro.33 In addition, Nav1.5 channels of the intercalated disk-pool were shown to not only co-localize with phosphotyrosin-β1, but also with connexin-43 (a well-known intercalated disk protein) and N-cadherin.34, 35 It has been suggested that mutations in Navβ1 may also disrupt channel-cytoskeletal interactions. The effect of Navβ1B (WT and R214Q) on other proteins may also plays a role in human health and disease. This crosstalk between Navβ1/Navβ1B and other proteins is an encrypted language that remains to be deciphered. (See more discussion in online supplement)
Limitations
Our data indicate that SCN1Bb/R214Q is a functional rare variant that occurs in all our patients together with 3 very common polymorphisms. The degree to which in vitro characteristics might be altered in the context of these polymorphisms is not known and will require further investigation. It is noteworthy that although the R214Q variant was not detected among 354 ethnically-matched healthy controls tested in Utica, NY, USA or Pavia, Italy, it was found in 4 of 453 presumably healthy controls tested in Rochester, Minn, USA (ECGs or clinical history not available). We are obliged to conclude that this variant may not be greatly over-represented in BrS or SIDS cases.
CONCLUSION
In summary, our study is the first to demonstrate that the Navβ1B subunit has a structural and functional association with both Nav1.5 and Kv4.3 and that a genetic variation in this subunit may further modulate INa and Ito so as to functionally modulate the expression of a BrS phenotype in adults as well as an arrhythmogenic substrate responsible for SIDS. Our study provides evidence in support of the hypothesis that a rare variant in the Navβ1B subunit leading to augmentation of net outward current via concomitant reduction in inward current and increase in outward current constitutes another mechanism contributing to the development of a BrS and SIDS phenotypes.
Supplementary Material
Acknowledgments
The authors are grateful to Judy Hefferon, Robert J. Goodrow, Susan Bartkowiak, Margherita Torchio, Rebellato Luca, Federica Dagradi, MD and Silvia Castelletti, MD for technical assistance.
FUNDING SOURCES
This work was supported by a grant HL47678 from NHLBI, grants from NYSTEM, the Masons of New York State and Florida for CA. Supported by the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program and the NIH, USA (HD42569) for MJA. Supported by MICINN grant (SAF2010-16120/) for OC.
ABBREVIATIONS
- AF
atrial fibrillation
- AP
action potential
- AV
atrioventricular
- AVNRT
atrioventricular nodal reentrant tachycardia
- BrS
Brugada syndrome
- CCD
cardiac conduction defect
- CPVT
catecholaminergic polymorphic ventricular tachycardia
- CAMs
cellular adhesion molecules
- DHPLC
denaturing high performance liquid chromatography
- ECD
extracellular domain
- ERS
early repolarization syndrome
- ICD
implantable cardioverter-defibrillator
- LQTS
long QT syndrome
- PCR
polymerase chain reaction
- RVOT
right ventricular outflow tract
- SQTS
short QT syndrome
- SSS
sick sinus syndrome
- INa
sodium channel current
- SCD
sudden cardiac death
- SIDS
sudden infant death syndromes
- Ito
transient outward potassium current
- VT/VF
ventricular tachycardia/ventricular fibrillation
- WT
wild type
Footnotes
CONFLICT OF INTEREST STATEMENT
Michael J. Ackerman is a consultant for Transgenomic and their FAMILION™ genetic test for cardiac ion channel abnormalities. In addition, “cardiac channel gene screen” and “know-how relating to long QT genetic testing” license agreements, resulting in consideration and royalty payments, were established between Genaissance Pharmaceuticals (then PGxHealth and now Transgenomic, Omaha, Neb) and Mayo Medical Ventures (now Mayo Clinic Health Solutions, Rochester, Minn) in 2004. However, Transgenomic did not provide financial support for this study. The other authors have no financial or other considerations to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Antzelevitch C. Brugada syndrome. PACE. 2006;29:1130–59. doi: 10.1111/j.1540-8159.2006.00507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antzelevitch C, Yan GX. J wave syndromes. Heart Rhythm. 2010;7:549–58. doi: 10.1016/j.hrthm.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burashnikov E, Pfeiffer R, Barajas-Martinez H, et al. Mutations in the cardiac L-type calcium channel associated J wave sydnrome and sudden cardiac death. Heart Rhythm. 2010;7:1872–82. doi: 10.1016/j.hrthm.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giudicessi JR, Ye D, Tester DJ, et al. Transient outward current (Ito) gain-of-function mutations in the KCND3-encoded Kv4.3 potassium channel and Brugada syndrome. Heart Rhythm. 2011;8:1024–32. doi: 10.1016/j.hrthm.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kattygnarath D, Maugenre S, Neyroud N, et al. MOG1: a new susceptibility gene for Brugada syndrome. Circ Cardiovasc Genet. 2011;4:261–8. doi: 10.1161/CIRCGENETICS.110.959130. [DOI] [PubMed] [Google Scholar]
- 6.Van Norstrand DW, Ackerman MJ. Genomic risk factors in sudden infant death syndrome. Genome Med. 2010;2:86. doi: 10.1186/gm207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tester DJ, Ackerman MJ. Sudden infant death syndrome: how significant are the cardiac channelopathies? Cardiovasc Res. 2005;67:388–96. doi: 10.1016/j.cardiores.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Arnestad M, Crotti L, Rognum TO, et al. Prevalence of long-QT syndrome gene variants in sudden infant death syndrome. Circulation. 2007;115:361–7. doi: 10.1161/CIRCULATIONAHA.106.658021. [DOI] [PubMed] [Google Scholar]
- 9.Remme CA, Wilde AA, Bezzina CR. Cardiac sodium channel overlap syndromes: different faces of SCN5A mutations. Trends Cardiovasc Med. 2008;18:78–87. doi: 10.1016/j.tcm.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe H, Darbar D, Kaiser DW, et al. Mutations in sodium channel b1 and b2 subunits associated with atrial fibrillation. Circ Arrhythm Electrophysiol. 2009;2:268–75. doi: 10.1161/CIRCEP.108.779181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe H, Koopmann TT, Le Scouarnec S, et al. Sodium channel b1 subunit mutations associated with Brugada syndrome and cardiac conduction disease in humans. J Clin Invest. 2008;118:2260–8. doi: 10.1172/JCI33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olesen MS, Jespersen T, Nielsen JB, et al. Mutations in sodium channel {beta}-subunit SCN3B are associated with early-onset lone atrial fibrillation. Cardiovasc Res. 2011;89:786–93. doi: 10.1093/cvr/cvq348. [DOI] [PubMed] [Google Scholar]
- 13.Hu D, Barajas-Martinez H, Burashnikov E, et al. A mutation in the beta 3 subunit of the cardiac sodium channel associated with Brugada ECG phenotype. Circ Cardiovasc Genet. 2009;2:270–8. doi: 10.1161/CIRCGENETICS.108.829192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan BH, Pundi KN, Van Norstrand DW, et al. Sudden infant death syndrome–associated mutations in the sodium channel beta subunits. Heart Rhythm. 2010;7:771–8. doi: 10.1016/j.hrthm.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valdivia CR, Medeiros-Domingo A, Ye B, et al. Loss-of-function mutation of the SCN3B-encoded sodium channel b3 subunit associated with a case of idiopathic ventricular fibrillation. Cardiovasc Res. 2010;86:393–400. doi: 10.1093/cvr/cvp417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P, Yang Q, Wu X, et al. Functional dominant-negative mutation of sodium channel subunit gene SCN3B associated with atrial fibrillation in a Chinese GeneID population. Biochem Biophys Res Commun. 2010;398:98–104. doi: 10.1016/j.bbrc.2010.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medeiros-Domingo A, Kaku T, Tester DJ, et al. SCN4B-encoded sodium channel b4 subunit in congenital long-QT syndrome. Circulation. 2007;116:134–42. doi: 10.1161/CIRCULATIONAHA.106.659086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deschênes I, Tomaselli GF. Modulation of Kv4.3 current by accessory subunits. FEBS Lett. 2002;528:183–8. doi: 10.1016/s0014-5793(02)03296-9. [DOI] [PubMed] [Google Scholar]
- 19.Burrell C, Reddy S, Haywood G, Cunningham R. Cardiac arrest associated with febrile illness due to U.K. acquired Cyclospora cayetanensis. J Infect. 2007;54:e13–e15. doi: 10.1016/j.jinf.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 20.Makaryus JN, Verbsky J, Schwartz S, Slotwiner D. Fever associated with gastrointestinal shigellosis unmasks probable Brugada syndrome. Case Report Med. 2009;2009:492031. doi: 10.1155/2009/492031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumaine R, Towbin JA, Brugada P, et al. Ionic mechanisms responsible for the electrocardiographic phenotype of the Brugada syndrome are temperature dependent. Circ Res. 1999;85:803–9. doi: 10.1161/01.res.85.9.803. [DOI] [PubMed] [Google Scholar]
- 22.Kanter RJ, Pfeiffer R, Hu D, Barajas-Martinez H, Carboni M, Antzelevitch C. Brugada-like syndrome in infancy presenting with rapid ventricular tachycardia and intraventricular conduction delay. Circulation. 2011 doi: 10.1161/CIRCULATIONAHA.111.054007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin N, D'Andrea MR, Lubin ML, Shafaee N, Codd EE, Correa AM. Molecular cloning and functional expression of the human sodium channel beta1B subunit, a novel splicing variant of the beta1 subunit. Eur J Biochem. 2003;270:4762–70. doi: 10.1046/j.1432-1033.2003.03878.x. [DOI] [PubMed] [Google Scholar]
- 24.McCormick KA, Srinivasan J, White K, Scheuer T, Catterall WA. The extracellular domain of the beta1 subunit is both necessary and sufficient for beta1-like modulation of sodium channel gating. J Biol Chem. 1999;274:32638–46. doi: 10.1074/jbc.274.46.32638. [DOI] [PubMed] [Google Scholar]
- 25.Uebachs M, Opitz T, Royeck M, et al. Efficacy loss of the anticonvulsant carbamazepine in mice lacking sodium channel beta subunits via paradoxical effects on persistent sodium currents. J Neurosci. 2010;30:8489–501. doi: 10.1523/JNEUROSCI.1534-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malhotra JD, Chen C, Rivolta I, et al. Characterization of sodium channel a- and b-subunits in rat and mouse cardiac myocytes. Circulation. 2001;103:1303–10. doi: 10.1161/01.cir.103.9.1303. [DOI] [PubMed] [Google Scholar]
- 27.Qu Y, Isom LL, Westenbroek RE, et al. Modulation of cardiac Na+ channel expression in Xenopus oocytes by b1 subunits. J Biol Chem. 1995;270:25696–701. doi: 10.1074/jbc.270.43.25696. [DOI] [PubMed] [Google Scholar]
- 28.Yang JS, Bennett PB, Makita N, George AL, Barchi RL. Expression of the sodium channel b1 subunit in rat skeletal muscle is selectively associated with the tetrodotoxin-sensitive a subunit isoform. Neuron. 1993;11:915–22. doi: 10.1016/0896-6273(93)90121-7. [DOI] [PubMed] [Google Scholar]
- 29.Nuss HB, Chiamvimonvat N, Perez-Garcia MT, Tomaselli GF, Marban E. Functional association of the b1 subunit with human cardiac (hH1) and rat skeletal muscle (m1) sodium channel a subunits expressed in Xenopus oocytes. J Gen Physiol. 1995;106:1171–91. doi: 10.1085/jgp.106.6.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ko SH, Lenkowski PW, Lee HC, Mounsey JP, Patel MK. Modulation of Nav1.5 by b1- and b3-subunit co-expression in mammalian cells. Pflugers Arch. 2005;449:403–12. doi: 10.1007/s00424-004-1348-4. [DOI] [PubMed] [Google Scholar]
- 31.Makielski JC, Limberis JT, Chang SY, Fan Z, Kyle JW. Coexpression of b1 with cardiac sodium channel a subunits in oocytes decreases lidocaine block. Mol Pharmacol. 1996;49:30–9. [PubMed] [Google Scholar]
- 32.Deschenes I, Armoundas AA, Jones SP, Tomaselli GF. Post-transcriptional gene silencing of KChIP2 and Navbeta1 in neonatal rat cardiac myocytes reveals a functional association between Na and Ito currents. J Mol Cell Cardiol. 2008;45:336–46. doi: 10.1016/j.yjmcc.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McEwen DP, Meadows LS, Chen C, Thyagarajan V, Isom LL. Sodium channel b1 subunit-mediated modulation of Nav1.2 currents and cell surface density is dependent on interactions with contactin and ankyrin. J Biol Chem. 2004;279:16044–9. doi: 10.1074/jbc.M400856200. [DOI] [PubMed] [Google Scholar]
- 34.Malhotra JD, Kazen-Gillespie K, Hortsch M, Isom LL. Sodium channel beta subunits mediate homophilic cell adhesion and recruit ankyrin to points of cell-cell contact. J Biol Chem. 2000;275:11383–8. doi: 10.1074/jbc.275.15.11383. [DOI] [PubMed] [Google Scholar]
- 35.Malhotra JD, Thyagarajan V, Chen C, Isom LL. Tyrosine-phosphorylated and nonphosphorylated sodium channel beta1 subunits are differentially localized in cardiac myocytes. J Biol Chem. 2004;279:40748–54. doi: 10.1074/jbc.M407243200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.