Abstract
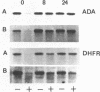
We have measured removal of pyrimidine dimers in defined DNA sequences in confluent and actively growing normal human and xeroderma pigmentosum complementation group C (XP-C) fibroblasts exposed to 10 J/m2 UV-irradiation. In normal fibroblasts 45% and 90% of the dimers are removed from the transcriptionally active adenosine deaminase (ADA) gene within 4 and 24 hours after irradiation respectively. Equal repair efficiencies are found in fragments located entirely within the transcription unit or partly in the 3' flanking region of the ADA gene. The rate and extent of dimer removal from the dihydrofolate reductase (DHFR) gene is very similar to that of the ADA gene. Repair of the transcriptionally inactive 754 locus is less efficient: 18% and 52% of the dimers are removed within 4 and 24 hours respectively. In spite of the limited overall repair capacity, confluent XP-C fibroblasts efficiently remove dimers from the ADA and DHFR genes: about 90% and 50% within 24 hours respectively. The 3' end of the ADA gene is repaired as efficiently as in normal human fibroblasts, but less efficient repair occurs in DNA fragments located in the DHFR gene and at the 5' end of the ADA gene. Repair of the inactive 754 locus does not exceed the very slow rate of dimer removal from the genome overall. Confluent and actively growing XP-C cells show similar efficiencies of repair of the ADA, DHFR and 754 genes. Our findings suggest the existence of two independently operating pathways directed towards repair of pyrimidine dimers in either active or inactive chromatin. XP-C cells have lost the capacity to repair inactive chromatin, but are still able to repair active chromatin.
Full text
PDF

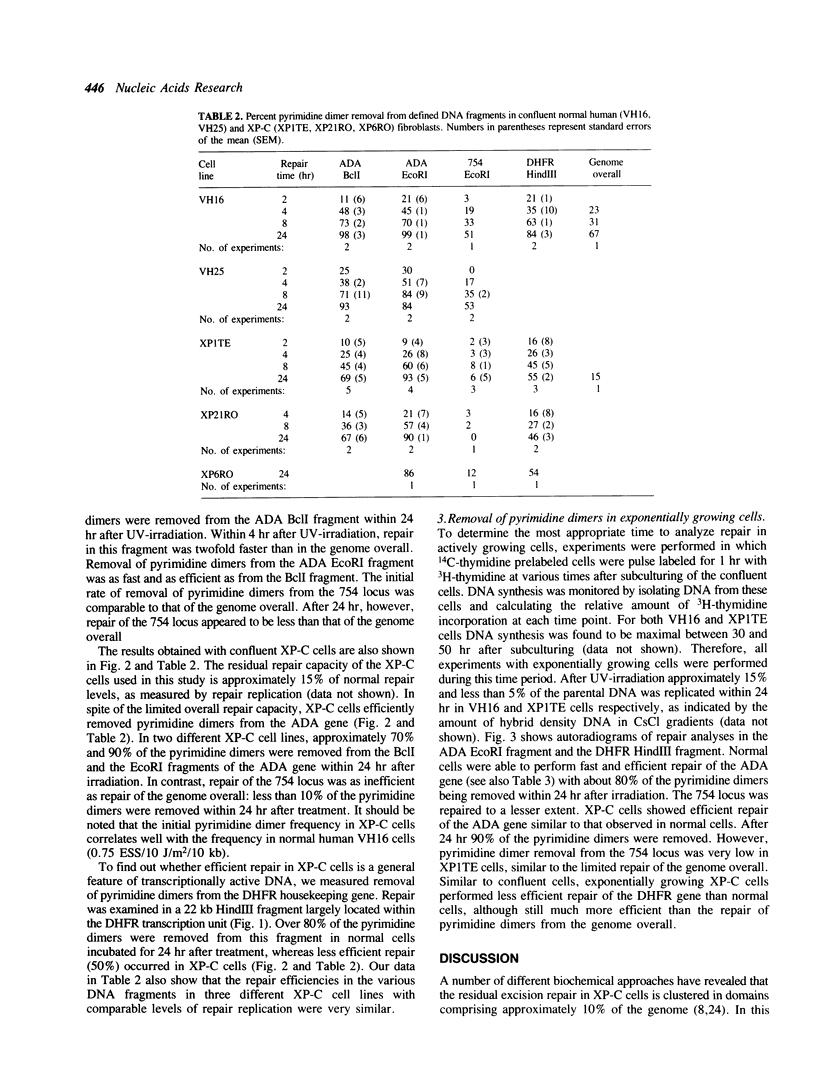


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berkvens T. M., Gerritsen E. J., Oldenburg M., Breukel C., Wijnen J. T., van Ormondt H., Vossen J. M., van der Eb A. J., Meera Khan P. Severe combined immune deficiency due to a homozygous 3.2-kb deletion spanning the promoter and first exon of the adenosine deaminase gene. Nucleic Acids Res. 1987 Nov 25;15(22):9365–9378. doi: 10.1093/nar/15.22.9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr V. A., Okumoto D. S., Hanawalt P. C. Survival of UV-irradiated mammalian cells correlates with efficient DNA repair in an essential gene. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3830–3833. doi: 10.1073/pnas.83.11.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr V. A., Smith C. A., Okumoto D. S., Hanawalt P. C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985 Feb;40(2):359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- Chan G. L., Little J. B. Resistance of plateau-phase human normal and xeroderma pigmentosum fibroblasts to the cytotoxic effect of ultraviolet light. Mutat Res. 1979 Dec;63(2):401–412. doi: 10.1016/0027-5107(79)90072-1. [DOI] [PubMed] [Google Scholar]
- Chen M. J., Shimada T., Moulton A. D., Cline A., Humphries R. K., Maizel J., Nienhuis A. W. The functional human dihydrofolate reductase gene. J Biol Chem. 1984 Mar 25;259(6):3933–3943. [PubMed] [Google Scholar]
- Ciejek E. M., Tsai M. J., O'Malley B. W. Actively transcribed genes are associated with the nuclear matrix. Nature. 1983 Dec 8;306(5943):607–609. doi: 10.1038/306607a0. [DOI] [PubMed] [Google Scholar]
- Cleaver J. E., Cortés F., Lutze L. H., Morgan W. F., Player A. N., Mitchell D. L. Unique DNA repair properties of a xeroderma pigmentosum revertant. Mol Cell Biol. 1987 Sep;7(9):3353–3357. doi: 10.1128/mcb.7.9.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver J. E. DNA repair in human xeroderma pigmentosum group C cells involves a different distribution of damaged sites in confluent and growing cells. Nucleic Acids Res. 1986 Oct 24;14(20):8155–8165. doi: 10.1093/nar/14.20.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver J. E. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968 May 18;218(5142):652–656. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- Dalton S., Younghusband H. B., Wells J. R. Chicken histone genes retain nuclear matrix association throughout the cell cycle. Nucleic Acids Res. 1986 Aug 26;14(16):6507–6523. doi: 10.1093/nar/14.16.6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Hirschhorn R., Beratis N. G., Martiniuk F. Adenosine deaminase. Alterations in activity and isozymes during growth of normal and genetically deficient fibroblasts. Exp Cell Res. 1978 Nov;117(1):103–109. doi: 10.1016/0014-4827(78)90432-9. [DOI] [PubMed] [Google Scholar]
- Hofker M. H., van Ommen G. J., Bakker E., Burmeister M., Pearson P. L. Development of additional RFLP probes near the locus for Duchenne muscular dystrophy by cosmid cloning of the DXS84 (754) locus. Hum Genet. 1986 Nov;74(3):270–274. doi: 10.1007/BF00282547. [DOI] [PubMed] [Google Scholar]
- Kantor G. J., Elking C. F. Biological significance of domain-oriented DNA repair in xeroderma pigmentosum cells. Cancer Res. 1988 Feb 15;48(4):844–849. [PubMed] [Google Scholar]
- Kantor G. J., Hull D. R. The rate of removal of pyrimidine dimers in quiescent cultures of normal human and xeroderma pigmentosum cells. Mutat Res. 1984 Jul-Aug;132(1-2):21–31. doi: 10.1016/0167-8817(84)90063-4. [DOI] [PubMed] [Google Scholar]
- Karentz D., Cleaver J. E. Excision repair in xeroderma pigmentosum group C but not group D is clustered in a small fraction of the total genome. Mutat Res. 1986 May;165(3):165–174. doi: 10.1016/0167-8817(86)90051-9. [DOI] [PubMed] [Google Scholar]
- Konze-Thomas B., Hazard R. M., Maher V. M., McCormick J. J. Extent of excision repair before DNA synthesis determines the mutagenic but not the lethal effect of UV radiation. Mutat Res. 1982 Jun;94(2):421–434. doi: 10.1016/0027-5107(82)90305-0. [DOI] [PubMed] [Google Scholar]
- Kraemer K. H., Lee M. M., Scotto J. Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch Dermatol. 1987 Feb;123(2):241–250. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- Lattier D. L., States J. C., Hutton J. J., Wiginton D. A. Cell type-specific transcriptional regulation of the human adenosine deaminase gene. Nucleic Acids Res. 1989 Feb 11;17(3):1061–1076. doi: 10.1093/nar/17.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loc P. V., Strätling W. H. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 1988 Mar;7(3):655–664. doi: 10.1002/j.1460-2075.1988.tb02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H. D., Bohr V. A., Hanawalt P. C. Differential DNA repair in transcriptionally active and inactive proto-oncogenes: c-abl and c-mos. Cell. 1986 May 9;45(3):417–423. doi: 10.1016/0092-8674(86)90327-2. [DOI] [PubMed] [Google Scholar]
- Maher V. M., Dorney D. J., Mendrala A. L., Konze-Thomas B., McCormick J. J. DNA excision-repair processes in human cells can eliminate the cytotoxic and mutagenic consequences of ultraviolet irradiation. Mutat Res. 1979 Sep;62(2):311–323. doi: 10.1016/0027-5107(79)90087-3. [DOI] [PubMed] [Google Scholar]
- Mayne L. V., Lehmann A. R. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne's syndrome and xeroderma pigmentosum. Cancer Res. 1982 Apr;42(4):1473–1478. [PubMed] [Google Scholar]
- Mellon I., Bohr V. A., Smith C. A., Hanawalt P. C. Preferential DNA repair of an active gene in human cells. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8878–8882. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I., Spivak G., Hanawalt P. C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987 Oct 23;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Mitchell D. L., Haipek C. A., Clarkson J. M. (6-4)Photoproducts are removed from the DNA of UV-irradiated mammalian cells more efficiently than cyclobutane pyrimidine dimers. Mutat Res. 1985 Jul;143(3):109–112. doi: 10.1016/s0165-7992(85)80018-x. [DOI] [PubMed] [Google Scholar]
- Mullenders L. H., van Kesteren A. C., Bussmann C. J., van Zeeland A. A., Natarajan A. T. Distribution of u.v.-induced repair events in higher-order chromatin loops in human and hamster fibroblasts. Carcinogenesis. 1986 Jun;7(6):995–1002. doi: 10.1093/carcin/7.6.995. [DOI] [PubMed] [Google Scholar]
- Mullenders L. H., van Kesteren A. C., Bussmann C. J., van Zeeland A. A., Natarajan A. T. Preferential repair of nuclear matrix associated DNA in xeroderma pigmentosum complementation group C. Mutat Res. 1984 Oct;141(2):75–82. doi: 10.1016/0165-7992(84)90014-9. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y., Yamashita K., Sekiguchi M. Purification and characterization of normal and mutant forms of T4 endonuclease V. J Biol Chem. 1982 Mar 10;257(5):2556–2562. [PubMed] [Google Scholar]
- Player A. N., Kantor G. J. The endogenous nuclease sensitivity of repaired DNA in human fibroblasts. Mutat Res. 1987 Sep;184(2):169–178. doi: 10.1016/0167-8817(87)90074-5. [DOI] [PubMed] [Google Scholar]
- Protić-Sabljić M., Kraemer K. H. One pyrimidine dimer inactivates expression of a transfected gene in xeroderma pigmentosum cells. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6622–6626. doi: 10.1073/pnas.82.19.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J. H., Kraemer K. H., Lutzner M. A., Festoff B. W., Coon H. G. Xeroderma pigmentosum. An inherited diseases with sun sensitivity, multiple cutaneous neoplasms, and abnormal DNA repair. Ann Intern Med. 1974 Feb;80(2):221–248. doi: 10.7326/0003-4819-80-2-221. [DOI] [PubMed] [Google Scholar]
- Sauerbier W., Hercules K. Gene and transcription unit mapping by radiation effects. Annu Rev Genet. 1978;12:329–363. doi: 10.1146/annurev.ge.12.120178.001553. [DOI] [PubMed] [Google Scholar]
- Small D., Nelkin B., Vogelstein B. The association of transcribed genes with the nuclear matrix of Drosophila cells during heat shock. Nucleic Acids Res. 1985 Apr 11;13(7):2413–2431. doi: 10.1093/nar/13.7.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder J., Groudine M., Dodgson J. B., Engel J. D., Weintraub H. Hb switching in chickens. Cell. 1980 Apr;19(4):973–980. doi: 10.1016/0092-8674(80)90088-4. [DOI] [PubMed] [Google Scholar]
- Wiginton D. A., Kaplan D. J., States J. C., Akeson A. L., Perme C. M., Bilyk I. J., Vaughn A. J., Lattier D. L., Hutton J. J. Complete sequence and structure of the gene for human adenosine deaminase. Biochemistry. 1986 Dec 16;25(25):8234–8244. doi: 10.1021/bi00373a017. [DOI] [PubMed] [Google Scholar]
- Yang J. K., Masters J. N., Attardi G. Human dihydrofolate reductase gene organization. Extensive conservation of the G + C-rich 5' non-coding sequence and strong intron size divergence from homologous mammalian genes. J Mol Biol. 1984 Jun 25;176(2):169–187. doi: 10.1016/0022-2836(84)90419-4. [DOI] [PubMed] [Google Scholar]
- van Zeeland A. A., Smith C. A., Hanawalt P. C. Sensitive determination of pyrimidine dimers in DNA of UV-irradiated mammalian cells. Introduction of T4 endonuclease V into frozen and thawed cells. Mutat Res. 1981 Jun;82(1):173–189. doi: 10.1016/0027-5107(81)90148-2. [DOI] [PubMed] [Google Scholar]