SUMMARY
Recognition of DNA by the innate immune system is central to anti-viral and anti-bacterial defenses, as well as an important contributor to autoimmune diseases involving self DNA. AIM2 (absent in melanoma 2) and IFI16 (interferon-inducible protein 16) have been identified as DNA receptors that induce inflammasome formation and interferon production, respectively. Here we present the crystal structures of their HIN domains in complex with double-stranded (ds) DNA. Non-sequence specific DNA recognition is accomplished through electrostatic attraction between the positively charged HIN domain residues and the dsDNA sugar-phosphate backbone. An intramolecular complex of the AIM2 Pyrin and HIN domains in an autoinhibited state is liberated by DNA binding, which may facilitate the assembly of inflammasomes along the DNA staircase. These findings provide novel mechanistic insights into dsDNA as the activation trigger and oligomerization platform for the assembly of large innate signaling complexes such as the inflammasomes.
INTRODUCTION
The innate immune system responds to the presence of cytosolic DNA molecules through the secretion of interferons and proinflammatory cytokines (Hornung and Latz, 2010), and the activation of antigen-presenting cells to induce potent adaptive immune responses (Kis-Toth et al., 2011). Multiple cytosolic innate DNA receptors/sensors have been reported, including DAI (DNA-dependent activator of IRFs) (Takaoka et al., 2007), LRRFIP1 (Leucine-rich repeat and flightless I interacting protein 1) (Yang et al., 2010) and DDX41 (DEAD box polypeptide 41) (Zhang et al., 2011). In addition, the RNA sensor RIG-I (retinoic acid inducible gene I) indirectly detects DNA transcribed by RNA polymerase III (Ablasser et al., 2009).
Recently, a family of DNA-recognizing innate receptors was identified among the HIN-200 proteins (hematopoietic interferon-inducible nuclear proteins with a 200-amino-acid repeat) (Goubau et al., 2010; Ludlow et al., 2005), such as AIM2 (Burckstummer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009) and IFI16 (Kerur et al., 2011; Unterholzner et al., 2010). A third DNA-binding protein p202 was reported to be an inhibitor of the AIM2 signaling (Roberts et al., 2009). Both AIM2 and IFI16 contain C-terminal DNA-binding HIN domain(s) and an N-terminal Pyrin (PYD) domain that belongs to the death domain superfamily of signaling modules, and thus were renamed as the PYHIN family of receptors (Hornung et al., 2009; Schattgen and Fitzgerald, 2011) or the AIM2-like receptors (Unterholzner et al., 2010). AIM2 is predominantly a cytosolic protein that responds to dsDNA from both host and pathogens to form large signaling platforms known as the inflammasomes (Davis et al., 2011; Schroder and Tschopp, 2010), which also contain the adapter protein ASC (apoptosis-associated speck-like protein containing a caspase recruitment domain) and effector enzyme procaspase-1. These macromolecular complexes control the activation of procaspase-1 and subsequent maturation and secretion of IL-1β and IL-18. Innate receptors such as NLRP1, NLRP3, NLRP6, NLRP7, NLRC4, NAIP, AIM2 and IFI16 are known to form inflammasomes that respond to ligands or stimuli from various microbial or host sources. A major challenge in the field has been the lack of concrete evidence of direct receptor:ligand association for many of the inflammasomes, thus the true identities of the respective ligands are still unknown. In contrast, cellular and biochemical evidence has confirmed that AIM2 and IFI16 (see below) directly interact with dsDNA (Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Unterholzner et al., 2010).
IFI16 was originally identified as an anti-proliferative and DNA damage response protein in the nucleus (Choubey et al., 2008). Recently IFI16 and its mouse homolog p204 were shown to be cytosolic dsDNA receptors that induce interferon production (Unterholzner et al., 2010). IFI16 was also reported to form inflammasomes sensing DNA viruses replicating in the nucleus (Kerur et al., 2011). The cytosolic signaling pathway for interferon induction downstream of IFI16 appears to require the ER resident protein STING (stimulator of interferon genes) (Ishikawa et al., 2009; Unterholzner et al., 2010), which itself was shown to be a nucleotide sensor that induces type I IFN production (Burdette et al., 2011). Both AIM2 and IFI16 respond to dsDNA from various sources irrespective of their sequences or GC contents (Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Unterholzner et al., 2010), consistent with the principal requirement of the innate immune responses to diverse microbial threats as well as cellular stress. As such, these innate receptors play crucial roles in host defense against intracellular pathogens such as Francisella tularensis, vaccinia virus and herpes simplex virus type 1 (Fernandes-Alnemri et al., 2010; Jones et al., 2010; Rathinam et al., 2010; Unterholzner et al., 2010), as well as in autoimmune disease such as systemic lupus erythematosus (SLE) in which DNA is a major autoimmune target (Veeranki and Choubey, 2010).
Despite the elucidation of innate RNA recognition by the structures of the TLR3:dsRNA complex (Liu et al., 2008) and RIG-I:RNA complex (Jiang et al., 2011; Kowalinski et al., 2011; Lu et al., 2010; Luo et al., 2011; Wang et al., 2010), the mechanism for innate recognition of dsDNA, particularly the common B-form dsDNA as the primary stimulator of the cytosolic sensors (Ishii et al., 2006), remains elusive. The crystal structures of the HINa and HINb domains from IFI16 (Liao et al., 2011) demonstrated that each HIN domain contains two tandem β barrels of ~80 residues previously characterized as the OB (oligonucleotide/oligosaccharide binding) fold (Albrecht et al., 2005), but the mode of HIN:DNA interaction remains unknown. Furthermore, the mechanisms of receptor oligomerization for the AIM2 and IFI16 inflammasomes are unclear. Neither AIM2 nor IFI16 contains an oligomerization domain, even though such domains were essential for other inflammasome receptors as evidenced by the majority of their dysfunctional mutations at these domains (Aksentijevich et al., 2007; Schroder and Tschopp, 2010).
To understand the mechanisms of dsDNA recognition, receptor activation and oligomerization, we determined the crystal structures of the HIN domains from both AIM2 and IFI16 in complex with the B-form dsDNA. Our findings establish electrostatic attraction as the basis for non-sequence specific DNA recognition, identify DNA as the ligand that releases the signaling domain PYD from its intramolecular complex with the HIN domain, and define the multivalent ligand dsDNA as the oligomerization platform for the inflammasome formation.
RESULTS
Overview of The HIN:DNA Complex Structures
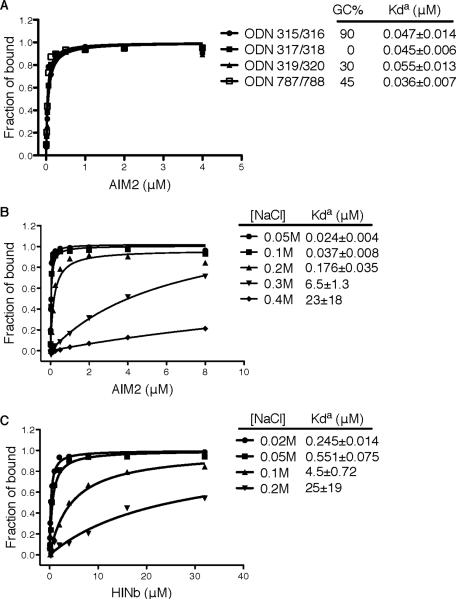
Previous reports demonstrated that DNA of various sequences or GC contents from host, microbial and synthetic sources are equivalent in their ability to stimulate AIM2 and IFI16 (Burckstummer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Roberts et al., 2009; Unterholzner et al., 2010). In agreement, here we showed quantitatively by a fluorescence polarization (FP) assay that different dsDNA bind the AIM2 HIN domain with comparable apparent affinities (Figure 1A and Table S1). The HIN:DNA binding was sensitive to salt concentrations (Figures 1B–C), suggesting essential contributions from ionic interactions. We chose to use the dsDNA derived from the vaccinia virus genomic repeat sequences (Baroudy and Moss, 1982) for further crystallographic studies (Table S1) and determined the crystal structures of the AIM2 HIN:DNA and IFI16 HINb:DNA complexes at 2.5–2.6 Å resolutions (Figures 2A–B and S1A–B, Table S2).
Figure 1. The HIN:DNA Interactions are Non-sequence Specific and Sensitive to Ionic Strength.
(A) Fluorescence polarization (FP) assays of the FAM-labeled dsDNAs of various sequences and GC contents upon binding to the AIM2 HIN domain. The apparent Kd values (Kda) are shown for each dsDNA.
(B) FP assays of the FAM-labeled dsDNA ODN787/788 upon binding to the AIM2 HIN domain in the presence of various concentrations of sodium chloride.
(C) FP assays of the FAM-labeled dsDNA ODN787/788 upon binding to the IFI16 HINb domain in the presence of various concentrations of sodium chloride.
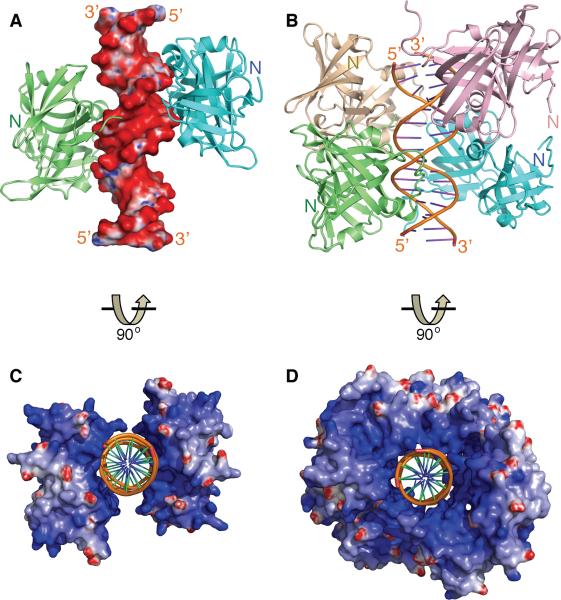
Figure 2. Overview of The HIN:DNA Complexes.
(A) The structure of the AIM2 HIN:DNA complex (crystal form I) is represented as lime and cyan-colored ribbons for each HIN domain and electrostatic charge surface for the dsDNA on a scale of -10 kT/e (red) to 10 kT/e (blue). Locations of the N-termini of the HIN domains are marked.
(B) Structure of the IFI16 HINb:DNA complex is shown as lime, cyan, lightpink and wheat ribbons for each HINb domain and orange ribbon for the dsDNA. Locations of the N-termini of the HIN domains are marked.
(C) Structure of the AIM2 HIN:DNA complex is rotated 90 degrees along the horizontal axis from the view in (A), and represented as electrostatic charge surface for the HIN domains and orange ribbon for the dsDNA.
(D) Structure of the IFI16 HINb:DNA complex is represented as electrostatic charge surface for the HIN domains and orange ribbon for the dsDNA. The view is rotated 90 degrees horizontally from that in (B).
See also Figure S1.
In spite of different crystal lattice packing, common modes of DNA binding by the HIN domains are evident upon inspection of the structures. The HIN:DNA complexes feature positively charged HIN domains embracing the dsDNA sugar-phosphate backbone in a concave surface of the protein (Figures 2C–D and S1C–F). Bonding between the HIN domain basic residues with the non-esterified phosphate oxygens dominates the HIN:DNA interface, in agreement with previous observations that nonbridging phosphoryl oxygens are critical for aligning DNA-binding proteins (Harrison, 1991). In addition, the N-termini of the HIN domains are all located distal to the DNA-binding interface (Figures 2A–B), potentially facilitating interaction of the N-terminal PYD domain with downstream adapter ASC at the periphery of the receptor assembly. Importantly, both AIM2 and IFI16 HIN domains bind both strands of the dsDNA, across both major and minor grooves (Figures 3A–B and S2A–C), in keeping with the requirement for dsDNA instead of ssDNA for innate signaling by AIM2 and IFI16 (Burckstummer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Unterholzner et al., 2010).
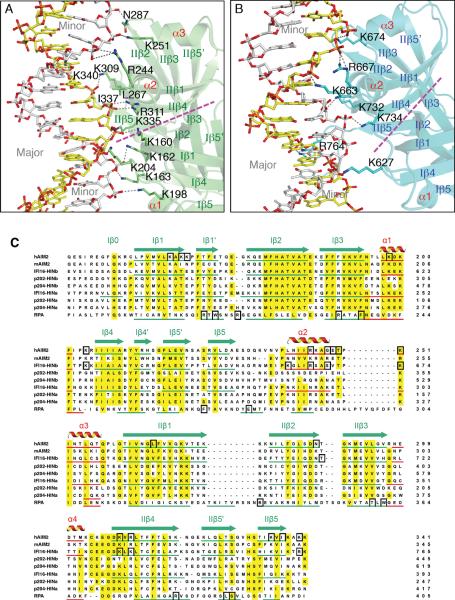
Figure 3. The HIN Domains Bind Both Strands of The dsDNA.
(A) Detailed HIN:DNA interactions for the AIM2 HIN:DNA complex. The hydrogen bonds are indicated as gray dotted lines. Secondary structures for the AIM2 HIN domain (lime) are labeled and the two DNA strands are colored yellow and silver, respectively. The approximate boundaries of the OB1-OB2 are marked with a magenta dotted line and the major and minor grooves of the dsDNA are marked in gray.
(B) Detailed HIN:DNA interactions are shown for the IFI16 HINb:DNA complex similar to (A), except the IFI16 HINb domain is colored cyan.
(C) Sequence alignment of the HIN domains. Sequences of selected dsDNA-binding HIN domains from human AIM2 (NP_004824), mouse AIM2 (NP_001013801), human IFI16 (Q16666), mouse p204 (NP_032355, a homolog of human IFI16), mouse p202 (NP_032353, an inhibitor of AIM2), as well as a ssDNA-binding OB superfamily protein RPA (NP_002936) were aligned by ClustalW (Thompson et al., 1994) with minor adjustments. The α helices are in red, and the β strands were underlined in green and marked with “I” and “II” for OB1 and OB2, respectively. Conserved residues are shaded in yellow, and DNA binding residues in black boxes.
See also Figure S2.
Both OB Folds and The Linker Between Them Engage The dsDNA Backbone
The DNA-binding surface of the AIM2 and IFI16 HIN domains consists of both OB folds (hereafter referred to as OB1 and OB2) and the linker between them (Figures 3 and S2, and Table S3). The DNA interface from the OB1 of AIM2 is centered at residues K162 and K163 between β1 and β1' strands, and K198 and K204 near the α1 helix. The OB1-OB2 linker contains amphipathic α2–α3 helices that contribute hydrogen bonds and van der Waals (vDW) contacts from R244, K251 or G247 and T249 for different AIM2 HIN domains. The OB2 of AIM2 HIN forms salt bridges and vDW contacts with DNA through residues R311 at the β4 strand and K335 and I337 at the β5 strand. R311 faces the minor groove of the dsDNA and forms bidentate hydrogen bonds with a DNA backbone phosphate (Figures 3A and S2A–B), unlike those from sequence-specific DNA-binding proteins that extend into the minor groove to contact the bases (Rohs et al., 2009). AIM2 HIN domains from two different crystal forms also employ distinctive DNA-binding residues bordering the above core DNA interface (Table S3), suggesting flexible interface with DNA. Overall, there is 1000–1200 Å2 of solvent accessible surface area buried between each AIM2 HIN domain and their DNA partners.
In comparison, the IFI16 HINb-DNA interfaces bury 700–800 Å2 of solvent accessible surface area, consistent with the lower DNA-binding affinity. Most of the IFI16 HINb DNA-binding residues are located at the OB1-OB2 linker and OB2 that form side chain and main chain hydrogen bonds as well as vDW contacts with the DNA backbone phosphates (Figures 3B and S2C, Table S3). These include residues K663, R667 at the linker α2 helix and K732, K734, and R764 near the β4 and β 5 strands. Even at the modest resolutions, many water molecules are visible along the DNA double strands, with some mediating protein-DNA interactions (Figure S2A–C).
Sequence alignment of the HIN domains illustrates that the DNA-binding residues are largely conserved between the AIM2 HIN and IFI16 HINb domains (Figure 3C), in particular those at the α1–β4 loops from OB1, the linker α2 helices, and the β4–β5 strands from OB2. In accord with its higher affinity for DNA, the AIM2 HIN domain also contributes additional DNA-binding residues at its β1 strands from both OB1 and OB2, which are conserved in mouse AIM2. The structures also explain the different DNA-binding affinities of the IFI16 HINa and HINb domains (Unterholzner et al., 2010). Residues K663 and R667 at the IFI16 HINb linker α2 helix are shifted in their positions compared with those at the IFI16 HINa domain, thus would locate the HINa residues away from the DNA interface. Additionally, the DNA-binding residues K734 and R764 from IFI16 HINb are not conserved in the HINa domains from either IFI16 or mouse p204, consistent with the lower DNA-binding affinities for the HINa domains (Unterholzner et al., 2010).
The HIN:DNA structures also demonstrates versatile modes of DNA binding: superposition of the HIN domains shows that their bound dsDNA can tilt and slide relative to the HIN domains (Figures 3C and S2D–F), partly due to the flexible lysine and arginine residues that dominate the HIN:DNA interface. Comparison of the IFI16 HINb structures in the presence and absence of dsDNA illustrates no major conformational changes (Figure S2G), suggesting that the DNA-binding surface of the HIN domains is pre-formed.
Mutations of Key Residues at The HIN:DNA Interface Compromises DNA Binding
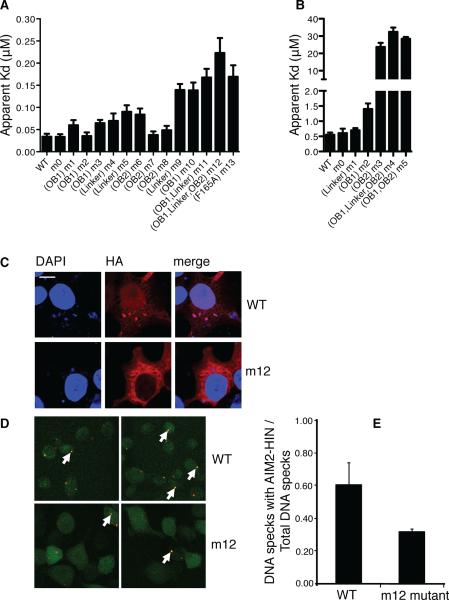
To probe the functional relevance of the observed HIN:DNA interactions, mutagenesis studies were carried out for the DNA-binding residues individually and in groups based on their locations at the OB1, linker and OB2 (Figures 4 and S3, Table S4). We first measured the apparent DNA-binding affinities of the wild type and mutant AIM2 HIN domains using the FP assay. Our data showed that several AIM2 HIN domain single-residue mutants had lower affinities for DNA (higher apparent Kd values), such as K204A at OB1 (m3), K251A at the OB1-OB2 linker (m5), and K309A at OB2 (m6), whereas point-mutation at other sites such as K198A at OB1 (m2) or R311A at OB2 (m7) did not significantly affect the HIN:DNA interaction (Figure 4A and S3A). In comparison, grouped mutations showed more prominent effects, with the mutant m12 harboring the most mutations retaining the weakest DNA-binding activity. To investigate if positively charged residues outside of the HIN:DNA interface are important for DNA binding, we mutated three basic residues K276A, K277A and K278A located at the opposite site of the AIM2 HIN domain DNA-binding surface. This mutant m0 possesses the same DNA-binding affinity as the wild type, suggesting that basic residues extraneous to the HIN:DNA interface are not essential for the interaction (Table S4). A mutation of F165A was previously reported to impair the association of AIM2 with DNA (Burckstummer et al., 2009). This residue is located at the OB1-OB2 interface to anchor the OB1 β1-β1' strands containing the DNA-binding residues K160, K162 and K163, as well as the adjacent K198 (Figure S3B). Consistent with the published result, the F165A mutant m13 possesses a diminished DNA-binding affinity comparable to that of the m10 or m11 mutant, in which the majority of the mutated DNA-binding residues reside in OB1 (Table S4).
Figure 4. Mutagenesis Studies of The HIN:DNA Interactions.
(A) Wild type and mutant AIM2 HIN domains were assayed for DNA binding and their apparent Kd values are plotted in a bar graph. Locations of the mutations for each mutant are marked below the graph. The “m0” mutants contain mutations of basic residues outside the DNA-binding surface. More details on the mutation sites are listed in Table S4. Panels of the binding curves for each mutant are presented in Figure S3A.
(B) Wild type and mutant IFI16 HINb domains were assayed for DNA binding and their apparent Kd values are plotted in a bar graph. Locations of the mutations for each mutant are marked below the graph. The “m0” mutants contain mutations of basic residues outside the DNA-binding surface. More details on the mutation sites are listed in Table S4. Panels of the binding curves for each mutant are presented in Figure S3D.
(C) Co-localization of DNA with wild type or mutant (m12) AIM2 HIN domains containing a hemagglutinin (HA) tag in HEK293T cells. Cells were stained 24 hours after transfection for AIM2 (anti-HA antibody with Alexa647, red) and DNA (DAPI, blue). Regions of colocalization are marked with white arrows.
(D) Co-localization of FITC-labeled dsDNA (red) with stably expressed wild type or mutant (m12) AIM2 HIN domains tagged with mCerulean (green) in AIM2 deficient macrophages. Regions of colocalization are marked with white arrows.
(E) The ratio of the number of dsDNA specks that colocalized with the wild type or mutant AIM2 HIN domains to the total number of dsDNA specks from (D) are shown for 2 full 1200×1200 pixel mosaic images, respectively.
See also Figure S3.
To further investigate the significance of the HIN:DNA interface in the context of the full-length AIM2 protein, we introduced some of the same mutations above in the intact AIM2 and measured the DNA-binding affinities. Similar to our data for the isolated HIN domains, mutations at the non-DNA binding surface (m0) or a single mutation at the linker region (m5) did not significantly affect DNA binding by the full-length AIM2 protein (Figure S3C and Table S4). By contrast, grouped mutants m10 (OB1) and m12 (OB1, linker and OB2) showed much diminished affinities for DNA, confirming that the DNA-binding residues are important for ligand association by the intact receptor.
In comparison to the AIM2 HIN domain, the IFI16 HINb has a much lower DNA-binding affinity (Figures 4B and S3D, Table S4), perhaps reflecting the presence of the HINa-b tandem in the full-length IFI16 receptor, and consistent with previous observations that the HINa-b tandem bound DNA much tighter than either HINa or HINb alone (Unterholzner et al., 2010). Mutation of the OB1-OB2 linker (m1) and OB1 (m2) only marginally reduced the HINb:DNA interaction. In contrast, DNA-binding was severely compromised by mutations at OB2 (m3), suggesting that it is essential for DNA association. Importantly, an irrelevant mutant m0 harboring mutations of five basic residues retained comparable DNA-binding affinity as the wild type protein, again suggesting that non-DNA contacting basic residues are not essential for binding (Table S4). Because all of the mutations are localized at the surface of the HIN domains, they are unlikely to disrupt the proper domain folding. This is illustrated by the structure of an IFI16 HINb mutant containing K663A and R667A mutations (Table S2, IFI16 crystal form III), which is essentially the same as the wild type protein (Figure S2G).
Intact DNA-binding Surface is Essential for Association with and Innate Response to DNA
To examine the HIN:DNA interaction in cells, confocal microscopy was employed to study the co-localization and clustering of the AIM2 HIN domains with DNA in HEK293T cells or AIM2-deficient mouse macrophages (Figures 4C–D). We show that the wild type but not mutant AIM2 HIN domain colocalized with DNA in HEK293T cells, and quantitative analysis in macrophages demonstrated diminished DNA colocalization for the mutant AIM2 HIN domain compared with the wild type (Figure 4E). Our attempts to analyze DNA colocalization with the full-length AIM2 protein were hampered by severe protein aggregation and cellular pyroptosis, perhaps due to the presence of the PYD domain that induced excessive protein oligomerization, and activation of the full-length receptor by the transfected plasmid DNA. Nevertheless, our data with the AIM2 HIN domain confirm the important contribution of the DNA-binding residues to the HIN:DNA association, and are consistent with our kinetic measurements of the HIN:DNA and full-length AIM2:DNA interactions (see above).
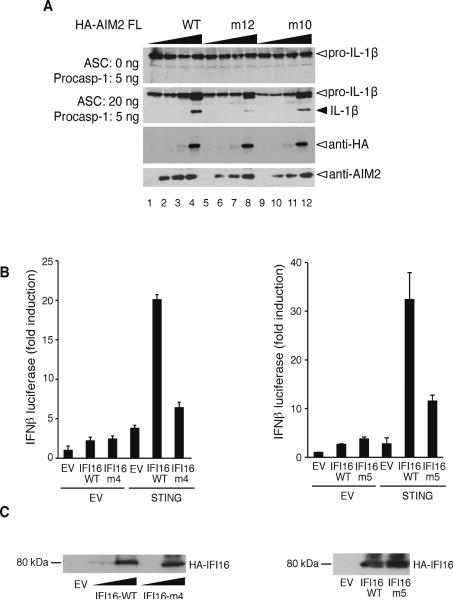
To further study the impact of the above mutations on the function of the full-length receptors in mammalian cells, we performed assays for inflammasome formation and interferon reporter with the full-length AIM2 and IFI16, respectively (Figure 5). Mutation of the DNA-binding residues for the AIM2 m10 and m12 mutants (Table S4) significantly diminished the maturation of IL-1β by the AIM2 inflammasome (Figure 5A, compare the IL-1β bands from lanes 4, 8 and 12). Similarly, when the IFI16/STING pathway was reconstituted in HEK293T cells, disruption of the IFI16 HINb DNA-binding residues impaired its ability to induce the IFN-β promoter in response to the transfected plasmid DNA (Figure 5B). The residual IFN-β induction by the IFI16 mutant may be partially due to the presence of the wild type HINa domain from the IFI16 receptor that is known to bind DNA (Unterholzner et al., 2010), as well as basal activation of the IFN-β promoter by the over-expressed STING. The reduction in IL-1β secretion or IFN-β induction was not due to differential expression of the wild type and mutant receptors, as they were expressed at comparable levels for both AIM2 and IFI16 in transfected HEK293T cells, with no observable endogeneous expression (Figures 5A bottom panels and 5C).
Figure 5. Innate immune Responses by The Full-length AIM2 and IFI16 Receptors.
(A) Reconstitution of the human AIM2 inflammasome using the wild type, or mutant (m12 and m10) full-length HA-AIM2 and ASC, procaspase-1 and luciferase-FLAG tagged pro-IL-1β. Maturation of the IL-1β is indicated with a black arrow. The expression levels of the wild type or mutant full-length HA-AIM2 are indicated at the bottom panels as probed by anti-HA or anti-AIM2 antibodies.
(B) Interferon-β promoter reporter assay for the wild type full-length IFI16 or IFI16 containing the HINb mutant m4 or m5 in HEK293T cells. EV indicates empty expression vector.
(C) The expression levels of the wild type or mutant full-length IFI16 proteins in (B) are detected by immunoblotting 48 hours after transfection.
DNA Displaces PYD Domain from Its Intramolecular Complex with The HIN Domain
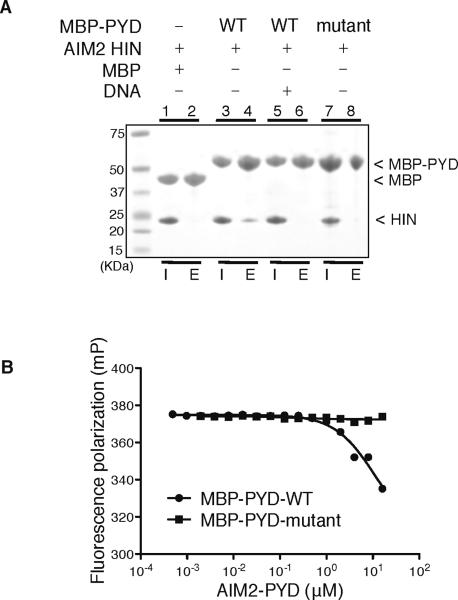
The HIN:DNA complex structures also suggested potential mechanisms of receptor activation. We note that the size of the AIM2 PYD domain (~10 Å radius), modeled using the ASC PYD structure (de Alba, 2009), is very similar to that of a B-DNA cylinder, and may be able to bind at the concave basic surface of the AIM2 HIN domain. We therefore tested if the AIM2 PYD and HIN domains form a protein complex, and if so, whether this interaction is affected by DNA binding. Transfection of the full-length AIM2 protein or its PYD domain caused severe protein aggregation and pyroptosis, as noted above, and has so far prevented us from analyzing the HIN:PYD domain interaction in a cellular context. As an alternative, we employed a pull-down assay using a maltose-binding protein (MBP) expression tag linked to the AIM2 PYD domain. We show that the wild type AIM2 PYD and HIN domains form a protein complex, which was disrupted by either the presence of dsDNA or mutation of acidic residues in PYD (Figure 6A). Conversely, the AIM2 PYD domain suppressed the HIN:DNA interaction (Figure 6B), even though it has no DNA-binding capacity (Figure S4A). In agreement, the full-length AIM2 receptor exhibited a decreased DNA-binding affinity in vitro compared with the isolated AIM2 HIN domain (Figure S4B). We therefore envision a model in which the PYD and HIN domains of AIM2 form an intramolecular complex in an autoinhibited “resting” state, with the PYD-binding and DNA-binding surface overlapping at the HIN domain. DNA binding by the HIN domain activates the receptor through displacing the PYD domain from this intramolecular complex, which facilitates the PYD domain downstream signaling to the adapter ASC (Figure S4C).
Figure 6. PYD domain Inhibits The HIN:DNA Interaction.
(A) Pull-down assay of the MBP, MBP-PYD wild type or mutant (containing mutations of E7A, L11A, D15A, D19A and E20A) with the AIM2 HIN domain. The wild type MBP-PYD (lane 4), but not mutant MBP-PYD (lane 8) or MBP (lane 2), was able to pull down the AIM2 HIN domain, which was significantly reduced by the addition of a 19mer dsDNA ODN 736/737 (lane 6). “I”: input; “E”: elution. (B) The AIM2 PYD domain inhibits the HIN:DNA interaction. Increasing concentrations of the wild type or mutant AIM2 PYD domain were incubated with an AIM2 HIN:DNA mixture, and the fluorescence polarization was measured and analyzed with program Prism. The IC50 for the wild type AIM2 PYD domain is 11 μM.
See also Figure S4.
DNA Serves as An Oligomerization Platform for The AIM2 Inflammasome
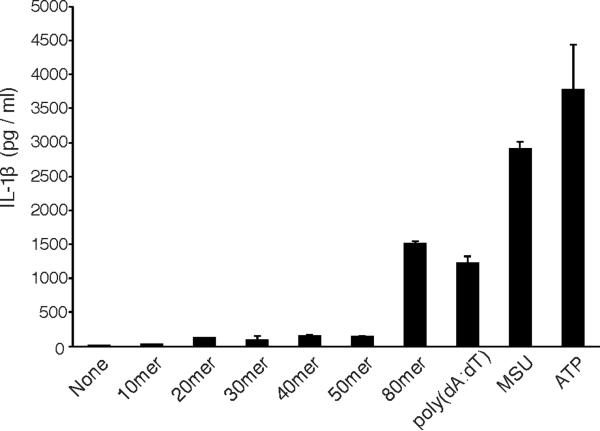
The dsDNA molecules in the HIN:DNA crystals form pseudo-continuous double helices through head-to-tail stacking, with the HIN domains decorated along the DNA staircases. An example from the AIM2 crystal form II is illustrated in figures S5A–B. Each HIN domain spans a spacing of 7–8 bp on each side of the dsDNA. In the IFI16 HINb crystal form I, a 16 bp dsDNA accommodates four HINb domains. It is possible that two of the HINb domains (i.e., C and D molecules with less interactions with DNA, Table S3) may mimic the HINa domains of IFI16, such that the four HINb domains may represent two HINa-b tandems from two IFI16 molecules. Previous studies demonstrated that 70 bp dsDNAs were required for optimal interferon induction by IFI16 Unterholzner et al., 2010). Based on the assumption of the HINa-b tandem in the IFI16 HINb crystal lattice, 70 bp may allow up to 9 IFI16 molecules with 18 HINa/b domains to oligomerize as a signaling complex. Similarly, we found that ~80 bp of dsDNA transfected into cells is required for optimal IL-1β induction (Figure 7), presumably through the activation of AIM2. A dsDNA of this length may be able to accommodate maximal 20 AIM2 HIN domains, although variable oligomerization states for the full-length AIM2 receptor are possible because of its larger size and the non-specific nature of DNA binding. One hypothetical model of such a multi-molecular complexes is presented in figures S5C–D, using the crystallographic asymmetric unit of the AIM2 crystal form II as a template to dock an AIM2 PYD domain model and the full-length ASC structure (2KN6) (de Alba, 2009). While the validity of the AIM2 inflammasome model awaits future structural and functional characterization, we note that oligomerization of the AIM2 receptors is unlikely to materialize in the absence of electrostatic charge neutralization through the HIN domain:DNA interaction. We therefore propose that instead of relying on a specialized oligomerization domain as in other inflammasomes, AIM2 may employ the multivalent ligand dsDNA as a platform to assemble large signaling complexes such as the inflammasomes.
Figure 7. Secretion of IL-1β induced by DNA of various lengths.
Human PBMCs were primed with LPS and transfected with the indicated dsDNA, or MSU and ATP as controls. The culture supernatants were assayed for IL-1β secretion 6 hours after transfection/stimulation.
See also Figure S5.
DISCUSSION
The structures reported here provide novel examples of non-sequence specific dsDNA recognition by innate immune receptors through electrostatic attraction. This appears to be one of the general principles of nucleic acid sensing by the innate immune system, as electrostatic attraction also plays important roles in RNA recognition by TLR3 (Liu et al., 2008) and RIG-I (Jiang et al., 2011; Luo et al., 2011; Kowalinski et al., 2011). The non-specific nature of sensing universal genomic materials allows the innate immune system to respond to the presence of threat from either infectious agents or host tissue damages. This type of interaction is indeed very common among DNA-binding proteins, as “non-specific” binding to DNA backbone also plays an important role in sequence-specific recognition by transcription factors and restriction enzymes (Luscombe et al., 2001), for example in the “facilitated diffusion” model (von Hippel and Berg, 1989). The non-specific DNA interface is highly flexible, which is reminiscent of the tilting and sliding of the dsDNA relative to the HIN domains in our structures. It is conceivable that electrostatic attraction may have evolved into an integral component of the innate nucleic acid recognition because of its non-specific and versatile nature.
Many signaling receptors reside in autoinhibited states in the absence of their ligands, and activation of the receptors is often accomplished by allosteric conformational changes (Pufall and Graves, 2002). For example, the apoptotic protease activating factor-1 (Apaf-1) contains a C-terminal WD-40 repeat domain that represses its ability to activate procaspase-9 in the absence of ligand (cytochrome C) binding, and ligand engagement by WD-40 releases this inhibition to facilitate the formation of a large macromolecular complex apoptosome (Hu et al., 1998; Srinivasula et al., 1998). More recently, an auto-repression model was proposed for the dsRNA receptor RIG-I, in which the RNA-binding helicase domain represses the CARD domains in the resting state, and dsRNA binding results in a structural change that facilitates downstream signaling by the CARD domains (Jiang et al., 2011; Kowalinski et al., 2011; Luo et al., 2011). For the DNA receptor AIM2, the apparently weak interaction between the HIN and PYD domains is likely significantly enhanced in the full-length receptor due to the high effective local concentrations from their covalent linkage, and can only be disrupted by high-affinity multivalent ligand such as dsDNA. The presence of cis-acting repressor domains in these receptors is an effective regulatory mechanism to prevent spurious activation of the signaling pathways: the autoinhibited state of the AIM2 receptor creates an elevated threshold for activation that prevents excessive immune response to trace amounts of nucleic acids, or to other negatively-charged cellular components. While the current work elucidated intramolecular interactions between the isolated HIN and PYD domains, future experiments will further examine the autoinhibition model of the AIM2 receptor using physiologically relevant cellular assays and animal models. These should further illustrate the importance of the exquisite control of innate receptor activation, as evidenced by diseases associated with inappropriate stimulation of the inflammasomes (Aksentijevich and Kastner, 2011; Schroder and Tschopp, 2010).
Regulation of the innate receptor oligomerization and inflammasome formation bears important physiological consequences. Mutations in the oligomerization domains of NLRP3, NLRP12, and NOD2 are associated with autoinflammatory disorders CAPS (cryopyrin-associated periodic syndromes) (Aksentijevich et al., 2007), FCAS2 (familial cold autoinflammatory syndrome 2) (Jeru et al., 2008), and Blau syndrome (Miceli-Richard et al., 2001), respectively. Such mutations are predicted to disrupt the inactive conformations of the receptors in the monomeric states, effectively lowering the threshold of inflammasome formation or receptor activation (Aksentijevich et al., 2007; Aksentijevich and Kastner, 2011). In comparison, AIM2 and IFI16 do not possess known oligomerization domains, which may be surrogated by the multivalent ligand dsDNA. Mutations in the receptors that diminish DNA binding may result in reduced oligomerization and signaling, and compromised immune response to microbial infections; whereas those that enhance DNA binding or destabilize the autoinhibited state may be linked to autoimmune disorders such as lupus and psoriasis. In this regard, the recent finding that the anti-microbial peptide LL-37 suppresses the AIM2 inflammasome activation in a psoriasis model suggests that this family of charged peptides may be exploited as anti-inflammatory agents (Dombrowski et al., 2011). In conclusion, our structural studies not only reveal the molecular basis of non-sequence specific dsDNA recognition and allow us to formulate a mechanism of the receptor autoinhibition and oligomerization, but also provide a framework for the design of therapeutic agents that modulate immune defense against infections or alleviate symptoms of autoimmune and autoinflammatory disorders.
EXPERIMENTAL PROCEDURES
A full description of the methods is in the Supplemental Information.
Protein Expression and Purification
The human AIM2 HIN (residues 144–343) and IFI16 HINb (residues 571–766) domains were cloned into a pET30a vector with a TEV cleavable N-terminal protein G β1 domain (GB1) tag or an MBP tag. Transformed BL21 (DE3) Codon Plus RIPL cells (Stratagene, Santa Clara, CA) were grown at 37 °C and then induced with 0.2 mM IPTG at 18 °C for 4 hours. Cells were lysed by sonication in buffer A (20 mM Tris-HCl, pH 8.0, 100 mM NaCl) plus 5 mM imidazole, DNase (Biomatik, Wilmington, DE) and protease inhibitors (Roche Applied Science, Indianapolis, IN). Soluble protein was purified from cell lysate by Hisprep IMAC column (GE Healthcare Bio-Sciences, Piscataway, NJ). Non-specific nucleic acid contaminants were removed by 0.1% polyethyleneimine (PEI) precipitation followed by 3M ammonium sulfate precipitation. The protein pellet was dissolved in buffer A before TEV protease cleavage. Further purification was carried out with a second IMAC column and size exclusion chromatography. The AIM2 PYD domain (residues 1–107) was purified using the same protocol as outlined above, without the PEI and ammonium sulfate precipitation steps.
Crystallization
DNA oligos were synthesized by IDT (Coralville, IA) and dissolved in buffer B (20 mM Hepes-Na, pH 7.4, 100 mM KCl and 5 mM DTT). The oligos were annealed by heating to 95 °C and slowly cooling to room temperature. Annealed dsDNA were added to protein solutions in buffer B and concentrated by centrifugal concentrators (Millipore, Billerica, MA) to 10–20 mg/ml before setting up hanging drop vapor diffusion method for crystallization. The AIM2 HIN:DNA complexes were crystallized with a well solution containing 8% PEG 1000, 100 mM KCl, 10 mM MgCl2, and 100 mM Mes-Na, pH 6.5. The IFI16 HINb:dsDNA complex was crystallized with a reservoir solution containing 20% PEG 3350, 100 mM potassium formate, and 100 mM Hepes-Na, 7.0. 20% ethylene glycol (v/v) was added to the reservoir solutions as the cryoprotectant to freeze the AIM2 HIN or IFI16 HINb crystals in liquid nitrogen for X-ray diffraction data collection.
X-ray Diffraction, Structure Determination and Refinement
X-ray diffraction data were collected at GM/CA-CAT at the Advanced Photon Source, Argonne National Laboratory (ANL). Data were processed with HKL2000 program suite (Zbyszek Otwinowski, 1997) and XDS (Kabsch, 2010). The IFI16 HINb-DNA and AIM2 HIN:DNA structures were determined by molecular replacement (MR) with Phaser (McCoy et al., 2007). A deposited IFI16 HINb structure (3B6Y) (Liao et al., 2011), and ideal dsDNA from Coot (Emsley and Cowtan, 2004) and make_na server (Lakshiminarayanan and Sasisekharan, 1970) were used as the initial search models. Structure building and refinement were carried out with Coot (Emsley and Cowtan, 2004) and Phenix (Adams et al., 2010). DNA base pair hydrogen bonding restraints from the PDBto3D restraints server (Laurberg et al., 2008) and TLS parameters generated by the TLSMD server (Painter and Merritt, 2006) or Phenix (Adams et al., 2010) were applied throughout the refinement. Twinned refinement protocols were applied for the AIM2 crystal forms I and II and IFI16 crystal form I as suggested by program Xtriage from Phenix (Adams et al., 2010). The crystal structures were validated by the Molprobity server (Chen et al., 2010) and RCSB ADIT validation server (Yang et al., 2004). Solvent accessible surface area was calculated with program Areaimol from the CCP4 suite (Lee and Richards, 1971; Potterton et al., 2003). Electrostatics surfaces were calculated with program Delphi (Honig and Nicholls, 1995) and displayed with program Pymol (Delano Scientific LLC, San Carlos, CA).
Fluorescence Polarization Assay
5'-Fluorescein (FAM) labeled (IDT, Coralville, IA) 20mer DNA oligo ODN 787 (Table S1) was dissolved in buffer A and annealed with its reverse complement ODN 788. 3 nM dsDNA were used for the AIM2 HIN and IFI16 HINb FP assays. Purified IFI16 HINb or AIM2 HIN samples were mixed with the FAM labeled dsDNA and diluted into assay buffers containing 20 mM Hepes-Na, pH 7.4 and 100 mM NaCl for the AIM2 HIN domains and 50 mM NaCl for the IFI16 HINb domains. The mixtures were then aliquoted in triplets into black 96 well plates and fluorescence polarization was measured with a Paradigm spectrometer (Molecular Devices, Sunnyvale, CA). For the inhibition assay, increasing concentrations of the AIM2 PYD domain or the KS-30 peptide was added to a mixture of 0.1 μM MBP-AIM2 HIN domain and 3 nM FAM-labeled 20mer dsDNA, and the fluorescence polarization was measured. Data were analyzed and plotted using program GraphPad Prism version 5.0, GraphPad Software, San Diego, CA.
Confocal Microscopy
Co-localization of the AIM2 HIN domains with DNA was studied with HEK293T cells and immortalized AIM2 deficient macrophages. Wild-type or mutant HA-AIM2-HIN constructs were transfected into HEK293T cells and the cells were stained with Alexa647-labeled anti-HA antibody (red) and DAPI (4,6-diamidino-2-phenylindole, blue). AIM2 deficient immortalized macrophages (Rathinam et al., 2010; Hornung et al., 2008) were transfected with mCerulean-fused AIM2 HIN domains (green) and FITC-labeled dsDNA (red). The ratio of the number of the dsDNA specks that colocalized with the AIM2 HIN domain to the total number of dsDNA specks was determined after setting a threshold on the fluorescence intensity of the AIM2 HIN mCerulean based on the average intensity of the cells.
Reconstitution of The AIM2 Inflammasomes
Reconstitution of the AIM2 inflammasome was carried out as previously described (Hornung et al., 2009), using HA-tagged AIM2 full-length expression constructs, Guassia-luciferase-FLAG tagged pro-IL-1β (~54 kDa), procaspase-1 and ASC expression constructs transfected into HEK293T cells. Cell lysates were probed with mouse anti-IL-1β monoclonal antibody (Clone 3ZD, National Cancer Institute, NIH). The expression levels for the AIM2 proteins were probed with anti-HA antibody (Roche Applied Science, Indianapolis, IN) and anti-AIM2 antibody (R. Johnstone, University of Melbourne, Australia).
Luciferase Reporter Gene Assay
Luciferase reporter gene assays were performed with HEK293T cells seeded in 96-well pates and transfected with firefly reporter construct under the control of the IFN-β promoter, GL3-Renilla control plasmid, STING expression vector and IFI16 expression vector. Cell lysates were assayed for the firefly luciferase activity and normalized to the renilla luciferase activity. The expression levels of the wild type or mutant full-length IFI16 were measured by immunoblotting with anti-HA antibody (Sigma, St. Louis, MO) 48 hours after transfection.
MBP Pull Down Assay
Purified AIM2 HIN domain and wild type or mutant MBP-AIM2-PYD domain (containing mutations of E7A, L11A, D15A, D19A and E20A) were mixed in buffer A with amylose beads (New England Biolabs, Inc., Ipswich MA) and rocked at room temperature for 2 hours. The beads were pelleted and washed twice with buffer A and the bound protein was eluted with the 25 mM maltose. To test the effects of the dsDNA ligand on the HIN:PYD domain interaction, a 19mer dsDNA (annealed from ODN 736 and ODN 737) was added to the HIN:PYD mixture upon binding to the amylose beads. Purified MBP protein was used as a control for non-specific interactions between the HIN domain and either the MBP tag or the amylose beads.
Stimulation of IL-1β Secretion with dsDNA
Stimulation of IL-1β secretion was carried out with human peripheral blood mononuclear cells (PBMCs) and dsDNA of various length. Cells were primed with 200 pg/ml LPS for 3 hours and transfected with indicated nucleic acids. ATP (5 mM) or monosodium urate (MSU, 250 μg/ml) was used as positive controls. 6 hours after stimulation supernatants were collected and assayed for IL-1β using ELISA. Data from one representative experiment out of two independent experiments (mean values + SEM) are depicted.
Modeling of The AIM2 Inflammasome
Construction of a hypothetical AIM2 inflammasome model was accomplished through sequential docking of the AIM2 PYD model and then the full-length ASC structure (de Alba, 2009) onto the HIN:DNA structures from the AIM2 crystal form II using program HEX (Ritchie et al., 2008). This DNA:AIM2:ASC model with a dsDNA footprint of ~16 bp was then propagated along a dsDNA staircase five times. The spacing among these five copies were chosen so they are well separated without steric clashes, but was otherwise arbitrary. The total footprint of the AIM2:ASC complexes at the dsDNA is ~100 bp. The resulting model was subjected to energy minimization using the relax mode of the Rosetta program (v3.2) (Kuhlman et al., 2003).
Supplementary Material
Highlights
Electrostatic attraction underlies innate dsDNA recognition by the HIN domains
Both OB folds and the linker between them engage the dsDNA backbone
An autoinhibited state of AIM2 is activated by DNA that liberates the PYD domain
DNA serves as an oligomerization platform for the inflammasome assembly
ACKNOWLEDGEMENTS
We thank the beam line scientists at the Argonne National Laboratory GM/CA-CAT (Argonne, IL) and the Brookhaven National Laboratory (Upton, NY) for their support on data collection. The authors would like to thank Abubakar Mian from LI, NIAID for technical support. We thank Ronald Schwartz, Michael Lenardo, and David Margulies for helpful discussions. We thank D. Eric Anderson at the Mass Spectrometry facility of NIDDK for technical support. T.S.X. is supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH. A.G.B. and L.U. are supported by Science Foundation Ireland (07/IN1/B934). K.A.F is supported by NIH grants AI083713 and AI067497. E.L. is supported by NIH grant AI067497. V.H. is supported by the European Research Council (ERC- 2009- StG 243046) and the DFG (SFB704 and SFB670). R.W.J. is a Principal Research Fellow of the National Health and Medical Research Council of Australia (NHMRC) and supported by NHMRC Program and Project Grants, the Susan G. Komen Breast Cancer Foundation, the Prostate Cancer Foundation of Australia, Cancer Council Victoria, The Leukemia Foundation of Australia, Victorian Breast Cancer Research Consortium and Victorian Cancer Agency.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS The PDB accession codes for the AIM2 HIN:DNA and IFI16 HINb:DNA structures listed in Table S2 are 3RN2, 3RN5, 3RNU, 3RLO, and 3RLN.
V.A.R. is supported by a NERCE Postdoctoral fellowship. The authors declare no financial conflict of interest.
REFERENCES
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nature immunology. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksentijevich I, C DP, Remmers EF, Mueller JL, Le J, Kolodner RD, Moak Z, Chuang M, Austin F, Goldbach-Mansky R, et al. The clinical continuum of cryopyrinopathies: novel CIAS1 mutations in North American patients and a new cryopyrin model. Arthritis Rheum. 2007;56:1273–1285. doi: 10.1002/art.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksentijevich I, Kastner DL. Genetics of monogenic autoinflammatory diseases: past successes, future challenges. Nat Rev Rheumatol. 2011;7:469–478. doi: 10.1038/nrrheum.2011.94. [DOI] [PubMed] [Google Scholar]
- Albrecht M, Choubey D, Lengauer T. The HIN domain of IFI-200 proteins consists of two OB folds. Biochem Biophys Res Commun. 2005;327:679–687. doi: 10.1016/j.bbrc.2004.12.056. [DOI] [PubMed] [Google Scholar]
- Baroudy BM, Moss B. Sequence homologies of diverse length tandem repetitions near ends of vaccinia virus genome suggest unequal crossing over. Nucleic Acids Res. 1982;10:5673–5679. doi: 10.1093/nar/10.18.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choubey D, Deka R, Ho SM. Interferon-inducible IFI16 protein in human cancers and autoimmune diseases. Front Biosci. 2008;13:598–608. doi: 10.2741/2705. [DOI] [PubMed] [Google Scholar]
- Davis BK, Wen H, Ting JP. The Inflammasome NLRs in Immunity, Inflammation, and Associated Diseases. Annu Rev Immunol. 2011;29:707–735. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alba E. Structure and interdomain dynamics of apoptosis-associated speck-like protein containing a CARD (ASC) J Biol Chem. 2009;284:32932–32941. doi: 10.1074/jbc.M109.024273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski Y, Peric M, Koglin S, Kammerbauer C, Goss C, Anz D, Simanski M, Glaser R, Harder J, Hornung V, et al. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci Transl Med. 2011;3:82ra38. doi: 10.1126/scitranslmed.3002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubau D, Rehwinkel J, Reis e Sousa C. PYHIN proteins: center stage in DNA sensing. Nat Immunol. 2010;11:984–986. doi: 10.1038/ni1110-984. [DOI] [PubMed] [Google Scholar]
- Harrison SC. A structural taxonomy of DNA-binding domains. Nature. 1991;353:715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- Hu Y, Ding L, Spencer DM, Nunez G. WD-40 repeat region regulates Apaf-1 self-association and procaspase-9 activation. The Journal of biological chemistry. 1998;273:33489–33494. doi: 10.1074/jbc.273.50.33489. [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeru I, Duquesnoy P, Fernandes-Alnemri T, Cochet E, Yu JW, Lackmy-Port-Lis M, Grimprel E, Landman-Parker J, Hentgen V, Marlin S, et al. Mutations in NALP12 cause hereditary periodic fever syndromes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1614–1619. doi: 10.1073/pnas.0708616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Patel SS, Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O'Rourke K, Chan S, Dong J, Qu Y, Roose-Girma M, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci U S A. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell host & microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kis-Toth K, Szanto A, Thai TH, Tsokos GC. Cytosolic DNA-Activated Human Dendritic Cells Are Potent Activators of the Adaptive Immune Response. J Immunol. 2011;187:1222–1234. doi: 10.4049/jimmunol.1100469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural Basis for the Activation of Innate Immune Pattern-Recognition Receptor RIG-I by Viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Lakshiminarayanan AV, Sasisekharan V. Stereochemistry of nucleic acids and polynucleotides. II. Allowed conformations of the monomer unit for different ribose puckerings. Biochim Biophys Acta. 1970;204:49–59. doi: 10.1016/0005-2787(70)90489-2. [DOI] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Liao JC, Lam R, Brazda V, Duan S, Ravichandran M, Ma J, Xiao T, Tempel W, Zuo X, Wang YX, et al. Interferon-Inducible Protein 16: Insight into the Interaction with Tumor Suppressor p53. Structure. 2011;19:418–429. doi: 10.1016/j.str.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, Segal DM, Davies DR. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou TY, Hu F, Herr AB, Strong RK, Kao CC, Li P. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XJ, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow LE, Johnstone RW, Clarke CJ. The HIN-200 family: more than interferon-inducible genes? Exp Cell Res. 2005;308:1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural Insights into RNA Recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, Hafner R, Chamaillard M, Zouali H, Thomas G, Hugot JP. CARD15 mutations in Blau syndrome. Nature genetics. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Pufall MA, Graves BJ. Autoinhibitory domains: modular effectors of cellular regulation. Annu Rev Cell Dev Biol. 2002;18:421–462. doi: 10.1146/annurev.cellbio.18.031502.133614. [DOI] [PubMed] [Google Scholar]
- Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie DW, Kozakov D, Vajda S. Accelerating and focusing protein-protein docking correlations using multi-dimensional rotational FFT generating functions. Bioinformatics. 2008;24:1865–1873. doi: 10.1093/bioinformatics/btn334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattgen SA, Fitzgerald KA. The PYHIN protein family as mediators of host defenses. Immunol Rev. 2011;243:109–118. doi: 10.1111/j.1600-065X.2011.01053.x. [DOI] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Molecular cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeranki S, Choubey D. Systemic lupus erythematosus and increased risk to develop B cell malignancies: role of the p200-family proteins. Immunol Lett. 2010;133:1–5. doi: 10.1016/j.imlet.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T, et al. Structural and functional insights into 5'-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2004;60:1833–1839. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nature immunology. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- Zbyszek Otwinowski WM. Processing of X-ray Diffraction Data Collected in Oscillation Mode In Methods Enzymol. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nature immunology. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.