Abstract
Iron can be involved in the pathogenesis of AMD through the oxidative stress because it may catalyze the Haber–Weiss and Fenton reactions converting hydrogen peroxide to free radicals, which can induce cellular damage. We hypothesized that genetic polymorphism in genes related to iron metabolism may predispose individuals to the development of AMD and therefore we checked for an association between the g.32373708 G>A polymorphism (rs867469) of the IRP1 gene and the g.49520870 G>A (rs17483548) polymorphism of the IRP2 gene and AMD risk as well as the modulation of this association by some environmental and life-style factors. Genotypes were determined in DNA from blood of 269 AMD patients and 116 controls by the allele-specific oligonucleotide-restriction fragment length polymorphism and the polymerase chain reaction-restriction fragment length polymorphism. An association between AMD, dry and wet forms of AMD and the G/G genotype of the g.32373708 G>A-IRP1 polymorphism was found (OR 3.40, 4.15, and 2.75). On the other hand, the G/A genotype reduced the risk of AMD as well as its dry or wet form (OR 0.23, 0.21, 0.26). Moreover, the G allele of the g.49520870 G>A-IRP2 polymorphism increased the risk of the dry form of the disease (OR 1.51) and the A/A genotype and the A allele decreased such risk (OR 0.43 and 0.66). Our data suggest that the g.32373708 G>A-IRP1 and g.49520870 G>A-IRP2 polymorphisms may be associated with increased risk for AMD.
Keywords: Age-related macular degeneration, AMD, IRP-1 and -2, Gene polymorphism, Iron, Oxidative stress, Reactive oxygen species, Iron-regulatory proteins
Introduction
Age-related macular degeneration (AMD) is an eye disease that is characterized by progressive vision loss of variable severity accompanied by distinct changes in the structure of the retina, retinal pigment epithelium (RPE), Bruch’s membrane and choriocapillaris [1]. Currently, AMD is estimated to affect approximately 30 million people globally and this number is expected to triple over the next 25 years [2]. AMD ranks third among the global causes of visual impairment with a blindness prevalence of 8.7%. It is the primary cause of visual deficiency in industrialized countries [3].
Although the pathophysiology of AMD is not clearly understood, it is well accepted that both oxidative stress and genetic factors in combination with environmental and life-style factors play an important role in the pathogenesis of the disease. The most consistent major risk factor for AMD is age. The development of AMD is a slow progressive process that occurs with aging and mainly affects people over age 60 [4]. While people in middle age have about a 2% risk of getting AMD, the risk increased to nearly 30% in those over age 75 [5]. Also, many other risk factors, including smoking, female sex, obesity, atherosclerosis, white race, exposure to sunlight and consumption of a non-balanced diet, have been proposed for AMD development [6, 7]. Results of several studies have shown that AMD affects Caucasians more than other populations [8–10].
Age-related disorders, including retinal diseases may result from cumulative oxidative damage resulting from reactive oxygen species (ROS) [11–13]. The macula is a source of high metabolic activity and is therefore exposed to high levels of ROS. With age, the balance between the production of ROS and local antioxidant levels can be disturbed, resulting in oxidative damage to outer segments of photoreceptors and lead to a progressive deterioration of the RPE [14]. One source of ROS generation is iron. Iron may catalyze the Haber–Weiss and Fenton reactions converting hydrogen peroxide to free radicals, which can cause oxidative tissue damage. AMD may be exacerbated by retinal iron overload and histopathology of eyes with macular degeneration has shown elevated levels of iron in RPE, Bruch’s membrane and within drusen [15, 16]. Moreover, the concentration of retinal iron increases with age [17]. Iron homeostasis is controlled by the regulation of the expression of iron-regulatory proteins (IRPs), which can bind iron-responsive elements (IREs) on the mRNA of target proteins.
A number of epidemiological studies have implicated AMD as an inherited disease showing that family members are at an increased risk of the disease [18–20]. Considerable evidence in family, twin and sibling studies exists, suggesting a genetic basis of AMD. Twin studies have demonstrated that genetic factors contribute to 46–71% of the overall variation in severity of macular degeneration. When one or both twins have AMD, concordance for AMD is 55% among monozygotic twins and 25% among dizygotic twins [21]. Several family studies have shown that patients with a positive family history of AMD are at increased risk of this disease [22–24]. Interestingly, Luo et al. [25] identified 4,764 patients with AMD and examined familial aggregation and risk of AMD in a Utah population using a population-based case–control study. The results showed that the population-attributable risk for AMD was calculated to be 0.34. Recurrence risks in relatives indicate increased relative risk in siblings (2.95), first cousins (1.29), second cousins (1.13), and parents (5.66) of affected individuals.
In recent years, the role of genetic factors in AMD development has been extensively investigated. Numerous case–control studies have confirmed the association between single nucleotide polymorphisms (SNPs) and AMD [26–28]. We hypothesized that genetic polymorphisms in the IRPs genes may be associated with development of AMD. To test our hypothesis, we evaluated the association between the g.32373708 G>A polymorphism (rs867469) of the IRP1 gene and the g.49520870 G>A (rs17483548) polymorphism of the IRP2 gene and AMD as well as the modulation of this association by some environmental and life-style factors. Both polymorphisms are SNPs with a minor allele frequency >3% in European population and are located in the 5′ flanking region of these genes. Polymorphism in this region can affect mRNA stability and degradation, and gene expression. [29].
Materials and methods
Study subjects and data collection
This case–control study included a total of 270 patients with AMD and 116 disease-free control subjects. Among AMD patients, 100 had dry AMD and the remaining 170—wet form of this disease. Eight patients with the dry form of the disease had geographic atrophy. The control subjects had no clinical evidence of AMD after undergoing the same comprehensive ophthalmic examination that was performed to confirm AMD in the patient group. Medical history was obtained from all subjects and no one reported current or previous cancer or any genetic disease. All patients and controls were examined in the Department of Ophthalmology, Medical University of Warsaw (Warsaw, Poland). They underwent ophthalmic examination, including best-corrected visual acuity, intraocular pressure, slit lamp examination, and fundus examination, performed with a slit lamp equipped with either non-contact or contact fundus lenses. Diagnosis of AMD was confirmed by optical coherence tomography (OCT) and, in some cases, by fluorescein angiography (FA) and indocyanin green angiography (ICG). OCT evaluated retinal thickness, the presence of RPE atrophy, drusen, or subretinal fluid and intraretinal edema; angiography assessed the anatomical status of the retinal vessels, the presence of choroidal neovascularization and leakage. The OCT examinations were performed with Stratus OCT model 3000, software version 4.0 (Oberkochen, Germany). The FA and ICG examinations were performed with a Topcon TRC-50I IX fundus camera equipped with the digital Image Net image system, version 2.14 (Topcon, Tokyo, Japan). An informed written consent was signed by all participants and the study design was approved by the Bioethics Committee of the Medical University of Warsaw. A structured questionnaire was used to determine demographic characteristics, family history of AMD (first-degree relatives), living environment (rural or urban areas) and life-style, including smoking and alcohol use. Each subject donated a venous blood sample of ~5 ml, 250 μl of which was used for genomic DNA extraction. Blood samples of all patients and controls were collected into ethylenediaminetetraacetic acid (EDTA) tubes and stored at −20°C until further use. All samples were coded after collection of blood and completing the questionnaire. Data on disease status, sex, age, smoking history, living environmental, and family history of AMD of the subjects enrolled in this study are presented in Table 1.
Table 1.
Characteristics of AMD patients and AMD-free control subjects enrolled in the study
| Characteristics | Controls | AMD | p | ||
|---|---|---|---|---|---|
| Number | Frequency | Number | Frequency | ||
| Disease status | 116 | ||||
| No AMD | |||||
| Dry AMD | 100 | 0.37 | |||
| Wet AMD | 170 | 0.63 | |||
| Sex | |||||
| Females | 82 | 0.78 | 179 | 0.66 | 0.0316 |
| Males | 23 | 0.22 | 92 | 0.34 | |
| Age | |||||
| ≤60 | 31 | 0.30 | 32 | 0.12 | <0.001 |
| ≥61 | 74 | 0.70 | 239 | 0.88 | |
| Mean ± SD | 68.20 ± 10.95 | 72.46 ± 8.51 | |||
| Range | 50–88 | 52–93 | |||
| Smoking | |||||
| Ever | 32 | 0.41 | 68 | 0.41 | 0.9251 |
| Never | 46 | 0.59 | 99 | 0.59 | |
| Living environment | |||||
| Rural | 32 | 0.38 | 50 | 0.30 | 0.2880 |
| Urban | 52 | 0.62 | 114 | 0.70 | |
| AMD in family | |||||
| Yes | 3 | 0.04 | 33 | 0.20 | <0.001 |
| No | 81 | 0.96 | 130 | 0.80 | |
p values for a two-sides χ2 test, p values <0.05 are in bold
SNP selection and primers design
There are many SNPs in the IRP1 and IRP2 genes listed in the dbSNP short genetic variations database in the NCBI http://www.ncbi.nlm.nih.gov/snp, but none of them has annotated any clinical significance and were not able to find any evidence for this. Our selection of polymorphisms was made on the basis of their location in the 5′ regulatory region, which usually contains the promoter. The phenotypic consequences of such DNA sequence changes may be associated with the difference in the transcription level, which can be quantitatively assessed in future research. Primers were designed using the IRP1 and IRP2 genomic sequence found at http://www.ensembl.org/Homo_sapiens/Gene/Sequence?g=ENSG00000122729;r=9:32384601-32450834 and http://www.ensembl.org/Homo_sapiens/Gene/Summary?g=ENSG00000136381;r=15:78730531-78793795;t=ENST00000258886 and Primer3Plus software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) for IRP2 SNP and Web-based allele-specific primer software (http://bioinfo.biotec.or.th/WASP/index/wasp ) for IRP1 SNP.
DNA preparation and storage
Genomic DNA was obtained from a 250 μl aliquot of blood using a commercially available AxyPrep™ Blood genomic DNA Miniprep Kit (Axygen Biosciences, Union City, CA, USA), according to the manufacturer’s instructions. Genomic DNA was directly isolated from the white blood cells. DNA purity and concentration were determined spectrophotometrically at 260 and 280 nm. Each DNA sample was stored in TE buffer (5 mM Tris–HCl, 0.1 mM EDTA, pH 8.5) at −20°C until analysis.
Genotyping
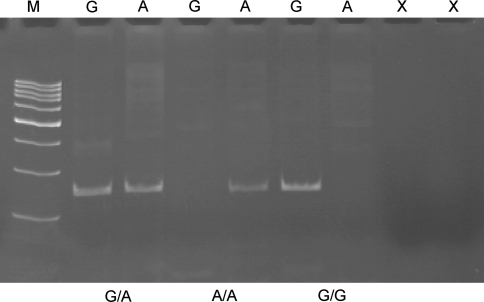
The genotypes of the g.32373708 G>A-IRP1 polymorphism were determined by the allele-specific oligonucleotide-PCR method. PCR was performed in a 25 μl reaction volume. The reaction mixture contained 50 ng of genomic DNA, 1 U Biotools DNA polymerase (Biotools, Madrid, Spain), 1 × reaction buffer [750 mM Tris–HCl, pH 9.0, 500 mM KCl, 200 mM (NH4)2SO4], 0.2 mM of each dNTP, 1.5 mM MgCl2, 0.25 μM of each primers (Metabion, Martinsried, Germany). The primers designed to detect the g.32373708 G>A SNP were as follows: allele-specific sense oligonucleotides 5′-TGCACACCTGCAAAGAAG-3′ for G variant and 5′-TGCACACCTGCAAAGAAA-3′ for A variant and antisense oligonucleotide 5′-CTAGATGAAAGGTGGTGAGG-3′. The allele-specific oligonucleotide-restriction fragment length polymorphism (ASO-PCR) conditions were as follows: 5 min of initial denaturation at 95°C, followed by 30 cycles of 30 s denaturation at 95°C, 30 s annealing at 56°C and 1 min extension at 72°C. The final extension step at 72°C for 5 min was also included. The PCR products (237 bp) were fractionated by electrophoresis on a 8% polyacrylamide gel, stained with ethidium bromide and viewed under UV light. Fig. 1 presents a representative gel from analysis of this polymorphism.
Fig. 1.
Genotypes of the g.32373708 G>A-IRP1 polymorphism (rs867469) determined by the allele-specific oligonucleotide-PCR (ASO-PCR) method and analyzed by a 8% polyacrylamide gel electrophoresis stained with ethidium bromide and viewed under UV light. Lane M displays GeneRuler ™ 100 bp molecular weight marker, lanes designated G or A show the results of amplification with primer specific to either the G or A allele, respectively and lane X shows a negative control comprising reaction mixture without target DNA. Genotypes are indicated in the lower part of the picture
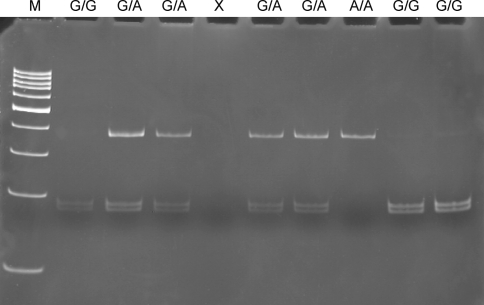
The polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method was used to determine the genotypes of the g.49520870 G>A-IRP2 polymorphism. PCR assay was performed in a total reaction volume of 25 μl containing the same chemicals as in the previous analysis except for primers. 360 bp length fragments containing polymorphic site were amplified using the following primers: sense 5′-CCCCCACTTGAAAACACG-3′ and antisense 5′-AGATCGTCGGACAGGAAAAC-3′. The PCR profile contained an initial denaturation step for 5 min at 95°C, 30 cycles at 95°C for 30 s, 30 s at 60°C annealing temperature and 60 s at 72°C and the final extension step for 5 min at 72°C. After amplification, the 360 bp PCR products were analyzed on a 3% agarose gel and digested with 3 U of BspTI (AflII) restriction endonuclease (Fermentas, Hanover, MD, USA) in a final volume of 15 μl for 16 h at 37°C. PCR products with a G at the polymorphic site were digested into two 183 and 177 bp fragments, while those with A were not because of the absence of a BspTI (AflII) restriction site. The G/G genotype produced two fragments (183, 177 bp), whereas the G/A genotype produced three fragments (360, 183, and 177 bp) and the homozygote A/A resulted in only one fragment 360 bp. Digested PCR products were separated by electrophoresis on a 8% polyacrylamide gel and visualized by ethidium bromide staining using a GeneRuler ™ 100 bp (Fermentas, Hanover, MD, USA) as a size marker. A representative gel for this polymorphism is presented in Fig. 2. All PCR amplifications were conducted in a C1000 Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA). Positive and negative (no template) controls were included in all sets. For quality control, we randomly selected 10% samples for each of the two SNPs to perform repeat assays and the results were 100% concordant.
Fig. 2.
Genotypes of the g.49520870 G>A-IRP2 polymorphism (rs17483548) determined by the PCR-RFLP detection and analyzed by a 8% polyacrylamide gel electrophoresis stained with ethidium bromide and viewed under UV light. Lane M shows GeneRuler ™ 100 bp molecular weight marker, lane X shows a negative control comprising reaction mixture without target DNA, all remaining lanes present genotypes indicated in the upper part of the picture
Statistical analysis
All statistical analyzes were performed using STATISTICA 9.0 software (Statsoft, Tulsa, OK, USA) and SigmaPlot v11.0 software (Systat Software, Inc., San Jose, CA, USA). To compare the distributions of demographic variables and selected risk factors between patients and controls Chi-square test was used. Hardy–Weinberg equilibrium (HWE) was checked using Chi-square test to compare the observed genotype frequencies with the expected frequencies among the case and control subjects. The χ2 analysis was also used to test the significance of the differences between distributions of genotypes and alleles in AMD patients and controls. The association between case–control status and each polymorphism, measured by the odds ratio (OR) and its corresponding 95% confidence interval (CI), was estimated using an unconditional multiple logistic regression model, both with and without adjustment for sex, age, smoking habit, living environment (rural versus urban areas), and family status of AMD. Smoking habit was categorized in terms of never smokers and smokers (including current and former). Stratified analysis according to age, sex, and family status of AMD was also conducted. To study a possible gene-environment interaction, the patients and controls were divided into subgroups depending on sex, family history and age. Unconditional logistic regression analyzes were also performed to assess the association between genotypes and risk for AMD after stratification of the individuals according to sex, age, and family status of AMD.
Results
Characteristics of the study population and relationship between demographic, environmental, life-style factors, and family history and the risk of AMD independent of genotype
The characteristics of the patients with AMD and AMD-free controls involved in this study are presented in Table 1. The mean ± SD age was 72.46 ± 8.51 years for the patients (range 52–93) and 68.29 ± 10.95 years for the controls (range 50–88) and 34% of the patients and 23% of the controls were men, whereas 66% of the patients and 82% of the controls were women. There were significantly more subjects with negative family history for AMD among the controls than patients (96 vs. 80%, p < 0.001). Therefore, these variables were further adjusted in the multivariate logistic regression model to control for possible confounding factors of the main effects of the polymorphisms. Moreover, we explored the relationships between age, sex, smoking, living environment and family history of AMD and the risk of AMD independently of genotype. We compared AMD patients and controls according to these parameters (Table 2). Our results suggest that male sex (OR 1.83, 95% CI 1.08–3.10; p = 0.024), age (7.42, 95% CI 2.62–21.02, p < 0.001) and first-degree relatives with AMD (OR 6.85, 95% CI 2.04–23.08; p = 0.002) significantly increased the risk of the disease.
Table 2.
Risk of AMD associated with age, sex, smoking, living environment and family history of AMD
| Characteristics | Controls | AMD | OR (95% CI) | p |
|---|---|---|---|---|
| Number (frequency) | Number (frequency) | |||
| Age | ||||
| From point | 1.05 (1.02–1.07) | <0.001 | ||
| From range | 7.42 (2.62–21.02) | <0.001 | ||
| Sex | ||||
| Females | 82 (0.78) | 179 (0.66) | 0.55 (0.32–0.92) | 0.024 |
| Males | 23 (0.22) | 92 (0.34) | 1.83 (1.08–3.10) | 0.024 |
| Smoking | ||||
| Ever (current, former) | 32 (0.41) | 68 (0.41) | 0.99 (0.57–1.71) | 0.964 |
| Never | 46 (0.59) | 99 (0.59) | 0.91 (0.53–1.58) | 0.748 |
| Living environment | ||||
| Rural | 32 (0.38) | 50 (0.30) | 0.71 (0.41–1.24) | 0.229 |
| Urban | 51 (0.62) | 114 (0.70) | 1.40 (0.81–2.44) | 0.229 |
| AMD in family | ||||
| Yes | 3 (0.04) | 33 (0.20) | 6.85 (2.04–23.08) | 0.002 |
| No | 81 (0.96) | 130 (0.80) | 0.15 (0.05–0.51) | 0.002 |
p values <0.05 along with corresponding ORs are in bold
OR odds ratio, 95% CI
Polymorphisms of the IRP1 and IRP2 genes, gene–gene interaction and AMD occurrence and progression
The genotype and allele distributions of the g.32373708 G>A-IRP1 and g.49520870 G>A-IRP2 polymorphisms in AMD patients and controls are summarized in Table 3. The observed genotype frequencies of the g.49520870 G>A-IRP2 SNP were all in agreement with the HWE calculated for the cases and controls (p > 0.05, data not shown). As shown in Table 3 the difference in the frequency distributions of genotypes of the g.32373708 G>A-IRP1 SNP between the cases (AMD, dry and wet forms of the disease) and controls was statistically significant (p < 0.05). An association between AMD, dry and wet forms of AMD and the G/G genotype of the g.32373708 G>A-IRP1 polymorphism was found (adjusted OR 3.40, 4.15, and 2.75, respectively). On the other hand, the G/A genotype reduced the risk of AMD as well as dry form of AMD or wet form of AMD, separately (adjusted OR 0.23, 0.21, and 0.26, respectively). Furthermore, in the dry form of AMD, the G allele of the g.49520870 G>A-IRP2 SNP increased the risk of the disease (OR 1.51) and the A/A genotype and the A allele decreased such risk (OR 0.43 and 0.66).
Table 3.
Distribution of genotypes, frequency of alleles of the g.32373708 G>A-IRP1 and g.49520870 G>A-IRP2 polymorphisms and odds ratio (OR) with 95% CI (95% CI) in patients with AMD, the disease in its dry and wet forms and individuals without visual disturbances (controls)
| Genotype/allele | Controls | AMD | Crude OR (95% CI) | Adjusted ORa (95% CI) | Dry AMD | Crude OR (95% CI) | Adjusted ORa (95% CI) | Wet AMD (95% CI) | Crude OR (95% CI) | Adjusted ORa (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|
| Number (frequency) | Number (frequency) | Number (frequency) | Number (frequency) | |||||||
| g.32373708 G>A, rs867469 | n = 98 | n = 269 | n = 99 | n = 170 | ||||||
| G/G | 22 (0.23) | 101 (0.38) | 2.07 (1.21–3.57) | 3.40 (1.71–6.78) | 35 (0.35) | 1.89 (1.01–1.69) | 4.15 (1.81–9.51) | 66 (0.38) | 2.19 (1.24–3.86) | 2.75 (1.31–5.76) |
| G/A | 67 (0.68) | 95 (0.35) | 0.25 (0.15–0.41) | 0.23 (0.23–0.43) | 36 (0.36) | 0.26 (0.14–0.48) | 0.21 (0.09–0.45) | 59 (0.35) | 0.25 (0.14–0.42) | 0.26 (0.13–0.52) |
| A/A | 9 (0.09) | 73 (0.27) | 3.68 (1.76–7.69) | 2.25 (0.96–5.28) | 28 (0.28) | 3.90 (1.73-8.80) | 1.97 (0.72–5.39) | 45 (0.27) | 3.56 (1.66-7.65) | 2.43 (0.97-6.13) |
| χ2 = 33.024, p < 0.0001 | χ2 = 22.047, p < 0.0001 | χ2 = 29.278, p < 0.0001 | ||||||||
| G | 172 (0.82) | 297 (0.55) | 0.94 (0.68–1.31) | 1.35 (0.89–2.05) | 106 (0.53) | 0.88 (0.59–1.31) | 0.16 (0.93–2.85) | 191 (0.56) | 0.98 (0.69–1.40) | 1.21 (0.75–1.97) |
| A | 38 (0.18) | 241 (0.45) | 1.06 (0.76–1.47) | 0.74 (0.49–1.12) | 92 (0.47) | 1.13 (0.76–1.69) | 0.62 (0.35–1.08) | 149 (0.44) | 1.01 (0.71–1.45) | 0.82 (0.51–1.33) |
| g.49520870 G>A, rs17483548 | n = 116 | n = 263 | n = 100 | n = 163 | ||||||
| G/G | 40 (0.34) | 107 (0.41) | 1.30 (0.83–2.05) | 0.93 (0.52–1.65) | 44 (0.44) | 1.50 (0.86–2.60) | 1.24 (0.61–2.53) | 63 (0.38) | 1.20 (0.73–1.97) | 0.81 (0.42–1.58) |
| G/A | 52 (0.45) | 121 (0.46) | 1.05 (0.68–1.63) | 1.16 (0.66–2.02) | 46 (0.46) | 1.05 (0.61–1.80) | 1.05 (0.52–2.11) | 75 (0.46) | 1.05 (0.65–1.70) | 1.21 (0.65–2.28) |
| A/A | 24 (0.13) | 35 (0.13) | 0.59 (0.33–1.04) | 0.87 (0.42–1.84) | 10 (0.10) | 0.43 (0.19–0.94) | 0.59 (0.21–1.65) | 25 (0.15) | 0.69 (0.37–1.29) | 0.99 (0.43–2.26) |
| χ2 = 3.640, p = 0.1620 | χ2 = 5.166, p = 0.0756 | χ2 = 1.445, p = 0.4855 | ||||||||
| G | 132 (0.57) | 335 (0.64) | 1.33 (0.97–1.81) | 1.00 (0.67–1.56) | 134 (0.67) | 1.51 (1.02–2.24) | 1.27 (0.77–2.11) | 201 (0.62) | 1.22 (0.87–1.70) | 0.91 (0.58–1.43) |
| A | 100 (0.43) | 191 (0.36) | 0.76 (0.56–1.04) | 0.99 (0.67–1.50) | 66 (0.33) | 0.66 (0.44–0.97) | 0.78 (0.47–1.30) | 125 (0.38) | 0.83 (0.59–1.15) | 1.10 (0.70–1.71) |
p values for a two-sides χ2 test, significant p and ORs are in bold
aOR adjusted for age, sex, AMD in family
–, not estimated
We also evaluated the associations between the occurrence of AMD and combined genotypes of the g.32373708 G>A-IRP1 and g.49520870 G>A-IRP2 polymorphisms. The distribution of combined genotypes of these polymorphisms is shown in Table 4.
Table 4.
Distribution of combined genotypes of the g.32373708 G>A-IRP1 and g.49520870 G>A-IRP2 polymorphisms and odds ratio (OR) with 95% confidence interval (95% CI) in patients with AMD as well as its dry and wet forms and individuals without visual disturbances (controls)
| Genotype | Controls (n = 79) | AMD (n = 254) | Crude OR (95% CI) | Adjusted ORa (95% CI) | Dry AMD (n = 99) | Crude OR (95% CI) | Adjusted ORa (95% CI) | Wet AMD (n = 155) | Crude OR (95% CI) | Adjusted ORa (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|
| Number (frequency) | Number (frequency) | Number (frequency) | Number (frequency) | |||||||
| G/G–G/G | 6 (0.07) | 38 (0.15) | 2.14 (0.87–5.27) | 2.94 (0.91–9.55) | 17 (0.17) | 2.52 (0.94–6.74) | 4.46 (1.21–16.41) | 21 (0.13) | 1.90 (0.74–4.91) | 2.03 (0.55–7.56) |
| G/G–G/A | 5 (0.06) | 45 (0.18) | 3.19 (1.22–8.34) | 3.52 (1.20–10.30) | 14 (0.14) | 2.44 (0.84–7.09) | 3.22 (0.91–11.40) | 31 (0.20) | 3.70 (1.38–9.93) | 3.05 (1.01–9.37) |
| G/G–A/A | 7 (0.09) | 14 (0.05) | 0.60 (0.23–1.54) | 1.05 (0.30–3.67) | 4 (0.04) | 0.43 (0.12–1.54) | 1.14 (0.26–5.08) | 10 (0.06) | 0.71 (0.26–1.94) | 1.01 (0.25–4.07) |
| G/A–G/G | 25 (0.32) | 36 (0.14) | 0.36 (0.21–0.64) | 0.23 (0.10–0.51) | 15 (0.15) | 0.39 (0.19–0.80) | 0.26 (0.09–0.68) | 21 (0.13) | 0.34 (0.17–0.65) | 0.25 (0.11–0.61) |
| G/A–G/A | 23 (0.30) | 40 (0.16) | 0.46 (0.25–0.82) | 0.48 (0.22–1.04) | 17 (0.17) | 0.50 (0.25–1.03) | 0.51 (0.19–1.36) | 23 (0.15) | 0.42 (0.22–0.82) | 0.51 (0.21–23) |
| G/A–A/A | 7 (0.09) | 12 (0.05) | 0.51 (0.20–1.34) | 1.25 (0.37–4.15) | 4 (0.04) | 0.43 (0.12–1.53) | 0.75 (0.15–3.82) | 8 (0.05) | 0.56 (0.19–1.60) | 1.55 (0.42–5.75) |
| A/A–G/G | 3 (0.04) | 31 (0.12) | 3.52 (1.05–11.85) | 2.38 (0.51–11.16) | 12 (0.12) | 3.49 (0.95–12.84) | 2.48 (0.46–13.24) | 19 (0.12) | 3.54 (1.01–12.35) | 2.40 (0.47–12.40) |
| A/A–G/A | 2 (0.02) | 31 (0.12) | 5.35 (1.25–22.90) | 3.78 (0.81–17.60) | 14 (0.14) | 6.34 (1.39–28.80) | 3.12 (0.55–17.55) | 17 (0.11) | 4.74 (1.06–21.07) | 4.30 (0.86–21.06) |
| A/A–A/A | 1 (0.01) | 7 (0.03) | 2.21 (0.27–18.25) | 0.23 (0.01–6.41) | 2 (0.02) | 1.61 (0.14–18.01) | – | 5 (0.03) | 2.60 (0.30–22.64) | 0.45 (0.02–11.61) |
Significant ORs are in bold; –, not estimated
aOR adjusted for age, sex, AMD in family
The presence of the A/A–G/A genotype of both polymorphisms increased the risk of AMD as well as its dry or wet form, separately (OR 5.35, 6.34, and 4.74, respectively), whereas the presence of the G/A–G/G genotype decreased such risk (OR 0.36, 0.39, and 0.34, respectively).
Furthermore, the G/A–G/A genotype may have a protective effect against wet AMD and AMD (OR 0.46 and 0.42, respectively). In the wet form of AMD and AMD, the G/G–G/A and A/A–G/G genotypes increased the risk of the disease (OR 3.70, 3.54, 3.19, and 3.52, respectively).
Additionally, we examined the distribution of genotypes/combined genotypes and alleles of the g.32373708 G>A-IRP1 and g.49520870 G>A-IRP2 polymorphisms in the group of wet AMD patients in comparison with dry AMD patients (data not shown). Such comparison was considered as a measure of AMD progression. We did not find any association between the progression of AMD and genotypes/combined genotypes and alleles of both polymorphisms.
Stratification and interaction analysis of IRP1 and IRP2 genotypes and the risk of AMD
The association between AMD and the tested SNPs was subjected to stratification analysis, but only data for the g.32373708 G>A-IRP1 are presented in Table 5 because such analysis for other polymorphism did not yield any significant result. In stratification analysis, the G/G genotype was associated with a significantly increased risk of AMD in subgroups of subjects of 61 years and older (adjusted OR 2.44) and women (adjusted OR 3.54). On the other hand, the G/A genotype reduced the risk of AMD in subgroups of subjects older than 61 years (adjusted OR 0.26), women (adjusted OR 0.20) and smokers (adjusted OR 0.19). Moreover, the occurrence of AMD was correlated with the presence of the A/A genotype in women (adjusted OR 2.87).
Table 5.
Distribution of genotypes of the g.32373708 G>A-IRP1 polymorphism stratified by age, sex and smokers in patients with dry AMD as compared with individuals without visual disturbances (controls)
| Genotype/allele | Controls | AMD | Adjusted ORa | p |
|---|---|---|---|---|
| Number (frequency) | Number (frequency) | |||
| Age >61 | n = 67 | n = 235 | ||
| G/G | 15 (0.22) | 88 (0.37) | 2.44 (1.13–5.25) | 0.022 |
| G/A | 46 (0.69) | 86 (0.37) | 0.26 (0.13–0.53) | <0.001 |
| A/A | 6 (0.09) | 61 (0.26) | 2.87 (1.03–7.95) | 0.043 |
| G | 76 (0.57) | 262 (0.56) | 1.09 (0.69–1.73) | 0.699 |
| A | 58 (0.43) | 208 (0.44) | 0.91 (0.57–1.44) | 0.699 |
| Women | n = 77 | n = 173 | ||
| G/G | 19 (0.25) | 68 (0.39) | 3.54 (1.60–7.84) | 0.002 |
| G/A | 52 (0.67) | 60 (0.35) | 0.20 (0.11–0.42) | <0.001 |
| A/A | 6 (0.08) | 45 (0.26) | 2.76 (0.95–7.98) | 0.060 |
| G | 90 (0.58) | 196 (0.57) | 1.36 (0.82–2.24) | 0.227 |
| A | 64 (0.42) | 150 (0.43) | 0.73 (0.44–1.21) | 0.227 |
| Smokers | n = 31 | n = 68 | ||
| G/G | 2 (0.06) | 24 (0.36) | 2.58 (0.77–8.59) | 0.122 |
| G/A | 24 (0.77) | 22 (0.32) | 0.19 (0.06–0.58) | 0.003 |
| A/A | 5 (0.16) | 22 (0.32) | 4.73 (0.94–23.69) | 0.059 |
| G | 34 (0.55) | 70 (0.48) | 0.94 (0.46–1.92) | 0.883 |
| A | 28 (0.45) | 66 (0.52) | 1.05 (0.51–2.14) | 0.883 |
p values <0.05 along with corresponding ORs are in bold
aOR adjusted for age, sex and smoking
Discussion
We showed that age, sex, and family status of AMD were significant risk factors in the development of AMD, which is in agreement with previous results obtained by others [30, 31]. However, we did not find any association between smoking and AMD. At present we have not any direct explanation of this fact except that smoking may be one of the factor of AMD pathogenesis and not the sole reason of AMD and the contribution of this factor depends on the population [32]. Smoking status in our questionnaire was defined as a smoker or non-smoker without taking into account passive smoking. It was shown that passive smoking is associated with an almost twofold increase in the risk of AMD for non-smokers having lived with smokers for 5 years or more. This might explain, at least in part, our apparently controversial data on association between AMD and smoking [33]. Therefore, more detailed characteristics of the population enrolled in the present study might have shed on this apparent lack of agreement with the established point of view.
Although the pathogenesis of AMD is not completely understood, a growing body of evidence suggests that oxidative stress and free radical damage may mediate or exacerbate macular degeneration [34]. Oxidative damage is implicated in several retinal diseases, including retinal degeneration and AMD [35–37]. In a large clinical trial, patients with dry AMD given dietary supplements of antioxidants and zinc had reduced progression to advanced AMD, suggesting that oxidative stress is somehow involved in its pathogenesis [38]. Iron is suggested as a potential source of oxidative radicals associated with degenerative processes affecting the central nervous system and the retina [34, 39, 40]. Hahn et al. [41] found that AMD-affected maculas had significantly increased total iron concentration compared with age-matched controls, suggesting that iron accumulation might play a role in this disease. Although iron can cause oxidative tissue damage through the Haber–Weiss and Fenton reactions, it is also an essential for several metabolic pathway. Homeostatic regulation of ferrous iron levels is critical for meeting physiologic demand while preventing the toxicity associated with iron overload [34, 42]. Several studies have confirmed that iron overload may play a crucial role in the pathogenesis of AMD [15, 34]. Balanced iron homeostasis is coordinated largely at the posttranscriptional level via the interaction of either of two IRPs with cis-regulatory RNA motifs—IREs located in the 5′ or 3′ untranslated regions (UTR) of mRNAs encoding proteins of iron uptake [transferrin receptor 1 (TfR1) and divalent metal transporter-1 (DMT-1)], export (ferroportin), utilization (5-aminolevulinate synthase) or storage (ferritin H- and L-chains) [43]. In mammals, two homologous IRPs have been identified. IRP1 also known as the cytosolic aconitase (ACO1) has two mutually exclusive functions. When iron is abundant, IRP1 assembles an Fe–S cluster and has aconitase activity, whereas—when iron is scarce, the apoprotein without its Fe–S cluster acquires IRE-binding activity [44]. IRP2 also called IREB2 (iron-responsive element binding protein 2) shares 79% homology with IRP1 but lacks aconitase activity [45]. Mice with combined total and constitutive deficiency of both IRPs show embryonic lethality, suggesting that the IRPs are fundamental for life [46–48]. In both iron deficiency and excess, IRP-mediated regulation rapidly restores the physiological cytosolic iron level. Under low-iron conditions, IRPs binds to IREs present in the 5′ UTR of mRNAs, such as in ferritin heavy and light chains and inhibits translation and concurrently IRPs bind to mRNAs containing IREs in the 3′ UTR, such as TfR1 and DMT1, resulting in increasing RNA’s stability and levels. And vice versa, under iron high conditions, IRP binding activity to IREs is reduced. Thus, under iron-poor condition, IRPs upregulate genes to increase iron uptake and under iron rich condition, IREs upregulate genes that are required for iron storage [49–51]. The importance of normal iron homeostasis is underlined by the fact that retinal dysfunction has been observed in some pathological conditions due to the lack of iron or an excess of iron [52–54].
Some results demonstrate that multiple genes and proteins are involved in development of AMD. AMD can be genetically associated with multiple susceptibility loci, i.a. 1q32 (complement factor H, CFH), 10q26 (ARMS2), 6p21.3 (complement factor B, BF; complement component 2, C2), 19p13.3–13.2 (complement component 3, C3) [55–61]. As mentioned, both genetic predispositions and environmental factors, such as smoking and ultraviolet rays, play important roles in pathogenesis of AMD. Interestingly, hereditary diseases of iron overload are caused by mutations in genes of both regulation and usage [62, 63]. Furthermore, there is a growing body of evidence that SNPs associated with inflammation, oxidative stress, angiogenesis and other pathological processes have been linked to AMD [61]. Because iron overload has been implicated in AMD, IRPs are a strong potential candidate for polymorphisms that could play a role in AMD. We assessed whether the g.32373708 G>A polymorphism in the IRP1 gene and the g.49520870 G>A polymorphism in the IRP2 gene increase the risk of AMD. To our knowledge, the g.32373708 G>A-IRP1 polymorphism and the g.49520870 G>A-IRP2 polymorphism have not been studied in AMD patients so far. We observed, for the first time, that the occurrence of AMD was positively correlated with the presence of the G/G genotype of the IRP1 SNP and of the G allele of the IRP2 SNP.
In conclusion, our work shows that genetic polymorphisms of the IRPs genes may be associated with development of AMD. These findings may be helpful in increasing our understanding of the etiology of AMD. Identification of genetic risk factors for AMD is the first step towards earlier detection and prevention, and in the future, better treatments.
Acknowledgments
This study was supported by the Grant number N N402 248336 of Ministry of Science and Higher Education.
Conflict of interest
The authors declare that there is no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
References
- 1.Meyer KJ, Davis LK, Schindler EI, Beck JS, Rudd DS, Grundstad AJ, Scheetz TE, Braun TA, Fingert JH, Alward WL, Kwon YH, Folk JC, Russell SR, Wassink TH, Stone EM, Sheffield VC. Genome-wide analysis of copy number variants in age-related macular degeneration. Hum Genet. 2010;129:91–100. doi: 10.1007/s00439-010-0904-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clemons TE, Milton RC, Klein R, Seddon JM, Ferris FL., 3rd Age-related eye disease study research group. Risk factors for the incidence of advanced age-related macular degeneration in the age-related eye disease study (AREDS) AREDS report no. 19. Ophthalmology. 2005;112:533–539. doi: 10.1016/j.ophtha.2004.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO). Priority eye diseases.http://www.who.int/blindness/causes/priority/en/index8.html. Accessed October 2009
- 4.Gehrs KM, Anderson DH, Johnson LV, Hageman GS. Age-related macular degeneration emerging pathogenetic and therapeutic concepts. Ann Med. 2006;38:450–471. doi: 10.1080/07853890600946724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Eye Institute, US National Institutes of Health. Facts about age-related macular degeneration. http://www.nei.nih.gov/health/maculardegen/armd_facts.asp. Accessed August 2010
- 6.Coleman HR, Chan CC, Ferris FL, 3rd, Chew EY. Age-related macular degeneration. Lancet. 2008;372:1835–1845. doi: 10.1016/S0140-6736(08)61759-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jager RD, Mieler WF, Miller JW. Age-related macular degeneration. N Engl J Med. 2008;358:2606–2617. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- 8.Weiter JJ, Delori FC, Wing GL, Fitch KA. Retinal pigment epithelial lipofuscin and melanin and choroidal melanin in human eyes. Invest Ophthalmol Vis Sci. 1986;27:145–152. [PubMed] [Google Scholar]
- 9.Kawasaki R, Wang JJ, Ji GJ, Taylor B, Oizumi T, Daimon M, Kato T, Kawata S, Kayama T, Tano Y, Mitchell P, Yamashita H, Wong TY. Prevalence and risk factors for age-related macular degeneration in an adult Japanese population: the Funagata study. Ophthalmology. 2008;115:1376–1381. doi: 10.1016/j.ophtha.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Ayala-Haedo JA, Gallins PJ, Whitehead PL, Schwartz SG, Kovach JL, Postel EA, Agarwal A, Wang G, Haines JL, Pericak-Vance MA, Scott WK. Analysis of single nucleotide polymorphisms in the NOS2A gene and interaction with smoking in age-related macular degeneration. Ann Hum Genet. 2010;74:195–201. doi: 10.1111/j.1469-1809.2010.00570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCall MR, Frei B. Can antioxidant vitamins materially reduce oxidative damage in humans? Free Radic Biol Med. 1999;26:1034–1053. doi: 10.1016/S0891-5849(98)00302-5. [DOI] [PubMed] [Google Scholar]
- 12.Ohia SE, Opere CA, Leday AM. Pharmacological consequences of oxidative stress in ocular tissues. Mutat Res. 2005;579:22–36. doi: 10.1016/j.mrfmmm.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 13.Khandhadia S, Lotery A. Oxidation and age-related macular degeneration: insights from molecular biology. Expert Rev Mol Med. 2010;12:e34. doi: 10.1017/S146239941000164X. [DOI] [PubMed] [Google Scholar]
- 14.Totan Y, Yağci R, Bardak Y, Ozyurt H, Kendir F, Yilmaz G, Sahin S, Sahin Tiğ U (2009) Oxidative macromolecular damage in age-related macular degeneration. Curr Eye Res 34:1089–1093 [DOI] [PubMed]
- 15.Wong RW, Richa DC, Hahn P, Green WR, Dunaief JL. Iron toxicity as a potential factor in AMD. Retina. 2007;27:997–1003. doi: 10.1097/IAE.0b013e318074c290. [DOI] [PubMed] [Google Scholar]
- 16.Blasiak J, Szaflik J, Szaflik JP. Implications of altered iron homeostasis for age-related macular degeneration. Front Biosci. 2011;16:1551–1559. doi: 10.2741/3804. [DOI] [PubMed] [Google Scholar]
- 17.Hahn P, Ying GS, Beard J, Dunaief JL. Iron levels in human retina: sex difference and increase with age. Neuroreport. 2006;17:1803–1806. doi: 10.1097/WNR.0b013e3280107776. [DOI] [PubMed] [Google Scholar]
- 18.Smith W, Mitchell P. Family history and age-related maculopathy: the blue mountains eye study. Aust N Z J Ophthalmol. 1998;26:203–206. doi: 10.1111/j.1442-9071.1998.tb01311.x. [DOI] [PubMed] [Google Scholar]
- 19.Hyman L, Neborsky R. Risk factors for age-related macular degeneration: an update. Curr Opin Ophthalmol. 2002;13:171–175. doi: 10.1097/00055735-200206000-00007. [DOI] [PubMed] [Google Scholar]
- 20.Scholl HP, Fleckenstein M, Charbel IP, Keilhauer C, Holz FG, Weber BH (2007) An update on the genetics of age-related macular degeneration. Mol Vis 13:196–205 [PMC free article] [PubMed]
- 21.Seddon JM, Cote J, Page WF, Aggen SH, Neale MC. The US twin study of age-related macular degeneration: relative roles of genetic and environmental influences. Arch Ophthalmol. 2005;123:321–327. doi: 10.1001/archopht.123.3.321. [DOI] [PubMed] [Google Scholar]
- 22.Klaver CC, Wolfs RC, Assink JJ, van Duijn CM, Hofman A, de Jong PT. Genetic risk of age-related maculopathy population-based familial aggregation study. Arch Ophthalmol. 1998;116:1646–1651. doi: 10.1001/archopht.116.12.1646. [DOI] [PubMed] [Google Scholar]
- 23.Seddon JM, Francis PJ, George S, Schultz DW, Rosner B, Klein ML. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA. 2007;297:1793–1800. doi: 10.1001/jama.297.16.1793. [DOI] [PubMed] [Google Scholar]
- 24.Ting AY, Lee TK, MacDonald IM. Genetics of age-related macular degeneration. Curr Opin Ophthalmol. 2009;20:369–376. doi: 10.1097/ICU.0b013e32832f8016. [DOI] [PubMed] [Google Scholar]
- 25.Luo L, Harmon J, Yang X, Chen H, Patel S, Mineau G, Yang Z, Constantine R, Buehler J, Kaminoh Y, Ma X, Wong TY, Zhang M, Zhang K. Familial aggregation of age-related macular degeneration in the Utah population. Vis Res. 2008;48:494–500. doi: 10.1016/j.visres.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Janik-Papis K, Zaras M, Krzyzanowska A, Wozniak K, Blasiak J, Szaflik J, Szaflik JP. Association between vascular endothelial growth factor gene polymorphisms and age-related macular degeneration in a Polish population. Exp Mol Pathol. 2009;87(3):234–238. doi: 10.1016/j.yexmp.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Fuse N, Mengkegale M, Miyazawa A, Abe T, Nakazawa T, Wakusawa R, Nishida K. Polymorphisms in ARMS2 (LOC387715) and LOXL1 genes in the Japanese with age-related macular degeneration. Am J Ophthalmol. 2011;151:550–556. doi: 10.1016/j.ajo.2010.08.048. [DOI] [PubMed] [Google Scholar]
- 28.Güven M, Görgün E, Unal M, Yenerel M, Batar B, Küçümen B, Dinç UA, Güven GS, Ulus T, Yüksel A. Glutathione S-transferase M1, GSTT1 and GSTP1 genetic polymorphisms and the risk of age-related macular degeneration. Ophthalmic Res. 2011;46:31–37. doi: 10.1159/000321940. [DOI] [PubMed] [Google Scholar]
- 29.Langaee T, Shin J. The genetics basis of pharmatogenomics. In: Zdanowicz MM, editor. Concepts in pharmacogenomics. Bethesda: American Society of Health-System Pharmacists; 2010. p. 29. [Google Scholar]
- 30.Geirsdottir A, Stefansson E, Jonasson F, Helgadottir G, Sigurdsson H. Age-related macular degeneration in very old individuals with family history. Am J Ophthalmol. 2007;143:889–890. doi: 10.1016/j.ajo.2006.11.057. [DOI] [PubMed] [Google Scholar]
- 31.Ting AY, Lee TK, MacDonald IM. Genetics of age-related macular degeneration. Curr Opin Ophthalmol. 2009;20:369–376. doi: 10.1097/ICU.0b013e32832f8016. [DOI] [PubMed] [Google Scholar]
- 32.Vingerling JR, Hofman A, Grobbee DE, de Jong PT. Age-related macular degeneration and smoking. The Rotterdam study. Arch Ophthalmol. 1996;114:1193–1196. doi: 10.1001/archopht.1996.01100140393005. [DOI] [PubMed] [Google Scholar]
- 33.Khan JC, Thurlby DA, Shahid H, Clayton DG, Yates JR, Bradley M, Moore AT, Bird AC, Genetic factors in AMD study Smoking and age related macular degeneration: the number of pack years of cigarette smoking is a major determinant of risk for both geographic atrophy and choroidal neovascularisation. Br J Ophthalmol. 2006;90:75–80. doi: 10.1136/bjo.2005.073643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunaief JL. Iron induced oxidative damage as a potential factor in age-related macular degeneration: the Cogan lecture. Invest Ophthalmol Vis Sci. 2006;47:4660–4664. doi: 10.1167/iovs.06-0568. [DOI] [PubMed] [Google Scholar]
- 35.Beatty S, Koh H, Phil M, Henson D, Boulton M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Surv Ophthalmol. 2000;45:115–134. doi: 10.1016/S0039-6257(00)00140-5. [DOI] [PubMed] [Google Scholar]
- 36.Shen J, Yang X, Dong A, Petters RM, Peng YW, Wong F, Campochiaro PA. Oxidative damage is a potential cause of cone cell death in retinitis pigmentosa. J Cell Physiol. 2005;203:457–464. doi: 10.1002/jcp.20346. [DOI] [PubMed] [Google Scholar]
- 37.Wu Z, Lauer TW, Sick A, Hackett SF, Campochiaro PA. Oxidative stress modulates complement factor H expression in retinal pigmented epithelial cells by acetylation of FOXO3. J Biol Chem. 2007;282:22414–22425. doi: 10.1074/jbc.M702321200. [DOI] [PubMed] [Google Scholar]
- 38.Age-related eye disease study research group (AREDS) A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E and beta carotene for age-related cataract and vision loss: AREDS report no. 9. Arch Ophthalmol. 2001;119:1439–1452. doi: 10.1001/archopht.119.10.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farkas RH, Chowers I, Hackam AS, Kageyama M, Nickells RW, Otteson DC, Duh EJ, Wang C, Valenta DF, Gunatilaka TL, Pease ME, Quigley HA, Zack DJ. Increased expression of iron-regulating genes in monkey and human glaucoma. Invest Ophthalmol Vis Sci. 2004;45:1410–1417. doi: 10.1167/iovs.03-0872. [DOI] [PubMed] [Google Scholar]
- 40.Moos T, Morgan EH. The metabolism of neuronal iron and its pathogenic role in neurological disease: review. Ann N Y Acad Sci. 2004;1012:14–26. doi: 10.1196/annals.1306.002. [DOI] [PubMed] [Google Scholar]
- 41.Hahn P, Milam AH, Dunaief JL. Maculas affected by age-related macular degeneration contain increased chelatable iron in the retinal pigment epithelium and Bruch’s membrane. Arch Ophthalmol. 2003;12:1099–1105. doi: 10.1001/archopht.121.8.1099. [DOI] [PubMed] [Google Scholar]
- 42.Hahn P, Qian Y, Dentchev T, Chen L, Beard J, Harris ZL, Dunaief JL. Disruption of ceruloplasmin and hephaestin in mice causes retinal iron overload and retinal degeneration with features of age-related macular degeneration. Proc Natl Acad Sci USA. 2004;10138:13850–13855. doi: 10.1073/pnas.0405146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muckenthaler MU, Galy B, Hentze MW. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu Rev Nutr. 2008;28:197–213. doi: 10.1146/annurev.nutr.28.061807.155521. [DOI] [PubMed] [Google Scholar]
- 44.Volz K. The functional duality of iron regulatory protein 1. Curr Opin Struct Biol. 2008;18:106–111. doi: 10.1016/j.sbi.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Viatte L, Gröne HJ, Hentze MW, Galy B. In vivo role(s) of the iron regulatory proteins (IRP) 1 and 2 in aseptic local inflammation. J Mol Med (Berl) 2009;87:913–921. doi: 10.1007/s00109-009-0494-8. [DOI] [PubMed] [Google Scholar]
- 46.Hahn P, Dentchev T, Qian Y, Rouault T, Harris ZL, Dunaief JL. Immunolocalization and regulation of iron handling proteins ferritin and ferroportin in the retina. Mol Vis. 2004;10:598–607. [PubMed] [Google Scholar]
- 47.Smith SR, Ghosh MC, Ollivierre-Wilson H, Hang Tong W, Rouault TA (2006) Complete loss of iron regulatory proteins 1 and 2 prevents viability of murine zygotes beyond the blastocyst stage of embryonic development. Blood Cells Mol Dis 36:283–287 [DOI] [PubMed]
- 48.Galy B, Ferring-Appel D, Kaden S, Gröne HJ, Hentze MW. Iron regulatory proteins are essential for intestinal function and control key iron absorption molecules in the duodenum. Cell Metab. 2008;7:79–85. doi: 10.1016/j.cmet.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 49.Hentze MW, Kühn LC. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc Natl Acad Sci USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 51.Goforth JB, Anderson SA, Nizzi CP, Eisenstein RS. Multiple determinants within iron-responsive elements dictate iron regulatory protein binding and regulatory hierarchy. RNA. 2010;16:154–169. doi: 10.1261/rna.1857210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lakhanpal V, Schocket SS, Jiji R. Deferoxamine (desferal)-induced toxic retinal pigmentary degeneration and presumed optic neuropathy. Ophthalmology. 1984;91:443–451. doi: 10.1016/s0161-6420(84)34267-1. [DOI] [PubMed] [Google Scholar]
- 53.Doly M, Bonhomme B, Vennat JC. Experimental study of the retinal toxicity of hemoglobinic iron. Ophthalmic Res. 1986;18:21–27. doi: 10.1159/000265409. [DOI] [PubMed] [Google Scholar]
- 54.Wang ZJ, Lam KW, Lam TT, Tso MO. Iron-induced apoptosis in the photoreceptor cells of rats. Invest Ophthalmol Vis Sci. 1998;39:631–633. [PubMed] [Google Scholar]
- 55.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 56.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanda A, Chen W, Othman M, Branham KE, Brooks M, Khanna R, He S, Lyons R, Abecasis GR, Swaroop A. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci USA. 2007;104:16227–16232. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montezuma SR, Sobrin L, Seddon JM. Review of genetics in age related macular degeneration. Semin Ophthalmol. 2007;22:229–240. doi: 10.1080/08820530701745140. [DOI] [PubMed] [Google Scholar]
- 59.Fritsche LG, Loenhardt T, Janssen A, Fisher SA, Rivera A, Keilhauer CN, Weber BH. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet. 2008;40:892–896. doi: 10.1038/ng.170. [DOI] [PubMed] [Google Scholar]
- 60.Patel N, Adewoyin T, Chong NV. Age-related macular degeneration: a perspective on genetic studies. Eye. 2008;22(6):768–776. doi: 10.1038/sj.eye.6702844. [DOI] [PubMed] [Google Scholar]
- 61.Ding X, Patel M, Chan CC. Molecular pathology of age-related macular degeneration. Prog Retin Eye Res. 2009;28:1–18. doi: 10.1016/j.preteyeres.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/S0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 63.Madsen E, Gitlin JD. Copper and iron disorders of the brain. Annu Rev Neurosci. 2007;30:317–337. doi: 10.1146/annurev.neuro.30.051606.094232. [DOI] [PubMed] [Google Scholar]