Abstract
The rhizarian amoeba Paulinella chromatophora harbors two photosynthetically active and deeply integrated cyanobacterial endosymbionts acquired ~60 million years ago. Recent genomic analyses of P. chromatophora have revealed the loss of many essential genes from the endosymbiont’s genome, and have identified more than 30 genes that have been transferred to the host cell’s nucleus through endosymbiotic gene transfer (EGT). This indicates that, similar to classical primary plastids, Paulinella endosymbionts have evolved a transport system to import their nuclear-encoded proteins. To deduce how these proteins are transported, we searched for potential targeting signals in genes for 10 EGT-derived proteins. Our analyses indicate that five proteins carry potential signal peptides, implying they are targeted via the host endomembrane system. One sequence encodes a mitochondrial-like transit peptide, which suggests an import pathway involving a channel protein residing in the outer membrane of the endosymbiont. No N-terminal targeting signals were identified in the four other genes, but their encoded proteins could utilize non-classical targeting signals contained internally or in C-terminal regions. Several amino acids more often found in the Paulinella EGT-derived proteins than in their ancestral set (proteins still encoded in the endosymbiont genome) could constitute such signals. Characteristic features of the EGT-derived proteins are low molecular weight and nearly neutral charge, which both could be adaptations to enhance passage through the peptidoglycan wall present in the intermembrane space of the endosymbiont’s envelope. Our results suggest that Paulinella endosymbionts/plastids have evolved several different import routes, as has been shown in classical primary plastids.
Keywords: Paulinella chromatophora, Endosymbiosis, Plastid, Pre-sequence, Targeting signal, Endosymbiotic gene transfer
Introduction
Primary plastid endosymbiosis and Paulinella chromatophora
Available data clearly demonstrate that plastids evolved from free-living cyanobacteria acquired by heterotrophic eukaryotic cells 1–2 billion years ago (Butterfield 2000; Douzery et al. 2004; Yoon et al. 2004; Kutschera and Niklas 2005; Kutschera 2009). This process, called primary endosymbiosis, resulted in plastids surrounded by two membranes. Such primary plastids are characteristic of three eukaryotic lineages: (i) glaucophytes, (ii) red algae, and (iii) green plants, including green algae and their land-plant descendants (Cavalier-Smith 2000; Palmer 2003; Gould et al. 2008; Archibald 2009). Although some authors still consider polyphyly of these photosynthetic groups and their plastids a reasonable hypothesis (Nozaki et al. 2007; Stiller 2007; Howe et al. 2008; Hampl et al. 2009), most now accept a monophyletic origin (Rodríguez-Ezpeleta et al. 2005; Reyes-Prieto et al. 2007; Burki et al. 2008). Consequently, glaucophytes, red algae, and green plants are grouped together in the kingdom of Archaeplastida or Plantae.
It has been common to assume that such an endosymbiosis was so difficult a process that it must be a unique evolutionary event, occurring only once in the common ancestor of archaeplastid lineages (Cavalier-Smith 2000; Keeling et al. 2005). Recently this view has been challenged by Paulinella chromatophora (Lauterborn 1895; Melkonian and Mollenhauer 2005), a thecate amoeba belonging to the supergroup Rhizaria (Bhattacharya et al. 1995; Yoon et al. 2009). P. chromatophora harbors two cyanobacterium-derived endosymbionts acquired independently of classical primary plastids ~60 million years ago (Marin et al. 2005, 2007; Archibald 2006; Yoon et al. 2006, 2009; Nowack et al. 2008) (Fig. 1). These photosynthetically active endosymbionts retain a peptidogylcan wall; like classical primary plastids they are surrounded by two membranes and are deeply integrated into the host cell’s metabolism and genetics.
Fig. 1.
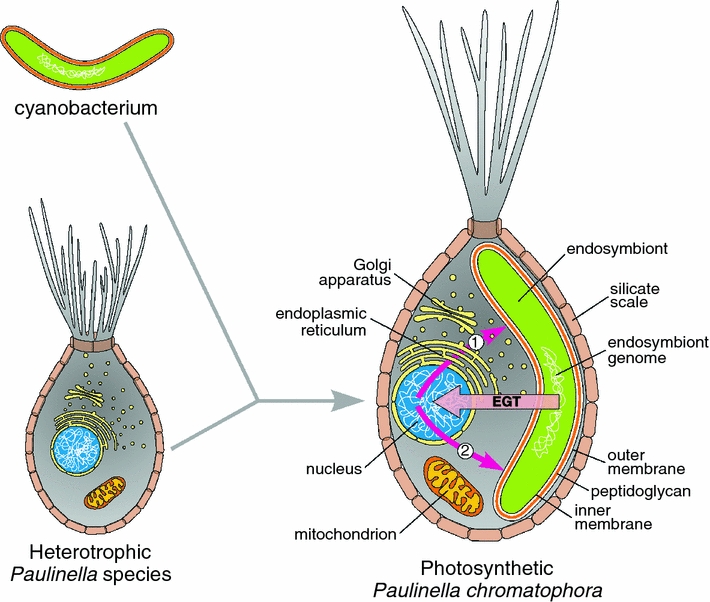
Primary plastid endosymbiosis in the rhizarian amoeba Paulinella chromatophora. About 60 million years ago, a heterotrophic and aplastidal ancestor of P. chromatophora engulfed a cyanobacterium, which was then stably integrated within the host cell as a photosynthetic endosymbiont. Today, the endosymbiont/plastid maintains the peptidoglycan wall but has a significantly reduced genome that has lost many essential genes. It is estimated that more than 30 endosymbiont genes were transferred to the host nuclear genome through the process known as the endosymbiotic gene transfer (EGT). This suggests that protein products of these genes must be imported into Paulinella endosymbionts. Such transport could proceed co-translationally in vesicles derived from the host endomembrane system (1) or by a post-translational pathway involving protein-conducting channels (2)
This tight host–endosymbiont relationship is especially well demonstrated by substantial reductions of the Paulinella endosymbiont genomes, which have been fully sequenced in two different strains, CCAC 0185 and FK01 (Nowack et al. 2008; Reyes-Prieto et al. 2010). The sizes and coding capacities of both genomes have decreased approximately three fold, down to ~1 Mb and ~900 genes compared to a ~3 Mb genome encoding ~3,500 genes in their closest free-living relative, the cyanobacterium Synechococcus WH5701 (see Nowack et al. 2008; Reyes-Prieto et al. 2010). This drastic genome reduction has been accompanied by the loss of many genes with products involved in essential biosynthetic pathways, such as the synthesis of amino acids (e.g., glutamine, arginine, methionine) and co-factors (e.g., riboflavine, biotin, coenzyme A) (Nowack et al. 2008; Reyes-Prieto et al. 2010). Moreover, and more importantly, individual genes were lost from vital and otherwise intact biosynthetic pathways (e.g., hemD encoding uroporphyrinogen III synthase), gene expression machinery (e.g., ligA encoding NAD-dependent DNA ligase), and subcellular structures (e.g., sulA encoding cell-division inhibitor that blocks FtsZ polymerization). Finally, Paulinella endosymbiont genomes are especially poor in genes coding for solute channels and membrane transporters (Nowack et al. 2008; Reyes-Prieto et al. 2010). All these features strongly suggest that Paulinella endosymbionts import nuclear-encoded proteins in ways similar to other true cell organelles such as classical primary plastids (Bhattacharya and Archibald 2006; Yoon et al. 2006; Bodył et al. 2007; Mackiewicz and Bodył 2010) (Fig. 1).
The best candidates for genes with protein products that are imported into Paulinella endosymbionts/plastids are those transferred from the endosymbiont genome to the host nuclear genome through a process called endosymbiotic gene transfer (EGT) (Timmis et al. 2004; Bock and Timmis 2008) (Fig. 1). Recent Paulinella genome and transcriptome analyses have identified more than 30 nuclear-encoded genes acquired via EGT (Nakayama and Ishida 2009; Reyes-Prieto et al. 2010; Nowack et al. 2011). The actual number of EGT-derived genes is likely much greater, perhaps between 40 and 125 genes as estimated by Nowack et al. (2011), although this still is much lower than the ~1700–2500 genes transferred from the genomes of classical primary plastids to their hosts’ nuclear genomes (for reviews see Bock and Timmis 2008; Kleine et al. 2009). Many of the Paulinella transferred genes are engaged in photosynthesis or photo-acclimation of thylakoid membranes and are transcriptionally regulated by the host cell.
Possible import routes of proteins into Paulinella endosymbionts
Most proteins imported into classical primary plastids carry N-terminal targeting signals known as plastid transit peptides (Bruce 2000, 2001; Lee et al. 2008). These peptides are sufficient for their translocation across the plastid envelope with the help of (i) the translocon at the outer chloroplast membrane (Toc) and (ii) the translocon at the inner chloroplast membrane (Tic) (for reviews see Inaba and Schnell 2008; Jarvis 2008; Agne and Kessler 2009; Benz et al. 2009). Each of these translocons consists of several specialized protein subunits. Toc involves three kinds of such subunits: (i) Toc34, Toc64, and Toc159 function as transit peptide receptors, (ii) Toc75 forms a protein-conducting channel, and (iii) Toc12 is responsible for delivering imported proteins to the Tic translocon. The Tic translocon is composed of (i) Tic20, Tic21, and Tic110 that probably constitute three independent protein-conducting channels, (ii) Tic32, Tic55, and Tic62 that form a redox regulon, (iii) Tic22 that is responsible for the coordination of the Toc and Tic translocons and/or intermembrane space protein targeting, and (iv) the Tic40 co-chaperone that, along with the scaffold-channel Tic110 subunit and the stroma-residing Hsp93 and Hsp70 chaperones, provides a motor machinery to pull imported proteins into the stroma.
Interestingly, searches of the Paulinella endosymbiont genome from the CCAC 0185 strain have identified homologs of plant toc12, tic32, and tic21 genes (Bodył et al. 2010). By analogy to classical primary plastids, the Paulinella Tic21 homolog may form a protein-conducting channel in the inner plastid membrane, while the Tic32 protein could be engaged in the regulation of the import process via redox sensing (Bodył et al. 2010). The Paulinella Toc12 homolog resembles its plant counterpart by the presence of DnaJ and transmembrane domains, but differs in secondary structure and in the location of its transmembrane domain (Bodył et al. 2010). Since the DnaJ domain is also typical of Hsp40 proteins, it is possible that Paulinella Toc12, anchored in the inner endosymbiont membrane as a result of its hydrophobic domain, operates as one of the subunits of the molecular protein import motor (Fig. 2).
Fig. 2.
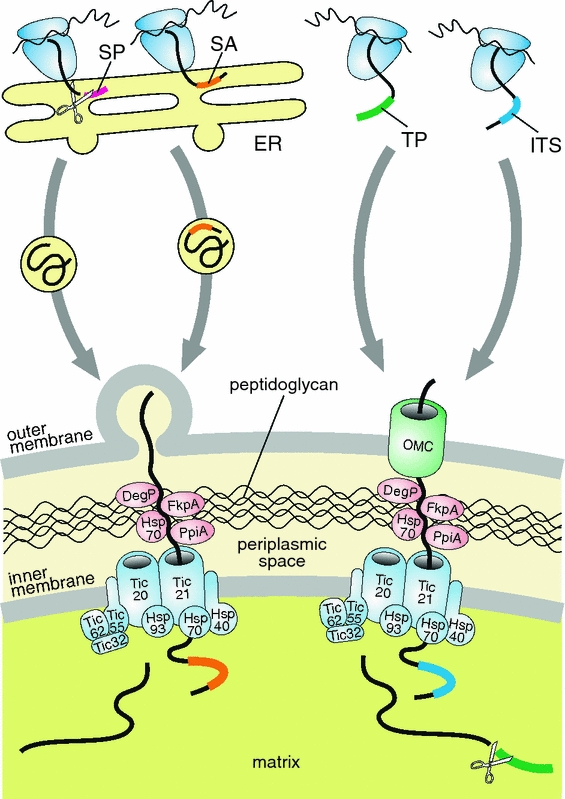
Hypothetical import pathways of nuclear-encoded proteins into the cyanobacterial endosymbionts of Paulinella chromatophora. One model postulates that these proteins carry an N-terminal signal peptide (SP) or signal anchor (SA) and are targeted to the endosymbionts in vesicles derived from the endoplasmic reticulum (ER) (left panel). However, by analogy with classical primary plastids and mitochondria, we cannot exclude the possibility that some proteins are equipped with a transit peptide (TP) or an internal targeting signal (ITS), which results in their translocation via some outer membrane channel (OMC) such as Omp85/Toc75, Tom40, or Tim22/OEP16 (right panel). Proteins released into the intermembrane space could migrate through the peptidoglycan wall freely, or with the assistance of molecular chaperones homologous to the higher plant Hsp70 (DnaK) as well as the bacterial DegP, FkpA, and PpiA, which are encoded in endosymbiont’s genome (Bodył et al. 2010). Protein translocation across the inner endosymbiont membrane could be mediated by a Tic-like translocon characteristic of classical primary plastids because the Paulinella endosymbiont genome encodes significant homologs to several Tic proteins (Bodył et al. 2010). The homologs of Hsp93, Hsp70, and Hsp40 could provide a pulling force to import proteins into the endosymbiont matrix
Although only three Toc and Tic homologs were found in the Paulinella endosymbiont genome, this does not exclude the possibility that other toc and tic genes were transferred to the host nuclear genome and their encoded proteins now are imported into the endosymbiont to create a Toc-Tic-like protein import apparatus. For example, the Paulinella Tic21, Tic32, and Toc12 homologs could form a translocon in the inner envelope membrane, together with other subunits (e.g., the endosymbiont-encoded Hsp93 and probably the nuclear-encoded Tic20, Tic55, and Tic62), that is very similar to the Tic system of classical primary plastids (Bodył et al. 2010). How proteins could pass across the outer membrane of Paulinella endosymbionts is less clear, however, because no homolog to the Toc75 pore (bacterial Omp85) is found in the endosymbiont’s genome (Bodył et al. 2010). At present, we cannot exclude the possibility that a gene encoding this protein was transferred to the host nuclear genome but has yet to be identified. Nevertheless, alternative ways for protein translocation across the outer membrane of Paulinella endosymbionts/plastids should be considered (Fig. 2).
As mentioned previously, the majority of proteins imported into classical primary plastids carry plastid transit peptides responsible for their Toc-Tic-dependent import, but some are equipped with typical endomembrane signal peptides (for reviews see Bhattacharya et al. 2007; Jarvis 2008; Bodył et al. 2009). It has been demonstrated experimentally that these proteins, specifically α-carbonic anhydrase, nucleotide pyrophosphatase/phosphodiesterase, and α-amylases (represented by αAmy3 and αAmy7), are trafficked to the plastids via the endomembrane system involving either the endoplasmic reticulum (ER) alone or in concert with the Golgi apparatus (Chen et al. 2004; Villarejo et al. 2005; Nanjo et al. 2006; Kitajima et al. 2009). Moreover, Armbruster et al. (2009) estimated that as many as 73 plastid-targeted proteins carry such signal peptides in Arabidopsis thaliana, constituting 5% of the plastid proteome. Therefore, it is reasonable that in P. chromatophora nuclear-encoded, endosymbiont-targeted proteins also could be delivered to the outer endosymbiont membrane in vesicles derived from the host’s endomembrane system. In support of this hypothesis, bioinformatics analyses of the sequence from Paulinella photosynthetic psaE gene, which was transferred to the host nuclear genome in the FK01 strain (Nakayama and Ishida 2009), revealed an upstream sequence with qualities of a typical signal peptide (Mackiewicz and Bodył 2010). Applied programs also showed the gene to encode an unambiguous cleavage site for this hypothesized peptide. These findings imply that the PsaE protein is first translocated into the ER lumen where it is processed, and then it is most likely targeted to the endosymbiont’s outer membrane in vesicles derived from the endomembrane system.
The above studies suggest that protein targeting to Paulinella endosymbionts/plastids proceeds via the endomembrane system, however, other bioinformatics analyses of nine EGT-derived genes from Paulinella CCAC 0185 strain did not yield evidence for a universal targeting signal (Nowack et al. 2011). Only products of two transferred genes showed some signal peptide predictions and in one case, PsaE, a weakly supported mitochondrial transit peptide was suggested. Because these analyses provided somewhat ambiguous results, we reanalyzed these sequences by considering more polypeptide variants based on potential translation initiation sites and applying additional bioinformatics tools that predict different targeting signals. We also performed statistical analyses of the basic properties of these hypothesized proteins, including molecular weights, charges, and amino acid compositions, as well as investigate the origins of their potential targeting signals.
Materials and methods
The sequence of psaE from the P. chromatophora FK01 strain was kindly supplied by Dr. Takuro Nakayama and Dr. Ken-ichiro Ischida (Nakayama and Ishida 2009), whereas other EGT candidates with sequenced 5′ ends from the P. chromatophora CCAC 0185 strain were obtained from Nowack et al. (2011). The set of 867 amino acid sequences encoded in the Paulinella endosymbiont genome was downloaded from Genbank (http://www.ncbi.nlm.nih.gov), and 1,762 sequences of proteins imported into classical primary plastids with annotated transit peptides were extracted from the Uniprot database (http://www.uniprot.org).
We applied 25 bioinformatics tools using default settings and all 35 eukaryotic models predicting potential N-terminal targeting signals, including (i) signal peptide, (ii) signal anchor, (iii) plastid transit peptide, and (iv) mitochondrial transit peptide (Table 1). Twelve of these distinguish three types of the targeting signals (i.e., signal peptide, plastid transit peptide, and mitochondrial transit peptide), while 18 algorithms were used to recognize signal peptides/signal anchors, two for plastid transit peptides, and three for mitochondrial transit peptides. To predict transmembrane α-helices in cyanobacterial homologs of Paulinella EGT-derived proteins, we used ConPred II with a selected prokaryote model (Arai et al. 2004).
Table 1.
Programs applied in this study that predict different kinds of N-terminal targeting signals including: signal peptide or signal anchor (SP/SA), plastid transit peptide (pTP), and mitochondrial transit peptide (mTP)
| Program name | References |
|---|---|
| Programs that distinguish SP, pTP, and mTP | |
| iPSORT | Bannai et al. (2002) |
| Predotar 1.03 | Small et al. (2004) |
| PredSL | Petsalaki et al. (2005) |
| PProwler 1.2 | Bodén and Hawkins (2005) |
| TargetLoc | Hoglund et al. (2006) |
| TargetP 1.1 | Emanuelsson et al. (2000) |
| Programs specializing in the prediction of SP/SA | |
| HECTARSEC | Gschloessl et al. (2008) |
| SignalP-HMM 3.0 | Nielsen and Krogh (1998) |
| Programs specializing in the prediction of SP | |
| DetecSig in ConPred II | Lao and Shimizu (2001) |
| MEMSAT 3.5 | Jones (2007) |
| MEMSAT-SVM | Nugent and Jones (2009) |
| Phobius | Käll et al. (2004) |
| PrediSi | Hiller et al. (2004) |
| Sigcleave in EMBOSS 3.0.0 | Rice et al. (2000) |
| SIGFIND 2.11 | Reczko et al. (2002) |
| Signal-3L | Shen and Chou (2007) |
| Signal-CF | Chou and Shen (2007) |
| SignalP-NN 3.0 | Bendsten et al. (2004) |
| SIG-Pred | Bradford (2001) |
| SOSUIsignal | Gomi et al. (2004) |
| SPEPLip | Fariselli et al. (2003) |
| Programs specializing in the prediction of pTP | |
| ChloroP 1.1 | Emanuelsson et al. (1999) |
| PCLR 0.9 | Schein et al. (2001) |
| Programs specializing in the prediction of mTP | |
| MitoProt II v1.101 | Claros and Vincens (1996) |
| MitoPred | Guda et al. (2004) |
Alignments of Paulinella sequences with their 10 top BLAST cyanobacterial homologs found in Genbank database (http://www.ncbi.nlm.nih.gov) were obtained in M-Coffee (Moretti et al. 2007) and prepared in Jalview (Waterhouse et al. 2009). Hydropathy plots of sequences were made assuming Kyte–Doolittle scale (Kyte and Doolittle 1982) and the sliding window length of 11 residues. Molecular weights and charges of proteins were calculated using pepstats from the EMBOSS 3.0.0 package (Rice et al. 2000). The non-parametric U Mann–Whitney test implemented in Statistica software (StatSoft, Inc., 2006) was used to estimate the statistical significance of differences in molecular weight, charge, and amino acid composition of analysed protein sets.
Numbers of amino acid substitutions per site between Paulinella CsoS4A and its 14 closest cyanobacterial homologs were estimated in TREEFINDER (Jobb et al. 2004) as maximum likelihood distances calculated under the best-fit model Dayhoff + Γ(5), whereas in the case of the Paulinella homolog to Synechococcus WH5701_13905, and its ten closest cyanobacterial sequences, the JTT + Γ(5) model was used. The relative numbers of non-synonymous (dN) and synonymous (dS) substitutions for these sequences were calculated according to the modified Nei–Gojobori method, assuming p-distance (Nei and Gojobori 1986) as implemented in MEGA 5.03 (Tamura et al. 2011). Protein domain searches were performed in NCBI CDD database (Marchler-Bauer et al. 2011).
Results
Prediction of targeting signals in Paulinella EGT-derived proteins
We analyzed upstream regions in nine newly identified EGT candidates with determined 5′-ends from the Paulinella CCAC 0185 strain (Nowack et al. 2011). All these proteins are listed in Table 2. The PsaE gene from Paulinella strain FK01 (Nakayama and Ishida 2009) also was included for comparison. Following other recent approaches (Mackiewicz and Bodył 2010; Nowack et al. 2011), we considered pre-sequences starting at all possible eukaryotic translation initiation sites: AUG, UUG, CUG, GUG, AUA, AUC, AUU, and ACG. When a translated sequence did not contain a stop codon in-frame in the upstream region of the mature protein, the longest possible peptide also was studied. A mature protein sequence was included in this analysis as a control. To predict potential N-terminal targeting signals, we used eukaryotic models that distinguish different N-terminal targeting signals, and algorithms specialized for prediction of signal peptides/signal anchors, plastid transit peptides, and mitochondrial transit peptides (Table 1).
Table 2.
Number of algorithms that predict a given targeting signal for the pre-sequences of Paulinella nuclear-encoded endosymbiont-targeted proteins considering all possible translation initiation sites (TIS)
| Protein namea | TISb | SP/SAc | pTP | mTP |
|---|---|---|---|---|
| PsaE (FK01 strain) | 32 | 27 | ||
| 22 | 24 | 1 | ||
| 15 | 5 | 1 | ||
| 12 | 2 | 1 | ||
| 0 | ||||
| PsaE (CCAC 0185 strain) | 13 (max) | 1 | 9 | |
| 12 | 8 | |||
| 3 | ||||
| 0 | ||||
| PsbN | 12 (max) | 21 | 1 | |
| 7 | 22 | |||
| 6 | 22 | |||
| 0 | 16 | |||
| Synechococcus WH5701_13415 homolog | 7 | 16 | 1 | |
| 4 | 6 | 1 | ||
| 2 | 4 | |||
| 1 | 2 | |||
| 0 | 3 | |||
| PsaK | 0 | 12 | ||
| PsaK, 2. copy | 1 | 8 | ||
| 0 | 10 | |||
| Hli | 8 | 1 | ||
| 2 | 1 | |||
| 0 | 1 | |||
| Synechococcus WH5701_06721 homolog | 7 (max) | 1 | 1 | |
| 0 | 2 | |||
| CsoS4A | 9 (max) | |||
| 0 | 1 | 1 | ||
| Synechococcus WH5701_13905 homolog | 15 | |||
| 3 | ||||
| 1 | ||||
| 0 |
Analyses of mature proteins (TIS = 0) are also shown. When a translated sequence did not contain a stop codon in the upstream region of the mature protein, the longest peptide was also analyzed (max). Maximum number of predictions amounts 30, 8, and 15 for signal peptide/signal anchor (SP/SA), plastid transit peptide (pTP), and mitochondrial transit peptide (mTP), respectively
aThe first listed PsaE is encoded in the nuclear genome of Paulinella FK01 strain, whereas the remaining proteins are encoded in the nuclear genome of Paulinella CCAC 0185 strain
bTranslation initiation site counted in codons upstream of mature protein
cSignal anchors were predicted for all PsbN polypeptides by two algorithms
Results of these analyses are presented in Table 2. As previously shown (Mackiewicz and Bodył 2010), 90% of algorithms predicted a signal peptide for the longest polypeptide that could be translated from a psaE open reading frame encoded in the nuclear genome of Paulinella FK01. In the case of CCAC 0185 strain, the signal peptide also was recognized confidently in 73% for Paulinella PsbN translation initiation site variants and in 53% for a Paulinella homolog of Synechococcus WH5701_13415; however, the mature polypeptides of the two Paulinella PsaK proteins reached only 40 and 33% predictability for signal peptides. For all these proteins with potential signal peptides, except for PsaE from the FK01 strain, there were only ambiguously predicted hypothetical cleavage sites, and these were scattered within regions of their mature protein sequences. These results suggest that the N-terminal part of these proteins functions as a signal peptide enabling their co-translational translocation into the ER lumen, but that this region is not processed, which resembles the signal anchor (see the next section).
Interestingly, in contrast to the PsaE from Paulinella FK01 strain, the PsaE encoded in the nuclear genome of Paulinella CCAC 0185 strain show no traits of a signal peptide. Its longest hypothetical polypeptide was predicted to have a mitochondrial transit peptide by 60% of the algorithms employed. We also note that the remaining proteins analyzed from the Paulinella CCAC 0185 strain did not show significant predictions of any targeting signals (Table 2).
Origin of potential targeting signals in Paulinella EGT-derived proteins
To reconstruct how the potential N-terminal targeting signals could have evolved, we compared sequences of Paulinella EGT-derived proteins with their ten most similar cyanobacterial homologs from BLAST searches (Fig. 3). The alignment of PsaE sequences shows that annotated cyanobacterial homologs wholly align only with the mature part of two PsaE proteins (Fig. 3a). To test whether Paulinella PsaE extensions likely evolved from sequences upstream of their cyanobacterial homologs, we compared these cyanobacterial regions with the Paulinella pre-sequences (data not shown). The respective sequences did not align with each other. Moreover, sensitive TBLASTN searches (with word size = 2 and other parameters adjusted for short sequences) using Paulinella PsaE pre-sequences as a query did not reveal any significant hits in cyanobacterial genomes. This indicates that the sequences encoding the potential signal peptide in the PsaE from FK01 strain, and the mitochondrial transit peptide-like signal of the homologous protein in the CCAC 0185 strain, were probably added after independent EGT events in their host nuclear genomes. The PsaE protein functions as subunit IV of the photosystem I (PSI) reaction center and is located on the stromal side of the thylakoid membrane (Barth et al. 1998; Klukas et al. 1999; Jordan et al. 2001; Jeanjean et al. 2008). Plant homologs of Paulinella PsaE are also equipped with N-terminal targeting signals but in these cases they represent typical plastid transit peptides responsible for import mediated by the Toc-Tic supercomplex.
Fig. 3.

Alignments of ten top BLAST cyanobacterial homologs with the following Paulinella sequences: a two PsaE proteins from FK01 (Pau_FK01) and CCAC0185 (Pau_CCAC0185) strains, b the homolog to Synechococcus WH5701_13415 protein (Pau_13415), c PsbN protein (Pau_PsbN), and d two PsaK proteins (Pau_PsaK_1, Pau_PsaK_2). Alignments were prepared in Jalview (Waterhouse et al. 2009) assuming Clustal X color scheme (see online version). Abbreviations: Cya, Cyanobium sp.; Cyt, Cyanothece sp.; Nod, Nodularia spumigena; Nos, Nostoc sp.; Sye, Synechococcus elongatus; Syn, Synechococcus sp.; Pro, Prochlorococcus marinus; The, Thermosynechococcus elongatus. Rectangles under the alignments show transmembrane domains predicted in at least 50% sites of cyanobacterial homologs by ConPred II (Arai et al. 2004)
Interestingly, the Paulinella homolog of Synechococcus WH5701_13415 shows quite a good signal peptide prediction but, unlike its cyanobacterial homologs, does not have any significant N-terminal extension (Fig. 3b). We suggest that, in this case, the N-terminus of the Paulinella protein was adapted to play the role of a signal peptide. In support of this hypothesis, the Paulinella sequence has lost some positively charged and polar residues, which are conserved in cyanobacteria homologs. Consequently, the N-terminal region of the Paulinella sequence has a more hydrophobic character, as is clearly visible in hydropathy profiles (Fig. 4a).
Fig. 4.
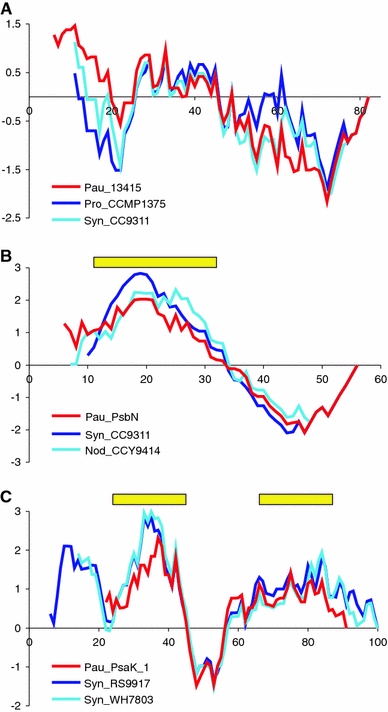
Hydropathy profiles of Paulinella EGT-derived proteins and their closest cyanobacterial homologs: a homolog to Synechococcus WH5701_13415 protein (Pau_13415), b PsbN protein (Pau_PsbN), and c PsaK protein (Pau_PsaK_1). The plots were made assuming Kyte–Doolittle scale (Kyte and Doolittle 1982) and the sliding window length of 11 residues. Rectangles over the profiles show transmembrane domains predicted in at least 50% of sites in cyanobacterial homologs by ConPred II (Arai et al. 2004). The x-axis corresponds to alignment positions. For the abbreviations of sequence names see legend to Fig. 3
Similarly, the Paulinella PsbN variant that shows high signal peptide predictability is similar in length to its cyanobacterial homologs and is not equipped with a substantial N-terminal extension (Fig. 3c). The function of the PsbN protein is still unknown. In photosynthetic eukaryotes, PsbN is encoded in plastid genomes. Some data suggest that this protein represents a component of photosystem II (PSII) (Ikeuchi et al. 1995; Zouni et al. 2001) but this has not been confirmed in other studies (Kashino et al. 2002a, b). Nevertheless, it cannot be ruled out that this protein can sometimes bind transiently to PSII (Plöscher et al. 2009). The presence of reliably predicted transmembrane α-helices in the N-termini of PsbN sequences (Fig. 3c), suggests that this protein is anchored in the thylakoid membrane. Because both signal peptides and transmembrane domains are enriched in hydrophobic residues, it is reasonable that the N-terminal region of Paulinella PsbN mimics a signal peptide and can be recognized by the SRP (signal recognition particle) during its targeting to the ER membrane. The SRP would enable co-translational translocation of this protein into the ER lumen and its subsequent transport in ER- or Golgi-derived vesicles to the Paulinella endosymbiont/plastid. However, in contrast to typical signal peptides, the N-terminal region of Paulinella PsbN most likely is not removed and is required to anchor this protein in its subcellular target, the endosymbiont’s thylakoid membrane. Actually, algorithms that predict cleavage sites of signal peptides gave ambiguous results and located them within the mature region of protein sequence, which suggests that the potential signal peptide is not processed. Furthermore, in contrast to most cyanobacterial homologs, Paulinella PsbN lacks several negatively charged, polar, and hydroxylated residues at its N-terminus. Consequently, it has a longer hydrophobic region than cyanobacterial proteins, which extends toward the N-terminus of the sequence (Fig. 4b).
We hypothesize that an adaptation of the transmembrane domain toward signal peptide function occurred in the two Paulinella PsaK proteins as well. PsaK is subunit X of photosystem I (PSI) (Jone et al. 1991; Jordan et al. 2001) and has two transmembrane α-helices responsible for its insertion into the thylakoid membrane (Kjaerulff et al. 1993; Mant et al. 2001; Düring et al. 2007). In higher plants, this protein is equipped with a typical transit peptide responsible for its import via the Toc-Tic supercomplex (Kjaerulff et al. 1993). Interestingly, the two Paulinella PsaK proteins, which are devoid of N-terminal extensions, are further shortened at their N-termini by at least seven amino acid residues compared with their cyanobacterial homologs (Fig. 3d). In some cyanobacterial PsaK sequences this region shows a weaker hydrophobic character than the more proximal main transmembrane domain (Fig. 4c). Consequently, the N-terminal transmembrane α-helix is located closer to the beginning of the sequence in Paulinella PsaK proteins, which could enable the evolution of signal peptide-like domains at their N-termini. Moreover, the N-terminal ends of Paulinella PsaK sequences do not contain positively charged residues that are conserved in cyanobacteria and are poorer in hydroxylated residues than their cyanobacterial homologs.
Molecular weights and charges of Paulinella EGT-derived proteins
It is notable that the majority of Paulinella EGT-derived genes identified to date encode small proteins (Nowack et al. 2011). Moreover, Mackiewicz and Bodył (2010) found that PsaE from Paulinella strain FK01 has the same number of positively and negatively charged residues. They proposed this as an adaptation to the passage of this protein through the negatively charged peptidoglycan wall located in the intermembrane space of the endosymbiont’s envelope (Kies and Kremer 1979). To check how generally representative these properties are for Paulinella EGT-derived proteins (i.e., nuclear-encoded, endosymbiont-targeted proteins), we compared their calculated molecular weights and charges against all proteins encoded in the Paulinella endosymbiont genomes, as well as proteins imported into classical primary plastids by means of transit peptides.
Results of these comparisons are summarized in Table 3. Paulinella EGT-derived proteins have, on average, four times lower molecular weight than Paulinella endosymbiont genome-encoded proteins and five times lower molecular weight than proteins targeted to classical primary plastids. Moreover, there are no plastid proteins smaller than the largest Paulinella EGT-derived protein and only 6% of Paulinella endosymbiont genome-encoded proteins fulfil this criterion. In addition, Paulinella EGT-derived proteins have more closely balanced numbers of positively and negatively charged residues than do proteins from the other two sets. All these differences in molecular weight and charge are statistically significant at P < 0.001 when analyzed by the U Mann–Whitney test.
Table 3.
Average and minimal-maximal range of molecular weight and absolute value of charge for three sets of plastid proteins
| Set of proteins | Number | Molecular weight (kDa) | Absolute value of charge |
|---|---|---|---|
| Nuclear-encoded proteins targeted to Paulinella endosymbiontsa | 28 | 7.6 (4.4–9.1) | 1.0 (0.0–5.5) |
| Proteins encoded in Paulinella endosymbiont genomes | 867 | 30.3 (3.5–151.2) | 5.5 (0.0–63.0) |
| Nuclear-encoded proteins targeted to primary plastids by means of pTP | 1,762 | 41.1 (10.0–202.1) | 5.0 (0.0–72.5) |
aWe included all TIS variants listed in Table 1 with the exception of one PsaK protein whose sequence was incomplete. In the case of PsaE from FK01 strain the sequence without the potential signal peptide and the sequence of mature protein were included because it is probable that its signal peptide is cleaved in the lumen of the endoplasmic reticulum and only the mature protein reaches the endosymbiont
The low-molecular weights and almost neutral charges of Paulinella EGT-derived proteins fit well with the properties of proteins that can pass freely through the peptidoglycan wall. In agreement with this hypothesis, Demchick and Koch (1996) demonstrated that globular, uncharged proteins up to 24 kDa in molecular weight pass freely through the isolated unstretched peptidoglycan sacculi of Escherichia coli (Gram-negative bacterium) and Bacillus subtilis (Gram-positive bacterium). Interestingly, the molecular weights of all variants of the Paulinella EGT-derived proteins analyzed are well below that threshold (Table 3), whereas only 38% of Paulinella endosymbiont genome-encoded proteins and 22% of plastid-targeted proteins have weights under 24 kDa.
Amino acid composition of Paulinella EGT-derived proteins
Because Paulinella EGT-derived proteins showed distinctive features in their molecular weights and charges, we also examined them for any peculiar characteristics in their amino acid compositions compared with their ancestral set, that is, proteins encoded in the Paulinella endosymbiont genome. We found that sequences from these two sets differ significantly in composition of 11 amino acid residues when analyzed in the U Mann–Whitney test (Table 4). Compared to those still encoded in the endosymbiont genome, Paulinella nuclear-encoded, endosymbiont-targeted proteins are richer in glycine, valine, methionine, and threonine, but poorer in arginine, serine, isoleucine, cysteine, leucine, glutamine, and histidine.
Table 4.
Average and quartile (Q1–Q2) range of amino acid percentages for Paulinella endosymbiont- and nuclear-encoded proteins
| Amino acid residue | Nuclear-encoded proteins targeted to endosymbiont | Proteins coded in endosymbiont genome | Bonferroni corrected P value |
|---|---|---|---|
| Glycine | 11.4 (10.2–12.9) | 7.1 (5.8–8.4) | 5.8E−15 |
| Valine | 8.0 (6.0–12.1) | 6.2 (4.9–7.5) | 0.0011 |
| Arginine | 4.2 (2.5–6.2) | 5.8 (4.5–7.3) | 0.0308 |
| Serine | 4.1 (2.6–6.9) | 7.0 (5.8–8.3) | 0.0058 |
| Methionine | 3.1 (2.6–5.3) | 2.2 (1.5–2.8) | 0.0010 |
| Isoleucine | 5.5 (2.4–6.3) | 7.5 (6.2–8.8) | 2.5E−07 |
| Threonine | 7.5 (6.1–8.6) | 5.0 (4.0–6.1) | 3.7E−09 |
| Cysteine | 0.0 (0.0–1.2) | 1.2 (0.6–1.7) | 1.6E−05 |
| Leucine | 9.2 (7.8–11.9) | 11.8 (9.4–14.2) | 0.0497 |
| Glutamine | 1.4 (0.0–3.6) | 3.9 (2.9–4.9) | 5.2E−05 |
| Histidine | 0.0 (0.0–1.9) | 1.8 (1.1–2.6) | 0.0004 |
Only amino acids are listed for which differences between these groups were statistically significant at P < 0.05 in the U Mann–Whitney test. We analyzed here the same set of Paulinella proteins as in the studies presented in Table 3
Discussion
Targeting signals of Paulinella EGT-derived proteins
Our analyses show that signal peptide-like sequences are the most commonly predicted N-terminal targeting signals in the Paulinella EGT-derived proteins studied, and were identified in five of the ten proteins analyzed (Table 2). This suggests that the signal peptide-carrying proteins are targeted to Paulinella endosymbionts/plastids via the host endomembrane system (Fig. 2) (see also Mackiewicz and Bodył 2010). The results obtained are rather unexpected because, by analogy to classical primary plastids, we might expect proteins imported into Paulinella endosymbionts/plastids to use N-terminal targeting signals resembling plastid transit peptides (Bruce 2000, 2001; Lee et al. 2008). Interestingly, a comparison of Paulinella EGT-derived proteins with their closest cyanobacterial homologs shows that the signal peptide-like sequences have different origins (Fig. 3). The signal peptide of PsaE protein from FK01 strain probably represents a typical cleavable signal peptide (see also Mackiewicz and Bodył 2010) that was added after transfer of this gene to the host nuclear genome. The Paulinella homolog of Synechococcus WH5701_13415 does not have such an extension, but its existing N-terminal sequence has acquired new properties of signal peptides and could play the same role (Table 2; Fig. 4). The N-terminal ends of PsbN and two PsaK proteins contain transmembrane domains that also show features of signal peptides and also could fulfil this function.
Some similarity of N-terminal transmembrane domains to signal peptides has been observed in plastid and mitochondrial outer membrane proteins (Kanaji et al. 2000; Lee et al. 2001, 2004; Horie et al. 2003; Waizenegger et al. 2003; Hofmann and Theg 2005). It was shown that several charged residues adjacent to the transmembrane domain play a crucial role in distinguishing these proteins from those directed to the endomembrane system. For example, experimental replacement of such residues with uncharged glycine, or their complete deletion, caused mistargeting of plastid AtOEP7 and AtToc64 proteins to the endoplasmic reticulum or the plasma membrane (Lee et al. 2001, 2004). Similar mutations that increase the hydrophobicity of the transmembrane domains and decrease the net positive charge within the flanking regions of mitochondrial outer membrane proteins Tom5 and Tom20 (Kanaji et al. 2000; Horie et al. 2003; Waizenegger et al. 2003) result in mistargeting to the endomembrane compartments. Interestingly, we observed similar changes and substitutions in the Paulinella PsbN and PsaK sequences compared with their cyanobacterial homologs (Fig. 3). This strongly suggests that these changes in the Paulinella transmembrane proteins represent the acquisition of signal peptide properties in their N-terminal transmembrane domains.
In contrast to the PsaE protein from Paulinella FK01 strain, its counterpart from the CCAC 0185 strain has a putative mitochondrial transit peptide (Table 2), which implies post-translational import perhaps involving a mitochondrial protein-conducting channel that was relocated to the outer endosymbiont membrane (Fig. 2). Good candidates for such a translocation channel are Tom40 and Tim22. In support of this hypothesis, the outer membrane of higher plant plastids contains the OEP16 channel for protochlorophyllide oxidoreductase A (Reinbothe et al. 2004; Pollmann et al. 2007), which probably evolved from a relocated mitochondrial Tim22 pore (Cavalier-Smith 2006). A third mitochondrial candidate could be the homolog of Omp85 (and, therefore, Toc75), but this gene has been identified to date only in trypanosomatid parasites (Pusnik et al. 2011). At present, we cannot exclude the possibility that the mRNA sequence of psaE obtained from CCAC 0185 strain is incomplete and could yet be found to contain upstream sequence encoding signal peptide properties. The published sequence is not limited by an in-frame stop codon upstream of the mature protein. Moreover, the available N-terminal extensions of the PsaE proteins from two Paulinella strains differ significantly (Fig. 3). This suggests that the PsaE proteins from each of these strains evolved distinct targeting signals when their genes were independently transferred to the hosts’ nuclear genomes. This possibility is supported by substantial differences between these two Paulinella psaE genes, including different intron positions, intron sequences, and 5′ and 3′ untranslated regions (Nowack et al. 2011).
The two-membrane envelope of Paulinella endosymbionts/plastids and the endomembrane system-mediated targeting of their nuclear-encoded proteins
Paulinella endosymbionts/plastids are surrounded by two membranes. Their inner membrane is certainly derived from the cyanobacterial plasmalemma, but origin of the outer membrane is less clear (Bodył et al. 2010; Mackiewicz and Bodył 2010). Because the cyanobacterial ancestor of Paulinella endosymbionts/plastids was surrounded by two membranes, it could be hypothesized that their outer membrane corresponds directly to the outer negibacterial membrane. In contrast, the cyanobacterial outer membrane could have been lost and replaced entirely by the host phagosomal membrane. It is also important to consider the possibility that the outer membrane of Paulinella endosymbionts/plastids has a chimeric origin, and contains components of both bacterial and eukaryotic membranes. The cyanobacterium initially engulfed by P. chromatophora was undoubtedly surrounded by three membranes, the host phagosomal membrane and the two envelope membranes of the endosymbiont, i.e., its plasma membrane and outer membrane (Bodył et al. 2010; Mackiewicz and Bodył 2010). In the initial stages of the endosymbiosis, it is reasonable that uncoordinated divisions of the cyanobacterium, and the phagosome containing it, resulted in regular escapes of these endosymbionts into the host cytosol. During these escapes the outer cyanobacterial membrane could have acquired some lipids and proteins from the phagosomal membrane, a kind of membrane mutation as termed by Cavalier-Smith (2000). This would have led to a chimeric bacterial–eukaryotic membrane (see also Bodył et al. 2009). The existence of clear signal peptides in Paulinella nuclear-encoded, endosymbiont-targeted proteins is compatible with this scenario.
The above evolutionary scenario is consistent with the process that led to classical primary plastids. As with Paulinella endosymbionts/plastids, they have a cyanobacterial origin and are surrounded by two membranes (Cavalier-Smith 2000; Palmer 2003; Gould et al. 2008; Archibald 2009). It was argued for many years that the outer plastid membrane was derived directly from the cyanobacterial outer membrane (Cavalier-Smith 2000); however, it contains lipids characteristic of negibacterial outer membranes (e.g., galactolipids), as well as those found in eukaryotic phagosomal membranes (e.g., phosphatidylcholine) (see Kilian and Kroth 2003). This discovery inspired the hypothesis that the outer plastid membrane has a chimeric bacterial–eukaryotic origin (Kilian and Kroth 2003; Bodył et al. 2009). The identification of many nuclear-encoded, plastid-targeted proteins with signal peptides in higher plants and green algae (Bhattacharya et al. 2007; Jarvis 2008; Armbruster et al. 2009; Bodył et al. 2009) provides additional strong support for this hypothesis.
Possible import routes of Paulinella proteins without N-terminal targeting signals
In contrast to the proteins discussed above, four Paulinella EGT-derived proteins show no evidence of any kind of N-terminal targeting signals (Table 2). It is possible that the sequences of Paulinella homologs to CsoS4A and Synechococcus WH5701_06721 are extended upstream and, therefore, could encode N-terminal targeting signals; none of their upstream sequences are constrained by in-frame stop codons. In contrast, the homologs to Hli and Synechococcus WH5701_13905 are most likely complete sequences.
The Paulinella Hli (high-light inducible protein), like its homologs, possesses a predicted α-helical transmembrane domain and probably is anchored in the thylakoid membrane (Funk and Vermaas 1999; Montané and Kloppstech 2000; He et al. 2001; Bhaya et al. 2002). The other three proteins do not have such recognizable transmembrane domains and, therefore, could reside in the intermembrane space or in the matrix. This indicates that the four proteins must pass one or two envelope membranes during their import into Paulinella endosymbionts/plastids. In such trafficking, they could be using some targeting signals that escaped detection by the algorithms used in this study, which are specialized for predicting classical N-terminal signals.
One such non-classical targeting signal is represented by the C-terminal cleavable region composed of positively charged residues, which was identified in the mitochondrial matrix-residing DNA helicase Hmi1 (Lee et al. 1999). Targeting signals that are not cleaved also were identified in proteins targeted to the inner membrane of classical primary plastids, including ceQORH (Miras et al. 2002, 2007) and Tic32 (Nada and Soll 2004). Their targeting signals appear to enable import into the plastid stroma from where they can be inserted into the inner membrane. The plastid-targeting signal in Tic32 probably is located in its first ten N-terminal residues, whereas in ceQORH it is encoded in an internal domain of 40 residues that is essential but not sufficient for correct plastid localization because it must act in concert with two adjacent domains required for import (Miras et al. 2007). It was demonstrated that import of ceQORH and Tic32 is mediated by a Toc-independent pathway because their translocation involves neither the Toc159 receptor nor the Toc75 channel (Miras et al. 2007). Interestingly, the gene for Toc75 was not identified in the Paulinella endosymbiont genome (Bodył et al. 2010) and is probably also absent from the host nuclear genome (Nowack et al. 2011). The lack of evidence for the presence of Toc75 and other Toc proteins, as well as the absence of recognizable plastid transit peptides in P. chromatophora, suggest that endosymbiont-directed proteins use an import route similar to the alternative pathways used by transit peptide-devoid proteins imported into higher plant plastids. Proteins with non-canonical signals targeted to classical primary plastids are likely to be more common than previously thought because proteomic studies of the A. thaliana plastid proteome revealed 142 (from 604) proteins without the N-terminal cleavable targeting pre-sequences (Kleffmann et al. 2004).
One very interesting aspect of Paulinella EGT-derived proteins is the nature of proteins located in the mitochondrial intermembrane space (IMS). They also are characterized by low molecular weight (7–15 kDa) and the absence of N-terminal transit peptides (Lutz et al. 2003; Herrmann and Hell 2005; Neupert and Herrmann 2007). Many contain conserved patterns of cysteine (and histidine) residues that enable them to bind cofactors or form disulfide bridges. Their translocation through the outer membrane translocons (Tom) requires them to be folded in the IMS, which is triggered by the acquisition of cofactors or by intramolecular disulfide bridges. However, the majority of Paulinella EGT-derived protein variants studied do not have any cysteine residue and eight have only one such residue, which is insufficient to form a disulfide bridge. These sequences also are histidine poor; ten have only one His residue and each of two translation initiation site variants of PsaK 2 copy has three histidines. The other class of IMS mitochondrial proteins that lacks classical transit peptides requires binding to affinity sites for translocation (Herrmann and Hell 2005; Neupert and Herrmann 2007). The targeting signal identified in a representative of this class, heme lyase, consists of a complex pattern of hydrophilic residues (Diekert et al. 1999). Concentration of such residues can be found in Paulinella EGT candidates lacking N-terminal signals but their importance for targeting should be verified experimentally. Similarly, some targeting information can be carried by several amino acids that are used more frequently in the EGT-derived proteins than in proteins encoded in the Paulinella endosymbiont genome (Table 4). They could, for example, facilitate protein import through some unidentified outer membrane channels (Fig. 2) or be involved in still unknown trafficking mechanisms.
Are some Paulinella nuclear-encoded proteins targeted to the endosymbiont as mRNAs?
There is one more possible route for the product of a nuclear-encoded gene to be expressed in an organelle that does not require any targeting signal in the protein product; it operates at the nucleic acid rather than the protein level. Gómez and Pallás (2010) showed recently that a viroid-derived ncRNA acting as a 5′UTR-end mediates the specific import of mRNA for Green Fluorescent Protein into plastids of the tobacco Nicotiana benthamiana. These results suggest the existence of an alternative transport pathway into plastids, where an ncRNA functions as a key regulatory molecule to control the import of plastid-directed, nuclear genes into this organelle. Such import of transcripts instead of proteins would explain the lack of targeting signals in some Paulinella EGT candidates.
An ongoing process of endosymbiotic gene transfer can explain the absence of targeting signals in some Paulinella nuclear-encoded proteins
Data from the above-discussed peculiar plastid and mitochondrial proteins suggest that the Paulinella EGT-derived proteins that lack recognizable targeting signals still could be imported into these endosymbionts; however, it is possible that some of them are not imported or that their import proceeds with low efficiency. Interestingly, copies of Paulinella homologs to both CsoS4A and Synechococcus WH5701_13905 that were transferred to the nucleus, both of which also still are retained in the endosymbiont’s genome, exhibit very high-substitution rates (Nowack et al. 2011). Estimated average numbers of amino acid substitutions per site between the nuclear copies and closest cyanobacterial sequences are 1.81 and 1.25 for Paulinella homologs to CsoS4A and Synechococcus WH5701_13905, respectively. The numbers for the corresponding endosymbiont-encoded copies are only 0.35 and 0.18 amino acid substitutions per site. Weaker purifying selection on nuclear copies also is evident at the DNA level. They show higher average dN/dS values compared to their cyanobacterial homologs. The ratios for Paulinella nuclear homologs to CsoS4A and Synechococcus WH5701_13905 are 0.40 and 0.50, whereas for the endosymbiont’s counterparts it is 0.20 and 0.15, respectively. In addition, an SH3 protein domain was not detected in the Paulinella nuclear homolog to Synechococcus WH5701_13905 although it was found in the endosymbiont’s copy and all cyanobacterial homologs. Domains typical for CsoS4A, such as ethanolamine utilization protein and carboxysome structural protein domain, were identified in the Paulinella nuclear gene copy but with 3 × 109 times higher E-value and 1.5 times lower bit score than in the endosymbiont and cyanobacteral homologs. These results suggest that the original functions of the Paulinella homologs to CsoS4A and Synechococcus WH5701_13905 could have changed after transfer to the nucleus. It is also possible that the Paulinella nuclear copies have not yet acquired efficient targeting signals and, therefore, the endosymbiont’s copies are still maintained.
Molecular weights and charges of Paulinella EGT-derived proteins
The most distinctive features of the EGT-derived proteins analysed are their low molecular weights and nearly neutral charges, which fit well with the properties of proteins known to cross peptidoglycan walls. The permeability limit of ~24 kDa we compared to Paulinella proteins is based on the peptidoglycan wall of E. coli, which is thinner (2–7 nm) than in cyanobacteria (e.g., 10 nm in the genus Synechococcus) (Hoiczyk and Hansel 2000). In some cases, however, it was found that the peptidoglycan wall can change in thickness locally, which could be also true in P. chromatphora. For example, 75–80% of the E. coli peptidoglycan surface is 2.5 nm thick while the remaining areas are ~7 nm in width (Labischinski et al. 1991). There is also the possibility of local lesions in the peptidoglycan wall, as was discovered at the contact sites of translocation pores in the outer and inner envelope membranes of glaucophyte plastids (Steiner and Löffelhardt 2005); however, if comparable import sites associated with local lesions have not evolved yet in the peptidoglycan wall in Paulinella endosymbionts/plastids, genes coding small proteins will be preferably transferred to the host’s nucleus. This could explain why the mass of nuclear-encoded proteins targeted to Paulinella endosymbionts/plastids (4.4–9.1 kDa) is several times lower than the benchmark limit of 24 kDa from E. coli. Moreover, the size of peptidoglycan pores, with diameters of ~2 nm, is probably more restrictive to protein passage than is the wall’s overall thickness. Interestingly, it was found that pores in the peptidoglycans from Gram-negative and -positive bacteria have similar average sizes and are relatively homogenous in size as well (Demchick and Koch 1996). If we assume that peptidoglycan pores in Paulinella endosymbionts/plastids are the same size as in other eubacteria, then the size limit determined for E. coli proteins should also be valid for Paulinella proteins.
Diffusion of charged molecules also can be influenced by peptidoglycan anionic groups (Steiner and Löffelhardt 2005). Consequently, it was suggested initially that the 1-carboxyl groups of d-glutamic acid of peptidoglycans in walls of glaucophyte plastids are amidated with N-acetylputrescine to lower their overall negative charge and polarity, thereby enhancing the passage of nuclear-encoded proteins into the plastid (Pfanzagl et al. 1996a, b; Pfanzagl and Löffelhardt 1999). Although the glaucophyte plastid-targeted proteins investigated to date turn out to be imported post-translationally, through a Toc-Tic supercomplex that bypasses the peptidoglycan wall (Steiner et al. 2005; Steiner and Löffelhardt 2005), problems associated with protein diffusion through the peptidoglycan wall still could be valid for Paulinella endosymbionts. So far, there is no evidence of a Toc-like translocon penetrating the endosymbiont envelope along with the Tic system (Bodył et al. 2010). Moreover, at least some of Paulinella EGT-derived proteins (with recognized signal peptides) are likely delivered to the endosymbionts in vesicles budding off from the host endomembrane system and are released into the endosymbiont intermembrane space, from where they must cross the peptidoglycan wall (Fig. 2) (Mackiewicz and Bodył 2010). It should be also pointed out that potential post-translational protein translocation through some protein-conducting channels in the outer endosymbiont membrane does not exclude release of the imported proteins directly to the intermembrane space. It is possible that such channels, even if present, have not developed a stable connection with the Tic-like translocons residing in the inner endosymbiont membrane.
Acknowledgments
We are very grateful to Dr. John W. Stiller for helpful comments and English editing and to two anonymous reviewers for their excellent comments and suggestions, which significantly improved the paper. A. B. is supported by funds from Wrocław University grant BS/1018/2010.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Agne B, Kessler F. Protein transport in organelles: the Toc complex way of preprotein import. FEBS J. 2009;276:1156–1165. doi: 10.1111/j.1742-4658.2009.06873.x. [DOI] [PubMed] [Google Scholar]
- Arai M, Mitsuke H, Ikeda M, Xia JX, Kikuchi T, Satake M, Shimizu T. ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 2004;32:W390–W393. doi: 10.1093/nar/gkh380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JM. Endosymbiosis: double-take on plastid origins. Curr Biol. 2006;16:R690–R692. doi: 10.1016/j.cub.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009;19:R81–R88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- Armbruster U, Hertle A, Makarenko E, Zühlke J, Pribil M, Dietzmann A, Schliebner I, Aseeva E, Fenino E, Scharfenberg M, Voigt C, Leister D. Chloroplast proteins without cleavable transit peptides: rare exceptions or a major constituent of the chloroplast proteome? Mol Plant. 2009;2:1325–1335. doi: 10.1093/mp/ssp082. [DOI] [PubMed] [Google Scholar]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;18:298–305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- Barth P, Lagoutte B, Sétif P. Ferredoxin reduction by photosystem I from Synechocystis sp. PCC 6803: toward an understanding of the respective roles of subunits PsaD and PsaE in ferredoxin binding. Biochemistry. 1998;37:16233–16241. doi: 10.1021/bi981379t. [DOI] [PubMed] [Google Scholar]
- Bendsten JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Benz JP, Soll J, Bölter B. Protein transport in organelles: the composition function and regulation of the Tic complex in chloroplast protein import. FEBS J. 2009;276:1166–1176. doi: 10.1111/j.1742-4658.2009.06874.x. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Archibald JM. The difference between organelles and endosymbionts. Response to Theissen and Martin. Curr Biol. 2006;16:R1017–R1018. doi: 10.1016/j.cub.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Helmchen T, Melkonian M. Molecular evolutionary analyses of nuclear-encoded small subunit ribosomal RNA identify an independent rhizopod lineage containing the Euglyphina and the Chlorarachniophyta. J Eukaryot Microbiol. 1995;42:65–69. doi: 10.1111/j.1550-7408.1995.tb01541.x. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Archibald JM, Weber AP, Reyes-Prieto A. How do endosymbionts become organelles? Understanding early events in plastid evolution. Bioessays. 2007;29:1239–1246. doi: 10.1002/bies.20671. [DOI] [PubMed] [Google Scholar]
- Bhaya D, Dufresne A, Vaulot D, Grossman A. Analysis of the hli gene family in marine and freshwater cyanobacteria. FEMS Microbiol Lett. 2002;215:209–219. doi: 10.1111/j.1574-6968.2002.tb11393.x. [DOI] [PubMed] [Google Scholar]
- Bock R, Timmis JN. Reconstructing evolution: gene transfer from plastids to the nucleus. Bioessays. 2008;30:556–566. doi: 10.1002/bies.20761. [DOI] [PubMed] [Google Scholar]
- Bodén M, Hawkins J. Prediction of subcellular localization using sequence-biased recurrent networks. Bioinformatics. 2005;21:2279–2286. doi: 10.1093/bioinformatics/bti372. [DOI] [PubMed] [Google Scholar]
- Bodył A, Mackiewicz P, Stiller JW. The intracellular cyanobacteria of Paulinella chromatophora: endosymbionts or organelles? Trends Microbiol. 2007;15:295–296. doi: 10.1016/j.tim.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Bodył A, Mackiewicz P, Stiller JW. Early steps in plastid evolution: current ideas and controversies. Bioessays. 2009;31:1219–1232. doi: 10.1002/bies.200900073. [DOI] [PubMed] [Google Scholar]
- Bodył A, Mackiewicz P, Stiller JW. Comparative genomic studies suggest that the cyanobacterial endosymbionts of the amoeba Paulinella chromatophora possess an import apparatus for nuclear-encoded proteins. Plant Biol. 2010;12:639–649. doi: 10.1111/j.1438-8677.2009.00264.x. [DOI] [PubMed] [Google Scholar]
- Bradford JR (2001) SIG-Pred. http://bmbpcu36.leeds.ac.uk/prot_analysis/Signal.html
- Bruce BD. Chloroplast transit peptides: structure function and evolution. Trends Cell Biol. 2000;10:440–447. doi: 10.1016/S0962-8924(00)01833-X. [DOI] [PubMed] [Google Scholar]
- Bruce BD. The paradox of plastid transit peptides: conservation of function despite divergence in primary structure. Biochim Biophys Acta. 2001;1541:2–21. doi: 10.1016/S0167-4889(01)00149-5. [DOI] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Pawlowski J. Phylogenomics reveals a new ‘megagroup’ including most photosynthetic eukaryotes. Biol Lett. 2008;4:366–369. doi: 10.1098/rsbl.2008.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield NJ. Bangiomorpha pubescens n. gen., n. sp.: implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology. 2000;26:386–404. doi: 10.1666/0094-8373(2000)026<0386:BPNGNS>2.0.CO;2. [DOI] [Google Scholar]
- Cavalier-Smith T. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 2000;5:174–182. doi: 10.1016/S1360-1385(00)01598-3. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Origin of mitochondria by intracellular enslavement of a photosynthetic purple bacterium. Proc R Soc B. 2006;273:1943–1952. doi: 10.1098/rspb.2006.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Huang LF, Li HM, Chen YR, Yu SM. Signal peptide-dependent targeting of a rice alpha-amylase and cargo proteins to plastids and extracellular compartments of plant cells. Plant Physiol. 2004;135:1367–1377. doi: 10.1104/pp.104.042184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou KC, Shen HB. Signal-CF: a subsite-coupled and window-fusing approach for predicting signal peptides. Biochem Biophys Res Commun. 2007;357:633–640. doi: 10.1016/j.bbrc.2007.03.162. [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Demchick P, Koch AL. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J Bacteriol. 1996;178:768–773. doi: 10.1128/jb.178.3.768-773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekert K, Kispal G, Guiard B, Lill R. An internal targeting signal directing proteins into the mitochondrial intermembrane space. Proc Natl Acad Sci USA. 1999;96:11752–11757. doi: 10.1073/pnas.96.21.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douzery EJ, Snell EA, Bapteste E, Delsuc F, Philippe H. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc Natl Acad Sci USA. 2004;101:15386–15391. doi: 10.1073/pnas.0403984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düring U, Ossenbühl F, Wilde A. Late assembly steps and dynamics of the cyanobacterial photosystem I. J Biol Chem. 2007;282:10915–10921. doi: 10.1074/jbc.M609206200. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Fariselli P, Finocchiaro G, Casadio R (2003) SPEPLip. http://gpcr.biocomp.unibo.it/cgi/predictors/spep/pred_spepcgi.cgi [DOI] [PubMed]
- Funk C, Vermaas W. A cyanobacterial gene family coding for single-helix proteins resembling part of the light-harvesting proteins from higher plants. Biochemistry. 1999;38:9397–9404. doi: 10.1021/bi990545+. [DOI] [PubMed] [Google Scholar]
- Gómez G, Pallás V. Noncoding RNA mediated traffic of foreign mRNA into chloroplasts reveals a novel signaling mechanism in plants. PLoS One. 2010;5:e12269. doi: 10.1371/journal.pone.0012269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi M, Sonoyama M, Mitaku S. High performance system for signal peptide prediction: SOSUIsignal. Chem-Bio Inf J. 2004;4:142–147. [Google Scholar]
- Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- Gschloessl B, Guermeur Y, Cock JM. HECTAR: a method to predict subcellular targeting in heterokonts. BMC Bioinformatics. 2008;9:393. doi: 10.1186/1471-2105-9-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guda C, Guda P, Fahy E, Subramaniam S. MITOPRED: A web server for the prediction of mitochondrial proteins. Nucleic Acids Res. 2004;32:W372–W374. doi: 10.1093/nar/gkh374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AG, Roger AJ. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci USA. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He QF, Dolganov N, Björkman O, Grossman AR. The high light-inducible polypeptides in Synechocystis PCC6803 expression and function in high light. J Biol Chem. 2001;276:306–314. doi: 10.1074/jbc.M008686200. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Hell K. Chopped trapped or tacked–protein translocation into the IMS of mitochondria. Trends Biochem Sci. 2005;30:205–211. doi: 10.1016/j.tibs.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Hiller K, Grote G, Scheer M, Münch R, Jahn D. PrediSi: prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 2004;32:W375–W379. doi: 10.1093/nar/gkh378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann NR, Theg SM. Chloroplast outer membrane protein targeting and insertion. Trends Plant Sci. 2005;10:450–457. doi: 10.1016/j.tplants.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Hoglund A, Donnes P, Blum T, Adolph HW, Kohlbacher O. MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences sequence motifs and amino acid composition. Bioinformatics. 2006;22:1158–1165. doi: 10.1093/bioinformatics/btl002. [DOI] [PubMed] [Google Scholar]
- Hoiczyk E, Hansel A. Cyanobacterial cell walls: news from an unusual prokaryotic envelope. J Bacteriol. 2000;182:1191–1199. doi: 10.1128/JB.182.5.1191-1199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie C, Suzuki H, Sakaguchi M, Mihara K. Targeting and assembly of mitochondrial tail-anchored protein Tom5 to the TOM complex depend on a signal distinct from that of tail-anchored proteins dispersed in the membrane. J Biol Chem. 2003;278:41462–41471. doi: 10.1074/jbc.M307047200. [DOI] [PubMed] [Google Scholar]
- Howe CJ, Barbrook AC, Nisbet RE, Lockhart PJ, Larkum AW. The origin of plastids. Philos Trans R Soc London Ser B. 2008;363:2675–2685. doi: 10.1098/rstb.2008.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Inoue Y, Vermaas W. Characterization of photosystem II subunits from the cyanobacterium Synechocystis sp. PCC6803. In: Mathis P, editor. Photosynthesis: from light to biosphere. Dordrecht: Kluwer Academic Publishers; 1995. pp. 297–300. [Google Scholar]
- Inaba T, Schnell DJ. Protein trafficking to plastids: one theme many variations. Biochem J. 2008;413:15–28. doi: 10.1042/BJ20080490. [DOI] [PubMed] [Google Scholar]
- Jarvis P. Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 2008;179:257–285. doi: 10.1111/j.1469-8137.2008.02452.x. [DOI] [PubMed] [Google Scholar]
- Jeanjean R, Latifi A, Matthijs HC, Havaux M. The PsaE subunit of photosystem I prevents light-induced formation of reduced oxygen species in the cyanobacterium Synechocystis sp. PCC 6803. Biochim Biophys Acta. 2008;1777:308–316. doi: 10.1016/j.bbabio.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jone CS, Kotani N, Aso K, Yang L, Enami I, Kondo K, Tsugita A. Amino acid sequence of the 8-kDa protein in photosystem I reaction center complex from a thermophilic cyanobacterium Synechococcus elongates. Protein Seq Data Anal. 1991;4:327–331. [PubMed] [Google Scholar]
- Jones DT. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics. 2007;23:538–544. doi: 10.1093/bioinformatics/btl677. [DOI] [PubMed] [Google Scholar]
- Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Krauss N. Three-dimensional structure of cyanobacterial photosystem I at 25 A resolution. Nature. 2001;411:909–917. doi: 10.1038/35082000. [DOI] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kanaji S, Iwahashi J, Kida Y, Sakaguchi M, Mihara K. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J Cell Biol. 2000;151:277–288. doi: 10.1083/jcb.151.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashino Y, Lauber WM, Carroll JA, Wang Q, Whitmarsh J, Satoh K, Pakrasi HB. Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry. 2002;41:8004–8012. doi: 10.1021/bi026012+. [DOI] [PubMed] [Google Scholar]
- Kashino Y, Koike H, Yoshio M, Egashira H, Ikeuchi M, Pakrasi HB, Satoh K. Low-molecular-mass polypeptide components of a photosystem II preparation from the thermophilic cyanobacterium Thermosynechococcus vulcanus. Plant Cell Physiol. 2002;43:1366–1373. doi: 10.1093/pcp/pcf168. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW. The tree of eukaryotes. Trends Ecol Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Kies L, Kremer BP. Function of cyanelles in the thecamoeba Paulinella chromatophora. Naturwissenschaften. 1979;66:578–579. doi: 10.1007/BF00368819. [DOI] [Google Scholar]
- Kilian O, Kroth PG. Evolution of protein targeting into “complex” plastids: the “secretory transport hypothesis”. Plant Biol. 2003;5:350–358. doi: 10.1055/s-2003-42710. [DOI] [Google Scholar]
- Kitajima A, Asatsuma S, Okada H, Hamada Y, Kaneko K, Nanjom Y, Kawagoem Y, Toyooka K, Matsuoka K, Takeuchi M, Nakano A, Mitsui T. The rice alpha-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell. 2009;21:2844–2858. doi: 10.1105/tpc.109.068288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaerulff S, Andersen B, Nielsen VS, Møller BL, Okkels JS. The PSI-K subunit of photosystem I from barley (Hordeum vulgare L) evidence for a gene duplication of an ancestral PSI-G/K gene. J Biol Chem. 1993;268:18912–18916. [PubMed] [Google Scholar]
- Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjölander K, Gruissem W, Baginsky S. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol. 2004;14:354–362. doi: 10.1016/j.cub.2004.02.039. [DOI] [PubMed] [Google Scholar]
- Kleine T, Maier UG, Leister D. DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol. 2009;60:115–138. doi: 10.1146/annurev.arplant.043008.092119. [DOI] [PubMed] [Google Scholar]
- Klukas O, Schubert WD, Jordan P, Krauss N, Fromme P, Witt HT, Saenger W. Photosystem I an improved model of the stromal subunits PsaC PsaD and PsaE. J Biol Chem. 1999;274:7351–7360. doi: 10.1074/jbc.274.11.7351. [DOI] [PubMed] [Google Scholar]
- Kutschera U. Symbiogenesis, natural selection, and the dynamic earth. Theory Biosci. 2009;128:191–203. doi: 10.1007/s12064-009-0065-0. [DOI] [PubMed] [Google Scholar]
- Kutschera U, Niklas KJ. Endosymbiosis, cell evolution, and speciation. Theory Biosci. 2005;124:1–24. doi: 10.1016/j.thbio.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Labischinski H, Goodell EW, Goodell A, Hochberg ML. Direct proof of a “more-than-single-layered” peptidoglycan architecture of Escherichia coli W7: a neutron small-angle scattering study. J Bacteriol. 1991;173:751–756. doi: 10.1128/jb.173.2.751-756.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao DM, Shimizu T. A method for discriminating a signal peptide and a putative 1st transmembrane segment. In: Valafar F, editor. Proceedings of the 2001 international conference on mathematics and engineering techniques in medicine and biological sciences (METMBS’01) Las Vegas: CSREA Press; 2001. pp. 119–125. [Google Scholar]
- Lauterborn R. Protozoenstudien II Paulinella chromatophora nov gen nov spec ein beschalter Rhizopode des Süßwassers mit blaugrünen chromatophorenartigen Einschlüssen. Z Wiss Zool. 1895;59:537–544. [Google Scholar]
- Lee CM, Sedman J, Neupert W, Stuart RA. The DNA helicase Hmi1p is transported into mitochondria by a C-terminal cleavable targeting signal. J Biol Chem. 1999;274:20937–20942. doi: 10.1074/jbc.274.30.20937. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Kim DH, Kim YW, Hwang I. Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell. 2001;13:2175–2190. doi: 10.1105/tpc.13.10.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Sohn EJ, Lee KH, Lee DW, Hwang I. The transmembrane domain of AtToc64 and its C-terminal lysine-rich flanking region are targeting signals to the chloroplast outer envelope membrane. Mol Cells. 2004;17:281–291. [PubMed] [Google Scholar]
- Lee DW, Kim JK, Lee S, Choi S, Kim S, Hwang I. Arabidopsis nuclear-encoded plastid transit peptides contain multiple sequence subgroups with distinctive chloroplast-targeting sequence motifs. Plant Cell. 2008;20:1603–1622. doi: 10.1105/tpc.108.060541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz T, Neupert W, Herrmann JM. Import of small Tim proteins into the mitochondrial intermembrane space. EMBO J. 2003;22:4400–4408. doi: 10.1093/emboj/cdg421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz P, Bodył A. A hypothesis for import of the nuclear-encoded PsaE protein of Paulinella chromatophora (Cercozoa Rhizaria) into its cyanobacterial endosymbionts/plastids via the endomembrane system. J Phycol. 2010;46:847–859. doi: 10.1111/j.1529-8817.2010.00876.x. [DOI] [Google Scholar]
- Mant A, Woolhead CA, Moore M, Henry R, Robinson C. Insertion of PsaK into the thylakoid membrane in a “Horseshoe” conformation occurs in the absence of signal recognition particle nucleoside triphosphates or functional albino3. J Biol Chem. 2001;276:36200–36206. doi: 10.1074/jbc.M102914200. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011;39(Database issue):D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin B, Nowack EC, Melkonian M. A plastid in the making: evidence for a second primary endosymbiosis. Protist. 2005;156:425–432. doi: 10.1016/j.protis.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Marin B, Nowack EC, Glockner G, Melkonian M. The ancestor of the Paulinella chromatophore obtained a carboxysomal operon by horizontal gene transfer from a Nitrococcus-like gamma-proteobacterium. BMC Evol Biol. 2007;7:85. doi: 10.1186/1471-2148-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkonian M, Mollenhauer D. Robert Lauterborn (1869–1952) and his Paulinella chromatophora. Protist. 2005;156:253–262. doi: 10.1016/j.protis.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Miras S, Salvi D, Ferro M, Grunwald D, Garin J, Joyard J, Rolland N. Non-canonical transit peptide for import into the chloroplast. J Biol Chem. 2002;277:47770–47778. doi: 10.1074/jbc.M207477200. [DOI] [PubMed] [Google Scholar]
- Miras S, Salvi D, Piette L, Seigneurin-Berny D, Grunwald D, Reinbothe C, Joyard J, Reinbothe S, Rolland N. Toc159- and Toc75-independent import of a transit sequence-less precursor into the inner envelope of chloroplasts. J Biol Chem. 2007;282:29482–29492. doi: 10.1074/jbc.M611112200. [DOI] [PubMed] [Google Scholar]
- Montané MH, Kloppstech K. The family of light-harvesting-related proteins (LHCs ELIPs HLIPs): was the harvesting of light their primary function? Gene. 2000;258:1–8. doi: 10.1016/S0378-1119(00)00413-3. [DOI] [PubMed] [Google Scholar]
- Moretti S, Armougom F, Wallace IM, Higgins DG, Jongeneel CV, Notredame C. The M-Coffee web server: a meta-method for computing multiple sequence alignments by combining alternative alignment methods. Nucleic Acids Res. 2007;35:W645–W648. doi: 10.1093/nar/gkm333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nada A, Soll J. Inner envelope protein 32 is imported into chloroplasts by a novel pathway. J Cell Sci. 2004;117:3975–3982. doi: 10.1242/jcs.01265. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Ishida K. Another acquisition of a primary photosynthetic organelle is underway in Paulinella chromatophora. Curr Biol. 2009;19:R284–R285. doi: 10.1016/j.cub.2009.02.043. [DOI] [PubMed] [Google Scholar]
- Nanjo Y, Oka H, Ikarashi N, Kaneko K, Kitajima A, Mitsui T, Muñoz FJ, Rodrı′guez-López M, Baroja-Fernández E, Pozueta-Romero J. Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-Golgi to the chloroplast through the secretory pathway. Plant Cell. 2006;18:2582–2592. doi: 10.1105/tpc.105.039891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Krogh A. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc Int Conf Intell Syst Mol Biol. 1998;6:122–130. [PubMed] [Google Scholar]
- Nowack EC, Melkonian M, Glöckner G. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr Biol. 2008;18:410–418. doi: 10.1016/j.cub.2008.02.051. [DOI] [PubMed] [Google Scholar]
- Nowack EC, Vogel H, Groth M, Grossman AR, Melkonian M, Glöckner G. Endosymbiotic gene transfer and transcriptional regulation of transferred genes in Paulinella chromatophora. Mol Biol Evol. 2011;28:407–422. doi: 10.1093/molbev/msq209. [DOI] [PubMed] [Google Scholar]
- Nozaki H, Iseki M, Hasegawa M, Misawa K, Nakada T, Sasaki N, Watanabe M. Phylogeny of primary photosynthetic eukaryotes as deduced from slowly evolving nuclear genes. Mol Biol Evol. 2007;24:1592–1595. doi: 10.1093/molbev/msm091. [DOI] [PubMed] [Google Scholar]
- Nugent T, Jones DT. Transmembrane protein topology prediction using support vector machines. BMC Bioinformatics. 2009;10:159. doi: 10.1186/1471-2105-10-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD. The symbiotic birth and spread of plastids: how many times and whodunit? J Phycol. 2003;39:4–11. doi: 10.1046/j.1529-8817.2003.02185.x. [DOI] [Google Scholar]
- Petsalaki EI, Bagos PG, Litou ZI, Hamodrakas SJ (2005) PredSL. http://hannibal.biol.uoa.gr/PredSL/ [DOI] [PMC free article] [PubMed]
- Pfanzagl B, Löffelhardt W. In vitro synthesis of peptidoglycan precursors modified with N-acetylputrescine by Cyanophora paradoxa cyanelle envelope membranes. J Bacteriol. 1999;181:2643–2647. doi: 10.1128/jb.181.8.2643-2647.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanzagl B, Allmaier G, Schmid ER, de Pedro MA, Löffelhardt W. N-acetylputrescine as a characteristic constituent of cyanelle peptidoglycan in glaucocystophyte algae. J Bacteriol. 1996;178:6994–6997. doi: 10.1128/jb.178.23.6994-6997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanzagl B, Zenker A, Pittenauer E, Allmaier G, Martinez-Torrecuadrada J, Schmid ER, De Pedro MA, Löffelhardt W. Primary structure of cyanelle peptidoglycan of Cyanophora paradoxa: a prokaryotic cell wall as part of an organelle envelope. J Bacteriol. 1996;178:332–339. doi: 10.1128/jb.178.2.332-339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plöscher M, Granvogl B, Zoryan M, Reisinger V, Eichacker LA. Mass spectrometric characterization of membrane integral low molecular weight proteins from photosystem II in barley etioplasts. Proteomics. 2009;9:625–635. doi: 10.1002/pmic.200800337. [DOI] [PubMed] [Google Scholar]
- Pollmann S, Springer A, Buhr F, Lahroussi A, Samol I, Bonneville JM, Tichtinsky G, von Wettstein D, Reinbothe C, Reinbothe S. A plant porphyria related to defects in plastid import of protochlorophyllide oxidoreductase. A Proc Natl Acad Sci USA. 2007;104:2019–2023. doi: 10.1073/pnas.0610934104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusnik M, Schmidt O, Perry AJ, Oeljeklaus S, Niemann M, Warscheid B, Lithgow T, Meisinger C, Schneider A. Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin. Curr Biol. 2011;21:1738–1743. doi: 10.1016/j.cub.2011.08.060. [DOI] [PubMed] [Google Scholar]
- Reczko M, Staub E, Fiziev P, Hatzigeorgiou A. Finding signal peptides in human protein sequences using recurrent neural networks. In: Gusfield D, Guigo R, editors. Algorithms in bioinformatics. Berlin/Heidelberg: Springer; 2002. pp. 60–67. [Google Scholar]
- Reinbothe S, Quigley F, Springer A, Schemenewitz A, Reinbothe C. The outer plastid envelope protein Oep16: role as precursor translocase in import of protochlorophyllide oxidoreductase. Proc Natl Acad Sci USA. 2004;101:2203–2208. doi: 10.1073/pnas.0301962101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Prieto A, Weber AP, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annu Rev Genet. 2007;41:147–168. doi: 10.1146/annurev.genet.41.110306.130134. [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A, Yoon HS, Moustafa A, Yang EC, Andersen RA, Boo SM, Nakayama T, Ishida K, Bhattacharya D. Differential gene retention in plastids of common recent origin. Mol Biol Evol. 2010;27:1530–1537. doi: 10.1093/molbev/msq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Löffelhardt W, Bohnert HJ, Philippe H, Lang BF. Monophyly of primary photosynthetic eukaryotes: green plants red algae and glaucophytes. Curr Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Schein AI, Kissinger JC, Ungar LH. Chloroplast transit peptide prediction: a peek inside the black box. Nucleic Acids Res. 2001;29:E82. doi: 10.1093/nar/29.16.e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HB, Chou KC. Signal-3L: a 3-layer approach for predicting signal peptides. Biochem Biophys Res Commun. 2007;363:297–303. doi: 10.1016/j.bbrc.2007.08.140. [DOI] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- StatSoft Inc. STATISTICA (Data Analysis Software System) Version. 2006;7:1. [Google Scholar]
- Steiner JM, Löffelhardt W. Protein translocation into and within cyanelles (review) Mol Membr Biol. 2005;22:1230–1232. doi: 10.1080/09687860500041411. [DOI] [PubMed] [Google Scholar]
- Steiner JM, Yusa F, Pompe JA, Löffelhardt W. Homologous protein import machineries in chloroplasts and cyanelles. Plant J. 2005;44:646–652. doi: 10.1111/j.1365-313X.2005.02559.x. [DOI] [PubMed] [Google Scholar]
- Stiller JW. Plastid endosymbiosis genome evolution and the origin of green plants. Trends Plant Sci. 2007;12:391–396. doi: 10.1016/j.tplants.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- Villarejo A, Burén S, Larsson S, Déjardin A, Monné M, Rudhe C, Karlsson J, Jansson S, Lerouge P, Rolland N, von Heijne G, Grebe M, Bako L, Samuelsson G. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat Cell Biol. 2005;7:1224–1231. doi: 10.1038/ncb1330. [DOI] [PubMed] [Google Scholar]
- Waizenegger T, Stan T, Neupert W, Rapaport D. Signal-anchor domains of proteins of the outer membrane of mitochondria: structural and functional characteristics. J Biol Chem. 2003;278:42064–42071. doi: 10.1074/jbc.M305736200. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2: a multiple sequence alignment and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol. 2004;21:809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Reyes-Prieto A, Melkonian M, Bhattacharya D. Minimal plastid genome evolution in the Paulinella endosymbiont. Curr Biol. 2006;16:R670–R672. doi: 10.1016/j.cub.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Nakayama T, Reyes-Prieto A, Andersen RA, Boo SM, Ishida KI, Bhattacharya D. A single origin of the photosynthetic organelle in different Paulinella lineages. BMC Evol Biol. 2009;9:98. doi: 10.1186/1471-2148-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouni A, Witt HAT, Kern J, Fromme P, Krauß N, Saenger W, Orth P. Crystal structure of photosystem II from Synechococcus elongatus at 38 Å resolution. Nature. 2001;409:739–743. doi: 10.1038/35055589. [DOI] [PubMed] [Google Scholar]


