Highlights
► Fermentation of xylose into ethanol is important in “biomass-to-biofuel” processes. ► Protein engineering has been extensively used to improve xylose fermentation. ► Published data on variants of XR and XDH are explored as predictors of strain performance. ► Applying internal reactant concentrations to bi-substrate kinetic equations give a reliable forecast on xylitol formation. ► A criterion of physiological fitness of engineered forms of XR is developed.
Keywords: Xylose fermentation, Xylose reductase, Xylitol dehydrogenase, Coenzyme specificity, Redox imbalance, Enzyme kinetics, Physiology
Abstract
An advanced strategy of Saccharomyces cerevisiae strain development for fermentation of xylose applies tailored enzymes in the process of metabolic engineering. The coenzyme specificities of the NADPH-preferring xylose reductase (XR) and the NAD+-dependent xylitol dehydrogenase (XDH) have been targeted in previous studies by protein design or evolution with the aim of improving the recycling of NADH or NADPH in their two-step pathway, converting xylose to xylulose. Yeast strains expressing variant pairs of XR and XDH that according to in vitro kinetic data were suggested to be much better matched in coenzyme usage than the corresponding pair of wild-type enzymes, exhibit widely varying capabilities for xylose fermentation. To achieve coherence between enzyme properties and the observed strain performance during fermentation, we explored the published kinetic parameters for wild-type and engineered forms of XR and XDH as possible predictors of xylitol by-product formation (Yxylitol) in yeast physiology. We found that the ratio of enzymatic reaction rates using NADP(H) and NAD(H) that was calculated by applying intracellular reactant concentrations to rate equations derived from bi-substrate kinetic analysis, succeeded in giving a statistically reliable forecast of the trend effect on Yxylitol. Prediction based solely on catalytic efficiencies with or without binding affinities for NADP(H) and NAD(H) were not dependable, and we define a minimum demand on the enzyme kinetic characterization to be performed for this purpose. An immediate explanation is provided for the typically lower Yxylitol in the current strains harboring XR engineered for utilization of NADH as compared to strains harboring XDH engineered for utilization of NADP+. The known XDH enzymes all exhibit a relatively high Km for NADP+ so that physiological boundary conditions are somewhat unfavorable for xylitol oxidation by NADP+. A criterion of physiological fitness is developed for engineered XR working together with wild-type XDH.
1. Introduction
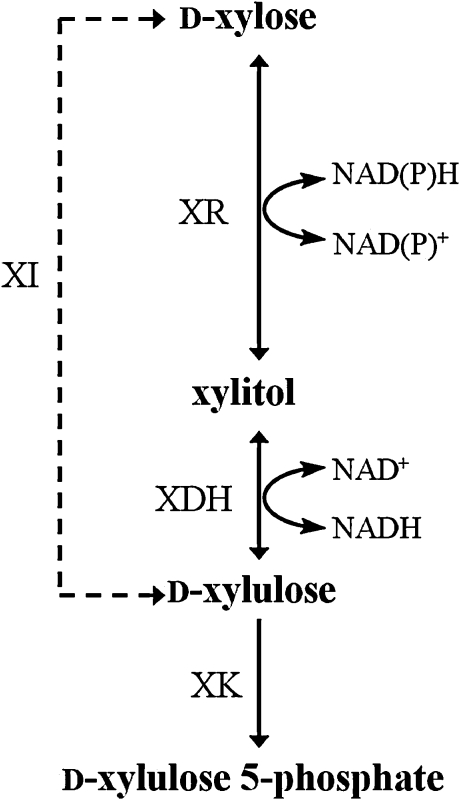
Xylose is the most abundant pentose sugar on earth. Its use for ethanol fermentation is a central step in many “biomass-to-biofuel” processes that are currently advancing to commercial production scale [http://biofuels.abc-energy.at/demoplants/ (April 16th, 2011)]. A microorganism capable of efficient fermentation of the main sugars present in the hydrolyzates of lignocellulosic biomass (glucose, xylose) would be highly desirable in the industrial process. Considering strain robustness and efficiency of glucose fermentation, the yeast Saccharomyces cerevisiae is a top candidate to be applied for biofuel production (Hahn-Hägerdal et al., 2007a). Metabolic engineering has been widely used to overcome the major drawback of S. cerevisiae that in its wild-type form, the organism is not able to utilize xylose (Chu and Lee, 2007; Hahn-Hägerdal et al., 2007b; Matsushika et al., 2009a). Utilization of xylose in other yeasts (e.g. Pichia stipitis, recently renamed to Scheffersomyces stipitis) (Kurtzman and Suzuki, 2010) occurs through a two-step oxidoreductive pathway, consisting of xylose reductase (XR) and xylitol dehydrogenase (XDH), that achieves net isomerization of xylose into xylulose via xylitol as intermediate (Scheme 1). Heterologous expression of the XR–XDH pathway has therefore been a prime strategy of development of xylose-fermenting strains of S. cerevisiae. An alternative strategy, inspired by the bacterial metabolism that involves direct isomerization of xylose into xylulose, has been pursued with notable success in the recent past (Brat et al., 2009; Kuyper et al., 2005; Madhavan et al., 2009). Examination of strain performance during fermentation of the real substrate, that is the lignocellulose hydrolyzate, has revealed the importance of physiological resistance to the inhibitors released during upstream processing of the feedstock (Almeida et al., 2011). Moreover, these studies show the possible role of reductive enzymes such as XR in conferring this resistance (Almeida et al., 2008; Liu, 2006). Strains constructed on the basis of the XR–XDH pathway therefore present a strong option for process development.
Scheme 1.
Pathways for utilization of xylose. XR, xylose reductase; XDH, xylitol dehydrogenase; XK, xylulose kinase; XI, xylose isomerase.
A common observation for “first generation” strains of S. cerevisiae expressing XR and XDH in their respective wild-type form (e.g. from P. stipitis) was production of fermentation by-products, especially xylitol in large amounts (Yxylitol), resulting in low ethanol yields (Yethanol) (Hahn-Hägerdal et al., 2007b; Jeffries, 2006). Mismatch in coenzyme usage by XR, which prefers NADPH over NADH, and XDH, which is dependent on NAD+, implies that regeneration of NAD(P) coenzymes during conversion of xylose into xylulose is incomplete. This in turn causes imbalance in the NAD(H) and NADP(H) pools which ultimately is the reason for the high value of Yxylitol. The mechanism underlying xylose fermentation at low Yxylitol under conditions of the natural host (e.g. P. stipitis) is not fully understood (Delgenes et al., 1986; Dellweg et al., 1984; Jeffries and Van Vleet, 2009; Skoog and Hahn-Hägerdal, 1990).
The different suggestions made for improving the performance of XR–XDH strains of S. cerevisiae can be categorized according to whether their approach was removal of the imbalance at its source or mitigation of the effects caused by it (Chu and Lee, 2007; Hahn-Hägerdal et al., 2007b; Matsushika et al., 2009a). The first of the two approaches which would seem to be most compelling in principle, requires that coenzyme utilization by XR and XDH be adjusted one to another without compromising the overall flux. In the apparent absence of a naturally well matched pair of XR and XDH, protein engineering was extensively applied to prepare variants of XR and XDH that show enhanced specificity for NADH and NADP+, respectively. Structure-guided mutagenesis and directed evolution were used independently or in combination. Tables S1 and S2 in Supporting Information provide a compilation of the reported variants of XR and XDH. The variants shown in Tables 1 and 2 were selected on the basis of prior use in S. cerevisiae strain engineering.
Table 1.
XR mutants with altered coenzyme specificity.
| XR | Coenzyme | Kinetic constants |
NADH preference |
Ref. | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KNAD(P)H (μM) | Kxylose (mM) | KiNAD(P)H (μM) | kcat (s−1) | vmax (U/mg) | () | Rsel | |||||||
| Ct WTa | NADH | 38 | 142 | 19 | 11 | ||||||||
| NADPH | 3 | 96 | 1 | 13 | 0.7 | 0.54 | 0.8 | 0.07 | 0.05 | 0.03 | Petschacher et al. (2005) | ||
| Ct K274Ma | NADH | 380 | 229 | 352 | 19 | ||||||||
| NADPH | 75 | 506 | 55 | 36 | 1.4 | 1.5 | 0.5 | 0.1 | 0.23 | 0.18 | Petschacher et al. (2005) | ||
| Ct N276Da | NADH | 26 | 99 | 60 | 14 | ||||||||
| NADPH | 17 | 170 | 21 | 37 | 0.7 | 2.3 | 0.4 | 0.25 | 0.42 | 0.23 | Petschacher et al. (2005) | ||
| Ct K274R N276Da | NADH | 41 | 106 | 30 | 12 | ||||||||
| NADPH | 128 | 722 | 64 | 30 | 6.8 | 29 | 0.4 | 1.2 | 8.5 | 5.8 | Petschacher et al. (2005) | ||
| Ps WTb | NADH | 30.5 | 31.5 | na | 6.9 | ||||||||
| NADPH | 2.5 | 47 | na | 10.5 | 0.7 | na | 0.7 | 0.05 | 0.08 | na | Watanabe et al. (2007a) | ||
| Ps R276Hb | NADH | 17 | 45.5 | na | 6.8 | ||||||||
| NADPH | 1.7 | 53.2 | na | 0.3 | 26.6 | na | 25.5 | 2.6 | 2.98 | na | Watanabe et al. (2007a) | ||
| Ps K270Rb | NADH | na | na | na | na | 6.7c | |||||||
| NADPH | na | na | na | na | 7.7c | na | na | 0.9 | na | na | na | Watanabe et al. (2007a) | |
| Ps WTd | NADH | 28.7 | 59.2 | 26 | na | 0.21e | |||||||
| NADPH | 1 | 62.2 | 1.4 | na | 0.3e | 0.7 | 0.29 | 0.7 | 0.02 | 0.02 | 0.04 | Bengtsson et al. (2009) | |
| Ps K270Rd | NADH | 62.8 | 145 | 57 | na | 0.96e | |||||||
| NADPH | 25.8 | 468 | 23 | na | 2.13e | 1.5 | 3.8 | 0.5 | 0.19 | 0.6 | 0.58 | Bengtsson et al. (2009) | |
| Ps K270M | NADH | na | na | na | na | ||||||||
| NADPH | 185.3f (290)d |
624.9f (454)d |
na (293)d |
na na |
(0.91)d,e |
na | na | na | na | na | na |
Kostrzynska et al. (1998) Bengtsson et al. (2009) |
|
| Ps WTf | NADH | 9 | 70 | 3 | na | 0.21e | |||||||
| NADPH | 1.6 | 35 | 1 | na | 0.28e | 0.6 | 1.3 | 0.8 | 0.13 | 0.07 | 0.13 | Runquist et al. (2009b) | |
| Ps N272D P275Qf | NADH | 26 | 80 | 50 | na | 2e | |||||||
| NADPH | 40 | 50 | 100 | na | 3.51e | 1.1 | 7.9 | 0.6 | 0.88 | 0.55 | 0.71 | Runquist et al. (2009b) | |
| Ps N272Df | NADH | 4 | 36 | 30 | na | 1.09e | |||||||
| NADPH | 24 | 40 | 50 | na | 2.4e | 0.8 | 15 | 0.5 | 2.7 | 3.03 | 0.84 | Runquist et al. (2009b) | |
| Ps P275Qf | NADH | −2g | 160 | 6 | na | 0.38e | |||||||
| NADPH | 5.5 | 72 | −3g | na | 0.36e | 0.9i | 8.0g | 1.1 | 5.8g | 2.61g | 0.08g | Runquist et al. (2009b) | |
| Ps WTd | NADH | na | na | na | na | 7.2c,h | |||||||
| NADPH | na | na | na | na | 15.7c,h | na | na | 0.46 | na | na | na | Watanabe et al. (2007b) | |
| Ps N272Dd | NADH | na | na | na | na | ||||||||
| NADPH | na | na | na | na | na | na | 0.6h | na | na | na | Watanabe et al. (2007b) | ||
| Ps WTj | NADH | 40 | 97 | 30 | na | 57 | |||||||
| NADPH | 3.2 | 68 | 6.6 | na | 57 | 0.9 | 1.1 | 1.0 | 0.08 | 0.06 | 0.15 | Rizzi et al. (1988) | |
Ct = Candida tenuis; Ps = Scheffersomyces stipitis (Pichia stipitis); na = not available.
Measured at 25 °C, pH 7.0.
Measured at 35 °C, pH 7.0.
Specific activities of purified enzyme.
Measured at 30 °C, pH 7.0.
Specific activities in crude cell extract.
Measured at 22 °C, pH 7.0.
Values of 1 μM are assumed for KNADH and KiNADPH to calculate NADH-preferences.
Measured with 133 mM xylose, 150 μM NAD(P)H.
Measured at 37 °C, pH 7.0.
Measured at pH 7.0 (temperature not stated).
Table 2.
XDH mutants with altered coenzyme specificity.
| XDH | Coenzyme | Kinetic constants |
NAD+ preference |
Ref. | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| (μM) | Kxylitol (mM) | kcat (s−1) | vmax (U/mg) | vmax/ (U/mg/mM) | () | ||||||
| Ps WTa | NAD+ | 381 | 21.7 | 17.5 | |||||||
| NADP+ | 170,000 | nab | 1.833 | (0.9–4)×104c | 9.5 | 4259 | na | Watanabe et al. (2005) | |||
| Ps D207A I208R F209Sa | NAD+ | 1300 | 55.7 | 4 | |||||||
| NADP+ | 897 | 31.1 | 41.67 | 0.2–0.8c | 0.1 | 0.07 | 0.04 | Watanabe et al. (2005) | |||
| Ps D207A I208R F209S N211Ra | NAD+ | 17,300 | nab | 23.83 | |||||||
| NADP+ | 1380 | 72.6 | 64 | 0.1–1c | 0.4 | 0.03 | na | Watanabe et al. (2005) | |||
| Ps S96C S99C Y102Ca | NAD+ | 739 | 30.3 | 30.33 | |||||||
| NADP+ | 9560 | nab | 0.91 | (1–6)×103c | 33.3 | 431 | na | Watanabe et al. (2005) | |||
| Ps S96C S99C Y102C D207A I208R F209Sa | NAD+ | 23,500 | nab | 29.5 | |||||||
| NADP+ | 1180 | 111 | 210 | 0.04–0.2c | 0.1 | 0.007 | na | Watanabe et al. (2005) | |||
| Ps S96C S99C Y102C D207A I208R F209S N211Ra | NAD+ | 7600 | nab | 13.17 | |||||||
| NADP+ | 1040 | 119 | 183.3 | 0.05–0.3c | 0.1 | 0.010 | na | Watanabe et al. (2005) | |||
| Ps WTd | NAD+ | na | na | na | 1.654e | ||||||
| NADP+ | na | na | na | 0.005e | na | 330 | na | na | Hou et al. (2007) | ||
| Ps D207A I208R F209Sd | NAD+ | na | na | na | 0.271e | ||||||
| NADP+ | na | na | na | 0.782e | na | 0.35 | na | na | Hou et al. (2007) | ||
| Ps S96C S99C Y102C D207A I208R F209Sd | NAD+ | na | na | na | 0.136e | ||||||
| NADP+ | na | na | na | 0.698e | na | 0.19 | na | na | Hou et al. (2007) | ||
| Gm WTf,g | NAD+ | 514.3 | na | na | 0.194e | 0.38e | |||||
| NADP+ | na | na | na | 0.0007e | (1–6)×103h | na | 540 | na | Krahulec et al. (2009) | ||
| Gm D202A L203Δ V204Δ E205P S206Rf,g | NAD+ | na | na | na | 0.017e | ||||||
| NADP+ | na | na | na | 0.067e | 1–9h | na | 0.25 | na | Krahulec et al. (2009) | ||
| Gm WTf,i | NAD+ | 909.8 | na | na | 0.197e | 0.22e | |||||
| NADP+ | na | na | na | 0.0006e | (1–6)×103h | na | 370 | na | Krahulec et al. (2009) | ||
| Gm D202A L203Δ V204Δ E205P S206Rf,i | NAD+ | na | na | na | 0.006e | ||||||
| NADP+ | na | na | na | 0.028e | 1–7h | na | 0.21 | na | Krahulec et al. (2009) | ||
Ps = Scheffersomyces stipitis (Pichia stipitis); Gm = Galactocandida mastotermitis; na = not available.
Measured at 35 °C, pH 9.0.
Identical Kxylitol values with NAD+ and NADP+ are assumed in the calculation of veff.
Range of veff values for substrate concentrations of NAD+ 0.5–1.0 mM; NADP+ 0.03–0.1 mM; xylitol 10–150 mM.
Measured at pH 7.0.
Measured in crude S. cerevisiae extract.
Measured at 25 °C, pH 7.0.
Measured with 150 mM xylitol.
Range of veff values for substrate concentrations of NAD+ 0.5–1.0 mM; NADP+ 0.03–0.1 mM.
Measured with 50 mM xylitol.
A series of “second generation” strains of S. cerevisiae are now available in which an engineered form of XR or XDH was expressed and performance in xylose fermentation benchmarked against the relevant reference strain that expresses the corresponding wild-type enzyme. Table 3 summarizes the results from the literature.1 Looking at the published evidence as a whole, a major complication is immediately revealed: the various XR–XDH strains differ immensely among each other with respect to their fermentation capabilities, and the achieved improvement in the relative distribution of fermentation products from xylose as compared to the corresponding reference strain was anything but uniform across them (Table 3). Now, all of the chosen combinations of XR and XDH were expected from in vitro kinetic data to be much better matched in coenzyme usage than the corresponding pair of wild-type enzymes. Therefore, this raises strong concern about the validity of the presumed correlation between the biochemical properties of the XR–XDH pathway and the physiology of the resulting S. cerevisiae strain under xylose fermentation conditions.
Table 3.
S. cerevisiae strains expressing an engineered XR–XDH pathway and their performances in xylose fermentation.
| Strain | Parental yeast strain | Genetic background | Xylose (Glucose) (g/L) | Yethanol (g/g) | Yxylitol (g/g) | Yglycerol (g/g) | qxylose (g/g/h) | Increase ethanol (%) | Increase xylitol (%) | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| XR mutant | ||||||||||
| BP000 (XM) | CEN.PK 113-5D | G CtXR/GmXDH/XK | 20 | 0.24 | 0.39 | 0.048 | 0.06 | WT | WT | Petschacher and Nidetzky (2008) |
| BP10001 (XM) | CEN.PK 113-5D | G CtXR–K274RN276D/GmXDH/XK | 20 | 0.34 | 0.19 | 0.021 | 0.08 | 42 | −51 | Petschacher and Nidetzky (2008) |
| TMB3001 (XM) | CEN.PK 113-7A | G PsXR/PsXDH/XK | 50 | 0.31 | 0.29 | 0.052 | 0.145 | WT | WT | Jeppsson et al. (2006) |
| TMB3270 (XM) | CEN.PK 113-11C | G PsXR–K270M/PsXDH/XK | 50 | 0.36 | 0.17 | 0.072 | 0.155 | 16 | −41 | Jeppsson et al. (2006) |
| TMB3260a (XM) | CEN.PK 113-7A | G PsXR/PsXDH/XK | 50 | 0.30 | 0.13 | 0.161 | 0.245 | WT | WT | Jeppsson et al. (2006) |
| TMB3271a (XM) | CEN.PK 113-11C | G PsXR–K270M/PsXDH/XK | 50 | 0.31 | 0.09 | 0.181 | 0.226 | 3 | −31 | Jeppsson et al. (2006) |
| TMB3321 (ns) | CEN.PK 2-1C | G PsXR/PsXDH/XK/TAL/TKL/RKI/RPE/ΔGRE3 | 50 (20) | 0.33b | 0.26c | 0.095b | na | WT | WT | Bengtsson et al. (2009) |
| TMB3322 (ns) | CEN.PK 2-1C | G PsXR–K270M/PsXDH/XK/TAL/TKL/RKI/RPE/ΔGRE3 | 50 (20) | 0.38b | 0.09c | 0.067b | na | na | −65 | Bengtsson et al. (2009) |
| TMB3323 (ns) | CEN.PK 2-1C | G PsXR–K270R/PsXDH/XK/TAL/TKL/RKI/RPE/ΔGRE3 | 50 (20) | 0.38b | 0.09c | 0.079b | na | na | −65 | Bengtsson et al. (2009) |
| TMB3424d (XM) | CEN.PK 2-1C | G PsXR/PsXDH/XK/TAL/TKL/RKI/RPE/ΔGRE3 | 60 | 0.12 | 0.53 | 0.03e | 0.06 | WT | WT | Runquist et al. (2010) |
| TMB3421d (XG) | CEN.PK 2-1C | G PsXR–N272DP275Q/PsXDH/XK/TAL/TKL/RKI/RPE/ΔGRE3 | 60 | 0.35 | 0.26 | 0.03e | 0.57 | 192 | −51 | Runquist et al. (2010) |
| TMB3422d (XG) | CEN.PK 2-1C | G PsXR–N272D/PsXDH/XK/TAL/TKL/RKI/RPE/ΔGRE3 | 60 | 0.37 | 0.24 | 0.03e | 0.62 | 208 | −55 | Runquist et al. (2010) |
| TMB3423d (XM) | CEN.PK 2-1C | G PsXR–P275Q/PsXDH/XK/TAL/TKL/RKI/RPE/ΔGRE3 | 60 | 0.18 | 0.57 | 0.02e | 0.097 | 50 | 8 | Runquist et al. (2010) |
| Y-WT (ns) | D452-2 | P PsXR/PsXDH | 15 (5) | 0.26c | 0.37c | 0.029c | na | WT | WT | Watanabe et al. (2007a) |
| Y-R276H (ns) | D452-2 | P PsXR–R276H/PsXDH | 15 (5) | 0.41c | 0.25c | 0.033c | na | 58 | −32 | Watanabe et al. (2007a) |
| Y-WTf (ns) | D452-2 | P PsXR/PsXDH | 15 (5) | 0.20b | 0.24c | na | na | WT | WT | Watanabe et al. (2007b) |
| Y-K270Rf (ns) | D452-2 | P PsXR–K270R/PsXDH | 15 (5) | 0.21b | 0.18c | na | na | na | −25 | Watanabe et al. (2007b) |
| XDH mutant | ||||||||||
| MA-N4 (XG) | INVSc1 | G PsXR/PsXDH/XK | 45 | 0.34 | 0.10 | na | 0.19 | WT | WT | Matsushika et al. (2008) |
| MA-N5 (XG) | INVSc1 | G PsXR/PsXDH–D207AI208RF209SN211R/XK | 45 | 0.36 | 0.06 | na | 0.25 | 6 | −40 | Matsushika et al. (2008) |
| MA-R4 (ns) | IR-2 | G PsXR/PsXDH/XK | 45 | 0.34 | 0.048 | 0.101 | na | WT | WT | Matsushika et al. (2009b) |
| MA-R5 (ns) | IR-2 | G PsXR/PsXDH–D207AI208RF209SN211R/XK | 45 | 0.37 | 0.038 | 0.076 | g | 9h | −21 | Matsushika et al. (2009b) |
| Y-WTi (ns) | D452-2 | P PsXR/PsXDH | 15 (5) | 0.27c,f | 0.35c,f | na | na | WT | WT | Watanabe et al. (2007c) |
| Y-ARSdR (ns) | D452-2 | P PsXR/PsXDH–D207AI208RF209SN211R | 15 (5) | 0.50c,f | 0.05c,f | 0.02b,f | na | 85 | −86 | Watanabe et al. (2007c) |
| H1290 (ns) | H158 | P/G PsXR/PsXDH/XK | 50 | 0.325 | 0.394 | 0.034 | 0.185 | WT | WT | Hou et al. (2007) |
| H1291 (ns) | H158 | P/G PsXR/PsXDH–D207AI208RF209S/XK | 50 | 0.261 | 0.236 | 0.019 | 0.16 | −20 | −40 | Hou et al. (2007) |
| H1292 (ns) | H158 | P/G PsXR/PsXDH–S96CS99CY102CD207AI208RF209S/XK | 50 | 0.223 | 0.467 | 0.02 | 0.121 | −31 | 19 | Hou et al. (2007) |
| XR and XDH mutants | ||||||||||
| BP000j (XM) | CEN.PK 113-5D | G CtXR/GmXDH/XK | 20 | 0.23 | 0.36 | 0.073 | 0.09 | WT | WT | Krahulec et al. (2009) |
| BP11001j (XM) | CEN.PK 113-5D | G CtXR–K274RN276D/GmXDH–D202AL203ΔV204ΔE205PS206R/XK | 20 | 0.23 | 0.37 | 0.046 | 0.06 | 0 | 3 | Krahulec et al. (2009) |
| BP11002j (XM) | CEN.PK 113-5D | G CtXR–N276D/GmXDH–D202AL203ΔV204ΔE205PS206R/XK | 20 | 0.20 | 0.41 | 0.041 | 0.06 | −13 | 14 | Krahulec et al. (2009) |
| BP000k (XM) | CEN.PK 113-5D | G CtXR/GmXDH/XK | 20 | 0.24 | 0.39 | 0.048 | 0.06 | WT | WT | Petschacher and Nidetzky (2008) |
| BP11001k (XM) | CEN.PK 113-5D | G CtXR–K274RN276D/GmXDH–D202AL203ΔV204ΔE205PS206R/XK | 20 | 0.24 | 0.34 | 0.023 | 0.06 | 0 | −13 | Krahulec et al. (2009) |
WT = wild-type; na = not available; XM = xylose metabolizing (anaerobic); XG = xylose growing (anaerobic); ns = ability to grow anaerobically on xylose not stated; G = xylose pathway integrated into the genome; P = plasmid based expression of the pathway; P/G = XR and XK integrated into genome, XDH expressed with a high copy plasmid.
Overproduces XR (2 copies).
Yields refer to total amount of sugar consumed (glucose + xylose).
Yields refer to the consumed xylose.
Different promoters for XR used compared to TMB3321/3322/3323.
Yield calculated from xylose uptake rate and glycerol production rate.
Yields calculated from concentrations given in the article or inferred from time course graphs of the fermentations.
qxylose 39% higher than for MA-R4.
Authors state that ethanol yield is not significantly higher compared to the yield of the strain expressing wild-type XDH.
Yields estimated from the published figure showing the fermentation time course.
Shake flask (oxygen limited).
Bioreactor (anaerobic).
The aim of this work was to examine to what extent the relative strain performance can be predicted from the kinetic comparison of wild-type and engineered forms of XR or XDH. Note: Unless mentioned otherwise, strain performance refers to the yield coefficients of the xylose fermentation, considering that Yxylitol should be minimized to obtain an as high as possible Yethanol. We show that in addition to the requisite information about enzyme kinetic properties, knowledge about intracellular metabolite concentrations is essential in the development of a reliable predictor. We further show that assessment of enzyme function under the in vivo boundary conditions should be done on the basis of rate equations obtained in bi-substrate kinetic analysis. The results presented provide an immediate explanation for the superior performance of the current strains harboring XR engineered for utilization of NADH as compared to strains harboring XDH engineered for utilization of NADP+. Moreover, a criterion of physiological fitness is developed for engineered XR working together with wild-type XDH. The published evidence on different XR–XDH strains is therefore brought into a coherent whole.
2. Materials and methods
2.1. Kinetic analysis of coenzyme specificity
Enzyme preference for reaction with NAD(H) as compared to NADP(H) can be kinetically expressed in several ways, as shown in Eqs. (1)–(4) where the superscript r is used to indicate the ratio NAD(H)/NADP(H). Turnover numbers (kcat) of purified enzymes are applied although in the case that kcat was not available, they are replaced by specific activities (vmax) of the enzyme preparation. Different expressions of enzyme catalytic efficiency (keff) may be used, depending on the accessible data. Interpretation of the kinetic parameters is strongly dependent on the kinetic mechanism utilized, and it is assumed here in agreement with the literature that both XR (Neuhauser et al., 1997; Rizzi et al., 1988) and XDH (Lunzer et al., 1998) operate by an ordered bi-substrate mechanism in which the coenzyme binds first. Under these conditions, the efficiency for coenzyme (keffA) reflects binding of NAD(P)H or NAD(P)+ to the free XR or XDH, respectively. KNAD(P)(H) is an apparent Michaelis constant for coenzyme (NADH, NADPH, NAD+ or NADP+). In practice, keffA is often obtained from initial rates recorded at saturating substrate concentration. The parameter , which considers binding affinity of the second substrate, is therefore relevant. Kxyl is the apparent Michaelis constant for xylose or xylitol. Results of full kinetic studies yield a dissociation constant for coenzyme (Ki) and the Michaelis constant for substrate (Km). Contrary to Eqs. (1)–(3) that are all empirical expressions of the coenzyme preference, the parameter Rsel in Eq. (4) presents a true selectivity constant of the enzyme for utilization of NAD(H) and NADP(H). However, its application is strictly valid only under conditions in which the concentrations of coenzyme and substrate are both limiting and kinetics is not further complicated by inhibition.
| (1) |
| (2) |
| (3) |
| (4) |
2.2. Kinetic analysis based on metabolite data
Preference for utilization of NAD(H) as compared to NADP(H) under given (physiological) reaction conditions () is described by Eq. (5). The equation presents a simplification of the full rate equation for simultaneous use of NAD(H) and NADP(H) for conversion of a single substrate (Banta et al., 2002), assuming that the reaction is irreversible and no product and substrate inhibition occurs (Banta et al., 2002; Petschacher and Nidetzky, 2005). Considering that we were mainly interested in relative rates of reaction with NAD(H) compared to NADP(H), these simplifying assumptions appeared to be fully justified. The use of NAD(H) and NADP(H) is indicated in superscript. The concentrations of coenzyme and substrate are in italics.
| (5) |
Eq. (6) compares enzymatic reaction rates using NAD(H) and NADP(H) with the simplifying assumption that the two conversions proceed completely independent one of another. In other words, two forms of the enzyme are assumed to be present in equal amounts, one using NAD(H) and the other using NADP(H). Kinetically, the assumption is strictly valid only under conditions of limiting reactant concentrations. However, because the currently used metabolic models of xylose utilization are usually based on exactly this assumption, it is reasonable to investigate the potential use of Eq. (6) for predictive purpose.
| (6) |
The intracellular concentrations of NADH and NADPH in S. cerevisiae were previously determined under conditions of xylose fermentation as 290 μM and 29 μM, respectively (Klimacek et al., 2010). The intracellular concentration of xylose is not known precisely. However, with the reasonable assumption that import of xylose is not limiting during the rather slow xylose utilization by S. cerevisiae under conditions when no glucose is present (Gárdonyi et al., 2003; Hamacher et al., 2002; Kötter and Ciriacy, 1993; Krahulec et al., 2010; Olofsson et al., 2011), the intracellular xylose concentration was approximated by the external concentration and assumed as 133 mM (20 g/L) in the reported calculations. Note: There have been reports suggesting that xylose transport can become rate-limiting in S. cerevisiae (Hector et al., 2008; Jojima et al., 2010; Katahira et al., 2008; Runquist et al., 2009a). However, this applies mainly to situations when glucose and xylose are co-utilized, when xylose concentrations in the media are low, or when aerobic conditions are used. Taking into account uncertainty in the intracellular metabolite levels (Klimacek et al., 2010), especially that of xylose for which various assumptions needed to be made, sensitivity analysis for was performed for a broader range of NADPH and xylose concentrations (Fig. S1, Supporting Information).
Klimacek et al. (2010) have shown that the intracellular concentrations of NAD+ and NADP+ vary considerably depending on the substrate glucose or xylose utilized by S. cerevisiae (strain BP000). Moreover, the intracellular concentration of xylitol is to our knowledge not available. We therefore calculated of XDH for a representative range of concentrations: NAD+ (experimental: 0.5–1.0 mM); NADP+ (experimental: 30–100 μM); xylitol (assumed: 10–150 mM). In the case that the literature had reported only one of the and , it was assumed that both parameters are the same, consistent with the published evidence that (see Table S2).
2.3. Prediction of Yxylitol from the kinetically determined selectivities of XR
The different selectivities of XR calculated through use of Eqs. (1)–(6) were applied to obtain prediction of xylitol yields using Eq. (7), where Psel stands for selectivity.
| (7) |
Eq. (7) assumes that under conditions of a completely NAD+-dependent XDH, all of the xylose converted by NADPH accumulates as xylitol. was compared with the experimentally determined yield coefficient (). The resulting ΔYxylitol was obtained by Eq. (8) and subjected to statistical Box-plot analysis (NIST/SEMATECH e-Handbook of Statistical Methods, http://www.itl.nist.gov/div898/handbook/, April 2011) as well as to independent two-sample t-tests (Dean and Voss, 1999).
| (8) |
3. Results and discussion
3.1. Kinetic properties of mutated XR and XDH used in S. cerevisiae strain engineering
Both XR and XDH have previously been engineered for altered selectivity of use of NAD(H) and NADP(H) in their respective reactions. Structure–function and protein engineering studies of XR from C. tenuis (CtXR) have provided a molecular basis of XR coenzyme specificity (Kavanagh et al., 2002, 2003; Petschacher et al., 2005) that could subsequently be exploited in the re-design or targeted evolution of orthologs from P. stipitis, Candida boidinii, Hansenula polymorpha, and Talaromyces emersonii (see Table S1). The structure of CtXR has been particularly important because unlike XDH that contains the well known Rossmann-fold coenzyme binding domain (Habenicht et al., 1999; Meijers and Cedergren-Zeppezauer, 2004), XR displays an atypical coenzyme binding site derived from a (β/α)8 barrel fold (Kavanagh et al., 2003). Moreover, the site in the XR structure that accommodates the 2′-phosphate group in NADPH is highly flexible, complicating its optimization for preferred use of NADH (Kavanagh et al., 2003). Engineering of XDH for use of NADP+ followed a natural design strategy recognized within the protein superfamily of medium-chain dehydrogenase/reductases that includes XDH. Ketose reductase from silverleaf whitefly is closely related to XDH but differs from it in being a NADP+-dependent enzyme. Determinants of NADP+-dependent reductase activity were successfully grafted on XDH from P. stipitis.
Tables 1 and 2 show kinetic parameters of wild-type and mutated forms of XR and XDH, respectively that will subsequently be used in the analysis of “enzyme fitness” for function during xylose fermentation by the corresponding yeast strain. A complete summary of the kinetic data collected from the literature is given in Tables S1 and S2. It is somewhat unfortunate but also as expected from the different research groups involved that the kinetic parameters were not always obtained under exactly identical conditions. We are fully aware of the potential limitation caused. Depending on the availability of the requisite kinetic parameters, we used each applicable equation from Eqs. (1)–(6) for calculation of a coenzyme selectivity coefficient. The results can be found in Tables 1 and 2 as well as in Tables S1 and S2. Unfortunately, kinetic data for XDH did not allow determination of . An additional complication is that kinetic characterization of XDH was often performed at the optimum pH for the enzymatic oxidation of xylitol. Work done with Galactocadida mastotermitis XDH and P. stipitis XDH show that enzyme activity decreases strongly upon lowering the pH from the optimum value of about 9 or greater to the physiological range of around 7 (Klimacek et al., 2007; Rizzi et al., 1989). Apparent Km values may also be affected by the corresponding pH change. The reported substrate and coenzyme affinities are used in the discussion with these limitations in mind.
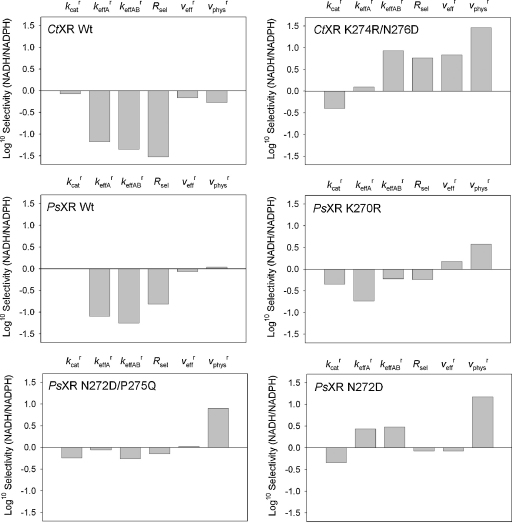
It becomes immediately clear from Table 1 and is further highlighted in Fig. 1 that the apparent preference for utilization of NAD(H) and NADP(H) can vary in a wide range, depending on the method used for its calculation. CtXR in the wild-type form (Ct WT) shows almost equal use of NADH and NADPH based on while according to Rsel the enzyme is almost completely specific for NADPH (Fig. 1). These results emphasize how important it is to have clearly defined the basis of the calculated selectivity parameter. Moreover, they raise the important question of which of the calculated selectivities is really informative with respect to yeast strain performance in xylose fermentation.
Fig. 1.
Comparison of wild-type and mutated forms of XR concerning usage of NADH and NADPH, using different expressions of coenzyme preference.
A general trend recognized within the series of XR enzymes is that selectivity parameters derived from kinetic and metabolite data (, ) indicated a higher preference for use of NADH than the corresponding parameters calculated from kinetic data alone (Fig. 1). This difference results mainly because the intracellular concentration of NADH exceeds that of NADPH by an order of magnitude. The opposite trend is seen across the series of XDH variants engineered for reaction with NADP+ (Table 2). The concentration of NADP+ is fivefold or more below the level of NAD+, implying that the physiological boundary conditions are somewhat unfavorable for xylitol oxidation by NADP+.
Besides showing a desired change towards improved utilization of NADH or NADP+ as stated by one of the calculated selectivity parameters, variants of XR and XDH can be further categorized according to whether the Michaelis constant for the relevant coenzyme was in the range of the physiological concentration or substantially above it. It turns out that whereas almost all of the shown XR variants (see Table 1) bind NADH tightly relative to the available intracellular concentration (NADH ≥5*KNADH), all of the mutated XDHs, listed in Table S2, display very low affinity for NADP+ as compared to coenzyme available ( >6*NADP+). XDH variants are therefore expected to be massively unsaturated by NADP+ under the physiological boundary conditions. In order to sustain flux through the XDH reaction, it would thus be necessary to compensate for the large fraction of enzyme not utilized, by a corresponding enhancement of the turnover number or an increase in the XDH expression level. However, either enhancement would have to be substantial, by one order of magnitude or greater. A prominent quadruple mutant of PsXDH (Ps D207A I208R F209S N211R) that has been used in the construction of several S. cerevisiae strains displays a approximately 3.7 times the of the wild-type enzyme. However, the predicted in vivo activity of this mutant with NADP+ () is below 35% of of the wild-type enzyme (Fig. S2, Supporting Information). The values of the above mentioned and another PsXDH mutant (Ps D207A I208R F209S, Table 2) were further enhanced by ∼3-fold through introduction a second zinc binding site in the protein structure (Ps S96C S99C Y102C D207A I208R F209S N211R and Ps S96C S99C Y102C D207A I208R F209S, respectively). However, these changes in were completely silent physiologically which was confirmed in two different strain backgrounds (Hou et al., 2007; Watanabe et al., 2007c).
In summary, therefore, XR has been engineered to a point where the best improved mutants show between 6-fold (Rsel, Ct K274R N276D) and 30-fold (, Ct K274R N276D) preference for NADH. Several of these mutants retain the original ability of the wild-type enzyme to catalyze reduction of xylose efficiently when physiological concentrations of NADH are present. In XDH, by contrast, an almost complete reversal of coenzyme specificity from NAD+ to NADP+ has been achieved as judged on the bases of catalytic efficiencies. Based on , the NADP+-dependent XDH would seem to be ideally matched with wild-type XR. Using , the situation is less clear and it could be that imbalance in coenzyme utilization has now been shifted towards NADPH and NAD+ instead of NADH and NADP+. Fitness of XDH mutants for function under physiological boundary conditions for the available NADP+ appears to be strongly compromised by their high Km for the coenzyme.
Using the ratio between the intracellular coenzyme concentration and the corresponding Km as indicator, it is quite interesting to note that wild-type XR (Ct WT and Ps WT, Table 1) binds both NADPH and NADH tightly (ratio ≥7) and can therefore be considered saturated with coenzyme under the in vivo reaction conditions. The XDH in its wild-type form (Ps WT and Gm WT, Table 2), by contrast, shows a Km for NAD+ that is roughly matched to the available concentration of coenzyme (ratio ≈1). Applying S. cerevisiae NAD+ levels to P. stipitis and G. mastotermitis, this property of XDH would be consistent with fundamental notions of optimized enzyme efficiency in a metabolic pathway that has been evolved for operation above a minimum level of flux (Fersht, 1999). The XR, which is required to catalyze the first step of the pathway, is primed to drive the conversion of xylose forward.
3.2. Benefits of engineered XR or XDH for xylose fermentation by S. cerevisiae
S. cerevisiae strain construction used various pairs of XR and XDH in which usually one of the two enzymes had been engineered for altered coenzyme specificity. XR–XDH combinations were chosen such that recycling of NAD(P)H during conversion of xylose into xylulose should have been improved as compared to the reference strain harboring XR and XDH in wild-type form. An enzyme pair in which both XR and XDH had been mutated has only been tried once (Krahulec et al., 2009). Table 3 summarizes results of fermentation studies conducted with these strains. Comparison of the different strains is somewhat complicated because the extent of global genetic manipulation varies among them. Moreover, the overexpression of XR and XDH was done from genome-integrated genes or yeast episomal plasmids. Finally, the substrate used for fermentation was xylose or a mixture of glucose and xylose. In several cases, yield coefficients are based on the total amount of sugar consumed which as a procedure is undoubtedly relevant for technological application but otherwise it makes further analysis of the data exceedingly difficult. The important aspect of how fluxes at key steps in the metabolism influence the yields of ethanol and xylitol from xylose is therefore not readily accessible from assessment of the available data as a whole.
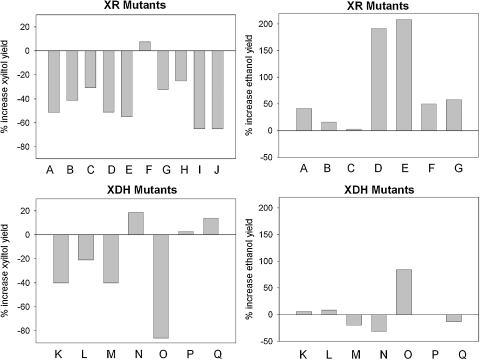
It was typical for the different strains although not true for all that a decrease in Yxylitol could be achieved by replacement of the original XR–XDH pair with an engineered combination of the two enzymes. However, in Fig. 2 it is clearly shown that strain performance varied substantially depending on whether it was XR or XDH that had been engineered. Except one, all enzyme combination derived from engineered XR and wild-type XDH gave a substantially decreased Yxylitol and generally it also brought about enhancement of Yethanol. In some strains (Fig. 2), the effect on Yxylitol appeared to have been fully translated into an increase in Yethanol. For XDH mutants, the effect on Yxylitol was mixed and there appears no immediate correlation between the relative changes in Yxylitol and Yethanol as compared to the respective reference (Fig. 2). Besides one case, XDH mutant strains gave little if any increase in Yethanol irrespective of the effect seen on Yxylitol. It is worth pointing out that positive effects on Yxylitol and Yethanol resulting from the best improved mutant of CtXR (Ct K274R N276D) were completely eliminated upon replacement of wild-type XDH (Gm WT; strain A in Fig. 2) by a mutated XDH (Gm D202A L203Δ V204Δ E205P S206R; strain P in Fig. 2), which according to the keffA-derived selectivity coefficient (Eq. (2)) would have been the much better companion enzyme for the XR mutant than wild-type XDH. It seems therefore that negative effects associated with the mutated XDH which we believe is introduction of a new kinetic bottleneck in the metabolism at the level of xylitol by far outweigh the benefits of the altered coenzyme specificity (Krahulec et al., 2009), also in line with observations of Hou et al. (2007). The idea receives further strong support from a study by Watanabe et al. (2007c) who used massive overexpression of NADP+-dependent XDH (Ps D207A I208R F209S N211R) from a high-copy plasmid. Under these conditions, little xylitol was produced (Yxylitol ∼0.05 g/g xylose) and the Yethanol calculated from the published data for mixed glucose/xylose fermentation was about 0.46 g/g total sugar consumed. Using the genome-integrated XDH, fermentation performance of the resulting strain was greatly deteriorated by comparison and essentially no improvement of Yethanol was achieved in relation to the reference strain harboring genes for the wild-type enzymes (Matsushika et al., 2008, 2009b). The strong requirement to compensate for catalytic deficiencies that were introduced in XDH upon mutation is plausibly indicated by these findings. The analysis that follows was therefore focused on mutated XR.
Fig. 2.
Changes in yield of xylitol and ethanol from xylose resulting from the use of an engineered XR or XDH as compared to the corresponding wild-type reference. Strains expressing XR mutants: A BP10001/BP000; B TMB3270/TMB3001; C TMB3271/3260; D TMB3421/TMB3424; E TMB3422/TMB3424; F TMB3423/TMB3424; G Y-R276H/Y-WT; H Y-K270R/Y-WT; I TMB3322/TMB3321; J TMB3323/TMB3321. Ethanol yields under H, I, and J are from mixed glucose/xylose fermentations and therefore not displayed. Strains expressing XDH mutants: K MA-N5/MA-N4; L MA-R5/MA-R4; M H1291/H1290; N H1292/H1290; O Y-ARSdR/Y-WT. Strains expressing XR and XDH mutants: P BP11001/BP000; Q BP11002/BP000. Table 3 lists all strains indicated in the figure and provides further details on them.
3.3. Prediction of Yxylitol from enzyme kinetic data for XR
Besides the fundamental interest in elucidating relationships between functional properties of the XR–XDH couple and the corresponding fermentation capabilities of the S. cerevisiae strain expressing the two enzymes, we considered the potential benefit for rational strain development if one had a reliable predictor (Psel), derived from enzyme kinetic data, to select promising variants of XR for further in vivo work. Experimental verification in the yeast is the most time-consuming step of the overall program. Knowledge-based reduction of the number of enzyme candidates to be evaluated in strain construction and fermentation studies would thus contribute most effectively to acceleration of the process development.
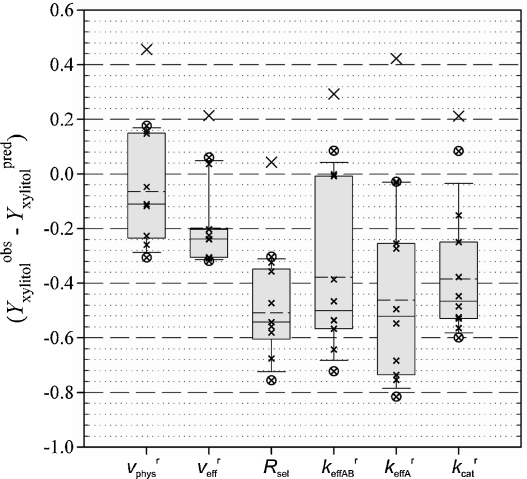
Using either one of the Psel values obtained with Eqs. (1)–(6), we applied Eq. (7) to calculate for 11 yeast strains that represent a diversity of CtXR and PsXR in the wild-type form and a total of seven enzyme variants derived from the two native enzymes. for S. cerevisiae expressing the K270M mutant of PsXR was not available due to insufficient kinetic characterization of the variant XR (see Table 1). We therefore applied kinetic data for the analogous mutant of CtXR (Ct K274M) in the calculation of . Comparison of the different values to the corresponding experimental using Eq. (8) gave a series of ΔYxylitol values for each enzyme, wild-type or variant. The results are shown as Box-plot in Fig. 3, and a complete summary of the data is given in Table S3 (Supporting Information). Quality of prediction of may be inferred from clustering of the data around zero (Fig. 3).
Fig. 3.
Box-plots for the differences between the observed and predicted xylitol yields. Boxes represent the 50% interquartile range. Solid and dashed lines within the boxes represent the sample median and the sample mean, respectively. Lower and upper bounds indicated by whisker caps display the 80% interquartile range. Small x and large X indicate data points obtained from the analysis and outliers, respectively. Open circles show data points outside the 80% interquartile range.
Variance analysis of the complete data set showed that at a p-level of 0.005, minimally two of the series of data analyzed were different from another. Normal distribution of the data was confirmed through their examination in a normal probability plot. Judging by distance from the sample median, a few outliers were identified, as indicated in Fig. 3, and excluded from the analysis (Table S3). Among the different expressions of Psel tested, only gave a near-perfect prediction of , that is ΔYxylitol ≈ 0, at the 99% confidence level. The standard deviation for ΔYxylitol () was 0.19 g/g which results in part from the significant statistical error in the kinetic parameters and the used metabolite concentrations. Values of ΔYxylitol () clustered around −0.2 g/g with a standard deviation of 0.14 g/g. The standard deviations of ΔYxylitol () and ΔYxylitol () were not different from another at a p-value of 0.18. Based on a two-sample t-test, the averages of ΔYxylitol () and ΔYxylitol () were statistically different at a p-value of 0.05. Psel values derived from kinetic parameters while ignoring intracellular metabolite concentrations (e.g. Rsel in Fig. 3) have the tendency to strongly overestimate xylitol production with an average ΔYxylitol between −0.38 g/g and −0.45 g/g. This result strongly emphasizes how important it is to combine kinetic and metabolite data in the evaluation of the fitness of XR for function under physiological reaction conditions. The comparison of predictions based on which used metabolite concentrations with those based on which did not use metabolite concentrations also supports this conclusion: ΔYxylitol () was clustered in a by far smaller range than ΔYxylitol () (see Fig. 3). The clearly superior performance of Psel () as compared to Psel () in predicting the experimental xylitol yield furthermore indicates that evaluation of the respective XR should involve bi-substrate kinetic characterization where the effect of substrate concentration on the steady-state rate is measured at several concentrations of coenzyme. Limitations of the kinetic shortcut where Km for substrate and coenzyme is determined at saturating concentrations of the respective other (see ) are pointed out in our analysis (Fig. 3). Note that enzymes need not be purified in order to carry out the suggested kinetic characterization in terms of . It is interesting that () was useful despite variation in strain genetic background, type of XR expression, and ability of the used yeast strain to grow under conditions of xylose fermentation (Table 3).
4. Conclusions
The analysis performed on the current diversity of engineered XR and XDH reveals limitations of the NADP+-utilizing XDH in terms of an affinity for binding NADP+ that is too low for efficient catalysis under in vivo boundary conditions. Further engineering strategies of XDH should address exactly this deficiency of existing mutants of this enzyme. The currently best improved variants of XR appear to be suitable for acting together with the wild-type form of XDH that is dependent on NAD+. The XR does so by making efficient use of the available NADH in the yeast cell. Additional optimization of XR by protein engineering should consider as target parameter to become maximally enhanced. This should be reflected in the experimental design of the study, in particular in the part dealing with the kinetic evaluation. Determination of the sensitivity of to changes in its constituent kinetic constants (see Eq. (6)) will assist in defining the most promising directions for the engineering of the XR used. Direct measurement of the enzyme preference for reaction with NADH as compared to NADPH was proposed by Petschacher and Nidetzky (2005). Applying the recently updated reaction conditions in S. cerevisiae could be a useful way to verify the kinetically determined . Application of XR optimized for in the context of a yeast strain in which bottlenecks elsewhere in the metabolism (e.g. the pentose phosphate pathway) have already been removed (Chu and Lee, 2007; Hahn-Hägerdal et al., 2007b; Jeffries, 2006; Matsushika et al., 2009a; Runquist et al., 2010), seems to be a promising strategy. Thereby full translation of the effect of XR engineering on decreasing Yxylitol into a corresponding enhancement of Yethanol might be achieved.
Summarizing, the aim of this work was to bring into a coherent whole the widely divergent evidence on the role of XR–XDH protein engineering on the fermentation capabilities of the resulting strains of S. cerevisiae. We show that it is possible to predict the strain performance upon introduction of a generated XR mutant. The conceptual approach should be useful to other strategies of metabolic engineering in which balance between NADP(H) and NAD(H) usage by a dehydrogenase/reductase is a key issue.
Acknowledgment
Financial support from the Austrian Science Fund FWF (P18275-B09 to BN) is gratefully acknowledged.
Footnotes
Our numbering of strain generation refers solely to XR-XDH pathway development and is not meant to reflect the overall advance in strain engineering that has previously targeted various other parts of the metabolism (Chu and Lee, 2007; Hahn-Hägerdal et al., 2007b; Jeffries, 2006; Matsushika et al., 2009a).
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jbiotec.2011.08.026.
Contributor Information
Mario Klimacek, Email: mario.klimacek@tugraz.at.
Bernd Nidetzky, Email: bernd.nidetzky@tugraz.at.
Appendix A. Supplementary data
References
- Almeida J.R., Modig T., Röder A., Lidén G., Gorwa-Grauslund M.F. Pichia stipitis xylose reductase helps detoxifying lignocellulosic hydrolysate by reducing 5-hydroxymethyl-furfural (HMF) Biotechnol. Biofuels. 2008;1:12. doi: 10.1186/1754-6834-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida J.R., Runquist D., Sànchez i Nogué V., Lidén G., Gorwa-Grauslund M.F. Stress-related challenges in pentose fermentation to ethanol by the yeast Saccharomyces cerevisiae. Biotechnol. J. 2011;6:286–299. doi: 10.1002/biot.201000301. [DOI] [PubMed] [Google Scholar]
- Banta S., Boston M., Jarnagin A., Anderson S. Mathematical modeling of in vitro enzymatic production of 2-keto-l-gulonic acid using NAD(H) or NADP(H) as cofactors. Metab. Eng. 2002;4:273–284. doi: 10.1006/mben.2002.0231. [DOI] [PubMed] [Google Scholar]
- Bengtsson O., Hahn-Hägerdal B., Gorwa-Grauslund M.F. Xylose reductase from Pichia stipitis with altered coenzyme preference improves ethanolic xylose fermentation by recombinant Saccharomyces cerevisiae. Biotechnol. Biofuels. 2009;2:9. doi: 10.1186/1754-6834-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brat D., Boles E., Wiedemann B. Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2009;75:2304–2311. doi: 10.1128/AEM.02522-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu B.C., Lee H. Genetic improvement of Saccharomyces cerevisiae for xylose fermentation. Biotechnol. Adv. 2007;25:425–441. doi: 10.1016/j.biotechadv.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Dean A.M., Voss D. Springer; New York: 1999. Design and Analysis of Experiments. [Google Scholar]
- Delgenes J.P., Moletta R., Navarro J.M. The effect of aeration on d-xylose fermentation by Pachysolen tannophilus, Pichia stipitis, Kluyveromyces marxianus and Candida shehatae. Biotechnol. Lett. 1986;8:897–900. [Google Scholar]
- Dellweg H., Rizzi M., Methner H., Debus D. Xylose fermentation by yeasts. 3. Comparison of Pachysolen tannophilus and Pichia stipitis. Biotechnol. Lett. 1984;6:395–400. [Google Scholar]
- Fersht A. Freeman and Company; New York: 1999. Structure and Mechanism in Protein Science. A Guide to Enzyme Catalysis and Protein Folding. [Google Scholar]
- Gárdonyi M., Jeppsson M., Lidén G., Gorwa-Grauslund M.F., Hahn-Hägerdal B. Control of xylose consumption by xylose transport in recombinant Saccharomyces cerevisiae. Biotechnol. Bioeng. 2003;82:818–824. doi: 10.1002/bit.10631. [DOI] [PubMed] [Google Scholar]
- Habenicht A., Motejadded H., Kiess M., Wegerer A., Mattes R. Xylose utilisation: cloning and characterisation of the xylitol dehydrogenase from Galactocandida mastotermitis. Biol. Chem. 1999;380:1405–1411. doi: 10.1515/BC.1999.180. [DOI] [PubMed] [Google Scholar]
- Hahn-Hägerdal B., Karhumaa K., Fonseca C., Spencer-Martins I., Gorwa-Grauslund M.F. Towards industrial pentose-fermenting yeast strains. Appl. Microbiol. Biotechnol. 2007;74:937–953. doi: 10.1007/s00253-006-0827-2. [DOI] [PubMed] [Google Scholar]
- Hahn-Hägerdal B., Karhumaa K., Jeppsson M., Gorwa-Grauslund M.F. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv. Biochem. Eng. Biotechnol. 2007;108:147–177. doi: 10.1007/10_2007_062. [DOI] [PubMed] [Google Scholar]
- Hamacher T., Becker J., Gárdonyi M., Hahn-Hägerdal B., Boles E. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology. 2002;148:2783–2788. doi: 10.1099/00221287-148-9-2783. [DOI] [PubMed] [Google Scholar]
- Hector R.E., Qureshi N., Hughes S.R., Cotta M.A. Expression of a heterologous xylose transporter in a Saccharomyces cerevisiae strain engineered to utilize xylose improves aerobic xylose consumption. Appl. Microbiol. Biotechnol. 2008;80:675–684. doi: 10.1007/s00253-008-1583-2. [DOI] [PubMed] [Google Scholar]
- Hou J., Shen Y., Li X.P., Bao X.M. Effect of the reversal of coenzyme specificity by expression of mutated Pichia stipitis xylitol dehydrogenase in recombinant Saccharomyces cerevisiae. Lett. Appl. Microbiol. 2007;45:184–189. doi: 10.1111/j.1472-765X.2007.02165.x. [DOI] [PubMed] [Google Scholar]
- Jeffries T.W. Engineering yeasts for xylose metabolism. Curr. Opin. Biotechnol. 2006;17:320–326. doi: 10.1016/j.copbio.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Jeffries T.W., Van Vleet J.R. Pichia stipitis genomics, transcriptomics, and gene clusters. FEMS Yeast Res. 2009;9:793–807. doi: 10.1111/j.1567-1364.2009.00525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppsson M., Bengtsson O., Franke K., Lee H., Hahn-Hägerdal B., Gorwa-Grauslund M.F. The expression of a Pichia stipitis xylose reductase mutant with higher KM for NADPH increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Biotechnol. Bioeng. 2006;93:665–673. doi: 10.1002/bit.20737. [DOI] [PubMed] [Google Scholar]
- Jojima T., Omumasaba C.A., Inui M., Yukawa H. Sugar transporters in efficient utilization of mixed sugar substrates: current knowledge and outlook. Appl. Microbiol. Biotechnol. 2010;85:471–480. doi: 10.1007/s00253-009-2292-1. [DOI] [PubMed] [Google Scholar]
- Katahira S., Ito M., Takema H., Fujita Y., Tanino T., Tanaka T., Fukuda H., Kondo A. Improvement of ethanol productivity during xylose and glucose co-fermentation by xylose-assimilating S. cerevisiae via expression of glucose transporter Sut1. Enzyme Microb. Technol. 2008;43:115–119. [Google Scholar]
- Kavanagh K.L., Klimacek M., Nidetzky B., Wilson D.K. The structure of apo and holo forms of xylose reductase, a dimeric aldo–keto reductase from Candida tenuis. Biochemistry. 2002;41:8785–8795. doi: 10.1021/bi025786n. [DOI] [PubMed] [Google Scholar]
- Kavanagh K.L., Klimacek M., Nidetzky B., Wilson D.K. Structure of xylose reductase bound to NAD+ and the basis for single and dual co-substrate specificity in family 2 aldo–keto reductases. Biochem. J. 2003;373:319–326. doi: 10.1042/BJ20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimacek M., Hellmer H., Nidetzky B. Catalytic mechanism of Zn2+-dependent polyol dehydrogenases: kinetic comparison of sheep liver sorbitol dehydrogenase with wild-type and Glu154→Cys forms of yeast xylitol dehydrogenase. Biochem. J. 2007;404:421–429. doi: 10.1042/BJ20061384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimacek M., Krahulec S., Sauer U., Nidetzky B. Limitations in xylose-fermenting Saccharomyces cerevisiae, made evident through comprehensive metabolite profiling and thermodynamic analysis. Appl. Environ. Microbiol. 2010;76:7566–7574. doi: 10.1128/AEM.01787-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrzynska M., Sopher C.R., Lee H. Mutational analysis of the role of the conserved lysine-270 in the Pichia stipitis xylose reductase. FEMS Microbiol. Lett. 1998;159:107–112. doi: 10.1111/j.1574-6968.1998.tb12848.x. [DOI] [PubMed] [Google Scholar]
- Kötter P., Ciriacy M. Xylose fermentation by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 1993;38:776–783. [Google Scholar]
- Krahulec S., Klimacek M., Nidetzky B. Engineering of a matched pair of xylose reductase and xylitol dehydrogenase for xylose fermentation by Saccharomyces cerevisiae. Biotechnol. J. 2009;4:684–694. doi: 10.1002/biot.200800334. [DOI] [PubMed] [Google Scholar]
- Krahulec S., Petschacher B., Wallner M., Longus K., Klimacek M., Nidetzky B. Fermentation of mixed glucose–xylose substrates by engineered strains of Saccharomyces cerevisiae: role of the coenzyme specificity of xylose reductase, and effect of glucose on xylose utilization. Microb. Cell Fact. 2010;9:16. doi: 10.1186/1475-2859-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman C.P., Suzuki M. Phylogenetic analysis of ascomycete yeasts that form coenzyme Q-9 and the proposal of the new genera Babjeviella, Meyerozyma, Millerozyma, Priceomyces, and Scheffersomyces. Mycoscience. 2010;51:2–14. [Google Scholar]
- Kuyper M., Hartog M.M., Toirkens M.J., Almering M.J., Winkler A.A., van Dijken J.P., Pronk J.T. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 2005;5:399–409. doi: 10.1016/j.femsyr.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Liu Z.L. Genomic adaptation of ethanologenic yeast to biomass conversion inhibitors. Appl. Microbiol. Biotechnol. 2006;73:27–36. doi: 10.1007/s00253-006-0567-3. [DOI] [PubMed] [Google Scholar]
- Lunzer R., Mamnun Y., Haltrich D., Kulbe K.D., Nidetzky B. Structural and functional properties of a yeast xylitol dehydrogenase, a Zn2+-containing metalloenzyme similar to medium-chain sorbitol dehydrogenases. Biochem. J. 1998;336:91–99. doi: 10.1042/bj3360091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan A., Tamalampudi S., Ushida K., Kanai D., Katahira S., Srivastava A., Fukuda H., Bisaria V.S., Kondo A. Xylose isomerase from polycentric fungus Orpinomyces: gene sequencing, cloning, and expression in Saccharomyces cerevisiae for bioconversion of xylose to ethanol. Appl. Microbiol. Biotechnol. 2009;82:1067–1078. doi: 10.1007/s00253-008-1794-6. [DOI] [PubMed] [Google Scholar]
- Matsushika A., Inoue H., Kodaki T., Sawayama S. Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Appl. Microbiol. Biotechnol. 2009;84:37–53. doi: 10.1007/s00253-009-2101-x. [DOI] [PubMed] [Google Scholar]
- Matsushika A., Inoue H., Watanabe S., Kodaki T., Makino K., Sawayama S. Efficient bioethanol production by a recombinant flocculent Saccharomyces cerevisiae strain with a genome-integrated NADP+-dependent xylitol dehydrogenase gene. Appl. Environ. Microbiol. 2009;75:3818–3822. doi: 10.1128/AEM.02636-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushika A., Watanabe S., Kodaki T., Makino K., Inoue H., Murakami K., Takimura O., Sawayama S. Expression of protein engineered NADP+-dependent xylitol dehydrogenase increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2008;81:243–255. doi: 10.1007/s00253-008-1649-1. [DOI] [PubMed] [Google Scholar]
- Meijers R., Cedergren-Zeppezauer E.S. Zn-dependent medium-chain dehydrogenases/reductases. In: Messerschmidt A., Bode W., Cygler M., editors. Handbook of Metalloproteins. Wiley; Chichester: 2004. pp. 5–33. [Google Scholar]
- Neuhauser W., Haltrich D., Kulbe K.D., Nidetzky B. NAD(P)H-dependent aldose reductase from the xylose-assimilating yeast Candida tenuis. Isolation, characterization and biochemical properties of the enzyme. Biochem. J. 1997;326:683–692. doi: 10.1042/bj3260683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson K., Runquist D., Hahn-Hägerdal B., Lidén G. A mutated xylose reductase increases bioethanol production more than a glucose/xylose facilitator in simultaneous fermentation and co-fermentation of wheat straw. AMB Express. 2011;1:4. doi: 10.1186/2191-0855-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petschacher B., Leitgeb S., Kavanagh K.L., Wilson D.K., Nidetzky B. The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and X-ray crystallography. Biochem. J. 2005;385:75–83. doi: 10.1042/BJ20040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petschacher B., Nidetzky B. Engineering Candida tenuis xylose reductase for improved utilization of NADH: antagonistic effects of multiple side chain replacements and performance of site-directed mutants under simulated in vivo conditions. Appl. Environ. Microbiol. 2005;71:6390–6393. doi: 10.1128/AEM.71.10.6390-6393.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petschacher B., Nidetzky B. Altering the coenzyme preference of xylose reductase to favor utilization of NADH enhances ethanol yield from xylose in a metabolically engineered strain of Saccharomyces cerevisiae. Microb. Cell Fact. 2008;7:9. doi: 10.1186/1475-2859-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzi M., Erlemann P., Bui-Thanh N.-A., Dellweg H. Xylose fermentation by yeasts 4. Purification and kinetic studies of xylose reductase from Pichia stipitis. Appl. Microbiol. Biotechnol. 1988;29:148–154. [Google Scholar]
- Rizzi M., Harwart K., Erlemann P., Bui-Thanh N.-A., Dellweg H. Purification and properties of the NAD+-xylitol-dehydrogenase from the yeast Pichia stipitis. J. Ferment. Bioeng. 1989;67:20–24. [Google Scholar]
- Runquist D., Fonseca C., Rådström P., Spencer-Martins I., Hahn-Hägerdal B. Expression of the Gxf1 transporter from Candida intermedia improves fermentation performance in recombinant xylose-utilizing Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2009;82:123–130. doi: 10.1007/s00253-008-1773-y. [DOI] [PubMed] [Google Scholar]
- Runquist D., Hahn-Hägerdal B., Bettiga M. Increased expression of the oxidative pentose phosphate pathway and gluconeogenesis in anaerobically growing xylose-utilizing Saccharomyces cerevisiae. Microb. Cell Fact. 2009;8:49. doi: 10.1186/1475-2859-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runquist D., Hahn-Hägerdal B., Bettiga M. Increased ethanol productivity in xylose-utilizing Saccharomyces cerevisiae via a randomly mutagenized xylose reductase. Appl. Environ. Microbiol. 2010;76:7796–7802. doi: 10.1128/AEM.01505-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog K., Hahn-Hägerdal B. Effect of oxygenation on xylose fermentation by Pichia stipitis. Appl. Environ. Microbiol. 1990;56:3389–3394. doi: 10.1128/aem.56.11.3389-3394.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Abu Saleh A., Pack S.P., Annaluru N., Kodaki T., Makino K. Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein-engineered NADH-preferring xylose reductase from Pichia stipitis. Microbiology. 2007;153:3044–3054. doi: 10.1099/mic.0.2007/007856-0. [DOI] [PubMed] [Google Scholar]
- Watanabe S., Kodaki T., Makino K. Complete reversal of coenzyme specificity of xylitol dehydrogenase and increase of thermostability by the introduction of structural zinc. J. Biol. Chem. 2005;280:10340–10349. doi: 10.1074/jbc.M409443200. [DOI] [PubMed] [Google Scholar]
- Watanabe S., Pack S.P., Saleh A.A., Annaluru N., Kodaki T., Makino K. The positive effect of the decreased NADPH-preferring activity of xylose reductase from Pichia stipitis on ethanol production using xylose-fermenting recombinant Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 2007;71:1365–1369. doi: 10.1271/bbb.70104. [DOI] [PubMed] [Google Scholar]
- Watanabe S., Saleh A.A., Pack S.P., Annaluru N., Kodaki T., Makino K. Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein engineered NADP+-dependent xylitol dehydrogenase. J. Biotechnol. 2007;130:316–319. doi: 10.1016/j.jbiotec.2007.04.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.