Abstract
The deregulation of microRNA (miRNA) is frequently associated with a variety of cancers, including hepatocellular carcinoma (HCC). In this study, we identified 10 upregulated miRNAs (miR-217, miR-518b, miR-517c, miR-520g, miR-519a, miR-522, miR-518e, miR-525-3p, miR-512-3p and miR-518a-3p) and 10 downregulated miRNAs (miR-138, miR-214, miR-214#, miR-27a#, miR-199a-5p, miR-433, miR-511, miR-592, miR-483-5p and miR-483-3p) by Taqman miRNAs array and quantitative real-time PCR (qRT–PCR) confirmation. Additionally, we investigated the expression and possible role of miR-138 in HCC. qRT–PCR results showed that miR-138 was downregulated in 77.8%(14/18) of HCC tissues compared with adjacent non-tumor tissues. Overexpression of miR-138 reduced cell viability and colony formation by induction of cell arrest in HCC cell lines and inhibited tumor cell growth in xenograft nude mice. The use of miR-138 inhibitor increased cell viability and colony formation in HCC cell lines and tumor cell growth in xenograft nude mice. Using TargetScan predictions, CCND3 was defined as a potential direct target of miR-138. Furthermore, CCND3 protein expression was observed to be negatively correlated with miR-138 expression in HCC tissues. The dual-luciferase reporter gene assay results showed that CCND3 was a direct target of miR-138. The use of miR-138 mimic or inhibitor could decrease or increase CCND3 protein levels in HCC cell lines. We conclude that the frequently downregulated miR-138 can regulate CCND3 and function as a tumor suppressor in HCC. Therefore, miR-138 may serve as a useful therapeutic agent for miRNA-based HCC therapy.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers worldwide (1). The development and progression of HCC is typical of a multistage process, which is believed to involve the deregulation of genes that are critical to cellular processes, such as cell cycle control, cell growth, apoptosis and cell migration and spreading. In the past decades, studies have focused on investigating the genes and proteins underlying the development and progression of HCC (2). Recently, an increasing number of reports have described a new class of small regulatory RNA molecules termed microRNAs (miRNAs) that are implicated in HCC progression (3).
MiRNAs are endogenous non-coding 20–22 nucleotide RNAs that have been identified as post-transcriptional regulators of gene expression (4). The miRNAs mainly bind to the 3′ untranslated regions (UTRs) of target messenger RNAs (mRNAs), resulting in mRNA degradation or the blockade of mRNA translation. Increasing evidence showed that miRNAs have significant roles in diverse biological processes (5). Meanwhile, deregulation of miRNAs has been observed in a wide range of human diseases, including cancer (6). In human cancer, miRNAs can function as oncogenes or tumor suppressor genes during tumor development and progression (7).
In this study, the expression profiles of 667 miRNAs were examined in human HCC and adjacent non-tumor tissues. A set of significantly differentially expressed miRNAs were identified in HCC tissues. Further investigation revealed that miR-138, a frequently downregulated miRNA in HCC, could induce cell cycle arrest in HCC cell lines. Moreover, cyclin D3 (CCND3) was characterized as a direct target of miR-138.
Materials and methods
Tissue specimens and cell lines
Eighteen pairs of human HCC and adjacent non-tumor tissues were obtained from surgical specimens immediately after resection from patients undergoing primary surgical treatment of HCC in the Eastern Hepatobiliary Surgery Hospital, Shanghai, China. No patient had received preoperative irradiation or chemotherapy. The samples were frozen in liquid nitrogen and stored at −80°C until use. Among these samples, three pairs were used for Taqman low density miRNA array (TLDA) analysis and all of them were used for quantitative real-time PCR (qRT–PCR) analysis. Clinical and pathological information was extracted from the patients’ medical charts and pathology reports (Table I). Written consent for tissue donation (for research purposes) was obtained from the patients before tissue collection and the protocol was approved by the Institutional Review Board of Eastern Hepatobiliary Surgery Hospital and Second Military Medical University.
Table I.
The data of patients (n = 18)
| Variable | Value |
| Age (years) | 48.5 ± 10.1 |
| Gender (male) | 14 (77.8%) |
| Tumor sizea (cm) | 8.73 |
| Tumor grade | |
| Well-differentiated (G1–2) | 7 |
| Moderately-differentiated (G3) | 11 |
| Poorly-differentiated (G4) | 0 |
| TNM stage | |
| I | 4 |
| II | 7 |
| III | 6 |
| IV | 1 |
TNM, tumor-node-metastasis.
Diameter of the biggest nodule.
Human embryonic kidney cells (HEK293T) and human HCC cell lines HepG2 and Huh7 were cultured in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum (Invitrogen, Carlsbad, CA) at 37°C in a humidified atmosphere containing 5% CO2.
Taqman low density miRNA array
Total RNA was isolated using mirVana miRNA isolation kit (Ambion, Austin, TX). For miRNA complementary DNA (cDNA) synthesis, RNA was reverse transcribed using the miRNA reverse transcription kit (Applied Biosystems, Foster City, CA) in combination with the stem-loop Megaplex primer pool (Applied Biosystems). Then, TLDA v2.0 was performed as the manufacturer’s protocol. For each cDNA sample, 667 small RNAs were profiled. All PCR reactions were performed on the 7900HT real-time PCR system (Applied Biosystems). Cycling conditions were as follows: 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Human U6 small RNA was used as an internal control to normalize RNA input. The data were analyzed by SDS v2.3 software. In details, the Ct value is defined as the fractional cycle number at which the fluorescence passes the fixed threshold. The fold change was calculated using the 2−ΔΔCt method and presented as the fold-expression change in tumors relative to their corresponding normal tissues after normalization to the endogenous control.
Quantitative real-time PCR
Synthesis of cDNA and qRT–PCR analysis of miRNA expression was carried out with TaqMan microRNA assay kits (Applied Biosystems) according to the manufacturer’s protocol. Briefly, total RNA was extracted using TRIzol Reagent (Invitrogen) from clinical samples or HCC cell lines and were used to synthesize cDNAs with gene-specific primers. Reverse transcriptase (RT) reactions contained 100 ng RNA, 50 nmol/l stem-loop RT primers, 1× RT buffer, 0.25 mmol/l each of the deoxyribonucleotide triphosphates, 3.33 U/μl MultiScribe reverse transcriptase and 0.25 U/μl ribonuclease inhibitor. The 15 μl reactions were incubated for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C and then held at 4°C. The cDNA product was used for the following qRT–PCR analysis directly. The 20 μl PCR reaction included 1.33 μl RT product, 1× TaqMan universal PCR master mix and 1 μl primers and probe mix of the TaqMan microRNA assay kit. Reactions were incubated in a 96-well optical plate at 95°C for 5 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. PCR reactions were run on a StepOne Plus real-time PCR machine (Applied Biosystems) and the data were analyzed by SDS v2.3 software same as in TLDA.
The construction of luciferase reporter plasmids
The fragment of 3′UTR of CCND3 (1054–2061 nt, Genbank accession no. NM_001136017.2) containing the two putative miR-138-binding sequences (1279–1285 nt and 1346–1352 nt) was amplified with the primers 5′-CCCTGGAGAGGCCCTCTGGA-3′ (forward) and 5′-TTCCAAGAAGCCAAAGCCAG-3′ (reverse). The PCR product was cloned into Firefly luciferase reporter vector pGL3 (Promega Corporation, Madison, WI), termed as pGL3-CCND3-3′UTR. The plasmid which carried the mutated sequence in the two complementary sites for the seed region of miR-138 (Figure 2A) was generated based on pGL3-CCND3-3′UTR plasmid by MutanBEST Kit (Takara Bio, Shiga, JP), termed as pGL3-CCND3-3′UTR-mut. Briefly, a forward miR-138 mutagenic primer F1-GAT ACATAGTGGTCGATTCCTTTTGAACGCCCCCCCCCACCCC and a reverse miR-138 mutagenic primer R1-CAATTCTGTCCCATCAGCCTGGCCCSCCCCCSGCTSGSGTTG or a forward miR-138 mutagenic primer F2-CTGCATCTGTGGTCGAGCGCCTTTCCCAACT CTAGCTGGGG and a reverse miR-138 mutagenic primer R2-GGAGGAGGAGCTTGACTAGCCACCGAAATGCAGACATGG, were synthesized and utilized in PCR experiment as described by the manufacturer. Two point mutations along the miR-138 seed binding sites were confirmed by sequencing analyses.
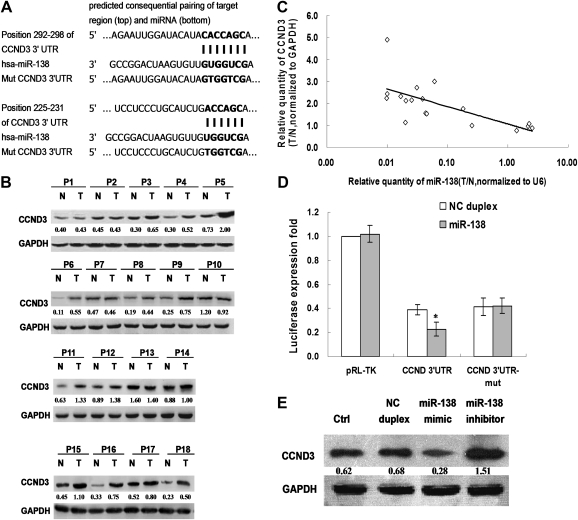
Fig. 2.
CCND3 is a direct target of miR-138. (A) The putative miR-138 binding sequence in the 3′UTR of CCND3 mRNA. Mutation was generated on the CCND3 3′UTR sequence in the complementary site for the seed region of miR-138. (B) The expression of CCND3 in 18 paired HCCs tumor tissues (T) and adjacent non-tumor tissues (N). (C) The negative correlation between the expression of miR-138 and CCND3 in paired HCC tissues. The relative quantity of miR-138 in HCC tumor tissues compared with adjacent non-tumor tissues (T/N) was showed after normalized to U6. The relative quantity of CCND3 protein in HCC tumor tissues compared with adjacent non-tumor tissues (T/N) was showed after normalized to glyceraldehyde-3-phosphate dehydrogenase. (D) Suppressed luciferase activity of wild-type 3′UTR of CCND3 by miR-138 mimic. HEK293T cells were cotransfected pGL3-CCND3-3′UTR or pGL3-CCND3-3′UTR-mut and miR-138 mimic or NC duplex. Firefly luciferase activity of each sample was measured 48 h after transfection and normalized to Renilla luciferase activity. (E) The expression of endogenous CCND3 regulated by miR-138. The expression level of endogenous CCND3 in HepG2 cells was analyzed 48 h after transfection with miR-138 mimic, miR-138 inhibitor or NC duplex by western blotting. Glyceraldehyde-3-phosphate dehydrogenase was used as an internal control. Column, mean of three independent experiments; bars, SD; *P < 0.01.
Transfection
The transfections were carried out using FuGene HD transfection reagent (Roche, Indianapolis, IN) following the manufacturer’s protocol. In brief, 2 × 104 HepG2 and Huh7 cells or 5 × 104 HEK293T cells in 24-well plate were transfected with indicated plasmid DNA, miRNA duplex (GenePharma, Shanghai, China), small interfering RNA (siRNA) (GenePharma) and collected 24–48 h after transfection for assay.
Cell viability assay
Twenty-four hours after transfection, 1000 transfected HepG2 and Huh7 cells were placed in a fresh 96-well plate in triplicate and maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum for 5 days. Cells were tested for proliferation per 24 h using Cell Titer-Blue cell viability assay (Promega Corporation) according to the manufacturer's instructions and the fluorescence value was recorded by multi-plate reader (Synergy 2; BioTek, Winooski, VT).
Cell cycle analysis
Forty-eight hours after transfection, 1 × 105 transfected HepG2 and Huh7cells were harvested, washed once in phosphate-buffered saline and fixed in 70% ethanol at 4°C overnight. Staining for DNA content was performed with 50 mg/ml propidium iodide and 1 mg/ml ribonuclease A at room temperature for 30 min. Populations in G0–G1, S and G2–M phase were measured by Cell Lab Quanta SC flow cytometry (Beckman Coulter, Fullerton, CA) and the data were analyzed by FlowJo v7.6 Software.
Colony formation assay
Twenty-four hours after transfection, 2000 transfected HepG2 and Huh7cells were placed in a fresh six-well plate in triplicate and maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum for 2 weeks. Cell colonies were fixed with 20% methanol and stained with 0.1% coomassie brilliant blue R250 at room temperature for 15 min. The colonies were counted by ELIspot Bioreader 5000 (BIO-SYS, Karben, GE).
Tumorigenicity assays in nude mice
Male BALB/c nude mice (5–6 weeks of age) were obtained from Shanghai Experimental Animal Center (Shanghai, China). Animal handling and experimental procedures were approved by the Animal Experiments Ethics Committee of Second Military Medical University. For in vivo tumorigenicity assay, all pyrimidine nucleotides in the miR-138 mimic, miR-138 inhibitor or nonrelative control (NC) duplex were substituted by their 2′-O-methyl analogues to improve RNA stability. MiR-138 mimic-or miR-138 inhibitor-transfected HepG2 cells (1 × 105) were suspended in 100 μl phosphate-buffered saline and then injected subcutaneously into left side of the posterior flank of six BALB/c nude mice, respectively. NC transfected or non-transfected HepG2 cells (1 × 105) were injected subcutaneously into right side of same 12 mice. Tumor growth was examined daily and the tumor volumes were calculated every week using the formula for hemi-ellipsoids: V =length (cm) × width (cm) × height (cm) × 0.5236. After 5 weeks, the mice were sacrificed and the tumors were dissected and photographed.
Dual-luciferase reporter assay
HEK293T cells seeded in 24-well plate in triplicate were cotransfected with pGL3-CCND3-3′UTR or pGL3-CCND3-3′UTR-mut and miRNA-138 mimic or non-relative control RNA duplex (NC duplex; GenePharma) by using FuGene HD transfection reagent. The pRL-TK (Promega Corporation) was also transfected as a normalization control. Cells were collected 48 h after transfection, and luciferase activity was measured using a dual-luciferase reporter assay kit (Promega Corporation) and recorded by multi-plate reader (Synergy 2; BioTek).
Western blotting
Protein extracts from HCC tissuses or HepG2 cells were prepared by a modified RIPA buffer with 0.5% sodium dodecyl sulfate in the presence of proteinase inhibitor cocktail (Complete mini; Roche). Twenty-five micrograms of protein of HCC tissues and their adjacent non-tumorous tissues were electrophoresed in 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis mini gels and transferred onto polyvinylidene difluoride membranes (Immobilon P−SQ; Millipore, Billerica, MA). After blocking with 5% non-fat milk, the membranes were incubated with rabbit anti-cyclin D3 antibody (1:1000 dilution; Epitomics, Burlingame, CA) or mouse anti-glyceraldehyde-3-phosphate dehydrogenase antibody (1:5000 dilution; Epitomics) at 4°C overnight, followed by incubation with horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse antibody (1:10000 dilution; KPL, Gaithersburg, MA) for 1 h at room temperature. Finally, signals were developed with Super Signal West Pico chemoluminescent substrate (Pierce, Rockford, IL), visualized by the GeneGnome HR Image Capture System (Syngene, Frederick, MD) and analyzed by Gene tools (Syngene).
Prediction of miRNAs targets
To investigate the target genes of miRNAs and the conserved sites bound by the seed region of miR-138, the TargetScan (http://www.targetscan.org/), MiRanda (http://www.microrna.org/microrna/home.do) and PicTar (http://pictar.mdc-berlin.de/) programs were used.
Gene ontology and Kyoto encyclopedia of genes and genomes pathway analysis
The top 25% miRNA targets that had been assigned the highest numbers of miRNA interaction sites were collected and subjected to Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis. GO analysis was applied to analyze the main function of the differential expression genes according to the GO, which is the key functional classification of National Center for Biotechnology Information (8).Generally, Fisher’s exact test and χ2 test were used to classify the GO category, and the false discovery rate (FDR) (9) was calculated to correct the P-value, the smaller the FDR, the smaller the error in judging the P-value. The FDR was defined as , where Nk refers to the number of Fisher’s test P-values less than χ2 test P-values. P-values were computed for the GOs of all the differential genes. Enrichment provides a measure of the significance of the function: as the enrichment increases, the corresponding function is more specific, which helps us to find those GOs with more concrete function description in the experiment. Within the significant category, the enrichment Re was given by ,where nf is the number of differential genes within the particular category, n is the total number of genes within the same category, Nf is the number of differential genes in the entire microarray and N is the total number of genes in the microarray (10). Similarly, pathway analysis was used to find out the significant pathway of the differential genes according to KEGG, Biocarta and Reatome. Still, the Fisher’s exact test and χ2 test were used to select the significant pathway, and the threshold of significance was defined by P-value and FDR. The enrichment Re was calculated like the equation above (11–13).
Statistical analysis
Data are presented as mean ± SD. Comparisons were made by using a two-tailed t-test or one-way analysis of variance for experiments with more than two subgroups. Correlation analysis was made by using Spearman correlation coefficient. P <0.01 was considered statistically significant.
Results
Patient characteristics
HCC and adjacent non-tumor tissues were obtained from 14 male and 4 female patients and their profiles are shown in Table I. The average age of these patients was 48.5 years (ranged from 35 to 71 years). Eleven patients (61.1%) had tumors >5 cm, two (11.1%) had tumors <2 cm and five (27.8%) had 2–5 cm tumors. Seven (38.9%) cases were well differentiated (I + II) and 11(61.1%) cases were moderately differentiated (III). Four patients (22.2%) were diagnosed at invasion T1, seven (38.9%) at invasion T2, six (33.3%) at invasion T3 and one (5.6%) at invasion T4.
Differential expression of miRNAs in HCC
The TLDA were performed in three pairs of HCC and adjacent non-tumor tissues. Only miRNAs altered by at least 3-fold in all three pairs of the samples were considered significant candidates. Under these strict criteria, 11 upregulated miRNAs and 12 downregulated miRNAs were identified. To validate the miRNA array data, qRT–PCR was performed in 18 pairs of HCC tissues. Ten upregulated and 10 downregulated miRNAs showed consistent changes in >50% tumorous tissues (Table II), of which miR-217, miR-520g, miR-522 and miR-525-3p were upregulated in >70% tumorous tissues and miR-199a-5p, miR-138, miR-483-5p and miR-511 were downregulated in >70% tumorous tissues (Table II).
Table II.
The deregulated miRNA in Hepatitis B virus-associated HCC
| miRNA | Fold change | P-value | Validationa | Representative predicted targets |
| Upregulated miRNAs | ||||
| hsa-miR-520g | 9475.09 | 0.0023 | 13 (72.2%) | VAPA, BET1, MMP2, GNAI3, PLCB1, PTPRJ, ENPP1, BMP6, ENAH, ARHGEF11 |
| hsa-miR-519a | 8204.48 | 0.0034 | 10 (55.6%) | TGFBR2, MYLK, PAFAH1B1, ROCK2, PRDM4, VAPA, EDNRB, ERBB3, JAK1, RPS6KA3 |
| hsa-miR-522 | 6178.34 | 0.0005 | 13 (72.2%) | SORBS1, UBE2G1, RAB7A, PDCD6IP, WHSC1, KITLG, YWHAZ, UBE2Q2, CCND2, UBE2K |
| hsa-miR-518e | 4096.74 | 0.0016 | 11 (61.1%) | RAP1B, NPAS2, SPRY4, MAP4K4, OGDH, WHSC1, EHMT1, RPS6KA1, JAK1, TRIP10 |
| hsa-miR-525-3p | 1489.49 | 0.0004 | 14 (77.8%) | HES1, MAP4K4, CCND3, ITGA2B, FGFR1, EPHB4, NRAS, BAIAP2, CBL, WHSC1 |
| hsa-miR-217 | 1080.33 | 0.0018 | 11 (59.4%) | RPS6KA3, RAP1B, STK4, MAP4K4, WHSC1, CACNB1, SOCS5, NPAS2, OGDH, CUL5 |
| hsa-miR-518b | 982.85 | 0.0034 | 14 (77.8%) | ATP11C, ATP8A1, ICK, AAK1, SOS1, ANLN, COX18, BCL11A, ATP1B1, LMO7 |
| hsa-miR-512-3p | 197.21 | 0.0045 | 10 (55.6%) | SOCS3, WASL, AK1, RAP1A, PPAP2B, SSX2IP, ASH1L, MAPK1, VANGL2, EDNRA |
| hsa-miR-517c | 130.92 | 0.0006 | 11 (61.1%) | SMAD2, ABL2, UBE2D1, GPAM, RAB11FIP4, SH3GLB1, PPARGC1A, CCND3, PPP3CA, CTBP1 |
| hsa-miR-518a-3p | 46.84 | 0.0023 | 10 (55.6%) | RPS6KA3, RAP1B, STK4, MAP4K4, WHSC1, RAB22A, CACNB1, SOS2, OGDH, TFDP1 |
| Downregulated miRNAs | ||||
| hsa-miR-199a-5p | 0.00237 | 0.0005 | 13 (72.2%) | JAG1, NLK, CACNB2, GCNT2, ROCK1, VEGFA, PI4KA, ACACA, SERPINE1, PPP1R12A |
| hsa-miR-433 | 0.00310 | 0.0027 | 10 (55.6%) | SORBS1, KRAS, EPAS1, CLTC, YWHAZ, PP3R1, CUL5, SMC1A, MYH9, GRB2 |
| hsa-miR-592 | 0.01137 | 0.0035 | 11 (59.4%) | PLXNA2, FOXO1, SMAD4, STX16, FOXO3, SLC8A1, ACACA, CSNK1A1, ACVR2A, UBE2D3 |
| hsa-miR-214# | 0.01327 | 0.0061 | 11 (59.4%) | PDGFRA, DLST, SEMA4C, JAG1, SMAD4, CASP9, CRY2, VTI1A, ACACA, WASL |
| hsa-miR-27a# | 0.02207 | 0.0053 | 10 (55.6%) | ELK1, IGF1, SSX2IP, CACNA2D2, AP2M1, MYH14, BAIAP2, ARFGAP2, PI4KA, SMAD5 |
| hsa-miR-483-5p | 0.03310 | 0.0034 | 14 (77.8%) | MAP4K4, ARFGAP2, DAB2, ARHGEF12, SERPINE1, VPS4A, DTX2, STK3, ADRBK1, ARRB2 |
| hsa-miR-483-3p | 0.03323 | 0.0051 | 12 (66.7%) | ITPR2, PDGFB, PRKAR1A, KITLG, SMAD4, IGF1, ARRB2, VPS4A, MYH9, TGFBR2 |
| hsa-miR-138 | 0.03727 | 0.0019 | 14 (77.8%) | RMND5A, GPR124, CREB3L2, SLC35F1, SYT13, KLF11, AMMECR1, GTPBP1, SH2B3, NFIX |
| hsa-miR-214 | 0.04213 | 0.0053 | 11 (59.4%) | NOTCH2, ACOX1, RASSF5, RUNX1, CACNB1, CDK6, IRS2, ARHGEF12, VANGL2, VASP |
| hsa-miR-511 | 0.07957 | 0.0049 | 13 (72.2%) | SPRY1, MYH10, VTI1A, UBE2D1, SRF, NTRK2, SH3GLB1, IRS2, SMAD4, GPAM |
GTP, guanosine triphosphate, IGF, insulin-like growth factor; MAPK, mitogen-activated protein kinase, MMP, matrix metalloproteinase; PDGF, platelet-derived growth factor; TGF, transforming growth factor; VEGF, vascular endothelial growth factor.
qRT–PCR was performed in 18 pairs of tumor and non-tumor tissues in HCC.
GO and KEGG pathways analysis of these deregulated miRNAs
The targets of the above 20 deregulated miRNAs were predicted by TargetScan and the representative target genes were listed in Table II. To judge the most significant candidates and investigate the cellular function, the signaling pathway and GOs of these target genes were analyzed. The results showed that a wide variety of cellular processes were featured significantly in signaling pathways (Supplementary Table S1 is available at Carcinogenesis Online). Many of these signaling pathways, such as insulin, mitogen-activated protein kinase, transforming growth factor-beta and Wnt signaling pathway, have been shown to participate in the tumorigenesis (14–17). However, some other signaling pathways have never been reported to play a role in tumorigenesis, e.g. axon guidance. Among all these differentially regulated signaling pathways, ‘regulation of actin cytoskeleton’ and ‘pathway in cancer’ appeared to be the most enriched two in both upregulated and downregulated miRNA groups. A similar phenomenon was observed in GOs analysis. Many cellular functions were featured significantly, of which the ‘signal transduction’ appeared to be the most enriched one (Supplementary Table S2 is available at Carcinogenesis Online). Based on these results, the miRNAs which were involved in both signal transduction and the regulation of actin cytoskeleton or pathway in cancer might be the most significant candidates. MiR-520g, miR-483-5p, miR-138, miR-199a-5p, miR-217 and miR-518e were selected under the criteria for further studies. Here, the studies on miRNA-138 were presented.
MiR-138 induces cell cycle arrest
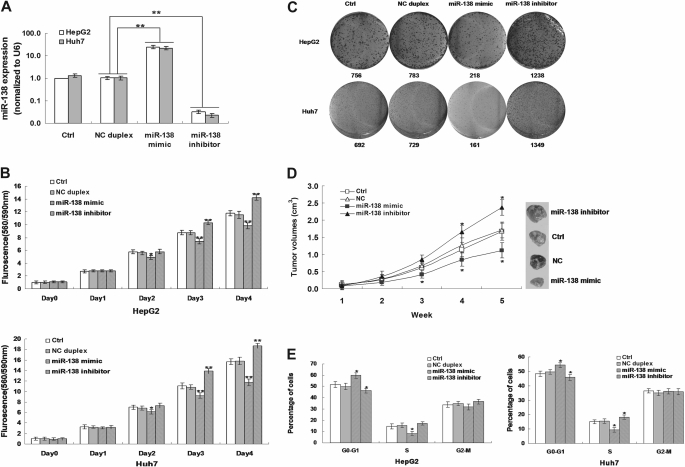
The significant reduction of miR-138 expression in HCC tissues indicated possible biological significance in tumorgenesis. At first, the effect of miR-138 on cell growth was evaluated in HepG2 and Huh7 cells transfected with or not, miR-138 mimic, miR-138 inhibitor or NC duplex. The expression of miR-138 was increased 25-folds (HepG2) and 22-folds (Huh7) in cells transfected with 20 nM miR-138 mimic but deceased 30-fold (HepG2) and 43-folds (Huh7) in cells transfected with 20 nM miR-138 inhibitor (Figure 1A). From second day (HepG2) or third day (Huh7) after the transfection, the viability of cells transfected with miR-138 mimic significantly decreased compared with that of NC duplex transfected or non-transfected cells, but the viability of cells transfected with miR-138 inhibitor significantly increased (Figure 1B). These results indicate miR-138 could inhibit cell growth.
Fig. 1.
The effect of miR-138 on cell viability (B), colony formation(C), xenograft tumor growth (D) and cell cycle (E). (A) The expression of miR-138 in HepG2 and Huh7 cells with or without transfection of miR-138 mimic, miR-138 inhibitor or NC duplex. (B) The effect of miR-138 on cell viability of HCC cell lines. (C) The effect of miR-138 on colony formation of HCC cell lines. (D) The effect of miR-138 on the tumor growth in xenograft nude mice. (E) The effect of miR-138 on cell cycle of HCC cell lines. Representative results (B, C and E) in HepG2 and Huh7 cells transfected with miR-138 mimic, miR-138 inhibitor, NC duplex or not. Column, mean of three independent experiments; bars, SD; *P < 0.01 and **P < 0.001. Representative results (D) in HepG2 transfected with miR-138 mimic, miR-138 inhibitor, NC duplex or not. *P < 0.01.
To validate the inhibitory effect of miR-138 on cell growth, the colony formation assay was performed in HepG2 and Huh7 transfected with or not, miR-138 mimic, miR-138 inhibitor or NC duplex. As showed in Figure 1C, HepG2 and Huh7 cells transfected with 20 nM miR-138 mimic displayed much fewer and smaller colonies (218 or 161 colonies) compared with NC duplex transfected (783 or 729 colonies) and non-transfected cells (756 or 692 colonies), but cells transfected with 20 nM miR-138 inhibitor displayed much more and larger colonies (1238 or 1349 colonies).
To further confirm the above findings, an in vivo mouse model was used. For the duration of the treatment with miR-138 mimic or miR-138 inhibitor for 5 weeks, tumor volume curves revealed a significant decrease in growth rates at the third, fourth and fifth week after treatment with miR-138 mimic and a significant increase in growth rates at the fourth and fifth week after treatment with miR-138 inhibitor whereas no significant differences in tumor growth rates were observed between the NC group and the control group (Figure 1D). These results indicate that introduction of miR-138 significantly inhibits tumorigenicity of HepG2 cells in xenograft nude mouse model.
To investigate the mechanism of inhibitory effect of miR-138, flow cytometry assay showed that the percentages of miR-138 mimic transfected HepG2 and Huh7 cells in the G0–G1 phase were 18% (HepG2) and 11% (Huh7) higher than that of NC duplex transfected or non-transfected cells, which paralleled with a 44% (HepG2) and 38% (Huh7) decrease in the S phase (Figure 1E). In miR-138 inhibitor-transfected cells, the percentages of cells in the G0–G1 phase were 8% (HepG2) and 6% (Huh7) less than that of NC duplex transfected or non-transfected cells, which paralleled with a 14% (HepG2) and 19% (Huh7) increase in the S phase (Figure 1E). These results indicate miR-138 could inhibit HepG2 and Huh7 proliferation by inducing cell cycle arrest at G1/S phase.
CCND3 is a direct target of miR-138
It is generally accepted that miRNAs exert their function through regulating the expression of their downstream target genes. CCND3 was predicted as a potential target of miR-138 by TargetScan and miRanda but not by PicTar. The 3′UTR of CCND3 mRNA contained a complementary site for the seed region of miR-138 (Figure 2A). CCND3 was found to be upregulated in 72.2% (13 of 18) HCC tissues compared with adjacent non-tumor liver tissues (Figure 2B). Furthermore, a negative correlation was found between the upregulated CCND3 protein and downregulated miRNA-138 (r = −0.730, P < 0.01) (Figure 2C). These results indicated that miR-138 may be associated with CCND3 and both of them may be involved in HCC tumorigenesis.
To validate whether CCND3 is a direct target of miR-138, a human CCND3 3′UTR fragment containing wild-type or mutant miR-138 binding sequence (Figure 2A) was cloned downstream of the firefly luciferase reporter gene in pGL3. In HEK293 cells cotransfected with the reporter plasmids and miR-138 mimic or NC duplex, the luciferase activity of the reporter that contained wild-type 3′UTR was significantly suppressed by miR-138 mimic, but the luciferase activity of mutant reporter was unaffected (Figure 2D), indicating that miR-138 may suppress gene expression through miR-138 binding sequence at the 3′UTR of CCND3. Furthermore, transfection of miR-138 mimic decreased CCND3 expression and transfection of miR-138 inhibitor increased CCND3 expression in HepG2 cells at protein (Figure 2E) but not mRNA level (data not shown), suggesting that CCND3 expression could be inhibited by miR-138 at post-transcriptional level. Together, the results show that miR-138 could regulate the expression of endogenous human CCND3 by directly targeting the 3′UTR of CCND3 mRNA and human CCND3 is a new target of miR-138.
CCND3 knockdown could induce cell cycle arrest
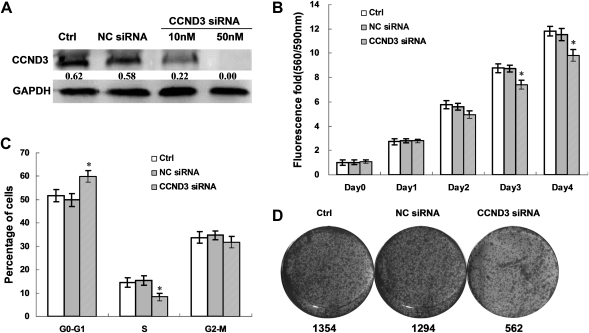
To identify whether inhibition of CCND3, just like miR-138 restoration, also resulted in HCC repression, the effects of knockdown of CCND3 on cell growth were examined. First, HepG2 cells were transfected with or not, CCND3 siRNA or control siRNA. Seventy-two hours after transfection, a dose-dependent knockdown of CCND3 was observed in HepG2 cells (Figure 3A). In cell viability assay and cell cycle analysis, in vitro knockdown of CCND3 repressed cell viability (Figure 3B), induced cell cycle arrest (Figure 3C) and inhibited the colony formation (Figure 3D). The similar data were obtained in Huh7 cells transfected with CCND3 siRNA (data not shown). These results indicate that CCND3 is most likely involved in the induction of cell cycle arrest by miR-138.
Fig. 3.
CCND3 could induce cell cycle arrest. (A) CCND3 siRNA efficiently inhibits the expression of CCND3. The expression of endogenous CCND3 was analyzed 48 h after transfection with CCND3 siRNA or NC siRNA by western blotting. Glyceraldehyde-3-phosphate dehydrogenase was used as an internal control. (B) CCND3 knockdown reduced cell viability of HCC cell lines. Representative results of cell viability in HepG2 cells transfected with CCND3 siRNA and NC siRNA or not. (C) CCND3 knockdown induced cell cycle of HCC cell lines. Representative results of cell cycle in HepG2 cells transfected with CCND3 siRNA NC siRNA or not. (D) CCND3 knockdown inhibited colony formation of HCC cell lines. Representative results of colony formation in HepG2 cells transfected with CCND3 siRNA and NC siRNA or not. Column, mean of three independent experiments; bars, SD; *P < 0.01.
Discussion
MiRNAs were found to be frequently deregulated in HCC, and some specific miRNAs were found to be associated with the clinicopathological features of HCC, such as metastasis, recurrence and prognosis (18–20). Moreover, compelling evidence demonstrates that miRNAs have important roles in HCC progression and directly contribute to the cell proliferation, avoidance of apoptosis and metastasis of HCC. Identifying the miRNAs and their targets that are essential for HCC progression may provide promising therapeutic opportunities.
In this study, with TLDA and real-time reverse transcription–PCR confirmation, 10 upregulated miRNAs (miR-217, miR-518b, miR-517c, miR-520g, miR-519a, miR-522, miR-518e, miR-525-3p, miR-512-3p and miR-518a-3p) and 10 downregulated miRNAs (miR-138, miR-214, miR-214#, miR-27a#, miR-199a-5p, miR-433, miR-511, miR-592, miR-483-5p and miR-483-3p) were identified in HCC. Important of these 20 deregulated miRNAs, only miR-199a-5p was reported to contribute to the increase of cell invasion by functional deregulation of DDR1 activity in HCC (21) and regulate Brm subunit of SWI/SNF in human cancers (22), and the other 19 deregulated miRNAs were first reported to be involved in HCC tumorigenesis. MiR-27a#, miR-214#, miR-518a-3p and miR-518e have never been reported in literature. The rest 15 miRNAs were reported in various cancer but not HCC. For example, the upregulated miR-512-3p and miR-525-3p were associated with a cisplatin-resistant phenotype in human germ cell tumors (23). The upregulated miR-519a and downregulated miR-511 and miR-485-5p were associated with histological subtypes in ovarian cancers (24). MiR-517c and 520g promotes in vitro and in vivo oncogenicity, modulates cell survival and robustly enhances growth of untransformed human neural stem cells in neuroectodermal brain tumors (25). MiR-433 could regulate tumor-associated proteins GRB2 in gastric carcinoma (26). MiR-592 was reported to be associated with the stepwise progression for transformation from normal colon to carcinoma (27). Thus, the role of these deregulated miRNAs in HCC tumorigenesis is pretty worthy to be investigated.
Compared with the previous version, the more newly found miRNAs (337 updated miRNAs) were profiled in TLDA v2.0, which may be the reason so many new differentially expressed miRNAs were identified in this study. Meanwhile, very strict criteria of selection defined in TLDA (significantly different in all three pairs of HCC tissues) led to the absence of some well-known HCC-related miRNAs. Such as miR-21 (28), miR-122 (29,30), miR-16 (31) and miR-29 (32,33) were excluded for their significantly differential expression only in two pairs of HCC tissues, and miR-181 (34), miR-221 (35,36), miR-125 (37) and miR-101 (38) were excluded for their differential expression in only one pair of HCC tissues or no differential expression at all. Using such strict criteria, we might miss some important candidates but catch some unique ones. Fortunately, qRT–PCR and follow-up studies proved that our strategy helped find the above new deregulated miRNAs.
All of HCC patients included in this study were HBsAg positive. Therefore, the expression patterns of identified miRNAs may mainly represent the alterations in Hepatitis B virus-positive HCC, which may partially account for the inconsistency between the others and our results. Although the HCC tissues from four tumor grades (G1–G4) were included, we were unable to perform statistical comparison on the profiles of differentially expressed miRNAs among four tumor grades due to the quantity restriction. Future studies including larger size of four tumor grades samples are required to elucidate the miRNAs associated with the clinicopathological features of HCC, which might severed as the biomarker for HCC.
KEGG pathway and GO enrichment analysis, which was based on the reported and predicted target genes of these deregulated miRNAs, were applied to identify which particular functions and pathways were enriched among genes controlling distinctive characters between HCC and adjacent non-tumor tissues. As a result, KEGG pathway analysis showed that proliferative (cell cycle, mitogen-activated protein kinase and Wnt), adhesive (actin cytoskeleton, adherens junction and focal adhension), survival (transforming growth factor-beta and ErbB) and oncogenic (renal cell carcinoma and pancreatic cancer) signaling pathways were abundant among the significantly enriched ones. Furthermore, the GOs related to signal transduction (signal transduction, small guanosine triphosphatase-mediated signal transduction, protein–amino acid phosphorylation and nerve growth factor receptor signaling pathway) and cell growth (cell differentiation, cell division, positive regulation of cell proliferation, cell cycle and multicellular organism development) represented up to 37% of the significantly enriched GOs. As expected, various cell process and signal pathways were involved in HCC tumorigenesis. To narrow down the scope of study and judge the most significant candidates, the intersection of signal transduction and regulation of actin cytoskeleton and pathway in cancer was selected for follow-up study.
Based on TargetScan search, miR-138 has 388 conserved targets, 32% of which belong to the intersection of signal transduction and regulation of actin cytoskeleton and pathway in cancer. In addition, we showed that miR-138, frequently downregulated in human HCC tissues, could suppress colony formation and induce cell cycle arrest in hepatoma cell lines and inhibit tumor growth in xenograft nude mice model. These results suggest the important role of miR-138 in HCC tumorigenesis. Reduced expression of miR-138 has been observed in different types of cancers (39–43) but not HCC. MiR-138 plays an important role in tongue squamous cell carcinoma cell migration and invasion by concurrently targeting RhoC and ROCK2 (39). MiR-138 could inhibit the expression of HIF-1a and regulate the apoptosis and migration of clear cell renal cell carcinoma cells (40). MiR-138 enhanced cell migration and invasion by targeting enhancer of zeste homologue 2 (EZH2) in squamous cell carcinoma cell lines (41). MiR-138 may play an important role in cancer initiation and progression by regulation of FOSL1 in squamous cell carcinoma (42) or G protein alpha-inhibiting activity polypeptide 2 (GNAI2) in tongue squamous cell carcinoma (43). We identified CCND3 as a target of miR-138 in HCC, which may provide new insights into the mechanisms underlying tumorigenesis. CCND3 is expressed in nearly all proliferating cells and could promote the cell cycle start (44). Liu et al. (45) reported miR-16 family (including miR-16, miR-195 and miR-424) could induce cell cycle arrest by targeting CCND3, CCNE1 and cyclin-dependent kinase (Cdk)-6. In TLDA analysis, miR-16 and miR-195 showed no difference in all three pairs of HCC tissues and miR-424 showed significant difference only in two of three pairs of HCC tissues, which suggest miR-16 family may not play important role in HCC tumorigenesis. Thus, we didn’t verify the regulation of miR-16 family on CCND3. The cell cycle is regulated by a family of the CDKs and their activating partners (cyclins). The G1/S phase transition is regulated primarily by D-type cyclins (D1, D2 or D3) in complex with CDK4/CDK6 and E-type cyclins (E1 or E2) in complex with CDK2. These complexes cooperate in phosphorylating and preventing Rb binding to E2F, thus activating E2F-mediated transcription and driving cells from G1 into S phase (46). Whether the CDK/pRb/E2F pathway is involved in the miR-138 induction of cell cycle arrest needs further study. On the other hand, Wang et al. (47) showed that CCND3–cdk4/cdk6 specifically phosphorylate C/EBP at Ser193 and support growth-inhibitory C/EBP–cdk2 and C/EBP–Brm complexes in differentiated cells , but we showed the promotion of growth of CCND3 regulated by miR-138 in HCC cell lines. This discrepancy may be due to different cell lines and different subjects of study. CCND3 is expressed abroad, suggesting that it has many different cell functions. CCND3 had been reported to be involved in the promotion of cell growth by several groups (48–52). Under the specific study design, the researchers could investigate only one or some aspects of CCND3 but all of it, which is needed to be further investigated.
In summary, we report the expression patterns of altered miRNAs in HCC and the potential role of miR-138 in tumorigenesis. Our data suggest the frequently downregulated miR-138 can regulate CCND3 and function as a tumor suppressor in HCC. Therefore, miR-138 may serve as a useful therapeutic agent for miRNA-based HCC therapy.
Supplementary material
Supplementary Table S1 and S2 can be found at http://carcin.oxfordjournals.org/.
Funding
This work was supported by Ministry of Science and Technology Key program (2008ZX10002-017 and 2012ZX10002009-004), Shanghai Leading Academic Discipline Project (B901) and Science Fund for Creative Research Groups, National Nature Science Foundation of China, China (30921006).
Supplementary Material
Acknowledgments
We thank research assistants at Orbital Instrument for their assistance in TDLA performance and Geminix for their assistance in GO and KEGG pathway analysis.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CDK
cyclin-dependent kinase
- cDNA
complementary DNA
- FDR
false discovery rate
- GO
Gene ontology
- HCC
hepatocellular carcinoma
- KEGG
Kyoto encyclopedia of genes and genomes
- miRNA
microRNA
- mRNA
messenger RNA
- NC
nonrelative control
- qRT–PCR
quantitative real-time–PCR
- RT
reverse transcriptase
- siRNA
small interfering RNA
- TLDA
Taqman low density miRNA array
- UTR
untranslated region
References
- 1.Parkin DM, et al. Global cancer statistics. CA Cancer J. Clin. 2002;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Aravalli RN, et al. Molecular mechanisms of hepatocellular carcinoma. Hepatology. 2008;48:2047–2063. doi: 10.1002/hep.22580. [DOI] [PubMed] [Google Scholar]
- 3.Nelson KM, et al. MicroRNAs and cancer: past, present, and potential future. Mol. Cancer Ther. 2008;7:3655–3660. doi: 10.1158/1535-7163.MCT-08-0586. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP, et al. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.He L, et al. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, et al. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Esquela-Kerscher A, et al. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 8.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupuy D, et al. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat. Biotechnol. 2007;25:663–668. doi: 10.1038/nbt1305. [DOI] [PubMed] [Google Scholar]
- 10.Schlitt T, et al. From gene networks to gene function. Genome Res. 2003;13:2568–2576. doi: 10.1101/gr.1111403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanehisa M, et al. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi M, et al. WholePathwayScope: a comprehensive pathway-based analysis tool for high-throughput data. BMC Bioinformatics. 2006;7:30. doi: 10.1186/1471-2105-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Draghici S, et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whittaker S, et al. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989–5005. doi: 10.1038/onc.2010.236. [DOI] [PubMed] [Google Scholar]
- 15.Min L, et al. Mitogen-activated protein kinases in hepatocellular carcinoma development. Semin. Cancer Biol. 2011;21:10–20. doi: 10.1016/j.semcancer.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 16.Mishra L, et al. Liver stem cells and hepatocellular carcinoma. Hepatology. 2009;49:318–329. doi: 10.1002/hep.22704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takigawa Y, et al. Wnt signaling in liver cancer. Curr. Drug Targets. 2008;9:1013–1024. doi: 10.2174/138945008786786127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braconi C, et al. MicroRNA expression profiling: a molecular tool for defining the phenotype of hepatocellular tumors. Hepatology. 2008;47:1807–1809. doi: 10.1002/hep.22326. [DOI] [PubMed] [Google Scholar]
- 19.Ladeiro Y, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 20.Mott JL. MicroRNAs involved in tumor suppressor and oncogene pathways: implications for hepatobiliary neoplasia. Hepatology. 2009;50:630–637. doi: 10.1002/hep.23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen QL, et al. Role of microRNA-199a-5p and discoidin domain receptor 1 in human hepatocellular carcinoma invasion. Mol. Cancer. 2010;9:227–239. doi: 10.1186/1476-4598-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakurai K, et al. MicroRNAs miR-199a-5p and -3p target the Brm subunit of SWI/SNF to generate a double-negative feed back loop in a variety of human cancers. Cancer Res. 2011;75:1680–1689. doi: 10.1158/0008-5472.CAN-10-2345. [DOI] [PubMed] [Google Scholar]
- 23.Port M. Micro-RNA expression in cisplatin resistant germ cell tumor cell lines. Mol. Cancer. 2011;10:52–59. doi: 10.1186/1476-4598-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim TH, et al. Deregulation of miR-519a, 153, and 485-5p and its clinicopathological relevance in ovarian epithelial tumours. Histopathology. 2010;57:734–743. doi: 10.1111/j.1365-2559.2010.03686.x. [DOI] [PubMed] [Google Scholar]
- 25.Li M, et al. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell. 2009;16:533–546. doi: 10.1016/j.ccr.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo H, et al. Down-regulated miR-9 and miR-433 in human gastric carcinoma. J. Exp. Clin. Cancer Res. 2009;28:82–90. doi: 10.1186/1756-9966-28-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oberg AL. miRNA expression in colon polyps provides evidence for a multihit model of colon cancer. PLoS One. 2011;6:e20465. doi: 10.1371/journal.pone.0020465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu S, et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 29.Tsai WC, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 30.Bai S, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J. Biol. Chem. 2009;284:32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsang TY, et al. P-glycoprotein enhances radiation-induced apoptotic cell death through the regulation of miR-16 and Bcl-2 expressions in hepatocellular carcinoma cells. Apoptosis. 2011;16:524–535. doi: 10.1007/s10495-011-0581-5. [DOI] [PubMed] [Google Scholar]
- 32.Wang CM, et al. miR-29c targets TNFAIP3, inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2011;411:586–592. doi: 10.1016/j.bbrc.2011.06.191. [DOI] [PubMed] [Google Scholar]
- 33.Fang JH, et al. MicroRNA-29b suppresses tumor angiogenesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology. 2011;54:1729–1740. doi: 10.1002/hep.24577. [DOI] [PubMed] [Google Scholar]
- 34.Arzumanyan A, et al. Does the hepatitis B antigen HBx promote the appearance of liver cancer stem cells? Cancer Res. 2011;71:3701–3708. doi: 10.1158/0008-5472.CAN-10-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon SO, et al. Deregulated expression of microRNA-221 with the potential for prognostic biomarkers in surgically resected hepatocellular carcinoma. Hum. Pathol. 2011;42:1391–1400. doi: 10.1016/j.humpath.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 36.Gramantieri L, et al. MicroRNA-221 targets Bmf in hepatocellular carcinoma and Correlates with tumor multifocality. Clin. Cancer Res. 2009;15:5073–5081. doi: 10.1158/1078-0432.CCR-09-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li W, et al. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int. J. Cancer. 2008;123:1616–1622. doi: 10.1002/ijc.23693. [DOI] [PubMed] [Google Scholar]
- 38.Chiang CW, et al. PKC alpha mediated induction of miR-101 in human hepatoma HepG2 cells. J. Biomed. Sci. 2010;17:35. doi: 10.1186/1423-0127-17-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang L, et al. Downregulation of the Rho GTPase signaling pathway is involved in the microRNA-138-mediated inhibition of cell migration and invasion in tongue squamous cell carcinoma. Int. J. Cancer. 2010;127:505–512. doi: 10.1002/ijc.25320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song T, et al. MiR-138 suppresses expression of hypoxia-inducible factor 1 (HIF-1) in clear cell renal cell carcinoma 786-O cells. Asian Pac. J. Cancer Prev. 2011;12:1307–1311. [PubMed] [Google Scholar]
- 41.Liu X, et al. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem. J. 2011;440:23–31. doi: 10.1042/BJ20111006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin Y, et al. Molecular characterization of the microRNA-138-Fos-like antigen 1 (FOSL1) regulatory module in squamous cell carcinoma. J. Biol. Chem. 2011;286:40104–40109. doi: 10.1074/jbc.C111.296707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang L, et al. Identification and experimental validation of G protein alpha inhibiting activity polypeptide 2 (GNAI2) as a microRNA-138 target in tongue squamous cell carcinoma. Hum. Genet. 2011;129:189–197. doi: 10.1007/s00439-010-0915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin J, et al. Cdk6-cyclin D3 complex evades inhibition by inhibitor proteins and uniquely controls cell's proliferation competence. Oncogene. 2001;20:2000–2009. doi: 10.1038/sj.onc.1204375. [DOI] [PubMed] [Google Scholar]
- 45.Liu Q, et al. The miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grillo M, et al. Validation of cyclin D1/CDK4 as an anticancer drug target in MCF-7 breast cancer cells: effect of regulated overexpression of cyclin D1 and siRNA-mediated inhibition of endogenous cyclin D1 and CDK4 expression. Breast Cancer Res. Treat. 2006;95:185–194. doi: 10.1007/s10549-005-9066-y. [DOI] [PubMed] [Google Scholar]
- 47.Wang GL, et al. Cyclin D3 maintains growth-inhibitory activity of C/EBPalpha by stabilizing C/EBPalpha-cdk2 and C/EBPalpha-Brm complexes. Mol. Cell. Biol. 2006;26:2570–2582. doi: 10.1128/MCB.26.7.2570-2582.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Q, et al. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin J, et al. Cdk6-cyclin D3 complex evades inhibition by inhibitor proteins and uniquely controls cell's proliferation competence. Oncogene. 2001;20:2000–2009. doi: 10.1038/sj.onc.1204375. [DOI] [PubMed] [Google Scholar]
- 50.Renate F, et al. Cdk6–cyclin D3 activity in murine ES cells is resistant to inhibition by p16INK4a. Oncogene. 2004;23:491–502. doi: 10.1038/sj.onc.1207133. [DOI] [PubMed] [Google Scholar]
- 51.Lu J, et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71:225–233. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, et al. Enhanced expression of cyclins and cyclin-dependent kinases in aniline-induced cell proliferation in rat spleen. Toxicol. Appl. Pharmacol. 2011;250:213–220. doi: 10.1016/j.taap.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.