Abstract
Transforming growth factor-β (TGF-β) signaling regulates many diverse cellular activities through both canonical (SMAD-dependent) and non-canonical branches, which includes the mitogen-activated protein kinase (MAPK), Rho-like guanosine triphosphatase and phosphatidylinositol-3-kinase/AKT pathways. Here, we demonstrate that miR-335 directly targets and downregulates genes in the TGF-β non-canonical pathways, including the Rho-associated coiled-coil containing protein (ROCK1) and MAPK1, resulting in reduced phosphorylation of downstream pathway members. Specifically, inhibition of ROCK1 and MAPK1 reduces phosphorylation levels of the motor protein myosin light chain (MLC) leading to a significant inhibition of the invasive and migratory potential of neuroblastoma cells. Additionally, miR-335 targets the leucine-rich alpha-2-glycoprotein 1 (LRG1) messenger RNA, which similarly results in a significant reduction in the phosphorylation status of MLC and a decrease in neuroblastoma cell migration and invasion. Thus, we link LRG1 to the migratory machinery of the cell, altering its activity presumably by exerting its effect within the non-canonical TGF-β pathway. Moreover, we demonstrate that the MYCN transcription factor, whose coding sequence is highly amplified in a particularly clinically aggressive neuroblastoma tumor subtype, directly binds to a region immediately upstream of the miR-335 transcriptional start site, resulting in transcriptional repression. We conclude that MYCN contributes to neuroblastoma cell migration and invasion, by directly downregulating miR-335, resulting in the upregulation of the TGF-β signaling pathway members ROCK1, MAPK1 and putative member LRG1, which positively promote this process. Our results provide novel insight into the direct regulation of TGF-β non-canonical signaling by miR-335, which in turn is downregulated by MYCN.
Introduction
Neuroblastoma is a highly enigmatic childhood cancer arising from precursor cells of the sympathetic nervous system with highly heterogeneous clinical behavior. Tumors in children under the age of 18 months are often successfully eliminated and will even sometimes spontaneously regress, whereas tumors in older children often become refractory to treatment (1). Metastasis is the overwhelming cause of mortality among neuroblastoma patients (2). Neuroblastoma is a cancer that primarily metastasizes to bone marrow and the survival rate of children with bone marrow metastasis is only ∼40% (3). Neuroblastoma is characterized by multiple recurrent genomic abnormalities (4), including amplification of the gene encoding the MYCN transcription factor, which is highly predictive of poor clinical outcome and metastatic disease (5). As a transcription factor, MYCN binds to the promoter regions of large numbers of genes and non-coding RNA sequences, modifying their transcriptional activation status (6–9).
The transforming growth factor-β (TGF-β) signaling pathway plays a pivotal and somewhat enigmatic role in the development and progression of cancer, acting both as a tumor suppressor and as a positive driver of cancer progression in later stages of various diseases, including breast, prostate and colon cancer (10,11). Initially, it was determined that TGF-β signaling transpires through TGF-β ligand binding to a type II receptor, which recruits and phosphorylates a type I receptor. The type I receptor then phosphorylates and activates SMAD transcription factors which subsequently translocate to the nucleus, thus regulating the transcription of specific genes. However, TGF-β is now understood to be considerably more intricate, with TGF-β receptors being able to activate additional signaling pathways, including the mitogen-activated protein kinase (MAPK), Rho-like guanosine triphosphatase and phosphatidylinositol-3-kinase (PI3K)/AKT pathways. These SMAD-independent non-canonical signaling pathways help to explain the diversity of phenotypic responses to TGF-β signaling (12). Ectopic overexpression of TGF-β in neuroblastoma cell lines inhibits cell proliferation (13) and overexpression of TGF-β type II receptor in LAN5 neuroblastoma cells suppresses their tumor-forming ability and induces differentiation (14).
Available evidence indicates that some genes forming part of the TGF-β signaling pathways are regulated by microRNAs (miRNA), which are short 19–22 nt RNA sequences that inhibit gene expression at a post-transcriptional level by targeting regions of sequence complementarity on the 3′untranslated regions (UTRs) of specific messenger RNAs (mRNAs) (15). MiRNAs can either cause degradation of mRNAs or can inhibit their translation and play major roles in normal growth and development. MiRNA dysregulation has oncogenic or tumor-suppressive functions in virtually all forms of cancer, including neuroblastoma (16,17). MiRNAs with neuroblastoma tumor-suppressive functions include proapoptotic miR-34a and miR-184 (18–23), anti-invasive miR-542-5p (24) and miR-10a/b, which induce neuroblastoma cell differentiation (25). On the other hand, the miR-17-5p-92 polycistronic cluster acts in an oncogenic fashion by targeting multiple members of the TGF-β canonical signaling pathway, including TGFBR2, SMAD2, SMAD4 and p21 (26,27). MiR-335 suppresses gastric cancer cell invasion and metastasis through directly targeting the specificity protein 1 (SP1) and Bcl-w (BCL2L2) mRNA transcripts (28). SP1 and Bcl-w form part of the PI3K/AKT pathway involved with TGF-β non-canonical signaling. The role of miR-335 in neuroblastoma has thus far never been investigated.
Here, we demonstrate that miR-335 regulation of TGF-β non-canonical signaling is substantially more pervasive than originally realized. In addition to targeting SP1 in the PI3K/AKT pathway, as previously demonstrated for gastric cancer, we have determined that miR-335 targets gene transcripts from the other two independent TGF-β non-canonical pathways, MAPK1 and Rho-like guanosine triphosphatase (ROCK1). MiR-335 targeting leads to a significant decrease in phosphorylation signaling within the respective pathways which culminates in inactivation of the motor protein myosin light chain (MLC) and therefore a decrease in the migratory and invasive potential of neuroblastoma cells. Additionally, we demonstrate that miR-335 targets the leucine-rich alpha-2-glycoprotein 1 mRNA transcript (LRG1), similarly leading to decreased migration and invasion of neuroblastoma cells by reducing the phosphorylation status of MLC thereby contributing to accumulating evidence of a possible involvement of LRG1 in TGF-β signaling. Finally, we demonstrate that miR-335 is directly repressed by the binding of MYCN to an upstream region of the miR-335 transcriptional start site, indicating a link between MYCN and the regulation of TGF-β signaling mediated through this miRNA. Thus, MYCN promotes disease progression by directly repressing miR-335, preventing this miRNA from targeting and post-transcriptionally inhibiting multiple genes, which promote cell migration and invasion.
Materials and methods
Cell lines and culture conditions
SK-N-AS and Kelly cell lines were purchased from the European Collection of Animal Cell Cultures. CHP-212 cells were purchased from the American Type Culture Collection. The MYCN repressible SHEP-TET21N cell line was obtained from Dr Louis Chesler and was treated with 100 ng/ml doxycycline to repress MYCN expression. All cell lines were validated for the presence of previously published genomic imbalances using array comparative genomic hybridization. Culture media was supplemented with 10% fetal bovine serum. For TGF-β stimulation studies, cells were starved overnight in 1% fetal bovine serum prior to stimulation with 5 ng/ml TGF-β (resuspended in 5 mM HCl) (AMS Biotechnology, Oxfordshire, UK) or 5 mM HCl (control cells).
Neuroblastoma tumors
MiR-335 expression profiling data was obtained from three published studies, which used the same TaqMan quantitative PCR (qPCR) platform (29–31).
Transfection procedures
MiRNA precursors, inhibitors and scrambled oligonucleotide controls (Ambion, Austin, TX and Exiqon, Copenhagen, Denmark) were transiently transfected into cells at a final concentration of 30 nM by reverse transfection using siPORT NeoFX reagent (Ambion). siGENOME smart pool small interfering RNAs (siRNAs) and negative controls (Dharmacon, Chicago, IL) were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) at a final concentration of 30 nM. ROCK1 expression vector, pCAG-myc-p160ROCK, was received as a gift from Dr Shuh Narumiya (Kyoto, Japan) (32). Two micrograms of expression plasmid or empty vector were transfected into Kelly cells using Lipofectamine 2000 (Invitrogen).
Quantitative real-time reverse transcription–PCR
MiRNA or total RNA was extracted from cells using either the miRNeasy Mini Kit or RNeasy Mini Kit (Qiagen, Crawley, UK). The expression of miR-335, ROCK1, MAPK1 and LRG1 was measured by TaqMan assays (Applied Biosystems, Foster City, CA). MiRNA and gene expression were normalized using endogenous RNU48 and 18S controls, respectively. For miR-335, expression levels of mature miRNA were quantified. See also Supplementary Methods, available at Carcinogenesis Online.
Microarray analysis
Kelly cells were transfected with either Pre-miR-335 or scrambled oligonucleotide and total RNA was extracted 48 h post-transfection. Array profiling began with initial creation and amplification of complementary DNA from total RNA using the WT-Ovation Pico RNA Amplification System (NuGEN, San Carlos, CA). Subsequent Cy3-labeling of complementary DNA using the NimbleGen One-Color DNA Labeling Kit was followed by hybridization to a 4 × 72 K human gene expression array (Roche NimbleGen, Madison, WI). Data analysis was performed using Roche NimbleScan software. The expression data was normalized using the Robust Multichip Average algorithm as described by Irizarry et al. (33) using quantile normalization as described previously (34). A 2-fold downregulation of genes compared with scrambled control was used as the threshold for the identification of potential miR-335 target genes. Potential target genes were subsequently cross-referenced against the TargetScan prediction algorithm (www.targetscan.org). Microarray data has been deposited at http://www.ebi.ac.uk/miamexpress/login.html (accession: E-MEXP-3274).
Cell migration and invasion assays
Cells transfected with miR-335 mimics, miR-335 inhibitor or a scrambled oligonucleotide were transferred into 8 μm pore size transwell inserts (BD Biosciences, San Jose, CA) or Matrigel Invasion Chambers (Invitrogen) 48 h post-transfection. For myosin inhibitor studies, cells were treated for 24 h with 10 μM blebbistatin (resuspended in dimethyl sulfoxide) (Tocris Bioscience, Bristol, UK) or dimethyl sulfoxide (control cells) and then transferred to migration and invasion chambers in blebbistatin or dimethyl sulfoxide. For TGF-β stimulation studies, cells were seeded into migration or invasion chambers for 24 h in either 5 ng/ml TGF-β or 5 mM HCl (control). Cells were seeded at a density of 2.5 × 104 for migration assays and 5 × 104 for invasion assays. Cells were suspended in serum-free media while media containing 10% fetal bovine serum was added to the lower chamber of the inserts. Inserts were subsequently incubated at 37°C for 24 h. Migratory or invasive cells were fixed using methanol, stained using 0.5% crystal violet and counted directly by light microscopy.
Significance testing
An unpaired Student’s t-test was used to determine statistical significance in all functional assays and densitometric analyses of western blot assays.
Cell proliferation assay
Proliferation rates of transfected cells were measured every 24 h over a 96 h period using the Cell Titre 96 Aqueous One Solution Cell Proliferation Assay (Promega, Madison, WI). Proliferation rates of Pre-miR-335-and miR-335 inhibitor-transfected cells were compared with scrambled oligonucleotide-transfected cells. siKinesin (Ambion) was used as a positive control for reduced cell proliferation.
Western blot analyses
Total protein was extracted from cells using RIPA lysis and extraction buffer (Fisher Scientific, Hudson, NH). Primary antibodies for ROCK1, MAPK1, MLC, pMLC and pMAPK1 (Cell Signalling Technology, Beverly, MA) were diluted 1:1000 in 5% bovine serum albumin in 1× Tris-buffered saline 0.1% Tween. LRG1 (Abnova, Taipei City, Taiwan) and α-tubulin (Abcam, Cambrdige, MA) were diluted 1:1000 in 5% milk 1× Tris-buffered saline 0.1% Tween followed by anti-rabbit or anti-mouse IgG horseradish peroxidase-conjugated secondary antibodies (Abcam). Densitometry was performed using GelEVal 1.22.
Immunofluorescence
MiR-335-, miR-335 inhibitor- or scrambled control-transfected Kelly cells were grown on glass coverslips for 48 h. Cells were then washed with phosphate-buffered saline and fixed using ice-cold 100% methanol. Cells were permeabilized using Triton-X 100 and blocked with 10% goat serum. Primary anti-ROCK1 antibody (1:300; Abnova) was incubated for 1 h at room temperature in a humidified chamber followed by incubation with secondary AlexaFluora 594 goat anti-rabbit antibody (1:500; Invitrogen). Cells were counter stained and mounted using Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA). For actin cytoskeleton staining, cells were stained using the Actin Cytoskeleton and Focal Adhesion Staining Kit (Millipore, Billerica, MA) as per manufacturer’s instructions. Images were visualized using an LSM 700 confocal microscope (Zeiss, Jena, Germany).
MYCN chromatin immunoprecipitation and microarrays
The details of the MYCN chromatin immunoprecipitation-chip experiments, along with validation of results, have been previously published (6).
Results
MiR-335 functions to suppress the migratory and invasive capacities of neuroblastoma cells
It has been previously demonstrated by miRNA profiling that miR-335 is significantly downregulated in MYCN-amplified (MNA) primary neuroblastoma tumors relative to non-MNA tumors (Supplementary Figure S1a is available at Carcinogenesis Online and ref. 29). Low expression of miR-335 was also correlated with poor overall patient survival in a cohort of 237 neuroblastoma patients using the R2 bioinformatic tool (http://r2.amc.nl) (Supplementary Figure S1b is available at Carcinogenesis Online). The significant repression of miR-335 in MNA tumors was intriguing, given the clinically aggressive phenotype of MNA tumors, and on this basis we selected this miRNA for further functional analysis in neuroblastoma cell lines.
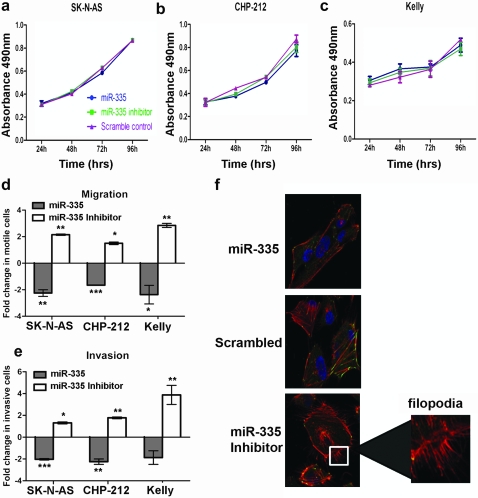
Transfection of the mature miR-335 mimics into two MNA (Kelly and CHP-212) and one non-MNA (SK-N-AS) cell lines resulted in ectopic overexpression (Supplementary Figure S1c is available at Carcinogenesis Online) but had no inhibitory affect on cell proliferation relative to the scrambled control (Figure 1a–c). Similarly, inhibition of endogenous miR-335 expression also had no significant impact on cell proliferation. However, ectopic overexpression of miR-335 significantly reduced cell migration across a non-matrigel membrane for all three cell lines relative to the scrambled control (Figure 1d). Conversely, miR-335 inhibition significantly enhanced migration relative to the scrambled control (Figure 1d). Similarly, ectopic overexpression of miR-335 significantly reduced the ability of CHP-212 and SK-N-AS cells to invade across a matrigel-coated membrane (Figure 1e). A reduction in cell invasiveness was not statistically significant in Kelly cells, which already have a low baseline level of invasive potential, however, a significant enhancement of invasion was evident for Kelly and the other cell lines following inhibition of endogenous miR-335 (Figure 1e.). These results indicate that miR-335 is involved in regulating the migratory and invasive potential of neuroblastoma cells. Furthermore, this alteration in migration and invasion potential is independent of cell proliferation in our experiments.
Fig. 1.
miR-335 regulates the migratory and invasive potential of neuroblastoma cell lines and does not affect cell viability. (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) cell viability assays following transfection of SK-N-AS (a), CHP-212 (b) and Kelly (c) cells with mature miR-335 mimics, miR-335 inhibitor or scrambled negative control. (d and e) Analysis of the ability of SK-N-AS, CHP-212 and Kelly cells to migrate across a non-matrigel membrane (d) and to invade across a matrigel membrane (e) following transfection with mature miR-335 mimics, miR-335 inhibitor or scrambled control. Asterisks indicate statistical significance obtained by unpaired Students’s t-test. *P < 0.05, **P < 0.005, ***P < 0.0005. (f) Staining of F-actin and focal adhesions in SK-N-AS cells with fluorescence-labeled phalloidin (red) and vinculin (green), respectively. Cell nuclei are stained with 4′,6-diamidino-2-phenylindole (blue). An enlarged image of the filopodia in miR-335 inhibitor-transfected cells is indicated by the white box.
Re-arrangements in the actin cytoskeleton provide the forces necessary for cancer cells to migrate and invade. Cell motion is defined by a complex and dynamic process, including actin stress fiber formation, contraction and membrane protrusion. In an effort to visualize any alteration in actin cytoskeleton dynamics that might accompany the observed changes in cell migration and invasion, SK-N-AS cells were stained with phalloidin and vinculin to visualize F-actin and focal adhesions, respectively. As illustrated in Figure 1f, cells transfected with miR-335 mimics displayed fewer and less intensely stained F-actin stress fibers and focal adhesions compared with scrambled control-transfected cells. This disorganization of the actin cytoskeleton is associated with the reduced migratory and invasive potential of the cells. In contrast, miR-335 inhibition induced a greater abundance of F-actin-rich filopodia protruding from the cell membrane compared with scrambled control-transfected cells. Abundant filopodia is a characteristic of highly invasive cancer cells and potentially contributes to the enhanced migratory and invasive potential of cells with reduced miR-335 expression.
Identification of miR-335 target genes
In order to identify potential miR-335 targets, microarray mRNA expression analysis was conducted on Kelly cells 48 h post-transfection with mature miR-335 mimics. A total of 252 genes were downregulated 2-fold or more across two biological replicates (Supplementary Table S1 is available at Carcinogenesis Online). Of these genes, 43 were computationally predicted targets for miR-335 as determined by the TargetScan miRNA prediction algorithm, representing a statistically significant enrichment for miR-335 target genes (P = 0.0009). Analysis of the genes that were upregulated 2-fold or more in response to miR-335 upregulation revealed a statistically significant underrepresentation of miR-335 target genes (P = 0.038). Of the 43 potential target genes identified, several are implicated in biological processes, such as cell motion, cell adhesion, angiogenesis and morphogenesis, which are relevant to the observed phenotypic effects of miR-335. Five potential target genes were selected for validation and further analysis: ASPH (aspartate beta-hydroxylase isoform a), ROCK1 (Rho-associated, coiled-coil containing protein), LRG1 (leucine-rich alpha-2-glycoprotein 1), MAPK1 and ETV1 (ets variant gene 1).
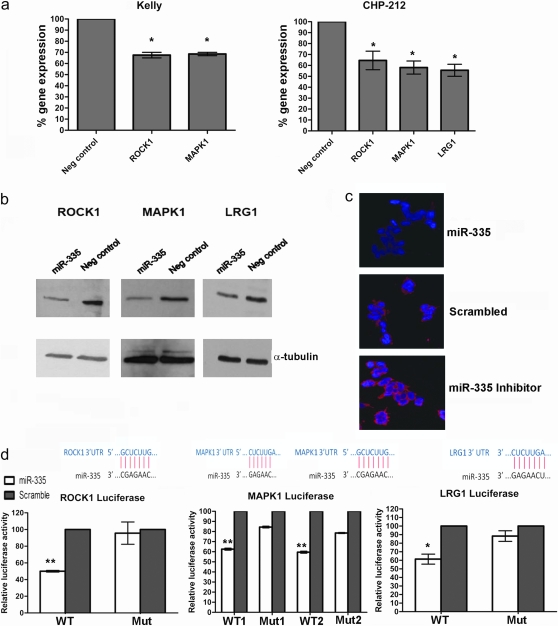
The downregulation of only three of these genes, ROCK1, MAPK1 and LRG1, following ectopic miR-335 upregulation, could be confirmed by real-time qPCR analysis. As illustrated in Figure 2a, ROCK1 and MAPK1 mRNA expression levels were significantly reduced 48 h post-transfection with miR-335 mimics in Kelly cells. LRG1 transcripts were undetectable in this cell line by reverse transcription–qPCR, however, significant reductions in mRNA transcripts for LRG1 and the other two genes (ROCK1 and MAPK1) were detected in CHP-212 cells within 48 h post-miR-335 transfection (Figure 2a). A corresponding reduction in protein level was also detectable by western blot for LRG1, ROCK1 and MAPK1 in response to miR-335 upregulation in CHP-212 cells (Figure 2b). Conversely, increased protein expression for the three genes was also demonstrated following transfection of cells with the miR-335 inhibitor (Supplementary Figure S2 is available at Carcinogenesis Online). In addition, miR-335 regulation of ROCK1 expression was also visualized by immunofluorescent staining of Kelly cells following transfection with miR-335 mimics, miR-335 inhibitor or a scrambled control. As indicated in Figure 2c, immunofluorescent staining of ROCK1 increased in Kelly cells transfected with miR-335 inhibitor and decreased in cells transfected with miR-335 mimics, relative to the scrambled control.
Fig. 2.
miR-335 target gene identification and validation. (a) Reverse transcription–qPCR expression analysis of ROCK1, MAPK1 and LRG1 in Kelly and CHP-212 cells following transfection with mature miR-335 mimics. LRG1 expression could not be detected in Kelly cells. (b) Western blot analysis revealed reduced expression of ROCK1, MAPK1 and LRG1 in CHP-212 cells transfected with miR-335. (c) Immunofluorescent staining of Kelly cells facilitated visualization of reduced ROCK1 protein expression in miR-335-transfected cells and increased expression of ROCK1 in miR-335 inhibitor-transfected cells. ROCK1 (red), 4′,6-diamidino-2-phenylindole (blue). (d) Luciferase reporter assays confirmed direct targeting of ROCK1, MAPK1 and LRG1 3′UTR regions by miR-335. A significant reduction in luciferase activity was measured for each of the wild-type miR-335 binding sites for each of the three genes compared with the mutated binding sites, normalized against scrambled control. The miRNA:mRNA base pairing for each gene is also illustrated. *P < 0.05, **P < 0.005.
To determine if miR-335 directly targets the 3′UTRs of ROCK1, MAPK1 and LRG1, luciferase reporter plasmids were constructed containing the miR-335 binding sites located within the 3′UTR for each of the genes (Supplementary Figure S3 is available at Carcinogenesis Online). Using the TargetScan database, single miR-335 binding sites were found within the 3′UTRs of ROCK1 and LRG1. Cotransfection of the reporter construct containing the wild-type binding sequence with mature miR-335 mimics resulted in a significant reduction in luciferase activity for ROCK1 and LRG1 (Figure 2d) in CHP-212 cells. In both cases, this effect was abrogated by a mutated target sequence, thereby confirming that ROCK1 and LRG1 are direct targets of miR-335. In the case of MAPK1, two miR-335 binding sites were predicted within its 3′UTR. The two binding sites are situated ∼1600 bp apart and therefore, a wild-type and mutant reporter construct was designed for each individual binding site (Supplementary Figure S3 is available at Carcinogenesis Online). Confirmation that MAPK1 is a direct target of miR-335 was provided by a significant reduction in luciferase activity for each wild-type binding site, which was abrogated by the mutated binding sites (Figure 2d). Therefore, ROCK1, MAPK1 and LRG1 are each direct targets of miR-335.
SiRNA-mediated inhibition of ROCK1, MAPK1 and LRG1 recapitulates the functional effects associated with miR-335 upregulation
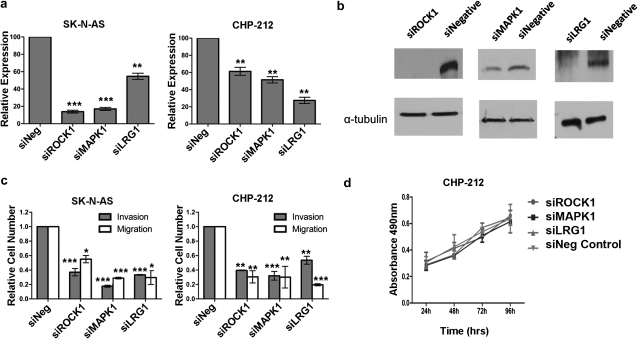
If miR-335 was regulating the cell invasive phenotype through targeting of ROCK1, MAPK1 and/or LRG1 mRNA transcripts, we reasoned that siRNA-mediated knockdown of these transcripts, either individually or collectively, should result in a decrease in cell invasion, similar to that observed following miR-335 overexpression. SiRNA-mediated knockdown of each of the genes was individually carried out, reducing mRNA levels by 50–90% in SK-N-AS cells and by 40–70% in CHP-212 cells, as determined by qPCR analysis of RNA isolated 48 h post-transfection (Figure 3a). A significant reduction in protein levels was validated by western blot analysis 72 h post-transfection (Figure 3b). Migration and invasion assays for CHP-212 and SK-N-AS cells following siRNA-mediated knockdown demonstrated a significant reduction in migration and invasion potential following independent knockdown of either ROCK1, MAPK1 and LRG1 expression (Figure 3c). Proliferation assays indicated that siRNA-mediated knockdown of the three genes had no impact on the rate of cell proliferation (Figure 3d). Therefore, independent knockdown of each of these three genes is capable of recapitulating the diminished cell migration and invasion observed upon upregulation of miR-335.
Fig. 3.
siRNA-mediated functional effects of ROCK1, MAPK1 and LRG1. (a) Reverse transcription–qPCR analysis demonstrated significant reductions in the expression levels of ROCK1, MAPK1 and LRG1 in SK-N-AS and CHP-212 cells 48 h post-transfection. (b) Western blot analysis also revealed significant reduction in protein levels of ROCK1, MAPK1 and LRG1 72 h post-transfection of CHP-212 cells. (c) SK-N-AS and CHP-212 cells demonstrated significant reductions in migration and invasion following knockdown of ROCK1, MAPK1 and LRG1. (d) Knockdown of ROCK1, MAPK1 and LRG1 did not alter the rate of proliferation of CHP-212 cells as compared with siNegative control transfected cells. *P < 0.05, **P < 0.005, ***P < 0.0005.
From the above described siRNA knockdown experiments, it is clear that the pleiotropic nature of miR-335, targeting multiple genes, does not easily allow for simple phenotypic rescue experiments to be performed. Nevertheless, we decided to determine if ectopic overexpression for one of the miR-335 target genes, ROCK1, would result in enhanced migration and invasion of neuroblastoma cells, providing further validation of our siRNA findings. A ROCK1 expression plasmid (pCAG-myc-p160ROCK) was transfected into Kelly cells, leading to ∼4-fold increased expression at mRNA level and at protein level (Supplementary Figure S4a and b is available at Carcinogenesis Online). Kelly cells were chosen for these experiments given the low migratory and invasive potential of these cells. Cells transfected with the ROCK1 plasmid demonstrated a significant enhancement in their migratory and invasive potential in comparison with cells transfected with the empty vector (Supplementary Figure S4c is available at Carcinogenesis Online). Furthermore, cells transfected with the ROCK1 plasmid demonstrated no change in the rate of proliferation as compared with empty vector-transfected cells (Supplementary Figure S4d is available at Carcinogenesis Online). Thus, the enhancement in migration and invasion that occurs following ROCK1 ectopic overexpression is consistent with the phenotypic effect following inhibition of endogenous miR-335.
MiR-335 regulation of TGF-β signaling
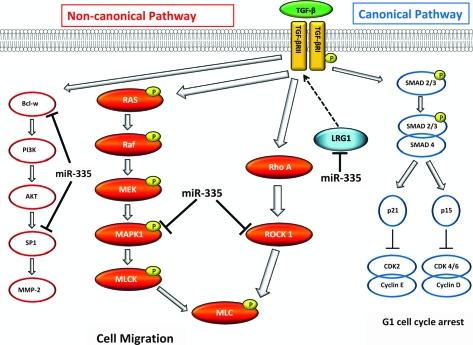
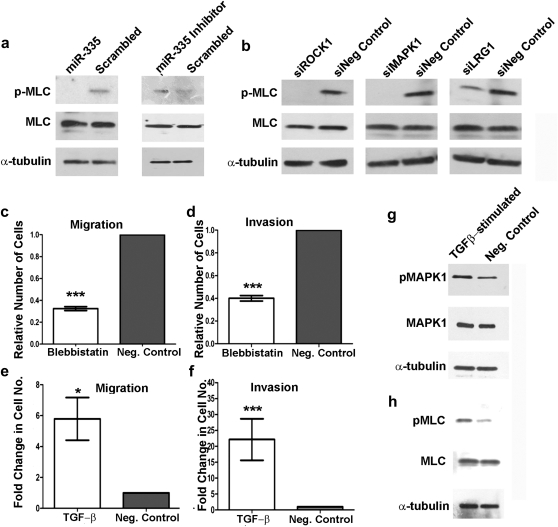
ROCK1 and MAPK1 form two independent branches of the non-canonical TGF-β signaling pathway that function to control cell migration (12). LRG1, although suspected of being involved with TGF-β signaling (35), has never been definitively placed in the pathway. The ROCK1 and MAPK1 signaling pathways converge to tightly regulate MLC phosphorylation or activation status, as illustrated in Figure 4. ROCK1 phosphorylates MLC at serine 19 (32); similarly MAPK1 phosphorylates MLC kinase, which in turn also phosphorylates serine 19 of MLC (36). Therefore, using a phospho-antibody specific to serine 19 of MLC, we demonstrate that transfection of CHP-212 neuroblastoma cells with miR-335 results in decreased levels of phosphorylated MLC protein (Figure 5a). Conversely, transfection with a miR-335 inhibitor resulted in increased levels of phosphorylated MLC protein, as analyzed by western blot 72 h post-transfection (Figure 5a). Furthermore, we demonstrate that miR-335 regulation of MLC phosphorylation manifests through direct targeting of ROCK1, MAPK1 and LRG1. Indeed, independent siRNA-mediated knockdown of each of these three genes induces a significant reduction in the phosphorylation status of MLC, also analyzed by western blot 72 h post-transfection (Figure 5b). MLC regulates the actin-binding activity of myosin and therefore the motile capacity of the cell (37). To confirm that MLC is the downstream target through which miR-335 ultimately influences cell motility in neuroblastoma cells, we demonstrate that the inhibitor of myosin activity, blebbistatin (38–40), significantly diminished neuroblastoma cell migration (Figure 5c) and invasion (Figure 5d).
Fig. 4.
Schematic illustration of TGF-β signaling pathway members and potential pathway member LRG1. TGF-β pathway comprises the canonical and non-canonical signaling pathways. miR-335 regulates cell migration and invasion through targeting of the three branches of the non-canonical TGF-β pathway. In this study, miR-335 has been demonstrated to directly target ROCK1, MAPK1 and putative TGF-β member LRG1. miR-335 inhibition of ROCK1 prevents phosphorylation of MLC thereby reducing actomyosin assembly and contraction and ultimately the motile potential of the cells. Similarly, inhibition of MAPK1 signaling also prevents phosphorylation of MLC.
Fig. 5.
MiR-335 regulation of TGF-β signaling. (a) Western blot analysis demonstrates decreased levels of phosphorylated MLC protein in CHP-212 transfected with miR-335 as compared with scrambled negative control. MiR-335 inhibitor-transfected cells displayed increased levels of phosphorylated MLC protein 72 h post-transfection. No change was evident in the levels of total MLC protein. (b) Individual siRNA-mediated knockdown of ROCK1, MAPK1 and LRG1 in CHP-212 cells resulted in a reduction in the level of phosphorylated MLC protein as compared with siNegative control-transfected cells, 72 h post-transfection. No change was evident in the levels of total MLC protein. Treatment of CHP-212 cells with 10 μM blebbistatin (myosin inhibitor) significantly reduced the migration (c) and invasion (d) potential of the treatment cells compared with control cells. Stimulation of CHP-212 cells with 5 ng/ml TGF-β induced a significant enhancement of the cells migratory (e) and invasive (f) capacities compared with control cells. (g) Stimulation of CHP-212 cells with 5 ng/ml TGF-β for 30 min induced an increase in pMAPK1 protein levels. (h) Increased levels of pMLC were observed following 24 h simulation with 5 ng/ml TGF-β. Asterisks indicate statistical significance obtained by unpaired Students’s t-test. *P < 0.05, **P < 0.005, ***P < 0.0005.
To demonstrate that the effect of miR-335 on migration and invasion is through TGF-β signaling, CHP-212 neuroblastoma cells were stimulated with TGF-β growth factor for 24 h in migration and invasion chambers. TGF-β-stimulated cells demonstrated significantly enhanced migratory (Figure 5e) and invasive (Figure 5f) capabilities compared with control cells. CHP-212 neuroblastoma cells stimulated with TGF-β had increased levels of phosphorylated MAPK1 (Figure 5g) and MLC proteins (Figure 5h) at 30 min and 24 h, respectively. The unavailability of a phospho-antibody for ROCK1 precluded the assessment of pROCK1 levels. These findings lead us to conclude that miR-335 regulates neuroblastoma cell migration and invasion by targeting the non-canonical branches of the TGF-β signaling pathway, which ultimately converge to tightly regulate MLC phosphorylation status. This is also the first report of a link between LRG1 and MLC, indicating that LRG1 also influences cell migration and invasion by altering the phosphorylation status of MLC, presumably by exerting its effect upstream of ROCK1 and MAPK1.
MYCN represses miR-335 expression in both neuroblastoma tumors and the SHEP-TET-21 MYCN-repressible cell line
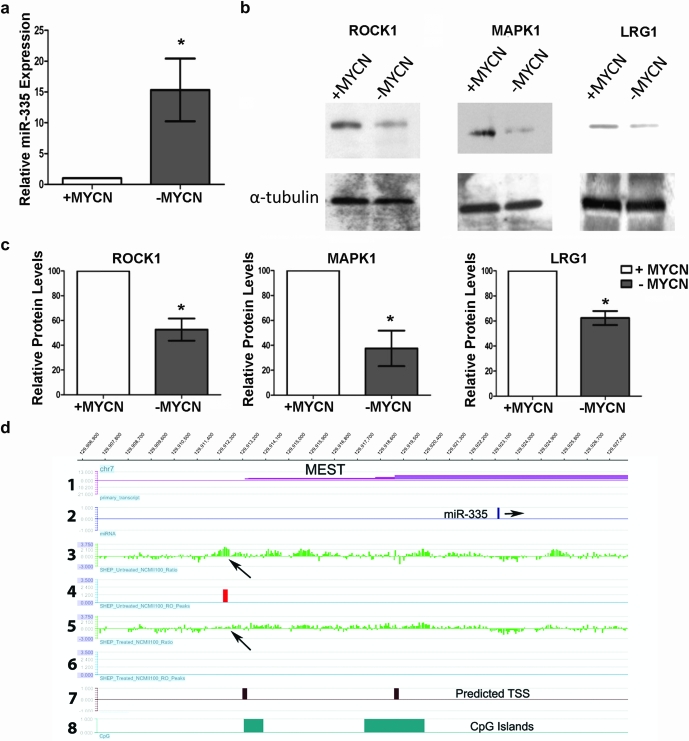
As previously demonstrated, miR-335 is significantly downregulated in MNA relative to non-MNA neuroblastoma tumors (Supplementary Figure S1a is available at Carcinogenesis Online). This expression pattern for miR-335 is also replicated in the neuroblastoma cell lines, with endogenous miR-335 expression being significantly lower in the MNA cell lines (CHP-212 and Kelly) compared with the non-MNA cell line SK-N-AS (Supplementary Figure S5 is available at Carcinogenesis Online). In order to experimentally confirm MYCN repression of miR-335, we used the well-characterized SHEP-TET21N cell line containing a repressible MYCN transgene (41), to determine if miR-335 levels changed when MYCN levels were repressed following the addition of doxycycline to the cell culture media. Treatment of these cells with doxycycline repressed MYCN expression (∼200-fold) and induced a resultant 15-fold increase in miR-335 expression (Figure 6a). Western blot analysis of SHEP-TET21N cells in the MYCN high and low expression states, corresponding to low and high miR-335 expression, respectively, revealed decreased levels of ROCK1, MAPK1 and LRG1 protein in cells with increased miR-335 activity (Figure 6b), consistent with their 3′UTRs being valid targets of this miRNA. Statistically significant differences in protein expression are illustrated in Figure 6c as determined by densitometric analyses.
Fig. 6.
MYCN regulates expression of miR-335 in neuroblastoma cell lines. (a) miR-335 expressional change in SHEP-TET21N containing a repressible MYCN transgene following doxycycline treatment. There is an average 15-fold increase in miR-335 expression in cells with low MYCN expression (treated) relative to those with high MYCN levels. (b) Western blot analysis of ROCK1, MAPK1 and LRG1 in SHEP cells reveal reduced protein expression when MYCN expression is repressed (treated with doxycycline) consistent with increased expression of miR-335. (c) Densitometric analysis of protein levels from western blots demonstrate statistically significant differences in protein expression in MYCN high and low states for all three genes, following normalization to the enodogenous control (α-tubulin). Densitometric data represent two biological repeat experiments. *P < 0.05. (d) An ∼20 kb tiled region upstream and downstream of miR-335 on chromosome 7 is displayed. MiR-335 is embedded within the MEST gene (panels 1 and 2). The third and fifth panels display the raw log2 ratios for MYCN in chromatin immunoprecipitation experiments from untreated (high MYCN expression) and doxycycline-treated (low MYCN expression) SHEP-TET21 cells, respectively. Peaks which are high confidence binding sites with a false discovery rate score of <0.05 are displayed as red (panels 4 and 6). Arrows point to a peak in the high MYCN-expressing cells that is lost in the low expressing state. Panels 7 and 8 show predicted potential miR-335 transcriptional start sites and positions of CpG islands, respectively.
To determine if miR-335 might be directly repressed by MYCN binding, we analyzed a previously published data set (6) of MYCN binding sites identified by chromatin immunoprecipitates applied to a custom tiling array containing 558 miRNA regions. Significant MYCN binding to a region ∼500 bp upstream of a predicted miR-335 transcriptional start site [based on the presence of a H3K4me3 mark (42)] was detected on the arrays (Figure 6d). MYCN binding at this site was abrogated in cells with low MYCN expression (i.e. treated with doxycycline). Notably, the MYCN binding site contained a non-canonical binding motif (CAGGTG), which has been demonstrated to bind MYCN (43).
Discussion
We have identified the ROCK1, MAPK1 and LRG1 gene transcripts as direct targets of miR-335 and have demonstrated that siRNA-mediated inhibition of each of these genes significantly reduces the migratory and invasive potential of neuroblastoma cells in vitro. ROCK1 and MAPK1 are members of two major branches of the TGF-β non-canonical signaling pathway (Figure 4). TGF-β signaling plays a fundamental role in the regulation of actin cytoskeleton dynamics during tumorigeneis, particularly during tumor progression and the development of metastases (11). The oncogenic functions of TGF-β signaling comprise three major branches of the non-canonical signaling pathway, the RhoA–ROCK1, extracellular signal-regulated kinase–MAPK and the PI3K/AKT pathways (12).
ROCK1 is a serine/threonine kinase that belongs to the Rho family of guanosine triphosphatase proteins that facilitate the reorganization of the actin cytoskeleton, a pivotal event during cell motion and invasion (44). Specifically, ROCK1 mediates stress fiber formation and actomyosin contractility by phosphorylation of MLC (32). The role of ROCK1 in tumor cell migration and invasion has been extensively studied and elevated ROCK1 expression has been associated with metastasis in many cancer types (32,45,46). Our study demonstrates that ROCK1 plays an important role in mediating migration and invasion in neuroblastoma.
MAPK1 is also a serine/threonine kinase and is a member of the MAPK family. A crucial role for MAPK signaling in the migration of many cancer cell types has been revealed in recent years (47). Among the downstream effectors of MAPK1 is MLC kinase, which phosphorylates MLC. By phosphorylation of MLC, MAPK1 stimulates focal adhesion turnover and actomyosin rearrangement (36). It appears that miR-335 functions to control neuroblastoma cell migration and invasion by the direct targeting of two genes involved in independent signaling pathways that converge on a common effector molecule, MLC, to tightly regulate control of actin cytoskeleton rearrangement (Figure 4). The diversity of MLC protein regulation is reflected by the temporal and spatial specificity of its different regulators, which facilitates localized contractility within the cell. Indeed, it has been demonstrated that MLC kinase and ROCK1 phosphorylate MLC at the cell periphery and cell centre, respectively (48).
Our study demonstrates that miR-335 also directly targets LRG1. Intriguingly, LRG1 is coordinately expressed with TGF-βRII, and it has been suggested that these two proteins could possibly interact, potentially connecting this gene to TGF-β signaling (35). Our finding that siRNA-mediated inhibition of LRG1 results in reduced levels of phosphorylated MLC protein further implicates LRG1 in TGF-β non-canonical signaling. Moreover, our study provides the first experimental confirmation for a direct role of LRG1 in cancer cell migration and invasion. LRG1 is a serum glycoprotein that has recently emerged as a gene of potential importance in cancer. Expression of LRG1 has been demonstrated to be upregulated in lung cancer, pancreatic cancer and ovarian cancer (49–51). LRG1 has previously been predicted to play a role in cell adhesion and migration given the leucine-rich repeat structure of the protein (52,53), its overexpression in high endothelial venules and its potential ability to bind extracellular matrix proteins (54).
A recent study in gastric cancer also demonstrated that miR-335 suppresses cell migration and invasion. Intriguingly, the target genes identified in this study are two members of the PI3K/AKT pathway, Bcl-w and SP1. The PI3K pathway is the third branch of the non-canonical TGF-β pathway, lending further evidence to the regulation of cancer cell migration and invasion by miR-335 through regulation of the non-canonical TGF-β pathway (Figure 4). Indeed, we have demonstrated that TGF-β stimulation of the neuroblastoma cells used in this study induces a significant enhancement of the cells migratory and invasive potential. Furthermore, we demonstrate that this enhancement in TGF-β-stimulated migratory potential is accompanied by an increase in the phosphorylated level of MLC.
Our study also demonstrates that miR-335 expression is repressed in MNA tumors, presumably as a consequence of direct MYCN binding to an upstream region of the predicted miR-335 transcriptional start site. Other unfavorable neuroblastoma tumor subtypes that have single copy MYCN status and loss of chromosome 11q material apparently tolerate higher levels of miR-335 transcripts than MNA tumors. Nevertheless, our experimental results on SK-N-AS, derived from a non-MNA 11q tumor, indicate that miR-335 also negatively impacts migration and invasion when levels are ectopically increased. Thus, miR-335 would appear to suppress migration and invasion potential in more than one genetic subtype of neuroblastoma. miR-335 can therefore be added to a growing list of miRNAs which have antitumorigenic effects on neuroblastoma cells (18,21–25,55,56).
Accumulating evidence now indicates that miR-335 plays a significant role in suppressing cell migration, invasion and metastasis in various cancer cell types by directly targeting a plethora of genes that modulate the cellular migratory machinery, including the aforementioned Bcl-w (BCL2L2) and SP1 genes in gastric cancer (28), tenacin c and SOX4 in breast cancer (57) and LRG1, ROCK1 and MAPK1, as reported here. Several of the target genes identified in this study and by Xu et al. (28) highlight significant regulation of the non-canonical TGF-beta pathway by miR-335. MiR-335 modulates cancer cell migration, invasion and metastasis by targeting all three branches of this oncogenic pathway and is clearly a master regulator of these processes in multiple forms of cancer.
Supplementary material
Supplementary Table S1 and Figures S1–S5 can be found at http://carcin.oxfordjournals.org/.
Funding
This work was supported in part by grants from Science Foundation Ireland (07/IN.1/B1776), Children’s Medical and Research Foundation, Cancer Research Ireland and National Institutes of Health (5R01CA127496).
Supplementary Material
Acknowledgments
We would like to thank Dr Shuh Narumiya for providing the ROCK1 expression plasmid.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- MAPK
mitogen-activated protein kinase
- miRNA
microRNA
- mRNA
messenger RNA
- MLC
myosin light chain
- MNA
MYCN amplified
- PI3K
phosphatidylinositol-3-kinase
- qPCR
quantitative PCR
- siRNA
small interfering RNA
- TGF-β
transforming growth factor-β
- UTR
untranslated region
References
- 1.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat. Rev. Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 2.Almeida MI, et al. MicroRNAs and metastases—the neuroblastoma link. Cancer Biol. Ther. 2010;9:453–454. doi: 10.4161/cbt.9.6.11215. [DOI] [PubMed] [Google Scholar]
- 3.Matthay KK, et al. Long-term results for children with high-risk neuroblastoma treated on a randomized trial of myeloablative therapy followed by 13-cis-retinoic acid: a children's oncology group study. J. Clin. Oncol. 2009;27:1007–1013. doi: 10.1200/JCO.2007.13.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stallings RL. Are chromosomal imbalances important in cancer? Trends Genet. 2007;23:278–283. doi: 10.1016/j.tig.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Brodeur GM, et al. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 6.Murphy DM, et al. Global MYCN transcription factor binding analysis in neuroblastoma reveals association with distinct E-box motifs and regions of DNA hypermethylation. PLoS One. 2009;4:e8154. doi: 10.1371/journal.pone.0008154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy DM, et al. Dissection of the oncogenic MYCN transcriptional network reveals a large set of clinically relevant cell cycle genes as drivers of neuroblastoma tumorigenesis. Mol. Carcinog. 2011;50:403–411. doi: 10.1002/mc.20722. [DOI] [PubMed] [Google Scholar]
- 8.Murphy DM, et al. Co-localization of the oncogenic transcription factor MYCN and the DNA methyl binding protein MeCP2 at genomic sites in neuroblastoma. PLoS One. 2011;6:e21436. doi: 10.1371/journal.pone.0021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westermann F, et al. Distinct transcriptional MYCN/c-MYC activities are associated with spontaneous regression or malignant progression in neuroblastomas. Genome Biol. 2008;9:R150. doi: 10.1186/gb-2008-9-10-r150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inman GJ. Switching TGFbeta from a tumor suppressor to a tumor promoter. Curr. Opin. Genet. Dev. 2011;21:93–99. doi: 10.1016/j.gde.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Meulmeester E, et al. The dynamic roles of TGF-beta in cancer. J. Pathol. 2011;223:205–218. doi: 10.1002/path.2785. [DOI] [PubMed] [Google Scholar]
- 12.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scarpa S, et al. Transforming growth factor beta regulates differentiation and proliferation of human neuroblastoma. Exp. Cell Res. 1996;229:147–154. doi: 10.1006/excr.1996.0352. [DOI] [PubMed] [Google Scholar]
- 14.Turco A, et al. Increased TGFbeta type II receptor expression suppresses the malignant phenotype and induces differentiation of human neuroblastoma cells. Exp. Cell Res. 2000;255:77–85. doi: 10.1006/excr.1999.4750. [DOI] [PubMed] [Google Scholar]
- 15.Kim VN. Small RNAs: classification, biogenesis, and function. Mol. Cells. 2005;19:1–15. [PubMed] [Google Scholar]
- 16.Stallings RL. MicroRNA involvement in the pathogenesis of neuroblastoma: potential for microRNA mediated therapeutics. Curr. Pharm. Des. 2009;15:456–462. doi: 10.2174/138161209787315837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stallings RL, et al. Therapeutic targeting of miRNAs in neuroblastoma. Expert Opin. Ther. Targets. 2010;14:951–962. doi: 10.1517/14728222.2010.510136. [DOI] [PubMed] [Google Scholar]
- 18.Foley NH, et al. MicroRNA-184 inhibits neuroblastoma cell survival through targeting the serine/threonine kinase AKT2. Mol. Cancer. 2010;9:83. doi: 10.1186/1476-4598-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tivnan A, et al. MicroRNA-184-mediated inhibition of tumour growth in an orthotopic murine model of neuroblastoma. Anticancer Res. 2010;30:4391–4395. [PMC free article] [PubMed] [Google Scholar]
- 20.Tivnan A, et al. MicroRNA-34a is a potent tumor suppressor molecule in vivo in neuroblastoma. BMC Cancer. 2011;11:33. doi: 10.1186/1471-2407-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole KA, et al. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol. Cancer Res. 2008;6:735–742. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei JS, et al. The MYCN oncogene is a direct target of miR-34a. Oncogene. 2008;27:5204–5213. doi: 10.1038/onc.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welch C, et al. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 24.Bray I, et al. MicroRNA-542-5p as a novel tumor suppressor in neuroblastoma. Cancer Lett. 2011;303:56–64. doi: 10.1016/j.canlet.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foley NH, et al. MicroRNAs 10a and 10b are potent inducers of neuroblastoma cell differentiation through targeting of nuclear receptor corepressor 2. Cell Death Differ. 2011;18:1089–1098. doi: 10.1038/cdd.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fontana L, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mestdagh P, et al. The miR-17-92 microRNA cluster regulates multiple components of the TGF-beta pathway in neuroblastoma. Mol. Cell. 2010;40:762–773. doi: 10.1016/j.molcel.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y, et al. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene doi: 10.1038/onc.2011.340. , doi: 10.1038/onc.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bray I, et al. Widespread dysregulation of MiRNAs by MYCN amplification and chromosomal imbalances in neuroblastoma: association of miRNA expression with survival. PLoS One. 2009;4:e7850. doi: 10.1371/journal.pone.0007850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mestdagh P, et al. MYCN/c-MYC-induced microRNAs repress coding gene networks associated with poor outcome in MYCN/c-MYC-activated tumors. Oncogene. 2010;29:1394–1404. doi: 10.1038/onc.2009.429. [DOI] [PubMed] [Google Scholar]
- 31.Schulte JH, et al. Accurate prediction of neuroblastoma outcome based on miRNA expression profiles. Int. J. Cancer. 2010;127:2374–2385. doi: 10.1002/ijc.25436. [DOI] [PubMed] [Google Scholar]
- 32.Itoh K, et al. An essential part for Rho-associated kinase in the transcellular invasion of tumor cells. Nat. Med. 1999;5:221–225. doi: 10.1038/5587. [DOI] [PubMed] [Google Scholar]
- 33.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolstad BM, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 35.Li X, et al. Expression of TGF-betas and TGF-beta type II receptor in cerebrospinal fluid of patients with idiopathic normal pressure hydrocephalus. Neurosci. Lett. 2007;413:141–144. doi: 10.1016/j.neulet.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 36.Huang C, et al. MAP kinases and cell migration. J. Cell Sci. 2004;117:4619–4628. doi: 10.1242/jcs.01481. [DOI] [PubMed] [Google Scholar]
- 37.Vicente-Manzanares M, et al. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duxbury MS, et al. Inhibition of pancreatic adenocarcinoma cellular invasiveness by blebbistatin: a novel myosin II inhibitor. Biochem. Biophys. Res. Commun. 2004;313:992–997. doi: 10.1016/j.bbrc.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 39.Pearson GW, et al. Real-time imaging reveals that noninvasive mammary epithelial acini can contain motile cells. J. Cell Biol. 2007;179:1555–1567. doi: 10.1083/jcb.200706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beadle C, et al. The role of myosin II in glioma invasion of the brain. Mol. Biol. Cell. 2008;19:3357–3368. doi: 10.1091/mbc.E08-03-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lutz W, et al. Conditional expression of N-myc in human neuroblastoma cells increases expression of alpha-prothymosin and ornithine decarboxylase and accelerates progression into S-phase early after mitogenic stimulation of quiescent cells. Oncogene. 1996;13:803–812. [PubMed] [Google Scholar]
- 42.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gualdrini F, et al. Addiction of MYCN amplified tumours to B-MYB underscores a reciprocal regulatory loop. Oncotarget. 2010;1:278–288. doi: 10.18632/oncotarget.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riento K, et al. Rocks: multifunctional kinases in cell behaviour. Nat. Rev. Mol. Cell Biol. 2003;4:446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- 45.Kidera Y, et al. Reduction of lung metastasis, cell invasion, and adhesion in mouse melanoma by statin-induced blockade of the Rho/Rho-associated coiled-coil-containing protein kinase pathway. J. Exp. Clin. Cancer Res. 2010;29:127. doi: 10.1186/1756-9966-29-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu S. The ROCK signaling and breast cancer metastasis. Mol. Biol. Rep. 2011;38:1363–1366. doi: 10.1007/s11033-010-0238-4. [DOI] [PubMed] [Google Scholar]
- 47.Inamdar GS, et al. Targeting the MAPK pathway in melanoma: why some approaches succeed and other fail. Biochem. Pharmacol. 2010;80:624–637. doi: 10.1016/j.bcp.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005;15:371–377. doi: 10.1016/j.tcb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 49.Li Y, et al. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis. 2011;32:1976–1983. doi: 10.1002/elps.201000598. [DOI] [PubMed] [Google Scholar]
- 50.Kakisaka T, et al. Plasma proteomics of pancreatic cancer patients by multi-dimensional liquid chromatography and two-dimensional difference gel electrophoresis (2D-DIGE): up-regulation of leucine-rich alpha-2-glycoprotein in pancreatic cancer. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007;852:257–267. doi: 10.1016/j.jchromb.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andersen JD, et al. Leucine-rich alpha-2-glycoprotein-1 is upregulated in sera and tumors of ovarian cancer patients. J. Ovarian Res. 2010;3:21. doi: 10.1186/1757-2215-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobe B, et al. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi N, et al. Periodicity of leucine and tandem repetition of a 24-amino acid segment in the primary structure of leucine-rich alpha 2-glycoprotein of human serum. Proc. Natl Acad. Sci. USA. 1985;82:1906–1910. doi: 10.1073/pnas.82.7.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saito K, et al. Gene expression profiling of mucosal addressin cell adhesion molecule-1+ high endothelial venule cells (HEV) and identification of a leucine-rich HEV glycoprotein as a HEV marker. J. Immunol. 2002;168:1050–1059. doi: 10.4049/jimmunol.168.3.1050. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y, et al. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 2007;67:976–983. doi: 10.1158/0008-5472.CAN-06-3667. [DOI] [PubMed] [Google Scholar]
- 56.Das S, et al. MicroRNA mediates DNA demethylation events triggered by retinoic acid during neuroblastoma cell differentiation. Cancer Res. 2010;70:7874–7881. doi: 10.1158/0008-5472.CAN-10-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tavazoie SF, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.