Abstract
A new study in Drosophila reports the genome-wide analysis of the maternal-to-zygotic transition in primordial germ cells, the progenitors of germline stem cells.
See research article http://genomebiology.com/2012/13/2/R11
In many species, the earliest stages of embryonic development occur in the absence of transcription and depend on maternally loaded mRNAs and proteins. During the maternal-to-zygotic transition (MZT), the control of development switches from the maternal genome to the zygotic genome. This is achieved by the combination of two processes: the massive degradation of maternal mRNAs and the activation of the zygotic genome (Figure 1). Over the past few years, genome-wide analyses of the MZT have been performed in whole embryos from several different species, including Drosophila [1-3].
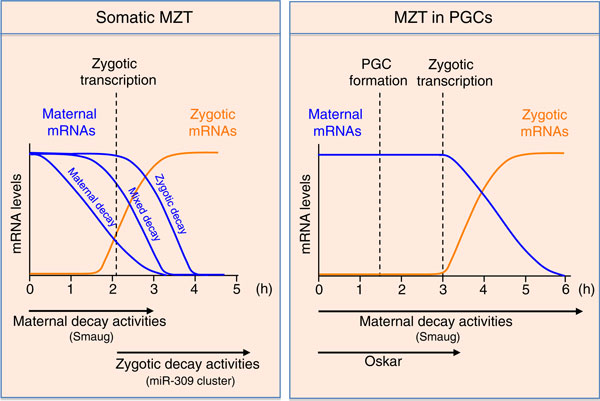
Figure 1.
Maternal-to-zygotic transition (MZT) in the soma and in primordial germ cells (PGCs). Profiles of unstable maternal mRNAs (blue) and of zygotic mRNAs (orange) are shown. In whole embryos, 14% of maternal mRNAs are degraded by maternal activities (Maternal decay), 22% by zygotic activities (Zygotic decay), and 25% by both (Mixed decay) [3]. Smaug is part of the maternal mRNA decay machinery and the miR-309 cluster is part of the zygotic machinery. Oskar might prevent Smg-dependent mRNA decay in the PGCs by binding to Smg. The dashed vertical lines indicate the time at which major zygotic transcription begins and the time of PGC formation. Time is indicated in hours after egg deposition (at 25°C).
Germ cells are specified and segregate from somatic lineages during early embryogenesis. In both Drosophila and Caenorhabditis elegans, the formation of germ cells depends on specific maternal components that accumulate in a specialized cytoplasm within the egg, called the germ plasm. An important function of the germ plasm is to protect germ cell progenitors from somatic differentiation. This is achieved in part by repressing transcription until the germ cell fate is established [4]. In Drosophila, the germ plasm (also called pole plasm) is localized at the posterior pole of the embryo. There, the germ cell progenitors, the primordial germ cells (PGCs), are the first cells to form. Just as in the somatic part of the embryo, MZT must occur in the PGCs to allow the switch from maternal to zygotic gene expression in these cells. Until recently, the low numbers of PGCs present in embryos have precluded large-scale analyses of their MZT; but in a new study published in this issue of Genome Biology, Siddiqui et al. [5] used flow cytometric sorting of GFP-labeled PGCs, proteomics and microarray-based gene-expression profiling to define the MZT in Drosophila PGCs.
In a first step, they identified proteins and mRNAs that were specific to or that were enriched in somatic cells or in PGCs. They found that different proteins and mRNAs were preferentially expressed in these two types of cells, consistent with the substantial differences in their biology. PGC-enriched proteins included ribosomal proteins and, not surprisingly, pole plasm components such as Oskar, Vasa, Tudor, Aubergine and Piwi. Between 5,000 and 6,000 mRNAs were found within each of the soma and PGC compartments, 1,700 and 1,300 of which were found to be enriched within PGCs and soma, respectively. PGC-enriched mRNAs were also associated with functions in the germ plasm and in the meiotic cell cycle, metabolism and energy production, whereas somatic-cell-enriched mRNAs had functions in development, cell fate and morphogenesis.
Maternal-to-zygotic transition in soma and primordial germ cells
A recent genome-wide analysis of the MZT in whole Drosophila embryos revealed that 60% of the 6,000 or so maternal transcripts that are preloaded into the egg are significantly degraded at 3 hours of development [3]. Slightly more than 2,000 mRNAs are zygotically transcribed at the same time point [3]. The decay of maternal mRNAs depends on two pathways, one involving maternal and the other zygotic activities. Transcripts can be degraded by one of the two pathways or by both (Figure 1).
Siddiqui et al. [5] profiled gene expression levels in purified PGCs at three time points, 1 to 3 hours, 3 to 5 hours and 5 to 7 hours after egg deposition, and found that 21% (about 1,200) of transcripts that were maternally preloaded in the PGCs were significantly degraded at 7 hours. These transcripts could be grouped into different classes according to their kinetics of decay. Interestingly, enriched Gene Ontology terms in each of these classes were largely non-overlapping, suggesting the co-regulation of distinct biological processes by mRNA decay. Among the transcripts that were destabilized at 3 to 5 hours were some that are related to post-transcriptional regulation in the pole plasm, consistent with a transition from post-transcriptional to transcriptional gene regulation in PGCs. Analysis of Gene Ontology terms enriched in transcripts that were destabilized at both time points identified transmembrane proteins involved in cell communication or adhesion, consistent with a possible need to reduce cell adhesion during the migration of PGCs to the somatic gonad.
In agreement with the relief of transcriptional repression in PGCs at 3 hours after egg deposition [4], about 800 mRNAs were found to be transcribed zygotically in the PGCs at the 3 to 5 hour time point. This group was enriched in mRNAs that function in transcription and in ribosomes/translation. Although maternal ribosomal proteins are more proportionally abundant in PGCs than in the soma when the cells form, the exclusion of maternal mRNAs encoding ribosomal proteins results in the requirement for zygotic production of new transcripts to generate ribosomes.
Zygotic transcripts can be grouped into several classes according to their patterns of synthesis, and the transcripts in each of these classes are involved in non-overlapping biological functions, again suggesting co-regulation of biological processes at the level of transcription.
Smaug is a major player in maternal mRNA degradation within both soma and primordial germ cells
The RNA-binding protein Smaug (Smg) is produced in the embryo for a short period during the first 3 hours of development. It has a major role in maternal mRNA decay in the somatic part of the embryo, contributing to the decay of two-thirds of unstable mRNAs [2,6]. Smg performs this function by recruiting the CCR4-NOT deadenylation complex to specific mRNA targets, thereby activating their poly(A) tail shortening, which in turn leads to their decay [7,8]. Importantly, Siddiqui et al. [5] found Smg to persist in PGCs for up to 7 hours of development, much longer than in somatic cells. They identified mRNAs whose decay depended on Smg by comparing PGC mRNAs in wild-type and smg mutant embryos. This comparison showed that one-third of unstable PGC mRNAs depend on Smg for their decay, revealing that Smg is a major factor in mRNA decay within PGCs, in addition to within the soma.
In whole embryos, Smg plays an indirect role in zygotic transcription through its effect on the degradation of mRNAs encoding transcriptional repressors [6]. Similarly, Smg has an indirect effect on zygotic transcription in the PGCs, which is likely mediated by the same mechanism as in the soma, although this has not yet been demonstrated. Consistent with its direct role in mRNA decay and indirect role in zygotic transcription within PGCs, mRNAs that depend on Smg for their decay are enriched in Smg-binding elements, whereas these recognition motifs are depleted in those mRNAs whose transcription is Smg-dependent. Interestingly, bioinformatic searches for other motifs that are relevant to mRNA decay, such as AU-rich elements, suggests that the corresponding binding proteins function independently of Smg in mRNA decay, as these motifs were not co-enriched with Smg-recognition elements on the same transcripts.
A consequence of the indirect role of Smg in zygotic transcription is its indirect role in the zygotic pathway of mRNA degradation in the somatic part of the embryo [6]. An important component of this pathway is the miR-309 cluster [9]. This cluster, together with a number of other microRNAs, is not expressed in PGCs when they form and thus is not involved in mRNA decay in these cells. However, whether the miR-309 cluster or other microRNAs are expressed in PGCs at later time points remains unknown.
Differences in the maternal-to-zygotic transition between soma and primordial germ cells
The data reported by Siddiqui et al. [5] emphasize that the major aspects of the MZT, namely general maternal mRNA decay and zygotic transcription, are similar between somatic cells and PGCs, although the specific mRNAs that are destabilized or synthesized do differ between the two cell types, consistent with their different developmental requirements.
Interestingly, their data also point out an important difference in the timing of MZT in the two cell types: the MZT is delayed in the PGCs in terms of both zygotic transcription and maternal mRNA decay (Figure 1). This is due, at least in part, to the repression of transcription in PGCs by specific factors, such as Polar granule component. This repression delays the onset of zygotic transcription in PGCs until 3 hours of development, compared to an onset at 2 hours in the soma. In turn, the delayed zygotic transcription results in a lack of zygotic mRNA decay machinery in newly formed PGCs. This could account for the slower kinetics of mRNA decay in PGCs, since the combined activities of the zygotic and maternal mRNA decay machineries have been shown to be more efficient than the action of the maternal machinery alone. The presence of Oskar in PGCs is also likely to contribute to delayed Smg-dependent mRNA decay in these cells. Oskar binds to Smg [10] and prevents Smg from binding to its mRNA target nanos, thus preventing the nanos transcript's decay specifically in the pole plasm and PGCs [8]. Although the application of this mechanism to the other targets has not yet been addressed, Oskar binding might prevent general Smg-dependent mRNA decay in PGCs.
The presence of Smg in the PGCs up to 7 hours after egg deposition raises the question of how mRNAs that are specifically stabilized in those cells, such as nanos, are prevented from undergoing Smg-dependent decay after the disappearance of Oskar. A possible mechanism that may explain the persistence of these Smg-dependent mRNAs in PGCs could be the slower kinetics of mRNA decay in the absence of zygotic decay components. Another intriguing possibility, however, is that mRNA-specific binding factors might selectively prevent decay in the PGCs. This awaits future investigation.
Abbreviations
MZT: maternal-to-zygotic transition; PGC: primordial germ cell; Smg: Smaug.
Competing interests
The authors declare that they have no competing interests.
Acknowledgements
I thank the members of our laboratory for their suggestions on this text. Work in the Simonelig laboratory is supported by the CNRS UPR1142, ANR (ANR-2010-BLAN-1201 01), FRM ("Equipe FRM 2007" and "Projets Innovants ING20101221078") and ARC (ARC Libre 2009 N°3192).
References
- De Renzis S, Elemento O, Tavazoie S, Wieschaus EF. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 2007;5:e117. doi: 10.1371/journal.pbio.0050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell. 2007;12:143–155. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Thomsen S, Anders S, Janga SC, Huber W, Alonso CR. Genome-wide analysis of mRNA decay patterns during early Drosophila development. Genome Biol. 2010;11:R93. doi: 10.1186/gb-2010-11-9-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Shirae-Kurabayashi M, Hanyu-Nakamura K. Repression of early zygotic transcription in the germline. Curr Opin Cell Biol. 2010;22:709–714. doi: 10.1016/j.ceb.2010.08.012. [DOI] [PubMed] [Google Scholar]
- Siddiqui N, Li X, Luo H, Karaiskakis A, Hou H, Kislinger T, Weswood JT, Morris Q, Lipshitz HD. Genome-wide analysis of the maternal-to-zygotic transition in Drosophila primordial germ cells. Genome Biol. 2012;13:r11. doi: 10.1186/gb-2012-13-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit B, He CH, Zhang F, Votruba SM, Tadros W, Westwood JT, Smibert CA, Lipshitz HD, Theurkauf WE. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development. 2009;136:923–932. doi: 10.1242/dev.031815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol. 2005;15:284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- Zaessinger S, Busseau I, Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133:4573–4583. doi: 10.1242/dev.02649. [DOI] [PubMed] [Google Scholar]
- Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18:501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Walker JA, Wharton RP. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol Cell. 1999;4:209–218. doi: 10.1016/S1097-2765(00)80368-8. [DOI] [PubMed] [Google Scholar]