Abstract
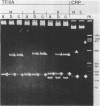
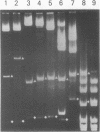
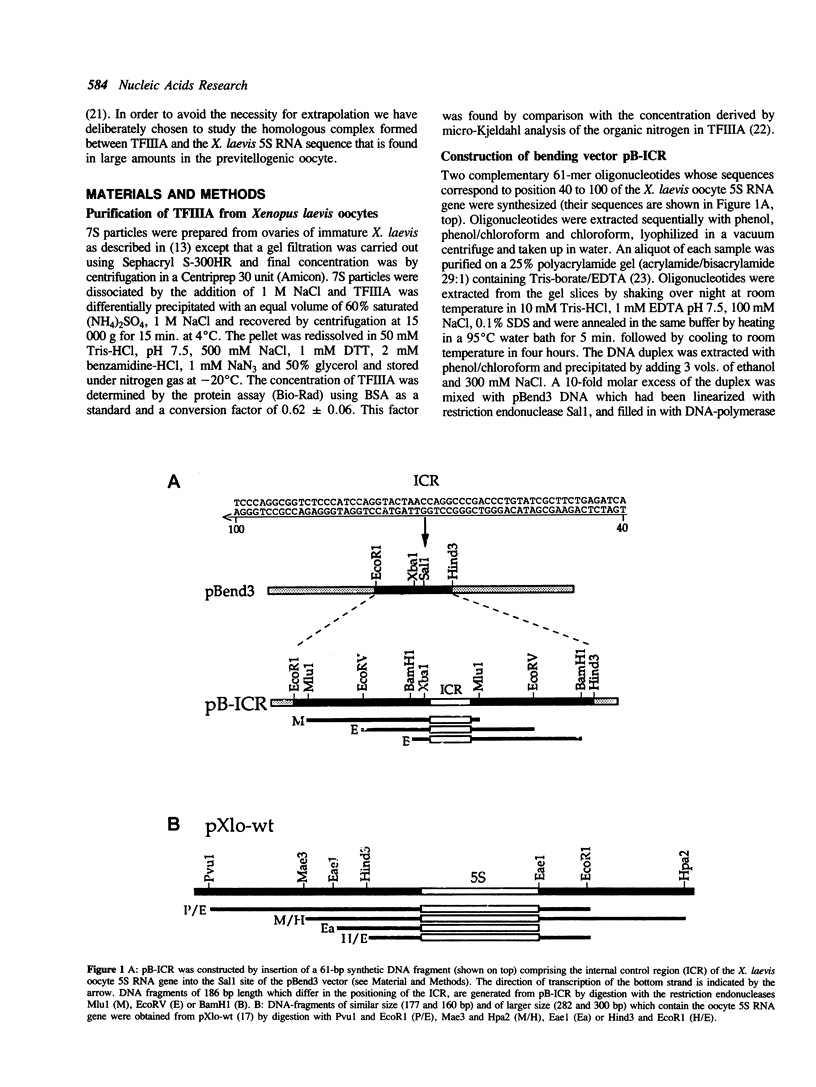
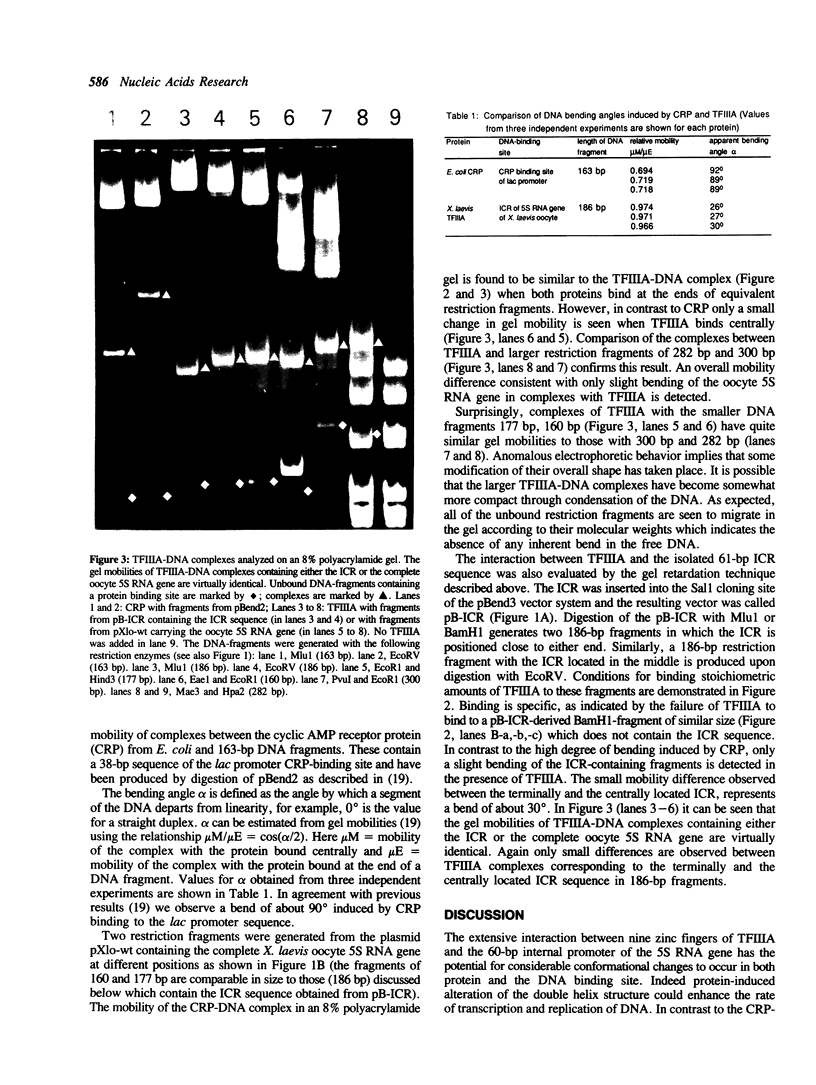
The extent and location of DNA-bending induced in the Xenopus laevis transcription factor IIIA-oocyte 5S RNA gene complex was determined by the gel retardation method. The electrophoretic mobilities of TFIIIA complexed with restriction fragments of 160, 177, 282 and 300 bp that contain the sequence of the major oocyte 5S RNA gene were compared. In these fragments the 120-bp gene is positioned either in the middle or at the end. Minor differences in the mobility of the complexes indicate that the degree of DNA bending is only slight. To determine the bending angle more precisely, a bending vector system, pBend3, was used to examine the complex of TFIIIA with the internal control region (ICR) of the 5S RNA gene. A 61-bp synthetic duplex corresponding to the ICR sequence was cloned into pBend3. Duplicated circular permuted restriction sites allow several 186-bp fragments to be generated in which the position of the ICR can be varied. Gel retardation of TFIIIA-DNA complexes with the ICR sequence contained in pBend3 indicates a bending angle of only 30 degrees and shows that interaction in the ICR could account for all of the bending found in the complete oocyte 5S RNA gene.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersen J., Delihas N., Hanas J. S., Wu C. W. 5S RNA structure and interaction with transcription factor A. 2. Ribonuclease probe of the 7S particle from Xenopus laevis immature oocytes and RNA exchange properties of the 7S particle. Biochemistry. 1984 Nov 20;23(24):5759–5766. doi: 10.1021/bi00319a014. [DOI] [PubMed] [Google Scholar]
- Bazett-Jones D. P., Brown M. L. Electron microscopy reveals that transcription factor TFIIIA bends 5S DNA. Mol Cell Biol. 1989 Jan;9(1):336–341. doi: 10.1128/mcb.9.1.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Brown R. S., Sander C., Argos P. The primary structure of transcription factor TFIIIA has 12 consecutive repeats. FEBS Lett. 1985 Jul 8;186(2):271–274. doi: 10.1016/0014-5793(85)80723-7. [DOI] [PubMed] [Google Scholar]
- Christiansen J., Brown R. S., Sproat B. S., Garrett R. A. Xenopus transcription factor IIIA binds primarily at junctions between double helical stems and internal loops in oocyte 5S RNA. EMBO J. 1987 Feb;6(2):453–460. doi: 10.1002/j.1460-2075.1987.tb04775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Fairall L., Martin S., Rhodes D. The DNA binding site of the Xenopus transcription factor IIIA has a non-B-form structure. EMBO J. 1989 Jun;8(6):1809–1817. doi: 10.1002/j.1460-2075.1989.tb03575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairall L., Rhodes D., Klug A. Mapping of the sites of protection on a 5 S RNA gene by the Xenopus transcription factor IIIA. A model for the interaction. J Mol Biol. 1986 Dec 5;192(3):577–591. doi: 10.1016/0022-2836(86)90278-0. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Blanco J., Tennant L. L. The 5S gene internal control region is B-form both free in solution and in a complex with TFIIIA. Nature. 1987 Oct 1;329(6138):460–462. doi: 10.1038/329460a0. [DOI] [PubMed] [Google Scholar]
- Huber P. W., Wool I. G. Identification of the binding site on 5S rRNA for the transcription factor IIIA: proposed structure of a common binding site on 5S rRNA and on the gene. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1593–1597. doi: 10.1073/pnas.83.6.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke L. A rapid micromethod for the determination of nitrogen and phosphate in biological material. Anal Biochem. 1974 Oct;61(2):623–627. doi: 10.1016/0003-2697(74)90429-1. [DOI] [PubMed] [Google Scholar]
- Liu-Johnson H. N., Gartenberg M. R., Crothers D. M. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 1986 Dec 26;47(6):995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk P. J. Characterization of the RNA binding properties of transcription factor IIIA of Xenopus laevis oocytes. Nucleic Acids Res. 1985 Jul 25;13(14):5369–5387. doi: 10.1093/nar/13.14.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk P. J., de Stevenson I. L., Wong H. H. Defining the binding site of Xenopus transcription factor IIIA on 5S RNA using truncated and chimeric 5S RNA molecules. Nucleic Acids Res. 1987 Mar 25;15(6):2737–2755. doi: 10.1093/nar/15.6.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroth G. P., Cook G. R., Bradbury E. M., Gottesfeld J. M. Transcription factor IIIA induced bending of the Xenopus somatic 5S gene promoter. Nature. 1989 Aug 10;340(6233):487–488. doi: 10.1038/340487a0. [DOI] [PubMed] [Google Scholar]
- Timmins P. A., Langowski J., Brown R. S. An elongated model of the Xenopus laevis transcription factor IIIA-5S ribosomal RNA complex derived from neutron scattering and hydrodynamic measurements. Nucleic Acids Res. 1988 Sep 12;16(17):8633–8644. doi: 10.1093/nar/16.17.8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrana K. E., Churchill M. E., Tullius T. D., Brown D. D. Mapping functional regions of transcription factor TFIIIA. Mol Cell Biol. 1988 Apr;8(4):1684–1696. doi: 10.1128/mcb.8.4.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P. Transcription fraction TFIIIC can regulate differential Xenopus 5S RNA gene transcription in vitro. EMBO J. 1988 Apr;7(4):1071–1079. doi: 10.1002/j.1460-2075.1988.tb02915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Xing Y. Y., Worcel A. The C-terminal domain of transcription factor IIIA interacts differently with different 5S RNA genes. Mol Cell Biol. 1989 Feb;9(2):499–514. doi: 10.1128/mcb.9.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C., Kim J., Adhya S. DNA bending by negative regulatory proteins: Gal and Lac repressors. Genes Dev. 1989 May;3(5):606–611. doi: 10.1101/gad.3.5.606. [DOI] [PubMed] [Google Scholar]