Expression of microRNAs (miRNAs) is a highly regulated process. This is more evident in the case of those forming part of clusters, where differential processing of individual miRNAs is observed. This paper addresses the influence of RNA tertiary structure on the differential processing of six miRNAs that form part of the pri-miR-17-92a cluster. The authors found that this cluster folds into a three-dimensional structure, which determines microRNA processing efficiency. This tertiary structure acts to create suboptimal protein recognition sites on the pri-miRNA cluster due to higher-order structure formation.
Keywords: primary microRNA, miRNA processing, tertiary structure, SHAPE analysis, hydroxyl radical footprinting, fluorescence anisotropy
Abstract
MicroRNAs control gene expression either by RNA transcript degradation or translational repression. Expressions of miRNAs are highly regulated in tissues, disruption of which leads to disease. How this regulation is achieved and maintained is still largely unknown. MiRNAs that reside on clustered or polycistronic transcripts represent a more complex case where individual miRNAs from a cluster are processed with different efficiencies despite being cotranscribed. To shed light on the regulatory mechanisms that might be operating in these cases, we considered the long polycistronic primary miRNA transcript pri-miR-17-92a that contains six miRNAs with diverse functions. The six miRNA domains on this cluster are differentially processed to produce varying amounts of resultant mature miRNAs in different tissues. How this is achieved is not known. We show, using various biochemical and biophysical methods coupled with mutational studies, that pri-miR-17-92a adopts a specific three-dimensional architecture that poses a kinetic barrier to its own processing. This tertiary structure could create suboptimal protein recognition sites on the pri-miRNA cluster due to higher-order structure formation.
INTRODUCTION
MicroRNAs (miRNAs) are small, 20- to 22-nt-long noncoding RNAs that regulate gene expression via diverse mechanisms ranging from mRNA cleavage and translational repression to transcriptional repression by heterochromatin formation (Ambros 2004; Chen and Rajewsky 2007). They have varied and unique expression patterns owing to their extensive control over the transcriptome and have been implicated in numerous biological processes including cellular differentiation, cell proliferation, apoptosis, synaptic plasticity, immunity, and metabolism (Bushati and Cohen 2007; Schratt 2009). Involvement of miRNAs across this spectrum of important biological phenomena requires stringent control over their expression. Recent findings have unraveled a plethora of post-transcriptional mechanisms which regulate miRNA biogenesis and activity (Filipowicz et al. 2008). Investigations on primary miRNAs have largely focused on transcripts incorporating a single pre-miRNA stem–loop that is processed sequentially into pre-miRNA and mature miRNA segments (Davis et al. 2008; Michlewski et al. 2008). These have shown that accessory proteins are required in addition to the microprocessor complex for proper primary transcript processing (Davis et al. 2008; Michlewski et al. 2008; Viswanathan and Daley 2010). Either the tissue-specific abundance of these accessory proteins or their signal-induced recruitment might explain the restricted expression of a few miRNAs. However, in vertebrates, it is known that 30% of miRNAs are found as part of a polycistronic cluster i.e., they are in quick succession on the same primary miRNA transcript (Megraw et al. 2007). Another level of complexity in regulation arises in such cases since it is known that the miRNAs that are part of the same cluster are present in different amounts in a given tissue (Thomson et al. 2006; Tang and Maxwell 2008). Further, their relative concentrations vary from tissue to tissue in the organism. It is not yet clear how this might be achieved.
Long noncoding RNA transcripts might act as scaffolds that self-orchestrate the binding of multiple proteins, where the binding of one protein on the RNA causes a structural remodeling of the latter that, in turn, either exposes a cryptic binding site or creates a new binding motif for the next protein in the cascade. This has been known to occur in noncoding RNAs such as ribosomal RNAs and ribosome biogenesis, 7SK RNA and 7SK RNP assembly, and snRNAs in spliceosome assembly (Talkington et al. 2005; Hogg and Collins 2007; Wahl et al. 2009). Thus, we considered a possibility where the primary miRNA cluster might self-orchestrate the binding of accessory proteins associated with processing different microRNA domains, thus bringing about their differential regulation. We, therefore, chose the intronic miRNA cluster called pri-miR-17-92a, which encompasses ∼800 nt on human chromosome 13, to address such differential regulation. Upon processing, this cluster produces six individual mature miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1) (Mendell 2008). Although it is widely regarded that the transcription of this cluster per se is regulated by c-Myc transcription factors, post-transcriptional regulation of this cluster has established that the miRNAs on this cluster are differentially expressed (O'Donnell et al. 2005; Thomson et al. 2006; Tang and Maxwell 2008). The molecular mechanisms that govern preferential enrichment of a given miRNA over others in the cluster are unclear, and we would like to address possible mechanisms that may be responsible. One could envisage that a primary miRNA cluster might adopt a higher-order structure, thereby creating a suboptimal display of recognition sites for the microprocessor complex. Structural studies indicate Drosha-DGCR8 binding requires a single-strand-double-strand junction (ss-ds) (Han et al. 2006). Thus, sequestration of such a recognition site could arise from tertiary structure formation from the helices of this cluster, thereby masking key ss-ds junctions. Tantalizing support for such a hypothesis is seen from the recruitment of hnRNP A1 which acts on a truncated form of the cluster, remodeling the local structure to facilitate the processing of pre-miR-18a (Guil and Cáceres 2007; Michlewski et al. 2008). In addition, the recent discovery of a higher molecular weight species of microprocessor complex that possesses RNA helicase activity suggests that processing might invoke restructuring of the RNA scaffold.
Studies on selected individual pre-miRNA hairpins of this cluster showed that they form helices. However, it is not known if, when these helices are part of a single primary miRNA transcript, they invoke tertiary interactions to adopt a well-defined higher-order structure. Using a battery of biophysical and biochemical assays, we have shown that pri-miR-17-92a, indeed, adopts a higher-order structure with solvent protected regions in a Mg2+-dependent manner. Further, this tertiary structure has a functional impact on the primary microRNA by posing a kinetic barrier to its processing.
RESULTS
In cellulis processing of native and truncated miRNA clusters
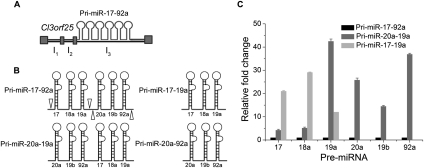
A pri-miR-17-92a cluster is usually schematically represented as six tandem stem–loops, implying the absence of tertiary structure (Mendell 2008). In order to test whether this was, indeed, the case, we made two constructs in addition to the native cluster, pri-miR-17-92a. These were a minicluster, pri-miR-17-19a, and pri-miR-20a-19a, which is a domain swapped or shuffled cluster, in the pcDNA3 vector that can be used for overexpression studies of the respective RNA in mammalian cells (Fig. 1A,B; also see Supplemental Fig. S1). In the absence of any tertiary interaction in pri-miR-17-92a, the shuffled transcript, which contains all six pre-miRNAs but in a different order, should be processed similarly in cells due to the availability of all the relevant binding sites to the processing proteins. To confirm this, we performed RT-PCR of all the relevant pre-miRNAs from pri-miR-17-92a and pri-miR-20a-19a from total small RNAs isolated from their overexpression in HeLa cells (see Supplemental Fig. S2A). This suggests that both the transcripts are processed into identical pre-miRNAs. Next, we performed quantitative real-time PCR analysis (qPCR) to look at the relative levels of the processed pre-miRNAs from both native and shuffled transcripts in this overexpression system. Total small RNAs isolated 24 h post-transfection from HeLa cells were reverse-transcribed using pre-miR specific primers and then quantified by qPCR using the ΔΔCT method (Livak and Schmittgen 2001). The pre-miRNAs from the pri-miR-17-92a transcript showed differential levels of expression (see Supplemental Fig. S2B). The qPCR analysis indicates that, with respect to pri-miR-17-92a, the individual pre-miRNAs from pri-miR-20a-19a show much higher levels and that this increase is not uniform for each pre-miR. Importantly, there is a marked increase in the levels of pre-miR-20a, pre-miR-19a, and pre-miR-92a from pri-miR-20a-19a, indicating a positional significance in the context of the entire transcript. As a control, the pre-miRNA level from the minicluster pri-miR-17-19a also undergoes processing to give significantly higher levels of all three pre-miRNAs relative to pri-miR-17-92a. Thus, a mere shuffling of discrete, pre-miR-containing hairpin domains is sufficient to alter the relative abundance of the processed pre-miRNAs. However, the in cellulis processing studies indicate structural differences between the native transcript and the shuffled transcript that impact their processing, which is beyond a simplistic secondary structure model (see Supplemental Fig. S2C,D for secondary structures). Further, the fact that the native cluster behaves differently from a simple minicluster implicates interaction between pri-miR-17-19a and pri-miR-20a-92a when they are fused to form the native transcript.
FIGURE 1.
The tertiary structure of pri-miR-17-92a regulates its processing. (A) Genomic structure of the C13orf25 gene, harboring the intronic pri-miR-17-92a cluster. (B) Diagrammatic representation of the pri-miR-17-92a transcript. Empty arrow heads indicate the transcript, pri-miR-17-19a, corresponding to the first three pre-miRNA domains (right, upper panel), and the last three pre-miRNA domains indicate the transcript, pri-miR-20a-92a (right, lower panel). The domains of the two constructs were swapped to create a shuffled transcript, pri-miR-20a-19a (left, lower panel). (C) Quantitative measurement of levels of the processed pre-miRNAs from total RNA isolated 24 h post-transfection from HeLa cells transfected either with pri-miR-17-92a-pcDNA3, or pri-miR-20a-19a-pcDNA3, or pri-miR-17-19a-pcDNA3. The relative expression levels of individual pre-miRNAs are determined using qPCR by the ΔΔCT method and shown. Error bars indicate SEM. The pre-miR-19a from the pri-miR-17-19a was quantified using Northern analysis.
Comparative sequence analysis of the pri-miR-17-92a cluster
Interactions between distinct hairpin domains could give rise to higher-order structure that might occur in the native transcript when the miniclusters are in the right orientation which would be missing in the individual miniclusters as well as in the shuffled transcript. So, we carried out comparative sequence analysis for preliminary indications of whether or not the native transcript could form tertiary structure.
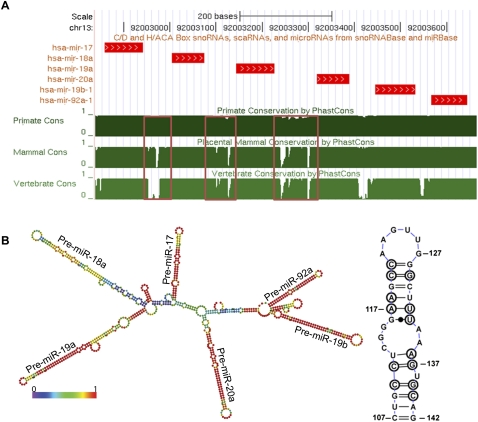
Phylogenetic or comparative sequence analysis is a powerful indicator of secondary and tertiary structure of RNA molecules (Gutell 1993). It extracts information about the RNA molecule from the conserved segments and covariations between homologous sequences across different phyla. When sequences from 19 species were aligned using the UCSC Genome browser (Fig. 2A), the sequence conservation histogram clearly revealed that the six individual stem–loops containing six pre-miRNAs were highly conserved as expected, as these regions are important for function and they are required to be helical for their processing. Notably, few inter-pre-miR regions, i.e., the intervening regions between the distinct pre-miRNA domains, such as regions between pre-miR-17 and pre-miR-18a (bases 107–142) or pre-miR-19a and pre-miR-20a (bases 388–480), are also found to be significantly conserved (Fig. 2A, boxed regions). Taking the first case as an example, a phylogenetic analysis of this region from the aligned sequences (see Supplemental Fig. S3 for alignment) showed that this sequence could fold back on itself to form a helix between pre-miR-17 and pre-miR-18a (Fig. 2B, right panel). This is further supported by the minimum free-energy structure using the structure prediction software RNAfold as well as “RNAstructure” (Hofacker et al. 1994; Reuter and Mathews 2010). Phylogenetic conservation of RNA segments for the pre-miR-containing domains might be attributed to their requirement for helicity. However, such an argument does not hold for the conservation of segments such as 107–142 or 388–480. A possible reason might be their importance in tertiary structure that might impact the function of the cluster as a whole.
FIGURE 2.
(A) Conservation pattern of human pri-miR-17-92a. The conservation patterns are based on the UCSC phastCons scores. The chromosomal regions of the miRNAs with an additional 20 nt flanking on both sides are presented. Sequence conservation of the region between the different pre-miRs shown in red box. It is clear that individual pre-miRNAs are highly conserved along with few inter-pre-miRNA regions. (B) The most stable thermodynamic structure of human pri-miR-17-92a is shown. This is predicted using the RNAfold algorithm and color-coded according to base-pairing probability. Base-pairing probability indicates the probability of a base pair existing throughout the ensemble of structures. This information is important because it gives an indication of structural remodeling or dynamics. Most of the pre-miRNAs are invariant throughout the ensemble, whereas pre-miR-18a can adopt alternate forms. A region between the pre-miR-17 and 18a flanking region is indicated by a box and shown to be conserved (see Supplemental Fig. S3A). The structure of this region found by covariation-based modeling is shown (right panel).
Indications of tertiary contacts
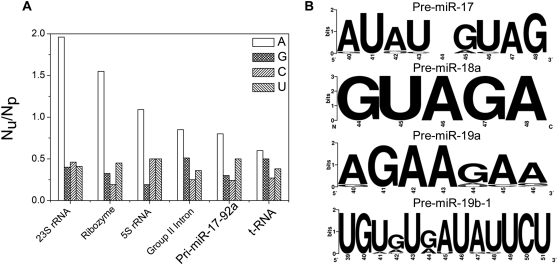
Prediction of tertiary contacts in an RNA molecule is not straightforward from phylogenetic analysis. However, the ratio of unpaired to paired adenosines from a secondary structure map has been a key indicator of tertiary structure in a given RNA (Gutell et al. 2000). This method has been applied to 16S rRNA and 23S rRNA, where it was found that the composition of single-stranded regions of the RNA, especially in the loops, was strongly biased toward adenosines (Gutell et al. 2000). Later, it was found that a vast majority of tertiary contacts are mediated by unpaired adenosines. We performed a similar analysis on the phylogenetic structure model of the pri-miR-17-92a cluster which showed a ratio of ∼1. This ratio for pri-miR-17-92a is encouraging and is comparable to the ratio obtained for the group II intron (Fig. 3A). t-RNA is also shown as a control. This analysis indicates that the pri-miRNA cluster might, indeed, possess tertiary structure.
FIGURE 3.
(A) Unpaired adenosine/paired adenosine ratio is shown for different large RNAs along with pri-miR-17-92a. For large structured RNAs, the unpaired/paired adenosine ratio is found to be >1, which qualitatively indicates adenosine-mediated tertiary contacts. Pri-miR-17-92a also has a Au/Ap ratio close to group II intron, indicating possible tertiary contacts mediated by adenosines. t-RNA serves as the control. (B) Sequence logo representation of terminal loop structures of different pre-miRNAs present in this cluster. Individual pre-miRNA structures are predicted from comparative sequence analysis, and terminal loop sequences are confirmed from known structures or phylogenetic sequence analysis. It is clear that pre-miR-18a, 19a, and 19b have very high sequence conservation.
In addition to identifying the helical regions of the cluster, we wanted to see if the loop regions showed any sequence conservation. The individual pre-miRNA sequences were retrieved from the miRNA registry and aligned. The loop regions of pre-miR-17, pre-miR-18a, and pre-miR-19a were taken from the published structures of the individual hairpins, while for pre-miR-20a, pre-miR-19b, and pre-miR-92a, the loop sequences were identified from the structures predicted by comparative sequence analysis (Krol et al. 2004). Sequence conservation of each of the terminal loops was converted into bits units and represented as a sequence logo (see Fig. 3B; also Supplemental Fig. S3B). It is evident that, for pre-miR-19a and pre-miR-18a, loop sequences are highly conserved. Such a high degree of loop sequence conservation could either be due to their importance in tertiary structure formation and/or for a protein-binding function. In fact, the loop of pre-miR-18a is known to be recognized by hnRNP A1 (Guil and Cáceres 2007). Further, although consecutive repeats of purine bases, GAA, are a well-known motif for protein binding due to their large aromatic faces, they also effectively mediate tertiary contacts via unpaired adenosines (Yeakley et al. 1996; Gutell et al. 2000). The loops of pre-miR-17 and pre-miR-19b-1 show conserved AU or GU repeats (Fig. 3B) that are also known to bind nuclear proteins that mediate RNA degradation or alternative splicing (Vakalopoulou et al. 1991; Marquis et al. 2006). Thus, the high sequence conservation of terminal loops in nearly all the pre-miRNA domains of this cluster is significant in the context of potentially acting as cryptic binding sites for different trans-acting protein factors that could mediate differential processing.
SHAPE analysis shows pri-miR-17-92a is structured
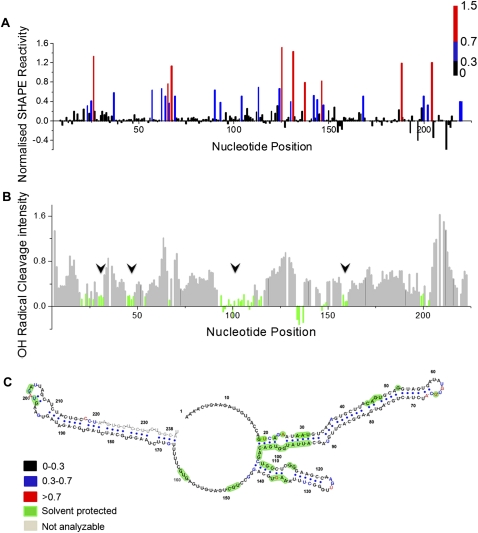
Given that sequence analysis of pri-miR-17-92a predicted its considerable potential to adopt tertiary structure, we proceeded to biochemically investigate its structure at the single nucleotide level using N-methyl isatoic anhydride (NMIA) and analysis by capillary electrophoresis (SHAPE, Selective 2′-hydroxyl acylation analyzed by primer extension) (Merino et al. 2005). The SHAPE technique has revolutionized RNA structural biology, as the analysis by high-throughput capillary electrophoresis precludes any associated band compression, thus enabling the study of long lengths of RNA. In a SHAPE reaction, the RNA is treated with the electrophilic reagent NMIA that selectively reacts with those ribose 2′-hydroxyl groups of nucleotides that are conformationally flexible (McGinnis et al. 2009). SHAPE reactivity thus reports on local nucleotide flexibility. Unconstrained nucleotides, i.e., nucleotides that are present in single-stranded structures, are likely to be more reactive to form the corresponding 2′-O-adduct. All four RNA nucleotides show nearly identical intrinsic reactivities when they are not constrained by base-pairing or tertiary interactions (Wilkinson et al. 2009). We applied this method to the 0.8-kb pri-miR-17-92a transcript and probed the structure of the 5′-terminal 220 nt, which span pre-miR-17 and pre-miR-18a, in the presence of Mg2+. We chose to focus on this region for the present study because protein-mediated structural remodeling has been reported for a 70-nt-long transcript corresponding to the pre-miR-18a domain. Normalized SHAPE reactivity of this 220-nt region in the context of the whole pri-miR-17-92a transcript is shown in Figure 4A and indicates the secondary structure of this region (see Supplemental Fig. S4A for raw trace). The structure of this region is color-coded according to SHAPE reactivity, where black, blue, and red indicate reactivity ranges of <0.3 (duplex), 0.3–0.7 (likely single-stranded), and ≥0.7 (single-stranded), respectively. SHAPE constraints were used to create the secondary structure model of this region (shown in Fig. 4C), using the RNAstructure program. This model is consistent with SHAPE data and color-coded according to reactivity. Notably, many regions are found to show low reactivity and also to be in the duplexed state, as shown in Figure 4C. Moderate and highly reactive nucleotides are found mainly in loops and internal bulges consistent with them being single-stranded. Importantly, a few nucleotides in loop regions, such as AUA in the pre-miR-17 loop, as well as in internal bulges showed low reactivity which points to their possible involvement in stacking interaction with neighboring base pairs or themselves base-pairing with other parts of the molecule. This is consistent with similar observations for other structured RNAs also analyzed by SHAPE (Watts et al. 2009). The accuracy of this SHAPE-based structure was compared with the structure predicted from phylogenetic analysis, as well as from the minimum free-energy model generated by the RNAstructure algorithm. This SHAPE directed model is consistent with the phylogenetic structure in that those nucleotides predicted to show evolutionarily conservation due to base-pairing possibility are also experimentally validated in the SHAPE model (see Supplemental Fig. S5A). A small 36-bp hairpin loop that is not a part of any pre-miR region, that is also predicted by phylogenetic analysis, was experimentally confirmed by the SHAPE directed model (see Fig. 2B). There are marginal inconsistencies in the case of a few loop residues of pre-miR-18a where the SHAPE-predicted structure, when it is part of the whole transcript, does not match the experimentally found structure when analyzed as an isolated domain.
FIGURE 4.
SHAPE and hydroxyl radical probing of pri-miR-17-92a. (A) Histogram of absolute nucleotide reactivities determined by SHAPE analysis of the pre-miR-17-18a region in the presence of Mg2+ ions. Color-coding is according to NMIA reactivity. (B) Histogram of cleavage intensity versus nucleotide position for the pri-miR-17-18a in the presence of Mg2+, determined using in situ–generated hydroxyl radicals. Solvent-inaccessible nucleotides in the folded RNA (intensities ≤ one-half the mean) are indicated in green (black arrows). (C) Secondary structure of the pre-miR-17-18a predicted by RNA structure with SHAPE constraints. Nucleotides are color-coded according to their reactivity toward NMIA. Solvent-inaccessible nucleotides are shown in green.
It is well-known that the adoption of tertiary structure by RNA simultaneously results in the burial of several tracts of nucleotides within the core of the folded structure (Rangan et al. 2003). The existence of such buried nucleotides may be revealed by probing solvent accessibility of nucleotides on the folded RNA transcript. This solvent accessibility of nucleotides on pri-miR-17-92a was probed by hydroxyl radical footprinting where in situ–generated hydroxyl radicals cleave locations on the RNA backbone directly proportional to their solvent exposure. Hydroxyl radicals were generated in situ using Fe(II)-EDTA, H2O2 in a solution of in vitro-transcribed pri-miR-17-92a in the presence and absence of Mg2+, followed by analysis using primer extension performed with fluorescently labeled primers that were then resolved by capillary electrophoresis. This yielded single-nucleotide-resolution cleavage information reporting on solvent exposure over a 225-nt tract at the 5′ terminus of pri-miR-17-92a (lower panel, Fig. 4B; Supplemental Figs. S4B, S6A). Cleavage intensities were normalized to a scale from 0 to 1.5, where 1.0 is defined as the average intensity of highly reactive nucleotides. On this scale, nucleotides with reactivities that are ≤ half the mean reactivity are generally considered solvent-inaccessible. Many regions indicated in green (15% of the sequence analyzed) showed tracts of solvent-inaccessible nucleotides according to the above mentioned scale. For example, the longest tract of nucleotides that were found to be solvent-inaccessible were A94–G106. Importantly, many symmetric and asymmetric internal bulges such as those present in the pre-miR-17 stem–loop were found to be solvent-inaccessible although they are reactive to NMIA. In many structured RNAs, such single-stranded regions often mediate tertiary interactions. Notably, G65 in the loop of pre-miR-17 and 4 nt (G198–U199 and G201–A202) located in the pre-miR-18a loop showed solvent inaccessibility. This indicates a burial of these regions due to further compaction, namely tertiary structure formation. Of particular interest, G198–U199 and G201–A202, which are known to function as hnRNP A1 binding sites, were found to be solvent-inaccessible when the pre-miRs are part of whole transcript pri-miR-17-92a. Thus, it might be possible that, in order to access these particular sites, hnRNP A1 would first have to bind the tertiary structured pri-miR-17-92a and unmask these sites for subsequent remodeling by further hnRNP A1 binding. There are also many sites which are solvent-protected on the inter-pre-miR stem–loop region between pre-miR-17 and pre-miR-18a. Thus, this hairpin loop also gets buried in the course of pri-miR-17-92a adopting its tertiary structure.
Pri-miR-17-92a undergoes folding transitions
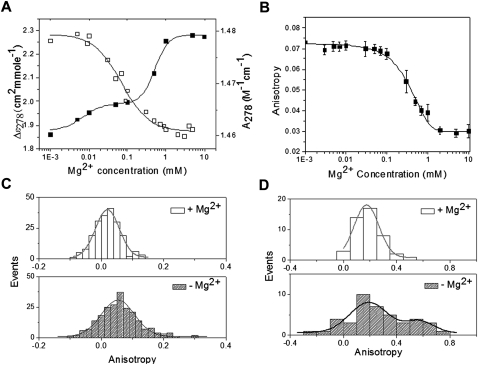
A combination of SHAPE analysis and hydroxyl radical footprinting on pri-miR-17-92a revealed that the transcript adopts a tertiary structure that required Mg2+ ions. The dramatic reactivity difference of a large tract of pri-miR-17-92a hinted at the possibility that this cluster might undergo a folding transition reminiscent of other structured RNAs where folding is essential to RNA functionality (Su et al. 2005; Baird et al. 2007). In order to see if the Mg2+-dependent structural changes could be captured by circular dichroism (CD) spectroscopy, a solution of 0.4 μM pri-miR-17-92a, in 10 mM sodium cacodylate buffer, pH 7, 150 mM NaCl, 0.1 mM EDTA, in the presence and absence of 10 mM Mg2+ were compared (see Supplemental Fig. S7A). Both samples showed CD profiles reminiscent of A-helical RNA. Importantly, upon addition of Mg2+, an increase in CD signal centered at 275 nm was observed, indicating extra base-pairing within the transcript facilitated specifically upon addition of Mg2+. When 8 M urea was added to 0.4 μM pri-miR-17-92a in 150 mM Na+, 10 mM Mg2+, it resulted in a substantial decrease in CD signal, characteristic of a urea-dependent denaturation of RNA structure (see Supplemental Fig. S7B). Taken together, these experiments present evidence that the pri-miR-17-92a cluster has the potential to fold in a Mg2+-dependent manner into a tertiary structure. If this cluster adopts a well-defined structure mediated by secondary and tertiary interactions, then it should undergo characteristic folding and unfolding transitions induced by Mg2+ addition and depletion. For this, 1 μM pri-miR-17-92a was dissolved in 10 mM sodium cacodylate buffer, pH 7, 150 mM NaCl, to which incremental amounts of Mg2+ were added, and circular dichroism of the sample was measured. As evident from Figure 5A, increasing Mg2+ showed a sigmoidal increase in signal at 278 nm, indicating that the pri-miRNA transcript undergoes a structural transition or folding mediated specifically by Mg2+. This is supported by its UV absorbance at 278 nm that concomitantly shows a characteristic decrease due to increased Mg2+-induced base stacking.
FIGURE 5.
Mg2+-dependent structural transition and compaction of pri-miR-17-92a. (A) Increase as well as decrease in CD and UV signals, respectively, at 278 nm as a function of Mg2+ concentration. RNA concentration was kept at 1 μM in 10 mM sodium cacodylate buffer, pH 7, 150 mM NaCl, and 0.1 mM EDTA. An increase in the CD signal at 278 nm is indicative of helix and tertiary structure formation. (B) Mg2+ ion-dependent collapse of 50 nM Alexa-488-labeled pri-miRNA 17-92a in 50 mM HEPES-KOH, pH 7.5, 100 mM KCl, and 1 mM EDTA is followed by anisotropy with incremental addition of Mg2+. The titration curve is fitted with the Hill equation. (C) Single molecule anisotropy histogram of folded pri-miR-17-92a (500 pM in 50 mM HEPES, KOH, 100 mM KCl, and 5 mM MgCl2) and unfolded pri-miRNA (with 5 mM EDTA, no Mg2+). Histograms are constructed from anisotropy values of freely diffusible Alexa-488 labeled pri-miRNA molecules in either of the states mentioned above. A threshold (Ip+Iperp) of 40 counts was used to discriminate single molecule peaks from background. (D) Pri-miRNA 17-92a conformational dynamics in the folded and unfolded state was captured using 70% glycerol. 1 nM folded and/or unfolded pri-miRNA was mixed with glycerol to have a 70% aqueous glycerol system, and anisotropy trajectories of single molecules were collected. A very large heterogeneous distribution from one such representative trajectory in the case of an unfolded sample indicates conformational plasticity in the absence of Mg2+ ions, as compared to the folded sample.
It is well-known that folded RNA molecules held together by tertiary interactions are more compact in size than in their unfolded state which is reflected in the change in their hydrodynamic radius (RH). To probe the change in RH upon addition of Mg2+, we probed the fluorescence anisotropy of fluorescently labeled pri-miR-17-92a. Fluorescence anisotropy is directly proportional to the hydrodynamic volume of the RNA given by the Perrin equation. First, ensemble Mg2+ titration experiments were performed on the labeled pri-miR-17-92a to (1) verify the Mg2+ concentration regime associated with the folding transition, and (2) provide results to compare with the subsequent single molecule experiments. To 50 nM Alexa-488-labeled pri-miR-17-92a in 50 mM HEPES-KOH, pH 7.5, 100 mM KCl, and 1 mM EDTA incremental amounts of Mg2+ were added, and the fluorescence anisotropy was monitored. As shown in Figure 5B, the mean fluorescence anisotropy of the RNA decreased with increasing Mg2+ concentrations, indicating lower RH and, therefore, compaction of the pri-miR-17-92a. Importantly, anisotropy of the pri-miR-17-92a decreases steadily until a 1 mM concentration of Mg2+, beyond which there is no further decrease. Bulk fluorescence anisotropy measurements indicate that in the presence of Mg2+ ions there is at least a 50% change in the hydrodynamic volume of the pri-miR-17-92a. This is consistent with the observations of other structured RNAs that become more compact in their completely folded state held together by tertiary interactions. In order to see whether this compaction is reversible or not, incremental additions of EDTA were made to 50 nM of the folded pri-miR-17-92a sample, and fluorescence anisotropy was monitored. This showed a profile consistent with a three-state behavior, with transitions occurring between 1–10 mM EDTA and another at 30 to 70 mM EDTA, recapitulating the folding curve (see Supplemental Fig. S8B). This strongly indicates at least two Mg2+ ions bound to the RNA, with one that is more strongly bound in the RNA requiring five- to sixfold more EDTA for sequestration. Thus, both Mg2+ and EDTA titration experiments probed by fluorescence anisotropy indicate Mg2+-specific folding behavior resulting in compaction of the pri-miRNA and gives an idea of the number the Mg2+ ions required to achieve this.
In order to see whether the introduction of fluorophores into the RNA influenced the above findings, we also monitored the change in hydrodynamic radius, RH, by Mg2+ titrations using a label-free technique such as dynamic light scattering. To 5 μM pri-miR-17-92a in 50 mM HEPES-KOH, pH 7.5, 100 mM KCl, and 1 mM EDTA, an incremental amount of Mg2+ was added, and the hydrodynamic radius was measured as a function of Mg2+ concentration. As expected, we observed a decreasing RH with Mg2+ concentration, as shown in Supplemental Figure S9A, which resembles the curve obtained by fluorescence anisotropy. Thus, incorporation of fluorophores does not significantly hamper pri-miRNA folding, and further, the sharp distribution of RH at such high (6.8 ± 0.5 nm) RNA strand concentrations is indicative of virtually no aggregation, suggesting a well-folded, highly structured RNA.
Pri-miR-17-92a transcript dynamics probed by single molecule fluorescence anisotropy
There are several examples where RNA function is synonymous with its specific secondary and tertiary structure and, in some cases, where its function is modulated by its structural plasticity or the ability to adopt stable alternate structures in response to specific stimuli, e.g., riboswitches or 7SK RNA (Smith et al. 2009; Herreweghe et al. 2007). Given that protein binding can drive an RNA folding path into an alternate structure, we wanted to quantitate the extent of inherent dynamicity in the folded state of the pri-miR-17-92a and assess its inherent capacity to sample multiple conformations in the absence of proteins, one of which might be stabilized upon interaction with protein partners. The contrary case would imply the need for specific factors that can open up the structure in order to drive processing.
Conformational distribution of an RNA molecule has been generally measured using single molecule fluorescence resonance energy transfer (FRET) that requires prior knowledge of the high resolution structure of the RNA for the introduction of a FRET pair at well-defined positions (Clegg 1992). However, a recently developed single molecule anisotropy-based method circumvents the need for a high resolution structure by measuring the hydrodynamic volumes of single RNA molecules, which also yields compaction heterogeneity or RNA dynamics within a given population (Sinha et al. 2006). Here, population distributions of conformations reporting on overall RNA dynamics are studied from a measure of anisotropy of single RNA molecules diffusing through a confocal volume at any instant. The pri-miR-17-92a is labeled by incorporating Alexa-488-labeled rUTP in an in vitro transcription mixture so that each of the transcripts incorporates at least two rUTPs at random uridine residues of the transcripts. This method works as long as (1) introduction of the fluorophore does not change the native structure and (2) structure formation does not change the lifetime of the fluorophore (see Supplemental Figs. S9B, S10). The fluorophore can rotate about its linker (local motion) as well as around the axis of the pri-miRNA cluster (global rotation). Both local and global rotation will depolarize the fluorescence which can be quantified by anisotropy. The hydrodynamic volume dependence on anisotropy is given by the Perrin equation: r = r0/[1 + (τRT/ηV)], where r is the anisotropy of the fluorophore, r0 is the anisotropy of the immobilized fluorophore, η is the viscosity of the medium, τ is the lifetime of the fluorophore, R is the gas constant, T is temperature, and V is the hydrodynamic volume of the RNA. The effective hydrodynamic volume of pri-miR-17-92a thus depends inversely on the extent of secondary structure and tertiary contacts that reflects the extent of RNA folding that leads to RNA compaction. This, in turn, dictates its translational and rotational dynamics in solution as well as the sampling times of various conformational states.
For the experiments, two samples were prepared at 0.5 nM concentration in 50 mM HEPES, KOH, pH 7.5, 100 mM KCl, and 1 mM EDTA with and without 10 mM Mg2+. Samples were excited by an Ar ion laser 488-nm line, and single molecule bursts were collected for each sample in two APDs located perpendicular to each other. Anisotropy of single molecules of pri-miRNAs was measured after filtering the data at a threshold of 40 counts. Anisotropy values for each sample were collected and presented as a histogram in Figure 5C. From both distributions of anisotropy values for pri-miR-17-92a it is evident that the width of the distribution of anisotropy in the presence of Mg2+ (w = 0.076) is much narrower than without Mg2+ (w = 0.106). This is consistent with the DLS studies that also indicate greater molecular compaction of the RNA in the presence of Mg2+ due to adoption of a specific tertiary structure by the pri-miR-17-92a. To investigate the conformational dynamics in the folded (+Mg2+, F state) and unfolded states (−Mg2+, U state), single molecule trajectories were recorded in 70% glycerol for both samples. It has been shown that the high viscosity of the glycerol water interface in a 70% glycerol solution of biomolecules functionally mimics relevant immobilized systems (Ha et al. 1999), enabling the assignment of distinct molecular states consistent with the observed distribution. The higher viscosity of 70% glycerol retains a given RNA molecule for a longer time period within the confocal spot, thus enabling the continuous measurement of anisotropy on an RNA molecule undergoing fluctuations. As shown in Figure 5D, the spread of the distribution in the case of the unfolded state (without Mg2+) indicates high conformational heterogeneity associated with a low degree of compaction, while the anisotropy distribution of the folded (+Mg2+) indicates a highly compact state which is relatively inflexible (see Supplemental Fig. S11A for trajectories). Thus, these experiments reinforce that, in the presence of Mg2+ ions, the pri-miR-17-92a cluster adopts the single folded state, with low conformational dynamics. They also reveal that the unfolded transcript (−Mg2+) is much more conformationally dynamic. This can be reconciled with Figure 2B, which shows the thermodynamically most stable secondary structure of pri-miR-17-92a as predicted by RNAfold (Hofacker et al. 1994). It is also color-coded according to base-pairing probability, i.e., the probability of occurrence of each base pair in an ensemble of structures calculated using partition functions which sum all the Boltzmann weighted free-energies of each base pair in all possible secondary structures (McCaskill 1990). When this base-pairing probability is grafted on the mean free-energy structure, it is apparent that barring pre-miR-18a, all the individual pre-miRNA hairpins are conserved and, therefore, invariant in the ensemble. However, pre-miR-18a shows a high amount of variability in its base-pairing probability, indicating the potential for structural plasticity. The base-paired regions between the individual pre-miRNA domains indicate relatively reduced base-pairing probabilities, too. These regions could contribute to the dynamicity observed in the single molecule experiments, in the absence of Mg2+ ions. Thus, the experimental observation of conformational dynamicity of the pri-miR-17-92a transcript under conditions that promote secondary structure formation is consistent with a theoretical prediction of flexibility based on energetics present in the secondary structure of the pri-miR transcript.
The shuffled transcript shows perturbed tertiary structure
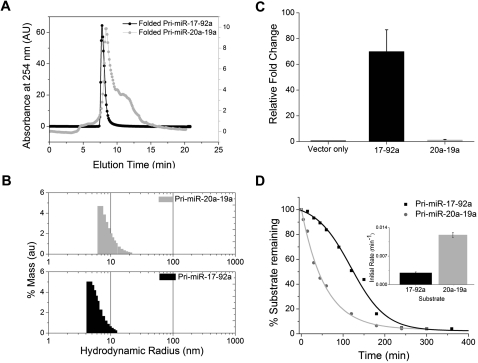
Using biochemical and biophysical methods, we established that native transcript pri-miR-17-92a undergoes a folding transition to adopt a well-defined tertiary structure. If this is true, then a shuffled transcript containing an altered order of hairpins might not have the potential to adopt the right folds to attain this tertiary structure. To investigate this, we used size exclusion chromatography (SEC) in conjunction with dynamic light scattering to compare the changes in RH of pri-miR-17-92a as well as the shuffled transcript pri-miR-20a-19a as a function of Mg2+ ions. Interestingly, it was found by SEC that, in conditions that promoted folding, pri-miR-17-92a showed a sharp peak in the chromatogram (Fig. 6A), while the shuffled transcript pri-miR-20a-19a showed a peak with a broad shoulder, indicating at least two structures that rapidly interconvert on SEC time scales. When the RH was measured by DLS, pri-miR-20a-19a showed a significantly higher radius (RH = 9.8 nm) with a broad distribution compared to pri-miR-17-92a (RH = 6.8 nm) (Fig. 6B). Thus, biophysical methods clearly indicate that the shuffled transcript pri-miR-20a-19a shows a distinct impairment in its ability to adopt tertiary structure, unlike pri-miR-17-92a. We also sought to obtain biochemical evidence of this by using an RNA aptamer, which has been shown to bind the apical loop of pre-miR-18a and A433-G441 in the native transcript in the presence of 5 mM Mg2+ ions (Lunse et al. 2010). The interaction between this aptamer and each of the pri-miR transcripts, pri-miR-17-92a and pri-miR-20a-19a, would indicate whether or not loss or alteration of tertiary structure could impact recognition of the RNA toward another biomolecule. We could, thus, use aptamer binding as a probe of RNA structure or dynamics or both. A careful analysis of the binding sites indicates two RNA motifs; the first one comprises the sequence 5′-AUCUAC complementary to the apical-loop domain of pre-miR-18a, while the second site (sequence 5′-CGACACAAU) is complementary to a region located between pre-miR-19a and pre-miR-20a. This aptamer is, thus, well placed to monitor the Mg2+-dependent changes in pri-miR-17-92a (see Supplemental Fig. S12). Upon incubation of pri-miR-17-92a with labeled aptamer for 30 min in different Mg2+ ion concentrations (data not shown), native PAGE indicated that binding was complete at 3 mM Mg2+ ion concentration. Importantly, pri-miR-17-92a did not bind the aptamer in the absence of Mg2+. The two aptamer binding sites are positioned 228 nt apart in the linear sequence of pri-miR-17-92a, the binding of which is possible only if the helices are proximal to each other in 3D space, which is facilitated by tertiary structure. Under similar conditions, pri-miR-20a-19a bound the aptamer efficiently independent of Mg2+.
FIGURE 6.
(A) Size-exclusion chromatogram of 1 μM pri-miR-17-92a and 20a-19a in folding buffer (composition: 50 mM HEPES, KOH, pH 7.5, 100 mM KCl, 5 mM Mg2+). The same buffer is used as the running buffer with a flow rate of 0.5 mL/min. (B) Dynamic light-scattering experiments with 1 μM pri-miR-20a-19a and pri-miR-17-92a in folding buffer. (C) Relative levels of pri-miRNA from native (pri-miR-17-92a) and shuffled (pri-miR-20a-19a) transcripts. Total RNA was isolated 24 h post-transfection from HeLa cells transfected either with empty pcDNA 3 vector, or pri-miR-17-92a-pcDNA3, or pri-miR-20-19a-pcDNA3 and was used for qRT-PCR using primers specific for each pri-miR indicated. 28S rRNA was used as an endogenous control, and levels were normalized to the empty vector. (D) Quantification of Northern blots to probe full-length native and shuffled transcripts remaining after in vitro processing shows that the kinetics of processing of native transcript is much slower. Initial rates were determined from the kinetic traces expressed as mean ± s.d. from three independent experiments, shown in the inset.
Effect of tertiary structure on processing in cellulis
We have shown that pri-miR-17-92a has a well-defined tertiary structure, and, though the shuffled transcript pri-miR-20a-19a contains the same sequence information, it does not possess a distinct higher-order structure. However, both the transcripts pri-miR-17-92a (N) and pri-miR-20a-19a (S) were processed into their respective pre-miRNA products, with the products from pri-miR-20a-19a showing much higher levels (see Fig. 1C; Supplemental Fig. S2B). In order to address the amount of substrate primary miRNA present at equilibrium in each case, we assessed the relative primary transcript levels in HeLa and HEK 293T cells transfected with pri-miR-17-92a and pri-miR-20a-19a. Total RNA in each case was isolated and reverse-transcribed using primers specific for pri-miR-17-92a and pri-miR-20a-19a, and then the cDNA was used for RT-PCR. This immediately gave a qualitative indication that, in both HeLa cells and HEK 293T cells, though the pri-miR-17-92a was present, pri-miR-20a-19a was not detectable (see Supplemental Fig. S13). In order to quantify the equilibrium levels of pri-miR-17-92a and pri-miR-20a-19a transcripts in cellulis, we performed qPCR using specific primers on total RNA isolated from HeLa cells 24 h post-transfection. The qPCR analysis confirmed that, in cellulis, the equilibrium levels of pri-miR-17-92a were nearly 70-fold greater than pri-miR-20a-19a. Given that (1) the in vitro lifetimes of both transcripts are similar (data not shown), (2) the rate of production of the transcripts in cellulis should be comparable as they utilize the same promoter, and (3) they give rise to the same product identities, and yet their equilibrium levels are so different, we were prompted to investigate the kinetics of processing of both transcripts under the same conditions. This was investigated in vitro by incubating either the pri-miR-17-92a or pri-miR-20a-19a transcripts in HeLa cell extract and detecting the unprocessed full-length transcript in a time-dependent manner, using a specific radio-labeled primer. Northern blot analysis showed that processing is, indeed, faster in the case of the shuffled transcript pri-miR-20a-19a than in the native, folded transcript pri-miR-17-92a (see Fig. 6D, also see Supplemental Fig. S14).
The time taken for 50% of the transcript to be processed in the case of pri-miR-17-92a and pri-miR-20a-19a was ∼120 min and 40 min, respectively. The rate of processing for pri-miR-17-92a and pri-miR-20a-19a was quantified by the initial rate method, given that the order of the reaction is not known, and this showed that the rate of processing of pri-miR-20a-19a is about fourfold higher than the rate of pri-miR-17-92a (Fig. 6D, inset). This shows that the tertiary structure adopted by the primary microRNA transcript has considerable influence on its processing in vitro and in cellulis.
DISCUSSION
Transcription is the major regulatory step in the biogenesis of mature miRNAs. Most primary miRNAs are transcribed by RNA Pol II, and consequently, their upstream regions possess canonical core promoters and enhancers similar to the promoters of protein-coding genes (Saini et al. 2007). Thus, the transcription factors that regulate primary miRNA transcription show significant overlap with those that control protein-coding genes, such as c-Myc and p53 (Simon et al. 2004). For example, pri-miR-17-92a transcription is activated by binding of c-Myc transcription factors to E-box elements of its gene promoter (O'Donnell et al. 2005). However, mature miRNA expression does not always correlate with expression of its corresponding pri-miRNA. In ES cells, though the primary transcript is well-expressed, its processing is initiated only at a specific time during differentiation (Thomson et al. 2006). In Xenopus, though the primary miRNA is transcribed to similar abundances in different tissues, the resultant mature miRNA products show very different relative abundances. Further, in ES cells, Thomson et al. have shown that, even within a given tissue, relative abundances of the distinct mature miRs present on the pri-miR-17-92a cluster are quite disparate and that those relative abundances also change as a function of cell differentiation status (Thomson et al. 2006). All of this points to the existence of mechanisms that regulate miRNA abundances post-transcriptionally, both in the nucleus and in the cytoplasm. Such post-transcriptional regulation is achievable by the collective action of a set of RNA-binding proteins. For example, pluripotency factor RNA-binding protein Lin28 binds the conserved terminal loop region of the pre-Let-7d and blocks let-7 production (Newman et al. 2008). KH-type splicing regulatory factor KSRP also binds the pri-let-7a loop region and promotes its processing (Trabucchi et al. 2009). Similarly, heterogeneous nuclear ribonucleoprotein (hnRNP) A1 binds specifically to human pri-miR-18a of the pri-miR-17-92a cluster and facilitates its processing by the Drosha-DGCR8 complex (Guil and Cáceres 2007). SF2/ASF, a prototypical SR protein splicing factor, promotes the biogenesis of mir-7 at the Drosha processing step by binding to the stem of pri-miR-7 RNA (Wu et al. 2010). The RNA-binding protein nuclear factor 90 (NF90) and its partner, NF45, are associated with the microprocessor complex (MPC) and act as negative regulators of Drosha processing (Sakamoto et al. 2009). The DEAD-box RNA helicases p68 and p72, which are associated with the large MPC, are required for the efficient Drosha-mediated processing of a subset of miRNAs (Fukuda et al. 2007). These are also known to promote the recruitment of Drosha itself or of auxiliary factors, such as estrogen-bound ERα, p53 and BMP-specific SMAD signal transducers that all modulate Drosha processing of subsets of miRNAs (Davis et al. 2008; Suzuki et al. 2009; Yamagata et al. 2009; Yan et al. 2009).
Here, we show that tertiary structure formation by primary miRNAs influences their processing into pre-miRNAs (Chaulk et al. 2011). The phylogenetic sequence analysis of the pri-miR-17-92a transcript revealed that many inter-pre-miR regions, previously presumed to be nonfunctional since they do not contain the mature miRNA sequence, are also conserved. SHAPE and hydroxyl radical footprinting assays confirmed that such inter-pre-miR regions, too, have well-defined secondary structure. In addition, specific nucleotides in these regions become solvent-inaccessible in the presence of Mg2+, indicative of tertiary structure in the RNA transcript. Folding of the RNA into its tertiary structure might be facilitated by the conserved terminal loops of the pre-miR domains, the inter-pre-miR regions, or both. The disruption of such structured inter-pre-miR elements, such as the region between pre-miR-19a and pre-miR-20a, yields a shuffled transcript pri-miR-20a-19a with impaired tertiary structure which shows altered processing efficiency in vitro and in cellulis. This suggests a model where tertiary structure formation by a primary miRNA transcript imposes a kinetic barrier to its processing where the conformation adopted is transparent to the MPC, leading to an inhibition imposed at the earliest stage of its processing into pre-miRs. The splicing machinery has been connected with the recruitment of the MPC. It is also known that splicing of those introns that harbor primary microRNA transcripts are retarded compared to those that do not, and the timescales of the latter are generally on the order of 5–10 min (Singh and Padgett 2009). This is sufficient time to allow the transcribed RNA to fold. Thus, processing of primary microRNAs could be envisaged to occur cotranscriptionally, yet post-folding into a tertiary structure.
One might now consider mechanistically how the interaction of a folded RNA with known processing proteins could impact its processing. The most simplistic case would be the difference in processing mechanisms prefolding and post-folding. It has been observed that premature cleavage of ribozyme elements that undergo fast cleavage that are engineered into introns results in the degradation of the pre-mRNA transcript (Fong et al. 2009). However, the inclusion of ribozyme elements that undergo cleavage at retarded rates allows for proper pre-mRNA splicing (Fong et al. 2009). Following a similar logic, we know that primary microRNA transcripts are processed, and thereby cleaved, cotranscriptionally by Drosha/DGCR8. Absence of a tertiary structure in a transcript such as pri-miR-20a-19a leads to a fourfold faster cleavage rate, due to easy accessibility of binding sites for the MPC. This could result in premature cleavage that could preclude proper splicing of the host pre-mRNA transcript. The folding of primary microRNA 17-92a into a well-defined tertiary structure that impedes processing would aid exon tethering that, in turn, facilitates appropriate pre-mRNA processing. Considering the abundance of several structured clusters of primary microRNA transcripts in intronic regions, this could serve as a high-value benefit of impeded primary microRNA processing. We are currently investigating this hypothesis experimentally in our laboratory.
In the context of the processing of the primary microRNA transcript itself into the relevant pre-microRNAs, the tertiary structure could sequester binding sites, impede processing, and result in a larger abundance of unprocessed transcripts. Such unprocessed transcripts have been shown to accumulate in subnuclear bodies rich in alternative splicing factors such as sc35. (Pawlicki and Steitz 2008). Further, we know that, although primary microRNAs are expressed in ES cells, the corresponding pre- and mature microRNAs are expressed only at a later developmental stage. This strongly suggests the binding of an alternate set of protein factors to folded, unprocessed transcripts that could mediate an alternate processing fate for the primary microRNA. In such a scenario, it is worthwhile to consider structurally how the primary microRNA might access such alternate fates due to differential protein binding. For example, the MPC is known to exist in two forms, one of which is a high molecular weight complex that contains a DEAD-box helicase such as DDX5 that has been implicated in RNA processing (Fukuda et al. 2007). The high molecular weight MPC could remodel the transcript by unwinding the helices, while the low molecular weight MPC could distort the transcript differently by binding key ss-ds junctions. These differential distortions by the high and low molecular weight MPCs could result in a completely different set of exposed processing protein-binding sites made available to the system that could result in alternate processing fates.
Given the accessibility of binding sites of the primary microRNA transcript to different processing proteins, we could also consider how alternate processing pathways might be regulated structurally. In this context, looking more broadly at microRNA regulation, many let-7 family members are known to exist in highly sequence-conserved clusters. It is known that different processing proteins such as hnRNPA1, Lin 28, and KSRP compete for the same RNA target, yet bind the target very differently. For example, binding of Lin28 to RNA helices in the pre-microRNA domain has been shown to substantially bend the helix which, in the context of primary microRNA transcript, could deform tertiary structure of the RNA (Nam et al. 2011). Interestingly, while both KSRP and hnRNPA1 bind specific loop sequences that remodel RNA secondary structure, hnRNPA1 has also been shown to bind stem regions and locally unfold the transcript (Michlewski and Cáceres 2010). It has been repeatedly shown that secondary structure is a key determinant of the resultant tertiary structure of the RNA (Brion and Westhof 1997). Different secondary structures naturally result in alternate higher-order structure that could differentially expose processing sites. Thus, one can envisage how KSRP might facilitate the processing of let-7, while hnRNPA1 down-regulates the same despite both being competitive binders of the same primary microRNA. Further, hnRNPA1 binding in a different context of primary microRNA, such as pri-miR-17-92a, has been shown to locally remodel RNA secondary structure and facilitate its processing (Michlewski et al. 2008). This indicates the importance of the RNA structural context of the binding of a given processing protein, indicating that the RNA is, indeed, an active regulator of its processing. The tertiary structure of pri-miR-17-92a might mask recognition sites for the MPC, thus creating the observed kinetic barrier for its processing. However, one can also envisage that the folded cluster may be distorted into alternate conformations with the aid of different RNA-binding proteins that render it a suitable substrate for alternate processing fates. The protein interaction hierarchy with the folded transcript would be dictated by the Kds and relative abundances of the RNA binders in a given environment.
In order to see whether such a mechanism might be used more generally by primary miRNA clusters, we used the UCSC genome browser and investigated phylogenetically several primary miRNA clusters. We checked, along with the functional pre-miR domains, the extent of sequence conservation of the “nonfunctional” inter-pre-miR regions by phylogenetic analysis. As shown in Supplemental Figure S15, we found as many as 10 different pri-miRNA clusters with multiple miRNAs (≥2) that clearly indicate conservation of their inter-pre-miR regions, strengthening the fact that tertiary structure formation by pri-miRNA clusters may be a widespread occurrence.
Thus far, primary miRNA clusters are generally represented as a series of connected hairpins devoid of tertiary structure that implies efficient processing of all the hairpins in a cluster, all equally accessible. Such post-transcriptional regulation of a primary miRNA cluster devoid of tertiary structure needs to reconcile with stochastic processing that can occur by the binding of any one of its many RNA processing factors, yet results in preferential or asymmetric processing. The present study shows that the folding of the transcript into a tertiary structure might function as the first step of post-transcriptional regulation by precluding equal accessibility of the constituent pre-miR domains to their relevant protein partners and/or a kinetic barrier that facilitates exon tethering for effective and appropriate splicing. The proposed model suggests autoregulation imposed early on by the RNA scaffold adopting a tertiary structure.
MATERIALS AND METHODS
Plasmids
All the sequences of the RNA transcripts used for in vitro transcription were cloned into TOPO-pCR 2.1 TA cloning vector (Invitrogen) and linearized with Sal I and Eco R1 before transcription. The primary sequences pri-miR-17-92a and its related miniclusters pri-miR-17-19a and pri-miR-20a-92a were obtained by PCR from human genomic DNA using the oligonucleotide primers specific for each cluster (see Supplemental Table S1). The shuffled pri-miR 20-19a cluster was derived from overlap PCR of 17-19a and 20a-92a. The forward primer used for making each construct for transcription contains the T7 promoter sequence, and the reverse primer contains a Sal I site. For overexpression of the pri-miR-17-92a and pri-miR-20a-19a in mammalian cell lines, the PCR-amplified DNA was cloned into the pcDNA3.1/V5-His-TOPO TA vector (Invitrogen). The identity of all the constructs was confirmed by sequencing. The sequences of the primers used for making different constructs are given in Supplemental Table S1.
Cell transfection
HeLa cells and HEK 293T cells were grown in Dulbecco's Modifed Eagle's Medium (DMEM) with 10% FBS, penicillin, and streptomycin until they reached 70% confluency. For overexpression of the native pri-miR-17-92a and shuffled pri-miR-20-19a transcripts, the expression plasmid was transfected into these cells using HeLa Monster (Mirus Bio). The transfected medium was replaced after 4 h of incubation and replaced with fresh medium. The cells were collected 24 h post-transfection and used either for total RNA isolation by using Trizol (Invitrogen) or Small RNA isolation (mirPremier RNA isolation kit, Sigma).
RT-PCR
For the expression analysis of the native and shuffled transcripts, total RNA was isolated from both HeLa and HEK 293T cells as indicated above. Briefly, 3 μg of total RNA was reverse-transcribed using SuperScript III Reverse Transcriptase (Invitrogen) with gene-specific primers. 4 μL of the cDNA was used for a standard PCR using Taq DNA Polymerase (New England Biolabs). As a control, 18S was reverse-transcribed and amplified. The RT-PCR products were analyzed on a 1% agarose gel.
Quantitative RT-PCR
Total RNAs from cultured HeLa cells were isolated using Trizol reagent (Invitrogen). RNA samples were reverse-transcribed into cDNA with Superscript III reverse transcriptase (Invitrogen). Real-time PCR was performed with gene-specific primers and Power SYBR Green PCR Master Mix (Applied Biosystems) using the 7500 Fast Real-Time PCR System (Applied Biosystems). For qPCR of pre-miRNAs, total small RNAs were isolated using miRVAna miRNA isolation kit (Ambion) according to the manufacturer's protocol.
Relative quantities of pri-miRNAs and pre-miRNAs were determined using the comparative CT method (ΔΔCT method) as provided by the manufacturer. Briefly, relative quantities of pri-miRNAs were determined by this method normalized to an endogenous reference (28S rRNA) and relative to the calibrator (empty vector). Similarly, the pre-miRNA levels of the shuffled transcript and minicluster 1 were normalized to an endogenous reference (5.8S) and relative to the native transcript.
Northern blots
Northern blotting of the in vitro processing reactions assesses the kinetics of the native and shuffled pri-miR transcripts. 5 μg of pri-miR-17-92a and shuffled pri-miR-20a-19a in vitro transcribed RNAs were incubated under processing conditions in the presence of HeLa cell extract in a 50-μL reaction at 37°C for the indicated times, followed by phenol-chloroform extraction and ethanol precipitation. The RNAs were mixed with gel loading buffer and resolved on a 6% denaturing polyacrylamide gel in 1× TBE and then transferred to Hybond XL Nylon membrane (GE Healthcare). The membranes were UV cross-linked at 1200 mJ using UV Stratalinker (Strategene), baked at 80°C for 30 min, and prehybridized for 1 h at 40°C in hybridization buffer (1× SSC, 1% SDS, and 200 μg/mL of salmon sperm SS DNA [Sigma]). Probes for pri-miR transcripts and U6 RNA (as a loading control) end-labeled using a [γ-P32] ATP (BRIT) and T4 Polynucleotide Kinase (New England Biolabs) were left to hybridize overnight in hybridization buffer at 37°C. Following hybridization, the membranes were washed twice in wash buffer I (1× SSC and 0.2% [w/v] SDS) for 30 min each and once in wash buffer II (0.2× SSC and 1% SDS) at 42°C. The blots were then analyzed using a Phosphor Imager (Typhoon Trio, GE Healthcare).
Kinetic assays
For kinetics of processing, the Northern blots of the in vitro processing experiment were quantified. The amount of substrate remaining after time t of processing was quantified from the blots (Image J, NIH) and normalized to the substrate at t = 0. For determining the initial rates, the traces from three independent processing experiments constructed for both the native and shuffled transcripts were taken and expressed as mean ± s.d.
RNA probing experiments
For the SHAPE reaction, 10 pmols of RNA was taken in folding buffer (50 mM HEPES-KOH, pH 7.5, 100 mM KCl, 5 mM Mg2+) and reacted with 3 mM NMIA in DMSO (+NMIA) and with only DMSO (−NMIA). Similarly, for hydroxyl radical cleavage experiments, RNA in folding buffer was reacted with 1 μL each of 150 mM ascorbate, and 0.3% H2O2 with and without 1 μL 3.75 mM Fe(II)-EDTA for 15 sec. After precipitation of the RNA-adduct complex and/or cleaved RNA, it was dissolved in water and used in reverse transcription with primers (binding site 286–257 nt), each labeled with a different fluorophore (6-FAM, +NMIA/+·OH; NED, −NMIA/−·OH; VIC, ddGTP). Sequencing reactions were done by incorporating dideoxynucleotides into reverse transcriptase mix. After completion, all four reactions were mixed and precipitated and dissolved in deionized formamide. 10 μL of such solution were loaded onto the capillary of the ABI 3130xl genetic analyzer. The resulting electrophoretogram was analyzed by SHAPE finder software (Vasa et al. 2008).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article and includes additional methods and results that support findings described in the main text.
ACKNOWLEDGMENTS
We thank Professors David Lilley, John Kuriyan, and Eric Westhof for several valuable discussions during the course of this study and for their suggestions and comments on the present manuscript. We thank Professor G. V. Shivashankar for his kind guidance in setting up the single molecule system. We also thank the Sequencing Facility at NCBS for capillary electrophoresis. S.C., S.M., and A.P. thank CSIR of the Government of India for research fellowships. Y.K. thanks DBT for the Innovative Young Biotechnologist Award and the Wellcome Trust DBT India Alliance for a Senior Research Fellowship.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.031039.111.
NOTE ADDED IN PROOF
During the submission of this work, Chaulk et al. (2011) also showed complementary evidence of tertiary structure formation by pri-miR-17-92a at 2 nm resolution that showed a globular structure by Cryo EM. They addressed the effect of tertiary structure on processing in vitro using purified MPC components.
REFERENCES
- Ambros V 2004. The functions of animal microRNAs. Nature 431: 350–355 [DOI] [PubMed] [Google Scholar]
- Baird NJ, Fang XW, Srividya N, Pan T, Sosnick TR 2007. Folding of a universal ribozyme: The ribonuclease P RNA. Q Rev Biophys 40: 113–161 [DOI] [PubMed] [Google Scholar]
- Brion P, Westhof E 1997. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomol Struct 26: 113–137 [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM 2007. MicroRNA functions. Annu Rev Cell Dev Biol 23: 175–205 [DOI] [PubMed] [Google Scholar]
- Chaulk SG, Thede JL, Kent OA, Xu Z, Gesner EN, Veldhoen RA, Khanna SK, Goping IS, MacMillan AN, Mendell JT, et al. 2011. Role of pri-miRNA tertiary structure in miR-17∼92 miRNA biogenesis. RNA Biol 8: 1–10 [DOI] [PubMed] [Google Scholar]
- Chen K, Rajewsky N 2007. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet 8: 93–103 [DOI] [PubMed] [Google Scholar]
- Clegg RM 1992. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol 211: 353–387 [DOI] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A 2008. SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454: 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N 2008. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Fong N, Öhman M, Bentley DL 2009. Fast ribozyme cleavage releases transcripts from RNA polymerase II and aborts cotranscriptional pre-mRNA processing. Nat Struct Mol Biol 16: 916–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, et al. 2007. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol 9: 604–611 [DOI] [PubMed] [Google Scholar]
- Guil S, Cáceres JF 2007. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol 14: 591–596 [DOI] [PubMed] [Google Scholar]
- Gutell RR 1993. Comparative structure studies of RNA: Inferring higher-order structure from patterns of sequence variation. Curr Opin Struct Biol 3: 313–322 [Google Scholar]
- Gutell RR, Cannone JJ, Shang Z, Du Y, Serra MJ 2000. A story: Unpaired adenosine bases in ribosomal RNAs. J Mol Biol 304: 335–354 [DOI] [PubMed] [Google Scholar]
- Ha T, Laurence TA, Chemla DS, Weiss S 1999. Polarization spectroscopy of single fluorescent molecules. J Phys Chem B 103: 6839–6850 [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee J-K, Sohn SY, Cho Y, Zhang B-T, Kim VN 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125: 887–901 [DOI] [PubMed] [Google Scholar]
- Herreweghe VE, Egloff S, Goiffon I, Jady BE, Froment C, Monsarrat B, Kiss T 2007. Dynamic remodeling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J 26: 3570–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, Fontana W, Stadler PF, Bonhoeffer S, Tacker M, Schuster P 1994. Fast folding and comparison of RNA secondary structures. Monatsh Chem 125: 167–188 [Google Scholar]
- Hogg JR, Collins K 2007. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA 13: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Sobczak K, Wilczynska U, Drath W, Jasinska A, Kaczynska D, Krzyzosiak WJ 2004. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J Biol Chem 279: 42230–42239 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using real time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lunse CE, Michlewski G, Hopp CS, Rentmeister A, Caceres J, Famulok M, Mayer G 2010. An aptamer targeting the apical-loop domain modulate pri-miRNA processing. Angew Chem Int Ed 49: 4674–4677 [DOI] [PubMed] [Google Scholar]
- Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le BEC C, Osborne HB 2006. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem J 400: 291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill JS 1990. The equilibrium partition function and base pair binding probabilities for RNA secondary structures. Biopolymers 29: 1105–1119 [DOI] [PubMed] [Google Scholar]
- McGinnis JL, Duncan CD, Weeks KM 2009. High-throughput SHAPE and hydroxyl radical analysis of RNA structure and ribonucleoprotein assembly. Methods Enzymol 468: 67–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw M, Sethupathy P, Corda B, Hatzigeorgiou AG 2007. miRGen: A database for the study of animal microRNA genomic organization and function. Nucleic Acids Res 35: 149–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT 2008. miRiad roles for the miR-17-92a cluster in development and disease. Cell 133: 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino EJ, Wilkinson KA, Coughlan JL, Weeks KN 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 127: 4223–4231 [DOI] [PubMed] [Google Scholar]
- Michlewski G, Cáceres JF 2010. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol 17: 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, Guil S, Semple CA, Cáceres JF 2008. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol Cell 32: 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P 2011. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 147: 1080–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM 2008. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14: 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT 2005. c-Myc regulated microRNAs modulate E2F1 expression. Nature 435: 839–843 [DOI] [PubMed] [Google Scholar]
- Pawlicki JM, Steitz JA 2008. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J Cell Biol 182: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Masquida B, Westhof E, Woodson SA 2003. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc Natl Acad Sci 100: 1574–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH 2010. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11: 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini HK, Griffiths-Jones S, Enright AJ 2007. Genomic analysis of human miRNA transcripts. Proc Natl Acad Sci 104: 17719–17724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S, Aoki K, Higuchi T, Todaka H, Morisawa K, Tamaki N, Hatano E, Fukushima A, Taniguchi T, Agata Y 2009. The NF90–NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol Cell Biol 29: 3754–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G 2009. microRNAs at the synapse. Nat Rev Neurosci 10: 842–849 [DOI] [PubMed] [Google Scholar]
- Simon C, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116: 499–509 [DOI] [PubMed] [Google Scholar]
- Singh J, Padgett RA 2009. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol 16: 1128–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha D, Sastry S, Shivashankar GV 2006. Probing messenger RNA conformational heterogeneity using single-molecule fluorescence anisotropy. Appl Phys Lett 88: 103901–103903 [Google Scholar]
- Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA 2009. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol 16: 1218–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LJ, Waldsich C, Pyle AM 2005. An obligate intermediate along the slow folding pathway of a group II intron ribozyme. Nucleic Acids Res 33: 6674–6687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K 2009. Modulation of microRNA processing by p53. Nature 460: 529–533 [DOI] [PubMed] [Google Scholar]
- Talkington MW, Siuzdak G, Williamson JR 2005. An assembly landscape for the 30S ribosomal subunit. Nature 438: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang GQ, Maxwell ES 2008. Xenopus microRNA genes are predominantly located within introns and are differentially expressed in adult frog tissues via post-transcriptional regulation. Genome Res 18: 104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM 2006. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 20: 2202–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG 2009. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459: 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakalopoulou E, Schaack J, Shenk T 1991. A 32-kilodalton protein binds to AU-rich domains in the 3′ untranslated regions of rapidly degraded mRNAs. Mol Cell Biol 11: 3355–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC 2008. ShapeFinder: A software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA 14: 1979–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GJ 2010. Lin28: A microRNA regulator with a macro role. Cell 140: 445–449 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Swanstrom R, Burch CL, Weeks KM 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460: 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Vasa SM, Deigan KE, Mortimer SA, Gidding MC, Weeks KM 2009. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA 15: 1314–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J 2010. A splicing independent function of SF2/ASF in microRNA processing. Mol Cell 38: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, Naitou M, Takeyama K, Minami Y, O'Malley BW, Kato S 2009. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol Cell 36: 340–347 [DOI] [PubMed] [Google Scholar]
- Yan HL, Xue G, Mei Q, Wang YZ, Ding FX, Liu MF, Lu MH, Tang Y 2009. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J 28: 2719–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeakley JM, Morfin JP, Rosenfeld MG, Fu XD 1996. A complex of nuclear proteins mediates SR protein binding to a purine-rich splicing enhancer. Proc Natl Acad Sci 93: 7582–7587 [DOI] [PMC free article] [PubMed] [Google Scholar]