High-throughput RNA sequencing (RNA-seq) was used to identify 178 exons in 168 genes that exhibit robust splicing level changes in response to stimulation of a human T-cell line. Most of these exons also exhibit differential splicing between naive and activated primary T cells. The exons are significantly enriched in genes with functions specifically related to immune response and represent at least three distinct networks of signal-induced alternative splicing events, each being specifically associated with distinct cis-features. Together, the results provide evidence that multiple independent regulatory mechanisms converge to control T-cell response to antigen at the level of alternative splicing.
Keywords: RNA-seq, T cells, alternative splicing, cell signaling, cell stimulation
Abstract
The formation and execution of a productive immune response requires the maturation of competent T cells and a robust change in cellular activity upon antigen challenge. Such changes in cellular function depend on regulated alterations to protein expression. Previous research has focused on defining transcriptional changes that regulate protein expression during T-cell maturation and antigen stimulation. Here, we globally analyze another critical process in gene regulation during T-cell stimulation, alternative splicing. Specifically, we use RNA-seq profiling to identify 178 exons in 168 genes that exhibit robust changes in inclusion in response to stimulation of a human T-cell line. Supporting an important role for the global coordination of alternative splicing following T-cell stimulation, these signal-responsive exons are significantly enriched in genes with functional annotations specifically related to immune response. The vast majority of these genes also exhibit differential alternative splicing between naive and activated primary T cells. Comparison of the responsiveness of splicing to various stimuli in the cultured and primary T cells further reveals at least three distinct networks of signal-induced alternative splicing events. Importantly, we find that each regulatory network is specifically associated with distinct sequence features, suggesting that they are controlled by independent regulatory mechanisms. These results thus provide a basis for elucidating mechanisms of signal pathway–specific regulation of alternative splicing during T-cell stimulation.
INTRODUCTION
A hallmark of the human adaptive immune system is the ability of T cells to detect foreign antigen and direct an appropriate functional response. The detection of foreign antigen occurs through the T-cell receptor (TCR) and triggers an extensive array of receptor-proximal signaling proteins that, in turn, initiate a broad network of signaling cascades (Smith-Garvin et al. 2009). These antigen-activated signaling pathways result in changes in cell morphology and migration, secretion of cytokines and/or cytotoxins, and other effector activities (Smith-Garvin et al. 2009). Not surprisingly, many of these antigen-induced changes in cellular activity are conferred through changes in gene expression (Naito et al. 2011). Thus, the activity of the human immune system relies on the ability of T cells to undergo extensive changes in protein expression in response to antigen-triggered signaling pathways.
Most previous studies regarding gene regulation in T cells have focused on transcription (Smith-Garvin et al. 2009; Beurel et al. 2010; Naito et al. 2011). However, despite increasing evidence that post-transcriptional gene control is also an important feature of T-cell activation (Lynch 2004; Sandberg et al. 2008), relatively little is known about the regulation and biological roles of alternative pre-mRNA processing following stimulation of the immune system. Alternative splicing, in particular, is a ubiquitous mechanism for regulating gene expression. The vast majority of human genes are spliced in more than one pattern to give rise to multiple distinct mRNA isoforms that differ either in their coding sequence or in the presence of cis-regulatory signals that control expression (Pan et al. 2008; Wang et al. 2008). Importantly, variant proteins expressed from a single gene via alternative splicing often act in competition or opposition to one another, such that even small changes in the ratio of protein isoforms expressed from a given gene can have a dramatic effect on cellular function (Clower et al. 2010; David and Manley 2010; Goehe et al. 2010; Nilsen and Graveley 2010). Moreover, there is a growing number of cell types in which various stimuli have been shown to alter global splicing patterns (Ip et al. 2007; Lee et al. 2007; Hartmann et al. 2009).
A few genes such as that encoding the transmembrane phosphatase CD45, have well-characterized alternative splicing events that undergo pronounced changes in response to T-cell activation, in a manner that has important physiological consequences for immune function (Jacobsen et al. 2000, 2002; Hermiston et al. 2002; Lynch 2004). In particular, the skipping of three variable exons of the CD45 pre-mRNA upon T-cell activation reduces the phosphatase activity of the molecule, which contributes to attenuation of T-cell signaling and prevention of autoimmune disease (Jacobsen et al. 2000, 2002; Hermiston et al. 2002). This and other precedents for the importance of alternative splicing in immune responses suggests that determining the full profile of alternative splicing changes induced by T-cell stimulation is critical for a complete understanding of the cellular consequences of antigen challenge.
Here we use high-throughput RNA sequencing (RNA-seq) to investigate global changes in alternative splicing in a cultured T-cell line and in primary human T cells. We identify 178 genes that are predicted to exhibit robust signal-induced changes in isoform expression in cultured T cells, and provide independent confirmation of such changes for about 40 of these genes. Importantly, at least 80% of these signal-induced splicing changes have not previously been described as regulatory events. A large percentage of these genes are enriched in functions related to immunobiology, including components of several of the critical signaling pathways that mediate T-cell effector function, thus underscoring the likely importance of these alternative splicing events for proper T-cell function. Finally, by comparing alternative splicing induced by different stimuli in the cultured cell line with that in primary cells, we provide evidence for at least four distinct signaling pathways from cell stimulation to alternative splicing. One of these pathways is shared between the primary and cultured T cells, while we also identify alternative splicing events that are unique to primary human peripheral T cells or the cultured T-cell line. Collectively, our results significantly expand knowledge of genes that are regulated at the level of alternative splicing in response to cellular signaling, and also the signals that lead to such changes. We further identify novel cases of altered isoform expression involving genes that have critical functions in response to TCR signaling in human peripheral T cells, and we provide insight into the relationship between TCR signaling in primary cells and in a commonly used cultured T-cell line.
RESULTS
Global analysis of signal-induced alternative splicing and gene expression changes in the T-cell line JSL1
Given previously reported examples of the physiological relevance of signal-induced changes in gene expression to T-cell activity (Lynch 2004; Sandberg et al. 2008; Smith-Garvin et al. 2009; Naito et al. 2011), it is important to gain a more comprehensive view of how alternative splicing is used to control protein expression downstream from TCR-mediated signaling. To accomplish this, we used RNA-seq to quantify differential expression of alternate mRNA isoforms using the JSL1 human T-cell line. Stimulation of JSL1 cells with PMA triggers signals that mimic aspects of mature TCR signaling (Lynch and Weiss 2000; and see below). Total RNA was harvested from three independent samples each of resting and PMA-stimulated JSL1 cells; these samples were pooled; poly(A)+ mRNA was then isolated and used to generate cDNA libraries for RNA-seq analysis (see Materials and Methods). Between 45 and 48 million reads were obtained from the resting and PMA-stimulated samples and were mapped to genomic DNA sequence. Nonmapping reads were then aligned to a reference database comprising a comprehensive set of cDNA/EST-supported splice junction sequences (Fig 1A; Pan et al. 2008). By counting the relative ratios of reads that map uniquely to junctions representing exon inclusion and exon skipping, estimates for the percentage of transcripts with alternative exon inclusion (%In) were derived.
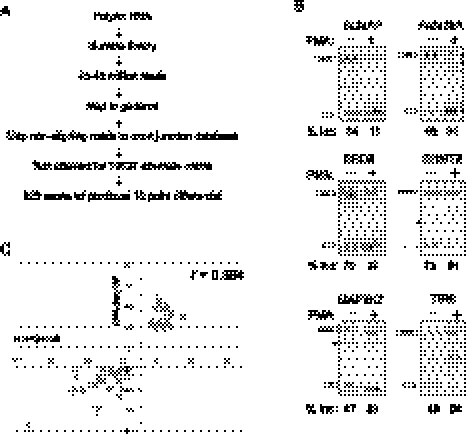
FIGURE 1.
RNA-seq and RT-PCR validation of alternative splicing in resting and PMA-stimulated JSL1 cells. (A) Work flow of RNA-seq analysis of isoform expression in JSL1 cells. (B) Representative RT-PCR of genes newly predicted by RNA-seq analysis to exhibit differential isoform expression in resting (−PMA) versus stimulated (+PMA) JSL1 cells. The percent inclusion of the variable exon shown is an average of at least three independent experiments. (*) Nonreproducible cryptic splice products that are not included in our quantification. (C) Plot of differential in percent exon inclusion in resting versus stimulated cells as calculated by RNA-seq (y-axis) and RT-PCR (x-axis). The Pearson coefficient is calculated as 0.894.
By applying minimal RNA-seq read coverage levels required to derive reliable estimates for alternative splicing levels, %In values could be determined for about 11,000 exons in both the resting and stimulated cell samples (Supplemental Table S1). Approximately 10% (929 of 10,637) of these exons exhibited at least a 10-point change in predicted %In between the resting and stimulated cells (Supplemental Table S1). This rate of predicted differential inclusion is consistent with a previous, smaller-scale analysis in which changes in %In were detected in 5%–10% of profiled alternative exons upon T-cell stimulation (Ip et al. 2007).
To focus on the most pronounced signal-dependent alternative splicing events, we identified a set of 178 exons in 168 genes that have at least a 20-point differential in %In between resting versus stimulated cells (Supplemental Table S1). Among these signal-induced changes in alternative splicing, 90 (51%) of the exons displayed increased inclusion, and 88 (49%) of the exons displayed increased skipping upon PMA stimulation. Notably, this list of “top candidates” captures previously defined and biologically important signal-responsive exons, including all three of the immune-regulated alternative exons in the CD45 gene, and also exons in the FYN, CUGBP2, and MAP4K2 genes (Lynch and Weiss 2000; Rothrock et al. 2003; Ip et al. 2007; Mallory et al. 2011). We next used low-cycle RT-PCR to validate predicted changes in the levels of inclusion of 46 additional exons among the list of 178. These events were chosen at random to represent a range from largest to smallest predicted changes in %In, and included sets of exons that display either increases or decreases in %In upon stimulation. For 32 of these exons, we observe differential %In values between resting and stimulated cells that are largely consistent with the RNA-seq predictions, while an additional five genes exhibited smaller yet statistically significant differential %In values (p < 0.05 for all; paired Student's t-test) (Fig 1B,C; Table 1). Together with the six exons validated in previous studies, the results from the RT-PCR validation experiments confirmed changes in %In levels between resting and stimulated cells for 80% (43/52) of the tested events (Table 1; Supplemental Table S1), with a Pearson correlation coefficient of 0.894 between %In values derived from the RNA-seq and RT-PCR-based measurements (Fig. 1C).
TABLE 1.
Signal-responsive exons in JSL1 cells validated by RT-PCR (n ≥ 3)
As further validation of the RNA-seq predictions, 11 of 12 exons predicted to have no notable change in %In (<5-point differential) between resting and activated JSL1 cells also showed a lack of signal responsiveness by RT-PCR analysis (Supplemental Table S2). Moreover, in general, predictions based on fewer than 20 reads per splice junction, or based on %In changes between resting and stimulated JSL1 cells of <15%, were not supported by RT-PCR results (data not shown). Therefore, we focused subsequent analyses on the set of 178 exons that are associated with a high rate of validation. Given the number and length of the RNA-seq reads analyzed in this study, we estimate that the list of 178 regulated exons we have detected are represented by the top ∼40% most highly expressed genes in JSL1 cells (Blencowe et al. 2009). However, given the unbiased nature of RNA-seq and our subsequent analysis (see below), we anticipate that these 178 alternative splicing events are representative of the larger set of signal-responsive cassette alternative exons in JSL1 cells.
To address whether signal-induced changes in mRNA expression levels may have affected our detection of %In changes, we next compared changes in total mRNA expression with changes in alternative splicing. In our previous, smaller-scale analysis of mRNA expression in JSL1 cells, we determined that upon PMA treatment, a similar percentage of genes exhibited alternative splicing changes as transcriptional changes. However, there was no significant overlap between the sets of genes affected by each process (Ip et al. 2007). In the present study, of 10,374 genes for which we obtained reliable sequence coverage in either resting or stimulated cells (10 or more reads per 50 bp of gene length), 312 exhibit a change of twofold or greater in mRNA expression between resting and stimulated cells (Supplemental Table S3). As observed in previous studies profiling gene expression in T cells (Lin et al. 2003; Ip et al. 2007), we observe that a larger number of genes are up-regulated upon stimulation than are repressed (207 vs. 115). Importantly, further consistent with our previous studies (Ip et al. 2007), none of the 178 differentially spliced exons are located within this set of 312 transcriptionally regulated genes. Therefore, while alternative splicing and transcriptional changes affect a similar percentage of genes, in general, these regulatory processes do not directly impact one another in the context of signal-induced changes in JSL1 cells.
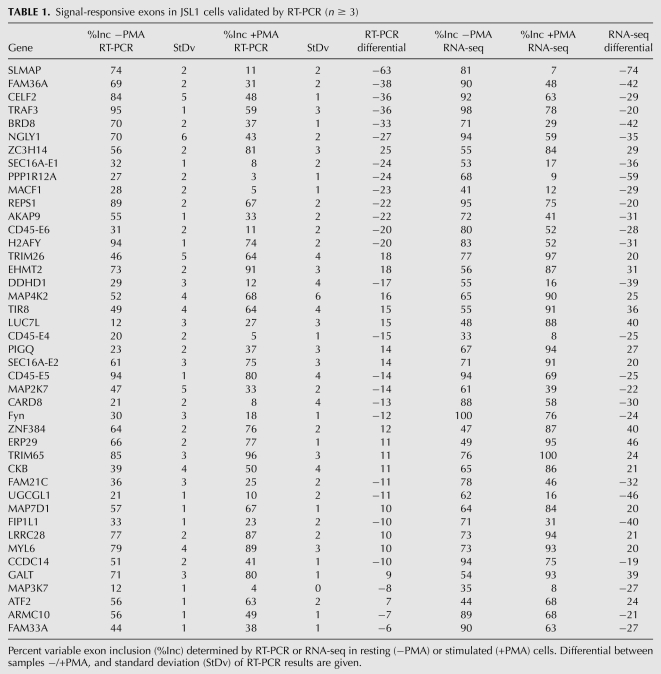
Interestingly, the signal-responsive exons we have identified are significantly enriched in genes involved in many aspects of cellular development and activity relevant to T-cell function, including cell signaling and gene expression, as well as functions that are specifically related to lymphocyte biology such as immune cell trafficking, inflammatory response, immunologic disease and cell-mediated immune response (p < 0.03; Fischer's exact test) (Fig. 2A; Supplemental Table S4A). Importantly, when we score the genes containing the set of 178 exons for involvement in specific signaling pathways, we observe enrichment in many of the signaling cascades that are triggered in response to T-cell activation and that are required for T-cell effector functions, such as NF-κB and JNK and MAP kinase signaling, as well as Rho-mediated changes in cell migration (Fig. 2B; Supplemental Table S4B; Smith-Garvin et al. 2009). Together, these results strongly suggest that alternative splicing likely plays a major role in shaping the T-cell response to stimulation. Moreover, in light of previous work demonstrating important functional roles for a few alternative splicing events in T cells (Jacobsen et al. 2000; Lynch 2004; Mallory et al. 2011), we predict that many of the new examples of regulated exons we identify here also contribute to an optimal immune response.
FIGURE 2.
Genes that undergo signal-responsive alternative splicing in JSL1 cells are enriched for activities required for T-cell function. (A) The 178 exons identified as signal-responsive in this study were analyzed by IPA (Ingenuity Systems; http://www.ingenuity.com) for functional categories. The categories graphed show above-threshold significance [−log(P-value) > 1.5] in enrichment relative to the 11,000 exons that were detectable in the RNA-seq analysis. The numbers above the bar graph represent the number of genes that score for each category. (B) Same as A, but genes were analyzed for their involvement in canonical signaling pathways.
Genes that displayed differential expression at the transcript level were also significantly enriched for functions related to T-cell biology, including almost all of the functional categories enriched among the genes with regulated alternatively splicing events (p < 0.03) (Supplemental Fig S1; Supplemental Table S5A,B). Interestingly, only “RNA trafficking” was identified as a function uniquely enriched among the alternatively spliced genes (Supplemental Fig. S1B). In contrast, transcription appears to regulate a broad spectrum of functions not impacted by alternative splicing, including general functions such as protein synthesis and trafficking, cellular morphology and mobility, and functions related to metabolism (Supplemental Fig. S1B; Supplemental Table S5A,B). Functions enriched among transcripts up-regulated by cell stimulation were largely overlapping those enriched among repressed genes (Supplemental Fig. S1A). Strikingly, however, the pathways enriched among the up-regulated and down-regulated genes are entirely distinct (Supplemental Fig. S1C; Supplemental Table S5C,D). Moreover, only one signaling pathway (NF-κB) includes genes regulated at both the level of transcription and alternative splicing (Supplemental Fig. S1D). Therefore, whereas both transcriptional and splicing regulation have the ability to influence T-cell biology, these mechanisms appear to do so through distinct signaling or functional pathways.
A subset of signal-responsive exons in JSL1 cells have similar behavior in primary T cells
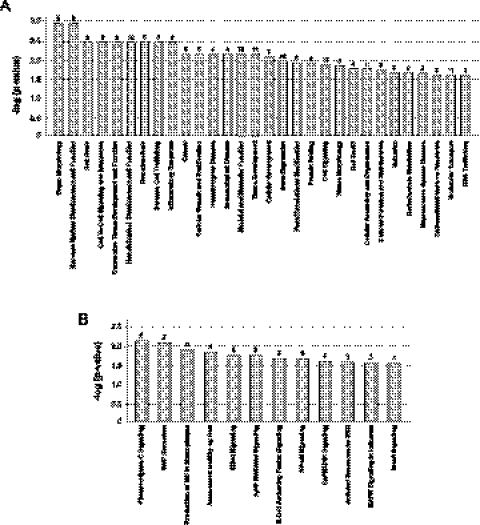
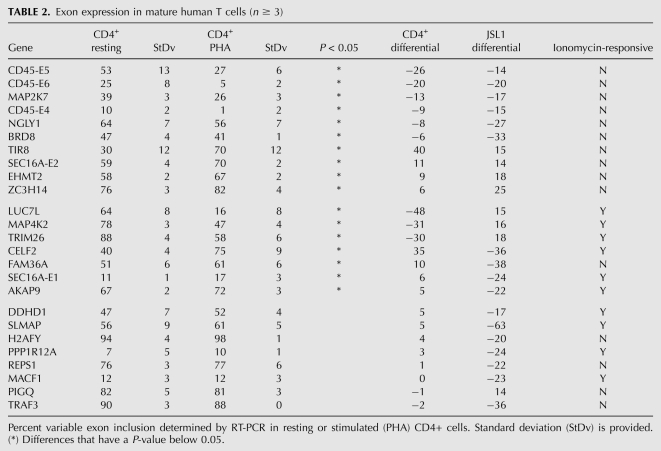
We next asked whether the signal-responsive exons detected in JSL1 cells are also regulated in response to antigen challenge of mature, peripheral T cells. CD4+ peripheral T cells were isolated from multiple donors and stimulated in culture with the lectin PHA, which functions as a surrogate antigen to stimulate the canonical T-cell receptor signaling pathway(s). Alternative splicing changes were then assayed for the top 25 genes in Table 1 by RT-PCR analysis of RNA harvested from resting or stimulated T cells from multiple independent donors. This revealed that 68% (17/25) of the tested exons show a statistically significant change in %In following PHA stimulation, whereas the remaining eight exons did not detectably respond to antigen stimulation (Fig 3A–D; Table 2). Similar results to those observed in the CD4+ T cells were obtained in CD8+ peripheral T cells, and also when either CD4+ or CD8+ T cells were stimulated with anti-CD3/CD28 antibodies instead of PHA (data not shown).
FIGURE 3.
Analysis of activation-induced splicing in CD4+ primary cells relative to JSL1 cells. (A) Graphical representation of the change in percent exon inclusion between resting and PHA-stimulated CD4+ cells (purple bars) compared with the change observed between resting and PMA-stimulated JSL1 cells (gray bars). The percent exon inclusion differential was calculated by RT-PCR and averaged from at least three independent donors (CD4+) and/or experiments (JSL1). Exons were further grouped in one of three categories: (I) responding similarly in both cell types (purple tint), (II) responding in an opposing manner in the two cell types (blue tint), or (III) showing no statistically significant change upon PHA treatment of CD4+ cells. See also Tables 1 and 2 for details. (B–D) Representative RT-PCR of isoform expression in resting (−PHA) versus stimulated (+PHA) CD4+ primary T cells, for genes in categories I (B), II (C), or III (D). The percent inclusion of variable exon shown is an average of at least three independent experiments.
TABLE 2.
Exon expression in mature human T cells (n ≥ 3)
Although the majority (10/17) of the PHA-dependent changes in alternative splicing in CD4+ primary cells were in the same direction (i.e., more or less exon inclusion) as the changes seen upon PMA treatment of JSL1 cells (Fig 3A,B; Table 2), some of the PHA-responsive exons (7/17) showed an opposite change in the PHA-stimulated primary cells compared with the PMA-stimulated JSL1 cells (Fig 3A,C; Table 2). Similar reciprocal changes in alternative splicing between JSL1 and CD4+ cells have been noted in previous studies (Ip et al. 2007; Mallory et al. 2011). The simplest explanation for this differential effect is that PMA triggers multiple distinct signaling pathways in JSL1 cells, some of which mimic canonical T-cell signaling, while others have opposing effects.
A PMA-sensitive pathway in JSL1 cells uniquely mimics TCR-signal-induced alternative splicing in primary T cells
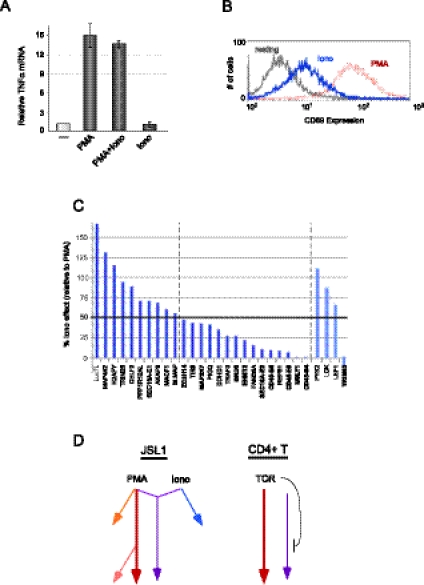
The classical model of T-cell signaling posits that antigen challenge and pre-TCR engagement activate Ras and calcium-dependent signaling pathways that synergize to achieve an optimal response (Smith-Garvin et al. 2009). PMA is thought to primarily trigger the Ras-dependent branch of this signaling cascade, while the calcium ionophore ionomycin induces calcium signaling. However, there is much evidence suggesting overlap or cross talk between these pathways in JSL1 cells. Most conspicuously, PMA alone triggers maximal expression of TNF-α in JSL1 cells with no synergy observed upon the addition of ionomycin (Fig. 4A). This result is striking since transcription from the TNF-α promoter has been well-documented to require Ca2+ signaling along with Ras stimulation (Tsai et al. 1996; Lopez-Rodriguez et al. 2001). Moreover, treatment of JSL1 cells with ionomycin results in at least partial expression of the cell-surface protein CD69 (Fig. 4B), which is traditionally considered a marker of Ras activation (D'Ambrosio et al. 1994). Therefore, PMA and ionomycin stimulation of JSL1 cells does not fit a clear-cut model of independent Ras and Ca2+ signaling, respectively.
FIGURE 4.
Analysis of signaling pathways in JSL1 cells and their effect on alternative splicing. (A) Induction of TNF-α expression in response to PMA and/or ionomycin treatment of JSL1 cells. TNF-α mRNA was measured by RT-PCR, and the average of two experiments is shown. (B) Cell-surface expression of CD69, as measured by flow cytometry, in JSL1 cells grown under resting (gray), PMA stimulated (red), or ionomycin-stimulated (blue) conditions. (C) Analysis of ionomycin-induced alternative splicing of the top 25 PMA-responsive genes. Ionomycin responsiveness was quantified by RT-PCR from at least three independent experiments, and the difference in exon inclusion between resting and Ionomycin-stimulated cells was graphed as a percent of splicing change observed upon PMA stimulation (PMA differential set to 100%). (D) Model of signaling pathways in JSL1 versus CD4 cells as indicated by alternative splicing responsiveness.
Given the apparent unconventional signaling in JSL1 cells, we wished to better delineate the pathways through which the various “PMA-responsive” genes are regulated. In particular, we sought to determine the relative contribution of ionomycin-induced signaling in the alternative splicing events we have identified. Notably, we find that 10 of the top 25 PMA-responsive genes show at least 50% of the maximal PMA response when stimulated with ionomycin alone (Fig. 4C, leftmost 10 genes; Table 2). In contrast, the other 15 genes did not display statistically significant or robust changes in alternative splicing upon ionomycin treatment (Fig. 4C, gene names between dotted lines). In no case did we observe any synergy following costimulation with PMA and ionomycin (data not shown). Therefore, we conclude that there are at least two distinct signaling pathways leading from PMA stimulation to alternative splicing, one of which is triggered by both PMA and ionomycin and another that is responsive only to PMA (Fig. 4D, purple and maroon arrows, respectively).
To ask whether differential signaling pathways might explain the distinct classes of alternative splicing response patterns observed when analyzing different cell types, we directly compared the ionomycin-response data with the data from CD4+ T cells. Strikingly, we observe that all (10/10) of the exons displaying the same directionality of change in splicing upon stimulation of CD4+ and JSL1 cells are responsive only to PMA, while six of the seven genes that had an opposite splicing pattern in CD4+ and JSL1 cells are also responsive to ionomycin. To investigate whether this correlation might be predictive, we next assayed three additional genes that we have previously found to be anti-correlated in response to stimulation of JSL1 and CD4+ cells (Lck, LEF1, Pyk2) (Ip et al. 2007). Remarkably, each of these genes displays a change in splicing in response to ionomycin that is similar to the response to PMA in JSL1 cells (Fig. 4C). In contrast, the one additional gene (TRIM65) we have thus far found to be spliced similarly in both PHA-stimulated primary cells and PMA-stimulated JSL1s, is entirely unresponsive to ionomycin (Fig. 4C).
Taken together, these results suggest that for 40% (10/25) of the genes initially characterized as “PMA-responsive” in JSL1 cells, the signaling pathway that leads to splicing modulation overlaps with an ionomycin-triggered cascade (Fig. 4D). Moreover, this ionomycin-triggered or “overlapping” pathway in JSL1 cells is distinct from signaling cascades triggered by antigen stimulation of CD4+ cells, and in some cases is even counter to the effects of TCR signaling, at least in terms of alternative splicing regulation (Fig. 4D). In contrast, we conclude that there is a PMA-induced pathway in JSL1 cells that mimics, at least at the level of alternative splicing, antigen stimulation of mature peripheral T cells. Such a model is further consistent with the fact that we observe full TNF-α expression with PMA alone, demonstrating that PMA is sufficient in JSL1 cells to induce at least some transcriptional events that typically require both a Ras and a Ca2+ signal. However, we note that not all PMA-only-induced alternative splicing events in JSL1s are seen in CD4+ T cells, suggesting that at least one PMA-induced mechanism in JSL1 cells is not functional in primary T cells (Fig. 3D; Table 2). This could be due to the involvement of a completely separate pathway or to the absence of one or more factors that are specifically required for a branch of the main PMA-only pathway (Fig. 4D, orange or pink arrows, respectively).
Distinct sequences correlate with discrete signal-responsive categories of alternative splicing
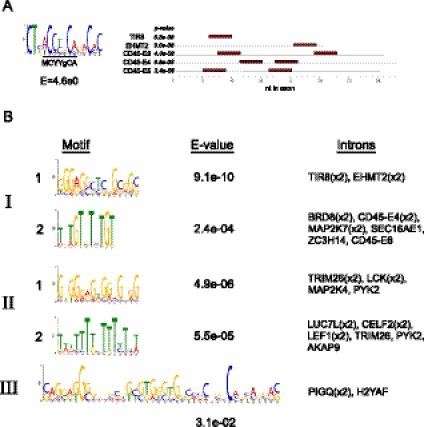
Finally, to begin to address whether the different sets of signal-induced alternative splicing events detected in our transcriptomic analysis are regulated by distinct factors, we investigated whether they are significantly enriched in specific sequence motifs indicative of binding sites of splicing regulators. An implication of the model in Figure 4D is that exons that are regulated by a common signaling pathway (i.e., one of the colored arrows in Fig. 4D) are likely regulated by similar or overlapping mechanisms. As most alternative splicing relies on the binding of regulatory proteins to cis-acting control sequences (Chen and Manley 2009), and in light of previous observations that coregulated sets of alternative exons tend to be regulated by one or more common regulatory factors, we reasoned that exons that fell into a given regulatory category might share specific regulatory motifs. We therefore queried each of the three categories of genes in Table 2 using the motif finder MEME (Bailey et al. 2009) to search for sequences that are significantly enriched in the regulated exons or their flanking intron sequences (see Materials and Methods). For the exons, we only found a single statistically enriched motif among the “category I” exons regulated similarly in JSL1 and CD4 cells (Fig. 5A). Strikingly, this enriched motif identified by MEME encompasses the “activation-response sequence” [ARS = (A/C)YYGCA] that we have previously demonstrated to be critical for the regulation of each of the three CD45 variable exons (Rothrock et al. 2003; Motta-Mena et al. 2011). MEME indeed identifies two copies of the enriched motif in each of the CD45 variable exons, as we have previously identified (Rothrock et al. 2003), and also identifies previously unrecognized occurrences of this sequence motif in two additional exons in category I (Fig. 5A). This motif is not found in categories II or III, nor is it enriched in the validated “nonresponsive” exons.
FIGURE 5.
Motif enrichment among signal-responsive alternative exons. (A) Logos of motif identified as enriched among category I exons and e-value of the enrichment (left) along with the location and P-value of the motif in each of the five exons in which it was identified (right). (B) Logos and corresponding e-values of the five motifs identified within the flanking introns of exons from categories I, II, and III, along with the names of the genes in which each motif is found. Detail of motifs and locations are provided in Supplemental Figure 1.
In contrast to these results, we observe significantly enriched sequence motifs within intronic sequences flanking the regulated exons belonging to each of the three categories (Fig. 5B; Supplemental Fig. S2). For categories I and II, two distinct sequence motifs are enriched. Matches to these motifs were found in the intron both upstream of and downstream from the variable exons. We also were able to identify a single motif in category III; however, this motif is present in only two of the eight genes in the category and has a relatively low e-value. Notably, the sequence motifs flanking the signal-responsive exons are similar but not identical between the three categories, suggesting functional differences between the regulatory factors that mediate signal-induced alternative splicing of these sets of genes. This observation is consistent with the distinct signaling requirements for the regulation of alternative exons belonging to classes I, II, and III. In sum, we conclude that there are likely multiple, distinct connections between signaling pathways and the regulatory factors that control splicing in T cells.
DISCUSSION
Alternative splicing is an important but under-investigated mechanism of regulation of T-cell function during immune responses. In this study, we use RNA-seq to identify genes that express distinct isoforms in response to stimulation of a human T-cell line. We identified 178 exons in 168 genes that are subject to signal-induced alternative splicing. The vast majority of these genes also exhibit differential isoform expression in naive and activated primary T cells.
Importantly, only a few of the 178 exons belong to genes previously described to undergo signal-induced alternative splicing in T cells or in any other system. For example, only 25 of these exons were among the high-confidence set of alternative splicing level predictions generated by our previous quantitative alternative splicing microarray profiling study (Ip et al. 2007). Conversely, many of the signal-induced alternative splicing changes predicted to occur based on our previous microarray profiling study were not detected by RNA-seq analysis. This can be attributed to the observations that a substantial number of the microarray-profiled exons lack sufficient read coverage required to generate reliable predictions for splicing changes, that the microarray platform is associated with a higher false-positive detection rate for splicing changes than is RNA-seq analysis, and possibly also because of false-negative detection in the RNA-seq analysis (data not shown). Thus, while both systems are capable of generating reliable predictions for alternative splicing changes on a large scale, RNA-seq has the main advantage of affording relatively unbiased detection of exons within expressed transcripts and, based on comparisons of relative RT-PCR validation rates (Ip et al. 2007; this study), is generally more quantitative. Regardless, our results in the present study greatly expand the repertoire of known signal-regulated splicing events. Moreover, because of the unbiased nature of RNA-seq analysis, these events are likely to be representative of the total spectrum of alternative exons that are regulated during T-cell activation. This is further reflected by the enrichment of the set of 178 exons in gene function categories that are specifically associated with the immune response.
Strikingly, the 178 exons are significantly enriched in genes involved in mediating critical T-cell effector functions, such as cell signaling, proliferation and development, and immune cell trafficking. For example, MAP2K7 controls the activity of the cJun transcription factor, which is essential for expression of cytokines and cell viability; EHMT2 has been implicated in controlling apoptosis; and TIR8 regulates Toll-like receptor signaling and the inflammation response (Hoffmann et al. 2008; Drexler et al. 2010; Huang et al. 2010; Wang et al. 2011). Therefore, although the precise functional impact of most of the splicing events we have uncovered remains unexplored, alternative splicing is predicted to play a significant role in shaping the ability of T cells to mount a robust and appropriate immune response.
Finally, in addition to providing novel examples of signal-induced alternative splicing, our data reveal important insights into the diversity of signaling pathways that control splicing. Specifically, comparison of the responsiveness of splicing to various stimuli in the cultured versus primary T cells enabled grouping of the signal-responsive exons into at least three distinct networks. Importantly, we find that each regulatory network is characterized by distinct sequence features, further suggesting independent regulatory mechanisms.
While directly testing the functionality of any of the intronic motifs in Figure 5 is beyond the scope of this study, the relevance of our motif analysis is strongly suggested by the unbiased identification of the known ARS regulatory motif among the category I genes. We have previously shown that the ARS motif functions by recruiting hnRNP L, LL, and PSF to regulate the category I exons in the CD45 gene (Rothrock et al. 2005; Tong et al. 2005; Melton et al. 2007; Topp et al. 2008; Motta-Mena et al. 2010). Our identification of the ARS motif in EHMT2 and TIR8 suggests that hnRNP L, LL, and/or PSF may also regulate at least a subset of additional category I exons. Interestingly, however, a G-rich motif was also identified in the introns of EHMT2 and TIR8, implying potential cross talk between signaling pathways and/or regulatory factors in mediating the final outcome of splicing of these genes. Moreover, the fact that the ARS motif was not enriched in exons outside category I underscores the likelihood that the category II and III exons are regulated by pathways and factors other than those described for the ARS-containing CD45 gene. Further identification of the regulatory factors will be required to fully delineate the signaling networks that control diverse alternative splicing events; however, our studies clearly demonstrate the presence of distinct regulatory pathways. Therefore, in summary, the data presented in this study provide a valuable basis for the future characterization of the function and mechanisms of alternative splicing networks controlled by distinct signal-induced pathways during T-cell stimulation.
MATERIALS AND METHODS
Cell culture
JSL1 cells (Lynch and Weiss 2000) were cultured in RPMI supplemented with 5% heat-inactivated fetal bovine serum (FBS). For stimulations, JSL1 cells were cultured with PMA (10 ng/mL; Sigma-Aldrich), ionomycin (1 μM; Sigma-Aldrich) or both for 60 h. DMSO was used as a vehicle control in all experiments.
CD4+ peripheral blood T cells were obtained from the University of Pennsylvania Human Immunology Core (IRB protocol #811028) with an average purity of 90%–95%. CD4+ cells were maintained in RPMI supplemented with 10% heat-inactivated FBS. CD4+ cells were collected from six donors and separately cultured under resting or stimulated (PHA; 10 μg/mL, Sigma-Aldrich L9017) conditions for 60 h.
RNA sequencing and analysis
Human UniGene sequences (build #208) and human genomic sequences (build #36) were downloaded from NCBI (ftp://ftp.ncbi.nih.gov/repository/UniGene/Homo_sapiens/, ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/). RefSeq data were downloaded from UCSC (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/database/refGene.txt.gz). Cassette alternative splicing events (n = 27,240) were mined from aligning EST/cDNA sequences to genomic sequence essentially as described previously (Pan et al. 2004, 2005; Saltzman et al. 2011).
The RNA-seq reads analyzed were 50-mers. For each splice site junction sequence aligned to sequence reads, a 90-nt-long sequence was extracted with 45 nt of overlap in each upstream and downstream exon. RNA-seq reads were mapped to exon–exon junction sequences, and the mapped reads were filtered as previously described (Pan et al. 2008). Percent exon inclusion values (%In) were calculated by counting the number of reads mapping uniquely (i.e., not mapping to another splice junction or genomic sequence) to the three junctions formed by alternative splicing of a single cassette skipping event. Specifically, the ratio of the average count for reads mapping to the two splice junctions formed by exon inclusion (C1:A, A:C2), over the total count for reads mapping both to the included exon junctions (C1:A, A:C2) and to the skipped exon junction (C1:C2), was determined. Percent exon inclusion values predicted for cassette AS events represented by 20 or more reads to at least one of these three junctions were shown to have a high correlation (r = 0.80) with independent AS microarray and RT-PCR measurements (Pan et al. 2008; Saltzman et al. 2011), and thus were considered high-confidence RNA-seq predictions for %In levels.
For gene expression measurements, reads uniquely mapping to representative RefSeq cDNAs were counted, and genes were filtered for those with 10 or more reads per 50 bp, which approximates the threshold for read coverage we used for the exon junction analysis. Reads per kilobase of cDNA per million reads (RPKM) values for each gene were determined. Ratios of RPKM values in resting and stimulated cells were then determined, and genes with a log base 2 ratio >2, or <−2, were considered to be genes highly regulated at the level of expression upon stimulation.
RT-PCR analysis
RNA was isolated using RNABee (Tel-Test, Inc.) according to the manufacturer's protocol. Reverse transcription-PCR (RT-PCR) was performed as previously described in detail (Lynch and Weiss 2000; Rothrock et al. 2003; Ip et al. 2007), using sequence-specific primers for individual genes and a cycle number chosen to provide a signal that is linear with respect to input RNA. Primer sequences for all genes analyzed are given in Supplemental Table S6. Quantification was done by densitometry using a Typhoon PhosphorImager. The percent inclusion was calculated as the percent of isoforms including the variable exon over total isoforms. A paired Student's t-test analysis was performed for all treatment groups relative to resting cells in JSL1 cells and CD4+ peripheral blood T lymphocytes using Microsoft Excel. Quantification of TNF-α gene expression was also determined by RT-PCR and normalized to GAPDH expression.
Ingenuity pathway analysis
Functional and canonical signaling pathway analyses were performed with Ingenuity Pathway Analysis software (Ingenuity Systems, http://www.ingenuity.com). For analysis of alternative splicing events, the approximately 11,000 genes that met our threshold of junction coverage by RNA-seq (Supplemental Table S1) were used as the reference set. The test set was the 178 genes that exhibited a ≥20 point differential in percent inclusion. For the gene expression data, we similarly compared the 312 genes that exhibited a ratio of change >2 upon stimulation [log(resting/stimulated) ≥ |2|] to the complete set of 10,374 genes with measurable expression in at least one of the data sets (Supplemental Table S3). Annotations with a −log(P-value) > 1.5 calculated by a Fischer's exact test were selected for functional analysis and canonical signaling pathway analysis.
CD69 cell-surface expression
JSL1 cells were stimulated under indicated conditions. 1 × 106 cells per condition were harvested and incubated with anti-CD69 antibody (BD Pharmingen 560968) for 15 min on ice. Cells were washed and resuspended in PBS supplemented with 1% FBS and analyzed by flow cytometry.
Motif analysis
For exons in each of the three categories, sequences of the alternative exons and the flanking 200 nt of intronic sequence were extracted from the human genome (NCBI36/hg18). These regions were scanned for enriched motifs using MEME v4.7.0 (Bailey et al. 2009), allowing for any number of repetitions of elicited motifs within the input sequences. Enriched motifs with an e-value below 5E0 were considered significant and are shown in Figure 5.
DATA DEPOSITION
RNA sequencing data have been deposited in GEO (GSE35871).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank members of the Lynch laboratory for advice and comments on this manuscript. This work was supported by R01 GM084034 to K.W.L. and by CIHR Operating Grants to B.J.B. N.M.M. was supported by GM084034S1, and C.Y. was supported by T32-GM07229.
Author contributions: F.H. prepared the RNAs for analysis; Q.P. and B.J.B. analyzed the RNA-seq data; N.M., C.Y., G.B., W.Z., S.A., and F.H. performed the RT-PCR validations in JSL1 cells and analyzed data; N.M. performed all experiments with the primary cells; and B.C. performed the motif analysis. K.W.L. directed the research, analyzed data, and wrote the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.032243.112.
REFERENCES
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS 2009. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurel E, Michalek SM, Jope RS 2010. Innate and adaptive immune responses regulated by glycogen synthase kinase-3 (GSK3). Trends Immunol 31: 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Ahmad S, Lee LJ 2009. Current-generation high-throughput sequencing: Deepening insights into mammalian transcriptomes. Genes Dev 23: 1379–1386 [DOI] [PubMed] [Google Scholar]
- Chen M, Manley JL 2009. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Natl Rev 10: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR 2010. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci 107: 1894–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ambrosio D, Cantrell DA, Frati L, Santoni A, Testi R 1994. Involvement of p21ras activation in T cell CD69 expression. Eur J Immunol 24: 616–620 [DOI] [PubMed] [Google Scholar]
- David CJ, Manley JL 2010. Alternative pre-mRNA splicing regulation in cancer: Pathways and programs unhinged. Genes Dev 24: 2343–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler SK, Kong P, Inglis J, Williams RO, Garlanda C, Mantovani A, Yazdi AS, Brennan F, Feldmann M, Foxwell BM 2010. SIGIRR/TIR-8 is an inhibitor of Toll-like receptor signaling in primary human cells and regulates inflammation in models of rheumatoid arthritis. Arthritis Rheum 62: 2249–2261 [DOI] [PubMed] [Google Scholar]
- Goehe RW, Shultz JC, Murudkar C, Usanovic S, Lamour NF, Massey DH, Zhang L, Camidge DR, Shay JW, Minna JD, et al. 2010. hnRNP L regulates the tumorigenic capacity of lung cancer xenografts in mice via caspase-9 pre-mRNA processing. J Clin Invest 120: 3923–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann B, Castelo R, Blanchette M, Boue S, Rio DC, Valcarcel J 2009. Global analysis of alternative splicing regulation by insulin and wingless signaling in Drosophila cells. Genome Biol 10: R11 doi: 10.1186/gb-2009-10-1-r11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermiston ML, Xu Z, Majeti R, Weiss A 2002. Reciprocal regulation of lymphocyte activation by tyrosine kinases and phosphatases. J Clin Invest 109: 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E, Ashouri J, Wolter S, Doerrie A, Dittrich-Breiholz O, Schneider H, Wagner EF, Troppmair J, Mackman N, Kracht M 2008. Transcriptional regulation of EGR-1 by the interleukin-1-JNK-MKK7-c-Jun pathway. J Biol Chem 283: 12120–12128 [DOI] [PubMed] [Google Scholar]
- Huang J, Dorsey J, Chuikov S, Perez-Burgos L, Zhang X, Jenuwein T, Reinberg D, Berger SL 2010. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem 285: 9636–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Tong A, Pan Q, Topp JD, Blencowe BJ, Lynch KW 2007. Global analysis of alternative splicing during T-cell activation. RNA 13: 563–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen M, Schweer D, Ziegler A, Gaber R, Schock S, Schwinzer R, Wonigeit K, Lindert RB, Kantarci O, Hemmer B, et al. 2000. A point mutation in PTPRC is associated with the development of multiple sclerosis. Nat Genet 26: 495–499 [DOI] [PubMed] [Google Scholar]
- Jacobsen M, Hoffmann S, Cepok S, Stei S, Ziegler A, Sommer N, Hemmer B 2002. A novel mutation in PTPRC interferes with splicing and alters the structure of the human CD45 molecule. Immunogenetics 54: 158–163 [DOI] [PubMed] [Google Scholar]
- Lee JA, Xing Y, Nguyen D, Xie J, Lee CJ, Black DL 2007. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biol 5: e40 doi: 10.1371/journal.pbio.0050040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Fillmore GC, Um TH, Elenitoba-Johnson KS, Lim MS 2003. Comparative microarray analysis of gene expression during activation of human peripheral blood T cells and leukemic Jurkat T cells. Lab Invest 83: 765–776 [DOI] [PubMed] [Google Scholar]
- Lopez-Rodriguez C, Aramburu J, Jin L, Rakeman AS, Michino M, Rao A 2001. Bridging the NFAT and NF-κB families: NFAT5 dimerization regulates cytokine gene transcription in response to osmotic stress. Immunity 15: 47–58 [DOI] [PubMed] [Google Scholar]
- Lynch KW 2004. Consequences of regulated pre-mRNA splicing in the immune system. Nat Rev Immunol 4: 931–940 [DOI] [PubMed] [Google Scholar]
- Lynch KW, Weiss A 2000. A model system for the activation-induced alternative-splicing of CD45 implicates protein kinase C and Ras. Mol Cell Biol 20: 70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory MJ, Jackson J, Weber B, Chi A, Heyd F, Lynch KW 2011. Signal- and development-dependent alternative splicing of LEF1 in T cells is controlled by CELF2. Mol Cell Biol 31: 2184–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton AA, Jackson J, Wang J, Lynch KW 2007. Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Mol Cell Biol 27: 6972–6984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta-Mena LB, Heyd F, Lynch KW 2010. Context-dependent regulatory mechanism of the splicing factor hnRNP L. Mol Cell 29: 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta-Mena LB, Smith SA, Mallory MJ, Jackson J, Wang J, Lynch KW 2011. A disease-associated polymorphism alters splicing of the human CD45 phosphatase gene by disrupting combinatorial repression by heterogeneous nuclear ribonucleoproteins (hnRNPs). J Biol Chem 286: 20043–20053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Tanaka H, Naoe Y, Taniuchi I 2011. Transcriptional control of T-cell development. Int Immunol 23: 661–668 [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, et al. 2004. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell 16: 929–941 [DOI] [PubMed] [Google Scholar]
- Pan Q, Bakowski MA, Morris Q, Zhang W, Frey BJ, Hughes TR, Blencowe BJ 2005. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet 21: 73–77 [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Rothrock C, Cannon B, Hahm B, Lynch KW 2003. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol Cell 12: 1317–1324 [DOI] [PubMed] [Google Scholar]
- Rothrock CR, House AE, Lynch KW 2005. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J 24: 2792–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Pan Q, Blencowe BJ 2011. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev 25: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Garvin JE, Koretzky GA, Jordan MS 2009. T cell activation. Annu Rev Immunol 27: 591–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A, Nguyen J, Lynch KW 2005. Differential expression of CD45 isoforms is controlled by the combined activity of basal and inducible splicing-regulatory elements in each of the variable exons. J Biol Chem 280: 38297–38304 [DOI] [PubMed] [Google Scholar]
- Topp JD, Jackson J, Melton AA, Lynch KW 2008. A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. RNA 14: 2038–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai EY, Yie J, Thanos D, Goldfeld AE 1996. Cell-type-specific regulation of the human tumor necrosis factor α gene in B cells and T cells by NFATp and ATF-2/JUN. Mol Cell Biol 16: 5232–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tang R, Lv M, Wang Q, Zhang X, Guo Y, Chang H, Qiao C, Xiao H, Li X, et al. 2011. Defective anchoring of JNK1 in the cytoplasm by MKK7 in Jurkat cells is associated with resistance to Fas-mediated apoptosis. Mol Biol Cell 22: 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]