During oocyte development, the cytoplasmic polyadenylation element-binding protein (CPEB) nucleates a set of factors on mRNA that controls cytoplasmic polyadenylation and translation. Because the precise stoichiometry between CPEB and CPE-containing RNA is responsible for the temporal order of mRNA polyadenylation during meiosis, the authors hypothesized that, if CPEB production exceeded the amount required to bind mRNA, the excess would be sequestered in an inactive form. They demonstrate that not only does CPEB form a dimer, but dimerization requires its RNA-binding domains. Dimer formation prevents CPEB from being UV cross-linked to RNA, which establishes a second pool of CPEB that is inert for polyadenylation and translational control. They propose that the balance between CPEB and its target mRNAs is maintained by CPEB dimerization, which inactivates spare proteins and prevents them from inducing polyadenylation of RNAs with low affinity binding sites. In addition, the dimers might serve as molecular hubs that release polyadenylation factors for translational activation upon CPEB dimer destruction.
Keywords: CPEB, polyadenylation, translation
Abstract
During oocyte development, the cytoplasmic polyadenylation element-binding protein (CPEB) nucleates a set of factors on mRNA that controls cytoplasmic polyadenylation and translation. The regulation of polyadenylation is mediated in part through serial phosphorylations of CPEB, which control both the dynamic integrity of the cytoplasmic polyadenylation apparatus and CPEB stability, events necessary for meiotic progression. Because the precise stoichiometry between CPEB and CPE-containing RNA is responsible for the temporal order of mRNA polyadenylation during meiosis, we hypothesized that, if CPEB production exceeded the amount required to bind mRNA, the excess would be sequestered in an inactive form. One attractive possibility for the sequestration is protein dimerization. We demonstrate that not only does CPEB form a dimer, but dimerization requires its RNA-binding domains. Dimer formation prevents CPEB from being UV cross-linked to RNA, which establishes a second pool of CPEB that is inert for polyadenylation and translational control. During oocyte maturation, the dimers are degraded much more rapidly than the CPEB monomers, due to their greater affinity for polo-like kinase 1 (plx1) and the ubiquitin E3 ligase β-TrCP. Because dimeric CPEB also binds cytoplasmic polyadenylation factors with greater affinity than monomeric CPEB, it may act as a hub or reservoir for the polyadenylation machinery. We propose that the balance between CPEB and its target mRNAs is maintained by CPEB dimerization, which inactivates spare proteins and prevents them from inducing polyadenylation of RNAs with low affinity binding sites. In addition, the dimers might serve as molecular hubs that release polyadenylation factors for translational activation upon CPEB dimer destruction.
INTRODUCTION
In early development, many maternal mRNAs are translationally repressed in oocytes that are arrested at the end of meiosis prophase I. In response to a hormonal cue, the oocytes reenter the meiotic divisions (indicated by germinal vesicle break down or GVBD) and commensurately activate the dormant mRNAs. A key regulator for both translational activation and meiotic progression is the cytoplasmic polyadenylation element-binding protein CPEB, which binds cytoplasmic polyadenylation elements (CPEs), U-rich structures (U4–6A1–2U1–2) in the 3′ untranslated regions (UTRs) of responding mRNAs (Richter 2007). CPEB not only binds RNA, but also recruits both the poly(A)-specific ribonuclease PARN and the noncanonical poly(A) polymerase Gld2 (defective in germline development 2) to the ribonucleoprotein (RNP) complex. PARN and Gld2 are both active yet their antagonistic activities result in short poly(A) tails and translational quiescence (Kim and Richter 2006). Another group of factors that interacts with the CPEB-containing complex includes symplekin, cleavage and polyadenylation specific factor (CPSF) complex, maskin, and eukaryotic initiation factor eIF4E (Richter 2007). Maskin binds eIF4E and inhibits the assembly of the initiation complex on the 5′ cap (Stebbins-Boaz et al. 1999; Cao et al. 2006). Upon reentry into meiosis, a signaling cascade initiated by progesterone binding to a surface-associated receptor results in activation of the kinase aurora A (Mendez et al. 2000a), which in turn phosphorylates CPEB. Phosphorylated CPEB induces the expulsion of PARN from the RNP (Kim and Richter 2006) and enhances the association with CPSF (Mendez et al. 2000b), leading to Gld2-catalyzed polyadenylation and translation (Barnard et al. 2004; Kim and Richter 2006).
In addition to polyadenylation-induced translation, progesterone also stimulates activation of cyclin-dependent kinase 1 (cdk1)/RINGO (rapid inducer of G2/M progression in oocytes) complex, which hyper-phosphorylates CPEB and elicits (i) binding of the embryonic poly(A)-binding proteins (ePAB) to the poly(A) tail to stabilize the tail (Kim and Richter 2007) and (ii) partial destruction of CPEB (Mendez et al. 2002). CPEB possesses a TSG motif (190TSGFSS195) in the PEST domain (a proline, glutamic acid, serine, and threonine-rich sequence typical of short-lived proteins) that resembles the binding site of the F-box protein of ubiquitin E3 ligase complex, β-transducin repeat-containing protein (β-TrCP). (The conventional β-TrCP binding site is a doubly phosphorylated DSG motif, DpSGΦXpS, where Φ represents a hydrophobic amino acid and X represents any amino acid [Fuchs et al. 2004].) A polo-like kinase homolog plx1 recognizes one of the cdk1 phosphorylation sites on CPEB and in turn phosphorylates the TSG motif, which then recruits β-TrCP for proteosome-mediated protein destruction (Setoyama et al. 2007). CPEB is therefore largely degraded when the oocytes undergo the prophase I to metaphase I transition (i.e., oocyte maturation), and remains relatively low in amount throughout the early embryonic stages. Paradoxically, the low level of CPEB is indispensible for the late maturation phase of CPEB-mediated translational activation. That is, if nondegradable CPEB mutants are introduced into the cell, it cannot proceed to meiosis II (Mendez et al. 2002), suggesting that a low CPEB to CPE ratio is critical to activate the translation of mRNAs for progression to meiosis II.
Protein dimerization or multimerization can be a regulatory mechanism to increase the diversity of catalytic dynamics without increasing the genetic burden. Several RNA-binding proteins are reported to form homo-dimers or multimers, including FMRP (fragile X mental retardation protein) (Adinolfi et al. 2003), quaking (Chen and Richard 1998), staufen 1 (Martel et al. 2010), ELAV (embryonic lethal abnormal visual) family proteins (Toba and White 2008), and She2p (Muller et al. 2009). Dimerization often involves heavily structured domains, among which are RNA-binding domains. A number of reports suggest that protein dimerization or multimerization is required for RNA binding, interaction with other proteins, translational regulation, or localization (Faller et al. 2007; Muller et al. 2009). These studies indicate that the α-helices of the RNA recognition motifs (RRMs) could form the basis of dimer formation yet still not interfere with the RNA binding that takes place on the opposite surface of β-sheet (Jang et al. 2006). In contrast, other investigators find that only the monomeric forms of RNA-binding proteins can bind RNA (Cole et al. 1993), and that the RRMs are used for protein–protein recognition instead of RNA recognition (Fribourg et al. 2003; Yao et al. 2007). Based on these studies, dimerization/multimerization is a common feature that introduces regulatory complexity, but the biochemical result, to bind or not bind RNA, may be protein-dependent.
Aplysia CPEB has been reported to harbor prion-like properties, that is, the ability to form amyloidogenic self-sustaining multimers, which is mediated by neuronal activity (Si et al. 2010). A Q-rich stretch (66 Q's from amino acid 53–139) at the N terminus is essential for the multimerization; however, vertebrate CPEB proteins contain no such Q-rich sequences. Instead, the C terminal half of all CPEB proteins is heavily structured, including two RRMs and two zinc fingers (ZFs), all of which are necessary for efficient RNA binding (Hake et al. 1998). Here, we provide evidence that CPEB forms dimers through the RNA-binding domains, which abrogates RNA binding; moreover, this dimerization is regulated during meiosis. We show that CPEB dimers have a strong affinity for polyadenylation factors and the protein destruction machinery, and thus may serve as a molecular reservoir in which its degradation may release components of the polyadenylation apparatus for translational activation. Finally, we demonstrate that excess CPEB monomers, but not dimers, are deleterious to meiotic progression. We hypothesize that CPEB dimers serve two purposes: to fine-tune the amount of CPEB that is required for oocyte maturation, and to act as a hub for polyadenylation factors that are released during maturation when the dimer is destroyed.
RESULTS
CPEB forms a dimer in oocytes in a cell cycle-dependent manner
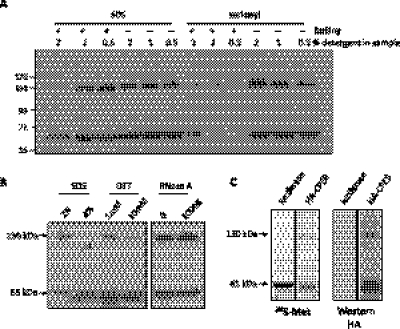
We noticed that Western blots of oocyte extracts probed for CPEB would sometimes yield two species of immunoreactivity, a fast migrating form that corresponded to the size of full-length CPEB (∼65 kDa) and a slower form that was double in size (∼130 kDa); these two species were particularly evident if the protein loading buffer contained reduced amounts of SDS (≤1%). Figure 1A shows this relationship over a concentration range of SDS in the loading buffer with and without boiling the sample. With the standard 2% SDS and boiling the sample, almost all the CPEB immunoreactivity had the expected mobility of ∼65 kDa. With reduced SDS or the elimination of sample boiling, the 130-kDa species became readily apparent. Substitution of the lysis buffer SDS with sarkosyl, a mild detergent, also yielded the two species (sample boiling with sarkosyl caused most of the protein to remain in the well). Treatment of the samples with double the amount (4%) of SDS reduced the 130-kDa CPEB immunoreactive species (Fig. 1B, left), suggesting that the 130-kDa band was not a result of nonspecific cross-reactivity. Further treatment of the samples with dithiothreitol (DTT, without β-mercaptoethanol) or RNase indicated that the 130-kDa CPEB immunoreactive species was stabilized by disulfide bonds but did not require RNA for the reduced electrophoretic mobility (Fig. 1B, middle and right). Finally, to determine whether the 130-kDa species was specific for the oocyte or involved proteins other than CPEB, a rabbit reticulocyte lysate was primed with mRNA encoding either luciferase or HA-tagged CPEB and supplemented with 35S-methionine. Figure 1C shows that the 65- and 130-kDa species were both evident when CPEB RNA was translated in the extract, which was further confirmed by Western blotting with the HA antibody. These results suggest that the 130-kDa species contains CPEB and has the electrophoretic migration expected of a dimer.
FIGURE 1.
CPEB migrates at 65 kDa and 130 kDa in a mild denaturing SDS-PAGE. (A) CPEB immunoreactive 130- and 65-kDa bands were observed when oocyte lysates were prepared with low SDS-containing sample buffer, with sarkosyl instead of SDS, or when the sample was not boiled. (B) Oocyte lysates were prepared with various amounts of SDS, sulfhydryl reducing agent, or RNase to characterize the 130-kDa CPEB immunoreactive band. Left two lanes: The 130-kDa band was sensitive to higher concentrations of SDS, suggesting that it is composed of two or more polypeptides. Middle two lanes: The 130-kDa band was sensitive to the DTT sulfhydryl reducing agent, suggesting that a disulfide bond is involved in protein stabilization. Right two lanes: The 130-kDa band was not sensitive to RNase treatment, suggesting that its formation is RNA-independent. (C) mRNAs encoding HA-CPEB and luciferase were translated in a rabbit reticulocyte lysate supplemented with 35S-methionine. The lysate was then directly applied to SDS-PAGE and visualized by both autoradiography and Western blotting. HA-CPEB migrates as two major species, one at 130 kDa and the other at 65 kDa.
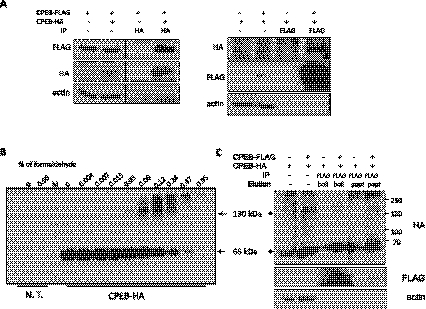
To determine whether the 130-kDa band is indeed a CPEB dimer, human embryonic kidney (HEK) 293T cells were co-transfected with constructs encoding CPEB-FLAG and CPEB-HA, which was followed by FLAG or HA immunoprecipitation (IP) and Western blotting for both epitopes. Figure 2A shows that IP with HA or FLAG antibody precipitated CPEB with both epitopes, indicating that the CPEB proteins interacted. In addition, treatment of CPEB-HA expressing HEK 293T cells with twofold increasing concentrations of formaldehyde followed by Western blotting for HA shows that high molecular weight species of CPEB corresponding in size to that of a predicted dimer gradually formed. Large aggregates were apparent at the higher formaldehyde concentrations (Fig. 2B). Similar formaldehyde cross-linking experiments were carried out with cells co-expressing CPEB-FLAG and CPEB-HA; again, dimer size CPEB-HA was co-immunoprecipitated with FLAG antibody even under very stringent washing conditions (Fig. 2C, also see Materials and Methods), suggesting that CPEB-FLAG and CPEB-HA were in close physical proximity to be covalently cross-linked. Taken together, the data in Figure 2 demonstrate that CPEB can dimerize.
FIGURE 2.
CPEB forms a dimer in mammalian cells. (A) CPEB-HA and CPEB-FLAG were co-expressed in HEK 293T cells followed by reciprocal co-immunoprecipitation (IP) with HA or FLAG antibodies and Western blotting with the same antibodies. These co-IPs demonstrate that CPEB forms a dimer. Actin served as an IP and loading control. (B) Extracts from CPEB-HA transfected HEK 293T were incubated with increasing amounts of formaldehyde, which was followed by immunoblotting for the HA epitope. Note that, with lower formaldehyde, a 130-kDa CPEB band was evident, which is the size of a CPEB dimer. N.T., nontransfected. (C) CPEB-HA and CPEB-FLAG were co-expressed in HEK 293T cells and cross-linked with 0.12% formaldehyde as in panel B. Stringent IP conditions were applied to minimize possible nonspecific adducts. The precipitate was then subjected to Western blotting with antibody for the HA and FLAG epitopes.
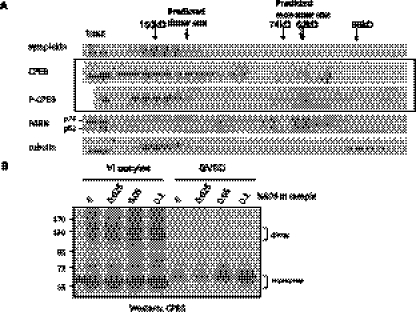
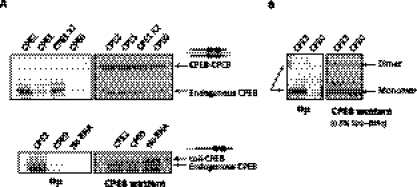
To determine the dimerization status of native CPEB, we performed gel filtration of lysates from dormant stage VI oocytes or mature oocytes that had undergone GVBD during the meiotic divisions. Using symplekin (150 kDa), PARN (74 kDa and 62 kDa), and tubulin (55 kDa) as size markers, Figure 3A shows that CPEB was predominant in high molecular complexes (>150 kDa); CPEB from the dormant oocytes was also evident in fractions that corresponded to dimer, but not monomer, in size. In contrast, CPEB from mature oocytes (P-CPEB), while still mostly in high molecular complexes, was mostly absent from the fractions corresponding to the dimer size but displayed a light peak at the monomer size. These data suggest that the CPEB dimers undergo a rearrangement or destruction during maturation. This possibility is also indicated by the data in Figure 3B, which show that, under conditions of low SDS, dormant oocytes contained both CPEB monomer and dimer as presented previously, but mature oocytes contained only CPEB monomers. These results suggest that CPEB dimerization is regulated during meiotic progression.
FIGURE 3.
Dimeric CPEB undergoes maturation-mediated destabilization in the native condition. (A) To examine the native status of CPEB, clarified lysate from dormant or GVBD oocytes were passed through a size exclusion column. Fractions of eluates were then assessed by Western blotting. Although most of CPEB was in complexes >150 kDa, in the dormant oocyte, there was a population of CPEB fractionating ∼130 kDa, potentially the dimers. In GVBD oocytes (P-CPEB), only a light peak ∼65 kDa, the monomer size, of CPEB could be detected, and the 130-kDa peak was not observed. These data suggest that, in the GVBD cells, CPEB undergoes an alteration such that the free dimeric CPEB is no longer stable. (B) Extracts from stage VI oocytes and GVBD oocytes were assessed by SDS-PAGE with low SDS in the sample buffer. The slow migrating CPEB species ∼130 kDa is evident from stage VI oocytes, but not from GVBD oocytes.
CPEB dimerization requires RNA-binding regions
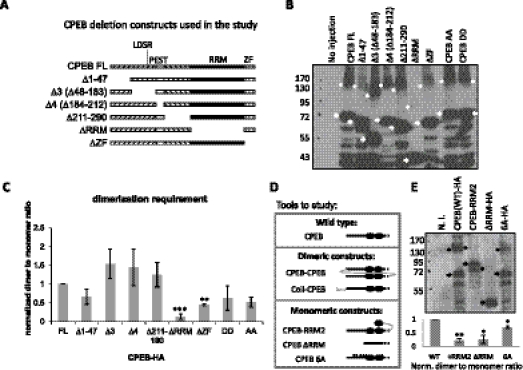
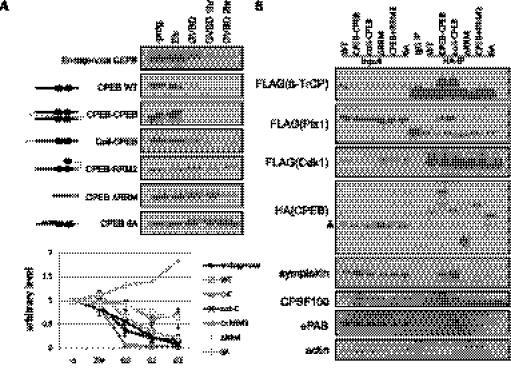
To determine the structural requirements for dimerization, serial deletions of CPEB were generated (Fig. 4A), expressed in oocytes by mRNA injection, and analyzed by low denaturing SDS-PAGE and Western blotting (Fig. 4B). The dimer to monomer ratio was quantified for four biological repeats (note that the asterisks refer to dimers and arrows to monomers); Figure 4C demonstrates that the RRMs and ZFs were particularly important for dimer formation. Indeed, the RRMs and ZFs are highly structured and surface-exposed, making them amenable for protein–protein interactions. Also note that, although the dimerization status of CPEB is regulated during meiotic progression, alanine mutations (CPEB-AA) and phosphomimetic mutations (CPEB-DD) of aurora A phosphorylation sites did not affect its dimerization.
FIGURE 4.
The CPEB RNA-binding domain is required for dimerization. (A) Serial deletions of CPEB used in this study. (B) mRNAs encoding the proteins noted (with HA epitopes at the carboxyl ends) in panel A were injected into oocytes, which was followed by Western blotting analysis. Arrows indicate monomers and asterisks indicate dimers. (C) Quantification of the results from four experiments similar to those presented in panel B. **: P-value < 5 × 10−3; ***: P-value < 5 × 10−4 as determined by paired t-test comparing to the full-length CPEB (FL). CPEB-AA and DD are alanine and phosphomimetic mutations of aurora A phosphorylation sites, respectively. (D) Depiction of dimer and monomer CPEB constructs. The solid line indicates N terminal half of CPEB; the closed circles indicate RRMs and the dashed line indicates ZFs. A curved gray line indicates the linker; a coiled-coil structure indicates the dimerization domain from GCN4, and small A's are alanine mutations. Dimeric CPEBs are (i) CPEB-CPEB: tandem CPEB in single peptide, and (ii) Coil-CPEB: CPEB fused with GCN4 dimerization domain (coiled-coil domain) at the N terminus. Monomeric CPEBs are (i) CPEB-RRM2: CPEB fused with an additional RRM to introduce intramolecular interaction and blocks intermolecular interaction, (ii) ΔRRM: deletion of the dimerization domain characterized in A–C, and (iii) 6A: six-point mutations of cdk1 phosphorylation sites that may change the surface charge and thus the conformation of CPEB to favor monomeric status. (E) Various CPEB proteins expressed in oocytes were assayed by mild denaturing SDS-PAGE (0.5% SDS with no reducing agent in the sample buffer), which shows that the monomeric constructs depicted in D indeed possess lower dimerization potential compared with CPEB WT. In the representative Western blot, arrows indicate monomers and asterisks indicate dimers. The lower panel shows the quantification of three replicates. * indicates statistical significance (**: P < 5 × 10−3; *: P < 0.05, Student's t-test).
We further generated various CPEB constructs that would be predicted to favor dimer or monomer formation (Fig. 4D). For CPEB dimers, the first one consisted of a single polypeptide containing two full-length CPEB proteins separated by a linker sequence (4 HA epitopes, a total of 40 amino acids, depicted by a gray line of CPEB-CPEB in Fig. 4D) that should allow for free rotation of the CPEB proteins (Robinson and Sauer 1998; Erickson 2009) (designated as CPEB-CPEB or C-C). The second was CPEB with an N-terminal coiled-coil dimerization domain that was modified from the yeast transcriptional factor GCN4 (designated as coil-CPEB) (Havranek and Harbury 2003; Wayne et al. 2010). To generate monomeric constructs, full-length CPEB was fused with three HA epitopes (a total of 31 amino acids) followed by RRM2 of CPEB. The HA epitopes constitute a flexible linker between full-length CPEB and RRM2 so that the isolated RRM could interact with the RRM of CPEB, thus forming an intramolecular interaction that should preclude an intermolecular interaction between two CPEB molecules (Wayne and Bolon 2007). CPEB-ΔRRM was identified as a monomer in the domain requirement for dimerization (Fig. 4A–C); CPEB-6A refers to CPEB with mutated cdk1 phosphorylation sites (Mendez et al. 2002), which was unexpectedly found to have reduced dimerization potential, possibly due to a change in the surface charge, thereby altering three-dimensional conformation. Analyzed by mild denaturing SDS-PAGE, Figure 4E (upper panel) demonstrates that indeed the monomeric constructs had lower potential to form an intermolecular dimer (marked by asterisks) compared with the monomeric species (marked by arrows); quantification from three replicates (lower panel) demonstrates that this difference is statistically significant (**: P < 5 × 10−3; *: P < 0.05) compared with wild type (WT). In summary, in Figure 4, we show that CPEB dimerizes through the RNA-binding region and, based upon this knowledge, CPEB dimeric and monomeric mutants are generated for detailed study.
CPEB dimers do not UV cross-link to RNA
CPEB dimerization occurs through the RNA-binding domain, which could preclude binding to RNA. To investigate this possibility, UV cross-linking with radiolabeled RNA and WT or dimeric CPEB was carried out. Extracts from oocytes injected with WT and CPEB-CPEB (Fig. 5A, upper panel) or WT and coil-CPEB constructs (Fig. 5A, lower panel) were primed with radiolabeled RNAs that contained or lacked CPEs, which was followed by UV irradiation, RNase digestion, and analysis by SDS-PAGE and phosphorimaging. Compared with the endogenous CPEB, which clearly cross-linked to CPE-containing RNAs, neither the tandem CPEB nor the coiled-coil domain-fused CPEB cross-linked to the RNA probes (Fig. 5A). Along the same lines, endogenous dimeric CPEB also showed no 32P-label transfer in the mild denaturing SDS-PAGE (Fig. 5B). These results indicate that CPEB dimerization occurs at the expense of RNA binding as assessed by UV cross-linking.
FIGURE 5.
Dimeric CPEBs do not cross-link to RNA. (A) Dimeric form of CPEB, i.e., tandem CPEB-CPEB and coiled-coil CPEB, were ectopically expressed in oocytes. The endogenous CPEB from the same lysate cross-linked to CPE-containing 32P-labeled RNA, while the dimeric CPEBs showed virtually no label transfer from the RNA. CPE2/1/0, radiolabeled RNA containing 2/1/0 CPE(s), CPE1 ×2, twice amount of CPE1 RNA that provides the same number of CPEs as CPE2 RNA in the cross-linking reaction. (B) Nontreated oocyte lysate was cross-linked to the radiolabeled RNA and analyzed on a mild denaturing SDS-PAGE. Although the dimeric CPEB was obvious on the Western blot, no corresponding cross-linking signal was detected by autoradiography.
CPEB dimers undergo rapid meiosis-dependent degradation
The data in Figure 3 suggest that CPEB dimers are degraded during oocyte maturation. To investigate this possibility further, a time course of GVBD-dependent CPEB degradation was performed. In this experiment, mRNAs encoding WT CPEB, CPEB-CPEB, coil-CPEB, CPEB-ΔRRM, CPEB-RRM2, or CPEB-6A were injected into oocytes followed by treatment with progesterone. A time course up to 2 h after GVBD showed that the dimeric CPEB (CPEB-CPEB and coil-CPEB) degraded faster than endogenous CPEB, while the monomeric CPEB (CPEB-ΔRRM, CPEB-RRM2, and CPEB-6A) degraded very little (Fig. 6A). Quantification of the Western blot from up to five biological repeats (lower panel of Fig. 6A) demonstrates that almost all dimeric CPEB degraded right after GVBD while still >50% of monomeric CPEB remained 2 h after GVBD. These results indicate that CPEB dimerization is indispensible for cell cycle-regulated protein degradation.
FIGURE 6.
Dimerization of CPEB facilitates GVBD-regulated protein destruction. (A) WT, dimeric form, or monomeric form of CPEB were expressed in dormant oocytes, which were then induced into GVBD by progesterone. Quantification from up to five independent experiments is shown in the lower panel. Compared with endogenous and WT CPEB, which were down-regulated after GVBD, the monomeric form of CPEBs, i.e., ΔRRM, CPEB-RRM2, and 6A, were stable along the time course. (B) HA-tagged CPEB and FLAG-tagged cdk1, plx1, and β-TrCP were co-expressed in oocytes, followed by IP with HA antibodies and Western blotting. Monomeric CPEBs were deficient of binding to destruction-related factors, including E3 ligase β-TrCP and its upstream kinase plx1, which explains the deficiency of the regulated destruction. In addition, the dimeric CPEBs bound to polyadenylation factors, symplekin, CPSF100, and ePAB, more efficiently. A triangle to the left of the HA(CPEB) blot indicates nonspecific reactivity of HA antibodies. Actin served as a negative control for IP.
To explore the mechanism of differential degradation between CPEB dimers and monomers, their association with the protein destruction machinery was examined. CPEB destruction is initiated by multiple cdk1-catalyzed phosphorylations (Mendez et al. 2002) following the resumption of oocyte maturation. Plx1 binds to one of the cdk1 phosphorylation sites in the PEST domain of CPEB, which in turn is recognized by β-TrCP of ubiquitin E3 ligase complex, which catalyzes ubiquitination and destruction by the proteasome (Setoyama et al. 2007). Co-IP experiments show that, although dimeric and monomeric CPEBs bound cdk1 similarly, the dimeric CPEB had significantly higher affinity for plx1 and β-TrCP relative to monomeric CPEB (Fig. 6B). Because these co-IPs were performed with RNase, these interactions are almost certainly RNA-independent. This enhanced binding of the protein destruction machinery by dimeric CPEB is probably due to a conformation that provides a particularly accessible binding surface for the degradation factors, resulting in robust GVBD-dependent degradation.
To our surprise, dimeric CPEB also bound many polyadenylation factors including symplekin, CPSF100, and ePAB more strongly than monomeric CPEB (Fig. 6B). It may be that the conformation of dimers is preferred for assembling the polyadenylation complex even though the dimeric CPEB complex lacks mRNA substrates. We propose that the rapid degradation of CPEB dimers during maturation releases the polyadenylation factors to efficiently induce polyadenylation and translation of CPE-containing mRNAs.
The deleterious effects of excess CPEB are reduced by dimerization
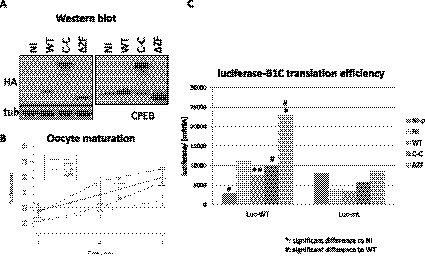
To further investigate the molecular function of the CPEB dimers, mRNAs encoding WT CPEB, tandem CPEB dimers (C-C), and CPEB-ΔZF were injected into oocytes and incubated overnight (protein expression shown in Fig. 7A). The next day, mRNAs encoding luciferase fused to the WT cyclin B1 3′ UTR (Luc-WT) or 3′ UTR with mutations in CPEs (Luc-mt) were injected; some of the oocytes were then treated with progesterone. The time course from three biological replicates shows that WT CPEB slowed the rate of maturation while the C-C CPEB did not (significant difference between WT and C-C injections, P < 5 × 10−4, as marked by ### in Fig. 7B). Furthermore, the luciferase activity normalized to the amount of mRNA determined by qRT-PCR shows a significant decrease of CPE-dependent translation upon GVBD when WT CPEB was overexpressed (cf. noninjected [NI] with WT of the left group in Fig. 7C, ** refers to P < 5 × 10−3). However, the C-C overexpression did not cause any adverse effect on translation; luciferase activity in this case was comparable with the noninjected (NI) and was significantly higher than the WT CPEB (# refers to P < 0.05 as in Fig. 7C). There was little effect of any of the CPEB proteins on the Luc-mt translation (Fig. 7C, right). These results suggest that CPEB dimerization may be a mechanism to protect cells from excess CPEB, which can potentially bind to U-rich but non-CPE sequences, ultimately leading to deregulated translation.
FIGURE 7.
CPEB dimerization ameliorates the deteriorating effect of CPEB overexpression. (A) Expression of exogenous CPEB WT, dimer CPEB-CPEB, and ΔZF mutant. Tubulin served as a loading control. (B) Exogenous CPEB (WT) slowed the rate of oocyte maturation while the CPEB dimer (C-C) had no effect. The maturation rate of dimer expression was significantly faster than that of WT expression (#). (C) Co-expression of luciferase with CPE-containing 3′ UTR shows that exogenous WT CPEB decreased its expression upon GVBD but not the dimeric CPEB. Luciferase expression with CPEB dimer (C-C) was significantly higher than that of CPEB WT (#), similar to noninjected oocytes (NI). On the right, various CPEB expressions did not affect luciferase expression with no CPE in its 3′ UTR (Luc-mt). NI-p, noninjected oocytes without progesterone treatment; NI, noninjected; WT, wild-type CPEB; C-C, tandem CPEB dimers; ΔZF, zinc finger deletion of CPEB. For the statistics, one mark represents P-value < 0.05; two marks represent P-value < 5 × 10−3; three marks represent P-value < 5 × 10−4 as determined by paired t-test.
DISCUSSION
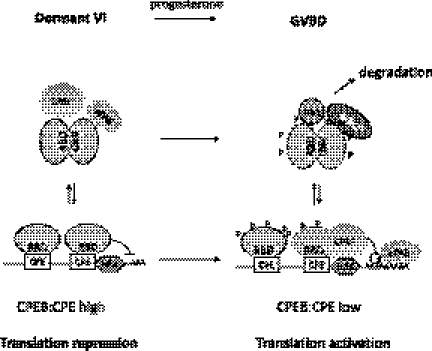
In this study, we provide evidence that CPEB forms dimers that (i) are dynamically regulated during oocyte maturation, (ii) are mediated by the RRMs, (iii) do not bind RNA, (iv) strongly associate with the protein degradation machinery, and (v) are necessary for translation and meiotic progression. Based on these results, Figure 8 proposes a model for CPEB dimer dynamics and function. CPEB proteins are translational repressors that bind to CPE-containing mRNAs and, in conjunction with Gld2 and PARN, maintain short poly(A) tails. In immature oocytes, most of the free CPEB form dimers through interactions with their RNA-binding domains; hence, this excess CPEB is unable to bind RNAs that do not contain bona fide CPEs. When the cells begin to mature, the dimeric CPEB is rapidly destroyed due to its preferential association with the kinase plx1 and the F-box protein β-TrCP of ubiquitin E3 ligase complex, which are activated upon GVBD. The destruction of the dimers releases their associated factors, including CPSF and ePAB, to possibly interact with the RNA-bound CPEB complex to facilitate polyadenylation and translation.
FIGURE 8.
The model of CPEB dimerization: CPEB dimerizes through the RNA-binding domain. The dimerization prevents excess CPEB from binding to mRNAs, which may cause translational deregulation. Moreover, CPEB dimers have higher affinity for degradation and some polyadenylation factors, which leads to rapid degradation of dimers. Dimer destruction releases the polyadenylation factors for translational activation of CPE-containing mRNA (also see the Discussion). Abbreviations: CPE, cytoplasmic polyadenylation element; HEX, AAUAAA hexanucleotide binding site for CPSF; RBD, RNA-binding domain; CPSF, cleavage and polyadenylation specific factor; ePAB, embryonic poly(A)-binding protein; plx1, polo-like kinase 1; β-TrCP, β-transducin repeat-containing protein–F-box protein of ubiquitin E3 ligase complex; GVBD, germinal vesicle break down.
Dimerization of RNA-binding proteins
Like many protein families, RNA-binding proteins have the potential to form homo- and/or hetero-dimers, as well as higher complexes. The active form of several RNA-binding proteins occurs when they homo-dimerize. For example, yeast She2p binds zip-code elements of ASH1 mRNA, directing it from the mother cell to the bud-tip of the daughter cell during mitosis. The locally translated Ash1p then acts as a repressor of mating-type switching exclusively in the daughter cell. Point mutations that abolish She2p dimerization/tetramerization inhibit RNA binding and its bud-tip localization, which is due to its deficiency of assembly into She3p-containing translocation mRNPs (Niessing et al. 2004; Muller et al. 2009). Therefore, the dimerization/tetramerization of She2p is essential for its RNA-binding and local translational control.
AUF1 (AU-rich element RNA-binding protein 1) is an RNA-binding protein that interacts with 3′ UTR AU-rich elements (AREs) to facilitate RNA destruction through deadenylation (Wilusz et al. 2001). This protein also contains two RRMs, both involved in RNA-binding. However, unlike CPEB, AUF1 dimerizes through an alanine-rich N terminal domain, as determined by a gel filtration assay. When the N terminal dimerization region is deleted, the RNA-binding affinity is ∼10-fold lower compared with the WT (DeMaria et al. 1997). Hence, the dimerization domain together with RRMs of AUF1 is essential for RNA binding.
Double-stranded RNA (dsRNA)-binding protein DGCR8 (DiGeorge syndrome critical region gene 8), the homolog of Pasha, assists the RNase III family enzyme Drosha to cleave primary microRNAs (pri-miRNAs) into precursor miRNAs. A crystal structure of the core DGCR8 suggests that the two tandem dsRNA-binding domains (dsRBDs) interact with a C-terminal helix and is arranged with pseudo twofold symmetry so that the two RNA-binding surfaces are exposed and point in opposite directions (Sohn et al. 2007). A gel filtration assay shows that the DGCR8 dimer further trimerizes using a helix downstream from dsRBDs, allowing cooperative RNA binding. In addition, only this trimeric form possesses the pri-miRNA processing ability (Faller et al. 2007), suggesting that the trimerization domain may be inhibitory or the trimeric structure can promote its enzymatic activity. Collectively, these examples demonstrate many RNA-binding proteins exert their function in the homo-dimeric/multimeric form. However, our studies indicate that dimeric CPEB, though active in the degradation pathway (Fig. 6), is dormant for RNA binding (Fig. 5). These results, together with the overexpression assay (Fig. 7), suggest that dimerization is a mechanism to keep the excess of CPEB inactive.
It may be intuitive to infer that the CPEB dimer should be deficient in RNA binding since the RNA-binding domain is masked by the protein–protein interaction. However, many examples show that the RNA-binding proteins can dimerize through the RNA-binding motifs but still maintain the RNA binding ability. First of all, there are usually multiple RNA binding motifs in an RNA-binding protein. In eukaryotic proteins, RRMs are often found as multiple copies within a protein (44%, two to six RRMs) and/or together with other RNA-binding domains (21%), among which ZFs are the most frequent (Lunde et al. 2007). The domain used for RNA binding and that used for dimerization may not overlap (Daher et al. 2001; Hitti et al. 2004; Martel et al. 2010). Secondly, even if the same motif is used for both RNA and protein interactions, the different surfaces may be used and thus these two functions do not interfere with each other (Jang et al. 2006; Sohn et al. 2007; Toba and White 2008). In the case of CPEB, probably the same surface for RNA binding is used for dimerization and hence the RNA-binding is blocked in the dimeric CPEB. This hypothesis needs to be further determined by detailed structural analysis. Unfortunately, CPEB is extremely difficult to purify en masse, which is necessary for crystallization and structural investigation.
Overall, dimerization/multimerization is a common mechanism to regulate the binding affinity and function of RNA-binding proteins. However, the impact of dimerization/multimerization is specific to each RNA-binding protein that needs to be determined experimentally.
Multimerization of ApCPEB and dimerization of xCPEB
It has been reported that ApCPEB has prion-like properties; that is, it acquires a distinct conformation that is self-sustaining and epigenetically inherited. In yeast, ApCPEB in this prion-like form has been reported to be the active form in that it binds mRNA and stimulates translation (Si et al. 2003; Heinrich and Lindquist 2011). In Aplysia sensory neurons, ApCPEB also can form self-sustaining multimers with amyloid characters; in addition, the multimerization can be induced by the neurotransmitter serotonin and injection of antibodies against the aggregate form of ApCPEB blocks long-term facilitation (Si et al. 2010). The N terminal Q-rich sequence of ApCPEB is required (yet not sufficient) to form aggregates in neurons, but this Q-stretch is not present in xCPEB. Hence, the biochemistry of ApCPEB multimerization and xCPEB dimerization is completely different. In our study, CPEB forms dimers through the C terminal RNA-binding domain and dimeric CPEBs are dormant in the RNA binding and the translation regulation. However, there are some common characters between ApCPEB multimers and xCPEB dimers. That is, either the multimerization or the dimerization is regulated by the extracellular stimulation and is critical for the cellular function. ApCPEB multimerization can be stimulated by serotonin and may contribute to the persistence of long-term facilitation, whereas xCPEB dimerization, which protects the cells from overloading CPEBs, is less prominent when cells are treated with progesterone and its degradation is indispensable for the meiotic progression.
MATERIALS AND METHODS
Antibodies
Antibodies used in this study include the following: rabbit anti-CPEB (Hake and Richter 1994), rabbit anti-HA (Sigma), mouse anti-HA (Covance), mouse anti-α-tubulin (Sigma-Aldrich), mouse anti-symplekin (BD Transduction Laboratories), rabbit anti-CPSF100 (Takagaki and Manley 2000; gift of J. Manley), rabbit anti-PARN (gift of M. Wormington), rabbit anti-ePAB (gift of J. Steitz), mouse anti-actin, rabbit anti-maskin (Stebbins-Boaz et al. 1999).
In vitro transcription and in vitro translation
RNAs used for oocyte injection were generated with mMESSAGE mMACHINE T3/T7 kit (Ambion). RNAs for protein expression were further polyadenylated with Escherichia coli poly(A) polymerase (New England Biolabs).
Labeled HA-CPEB protein was synthesized from in vitro transcribed mRNA using a rabbit reticulocyte lysate (Promega) following the manufacturer's instruction. The reaction was supplemented with 20 μCi methionine, L-[35S] to label the newly synthesized protein, and was applied directly to the gel. After electrophoretic transfer to nitrocellulose membrane, the protein was visualized by Western blotting or autoradiography.
Immunoprecipitation (IP)
Fifty to 100 injected oocytes or HEK 293T cells from a 10-cm dish were homogenized in IP buffer (150 mM NaCl, 25 mM HEPES-KOH pH 7.5, 10% glycerol, 1 mM MgCl2, 2 mM sodium orthovanadate, 2 mM β-glycerophosphate, 1 mM phenylmethylsulphonylfluoride [PMSF], 1 mM DTT, 2 mM EDTA, 0.5% Triton X-100, and protease inhibitor cocktail [Roche]). The clarified lysate was incubated with monoclonal anti-HA-agarose (Sigma-Aldrich) or anti–FLAG M2 affinity gel (Sigma-Aldrich) for 2 h to overnight at 4°C with 50 μg/mL RNase A (Sigma-Aldrich). The collected agarose beads or affinity gel were then washed five times with the IP buffer before boiling in SDS-sample buffer.
Formaldehyde cross-linking and IP
HEK 293T cells (confluent in a 10-cm dish) expressing both CPEB-HA and CPEB-FLAG or CPEB-HA only were homogenized in 1 mL IP buffer, incubated with 50 μg/mL RNase A at room temperature for 20 min. To avoid nonspecific aggregation, the lysate was diluted with PBS to 10 volumes before adding 32 μL 37% formaldehyde (Sigma-Aldrich) (final 0.12%). The cell lysate was incubated for 15 min at room temperature followed by an additional incubation with anti-FLAG M2 affinity gel (Sigma-Aldrich) at 4°C overnight. The collected beads were washed twice with wash buffer (1× PBS, 0.1% SDS, 0.5% sodium deoxycholate, and 0.5% NP-40) and three times with high-salt wash buffer (5× PBS, 0.1% SDS, 0.5% sodium deoxycholate, and 0.5% NP-40). Proteins were then eluted from the affinity gel following incubation with FLAG peptide at 30°C for 30 min. The supernatant was concentrated with a speed vacuum contractor to decrease the volume for loading. The affinity gel was further boiled with SDS-sample buffer. The eluates from both peptide elution and sample buffer boiling were analyzed by standard SDS-PAGE and Western blotting.
Gel filtration
Five milliliters of gravity packed control or mature oocytes were homogenized in equal volume of IP buffer (see above) supplemented with 1% sodium deoxycholate. The homogenate was centrifuged at 5K rpm for 20 min in SW 41 rotor (Beckman Coulter). The middle layer of the clarified lysate was extracted with an 18½G needle and centrifuged again at 36K for 30 min. The clear lysate was then passed through the glass wool to remove lipids and 0.45 μm PVDF membrane to remove large aggregates and cell debris. It was then loaded onto a Hiload 16/60 Superdex 200 prep grade column (Amersham Biosciences) attached to FPLC. The flow rate was adjusted to 1 mL/min and 1.5 mL fractions were collected for Western analysis.
UV cross-linking
To perform UV cross-linking, a sequence from the Xenopus cyclin B1 3′ UTR or ones that contained mutations in the CPEs (CPE2: GGGAGAUGUUUUUAAUGCGACUCUGGCGUUUUAAUAAAGCUCA; CPE1: GGGAGAUGUUUUUAAUGCGACUCUGGCGCGACAAUAAAGCUCA; CPE0: GGGAGAUGUCGACUUAGCGACUCUGGCGCGACAAUAAAGCUCA) were transcribed in vitro with 30 μCi UTP [α-32P], extracted with acid phenol/chloroform, and precipitated with isopropanol. 1 × 105 cpm of CPE2 probe was used in the cross-linking reaction; Based on the composition of U's, 0.75 × 105 cpm or 1.5 × 105 cpm of CPE1 probe was used to achieve the same molecules of probes or same number of CPEs in the reaction, respectively. Untreated or injected oocytes were lysed in IP buffer and then incubated with the RNA probes in gel retardation buffer (10 mM HEPES pH 7.6, 2 mM MgCl2, 0.1 mM ZnCl2, 50 mM KCl, 10% glycerol, and 1 mM DTT) supplemented with 2.5 mg/mL heparin, 50 μg/mL tRNA, 0.5 mM DTT, and 0.6 unit/μL RNaseOUT for 10 min on ice and then 10 min at room temperature. The protein–RNA mixture (20 μL) was applied per well on a Nunclon Δ Surface plate (Nunc) and UV-irradiated with 500 mJ (Stratalinker UV Crosslinker, Stratagene) on iced water. Following 100 ng RNase A treatment at 37°C for 30 min, the mixture was then boiled in SDS-sample buffer (omit boiling for low denaturing SDS-PAGE), followed by standard Western blotting and autoradiography.
Statistics
The raw GVBD percentage was preprocessed by quantile normalization (Bolstad et al. 2003), which normalizes the GVBD percentages from different experiments to an identical distribution so that one can compare the percentages across different experiments. To make a valid comparison of the GVBD percentage of various exogenous CPEB expressions along the time course, the normalized percentage was analyzed by Generalized Estimating Equation (GEE) (Zeger and Liang 1986), which is a standard statistical analysis for the correlated data by taking into consideration the interdependence in measurements across different time points. For other experiments with uncorrelated data, paired Student t-test was used to generate P-values between experiments.
ACKNOWLEDGMENTS
We thank J. Steitz (Yale University), J. Manley (Columbia University), and M. Wormington (University of Virginia) for gifts of antibodies, and D.N. Bolon (University of Massachusetts) for his advice on constructing dimers and monomers and sharing of GCN4 coiled-coil domain-containing plasmid. We are also grateful to all the current and previous lab members for the discussions and technical support, especially I. Alexandrov (Biomerix Corporation). This work is supported by NIH grant GM46779 (to J.D.R.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.031682.111.
REFERENCES
- Adinolfi S, Ramos A, Martin SR, Dal Piaz F, Pucci P, Bardoni B, Mandel JL, Pastore A 2003. The N-terminus of the fragile X mental retardation protein contains a novel domain involved in dimerization and RNA binding. Biochemistry 42: 10437–10444 [DOI] [PubMed] [Google Scholar]
- Barnard DC, Ryan K, Manley JL, Richter JD 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119: 641–651 [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Cao Q, Kim JH, Richter JD 2006. CDK1 and calcineurin regulate Maskin association with eIF4E and translational control of cell cycle progression. Nat Struct Mol Biol 13: 1128–1134 [DOI] [PubMed] [Google Scholar]
- Chen T, Richard S 1998. Structure-function analysis of Qk1: A lethal point mutation in mouse quaking prevents homodimerization. Mol Cell Biol 18: 4863–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JL, Gehman JD, Shafer JA, Kuo LC 1993. Solution oligomerization of the rev protein of HIV-1: Implications for function. Biochemistry 32: 11769–11775 [DOI] [PubMed] [Google Scholar]
- Daher A, Longuet M, Dorin D, Bois F, Segeral E, Bannwarth S, Battisti PL, Purcell DF, Benarous R, Vaquero C, et al. 2001. Two dimerization domains in the trans-activation response RNA-binding protein (TRBP) individually reverse the protein kinase R inhibition of HIV-1 long terminal repeat expression. J Biol Chem 276: 33899–33905 [DOI] [PubMed] [Google Scholar]
- DeMaria CT, Sun Y, Long L, Wagner BJ, Brewer G 1997. Structural determinants in AUF1 required for high affinity binding to A + U-rich elements. J Biol Chem 272: 27635–27643 [DOI] [PubMed] [Google Scholar]
- Erickson HP 2009. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol Proced Online 11: 32–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller M, Matsunaga M, Yin S, Loo JA, Guo F 2007. Heme is involved in microRNA processing. Nat Struct Mol Biol 14: 23–29 [DOI] [PubMed] [Google Scholar]
- Fribourg S, Gatfield D, Izaurralde E, Conti E 2003. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat Struct Biol 10: 433–439 [DOI] [PubMed] [Google Scholar]
- Fuchs SY, Spiegelman VS, Kumar KG 2004. The many faces of β-TrCP E3 ubiquitin ligases: Reflections in the magic mirror of cancer. Oncogene 23: 2028–2036 [DOI] [PubMed] [Google Scholar]
- Hake LE, Richter JD 1994. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell 79: 617–627 [DOI] [PubMed] [Google Scholar]
- Hake LE, Mendez R, Richter JD 1998. Specificity of RNA binding by CPEB: requirement for RNA recognition motifs and a novel zinc finger. Mol Cell Biol 18: 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havranek JJ, Harbury PB 2003. Automated design of specificity in molecular recognition. Nat Struct Biol 10: 45–52 [DOI] [PubMed] [Google Scholar]
- Heinrich SU, Lindquist S 2011. Protein-only mechanism induces self-perpetuating changes in the activity of neuronal Aplysia cytoplasmic polyadenylation element binding protein (CPEB). Proc Natl Acad Sci 108: 2999–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitti EG, Sallacz NB, Schoft VK, Jantsch MF 2004. Oligomerization activity of a double-stranded RNA-binding domain. FEBS Lett 574: 25–30 [DOI] [PubMed] [Google Scholar]
- Jang SB, Jeong MS, Carter RJ, Holbrook EL, Comolli LR, Holbrook SR 2006. Novel crystal form of the ColE1 Rom protein. Acta Crystallogr D Biol Crystallogr 62: 619–627 [DOI] [PubMed] [Google Scholar]
- Kim JH, Richter JD 2006. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell 24: 173–183 [DOI] [PubMed] [Google Scholar]
- Kim JH, Richter JD 2007. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes Dev 21: 2571–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G 2007. RNA-binding proteins: Modular design for efficient function. Nat Rev Mol Cell Biol 8: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Dugre-Brisson S, Boulay K, Breton B, Lapointe G, Armando S, Trepanier V, Duchaine T, Bouvier M, Desgroseillers L 2010. Multimerization of Staufen1 in live cells. RNA 16: 585–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD 2000a. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature 404: 302–307 [DOI] [PubMed] [Google Scholar]
- Mendez R, Murthy KG, Ryan K, Manley JL, Richter JD 2000b. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol Cell 6: 1253–1259 [DOI] [PubMed] [Google Scholar]
- Mendez R, Barnard D, Richter JD 2002. Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J 21: 1833–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Richter K, Heuck A, Kremmer E, Buchner J, Jansen RP, Niessing D 2009. Formation of She2p tetramers is required for mRNA binding, mRNP assembly, and localization. RNA 15: 2002–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessing D, Huttelmaier S, Zenklusen D, Singer RH, Burley SK 2004. She2p is a novel RNA binding protein with a basic helical hairpin motif. Cell 119: 491–502 [DOI] [PubMed] [Google Scholar]
- Richter JD 2007. CPEB: A life in translation. Trends Biochem Sci 32: 279–285 [DOI] [PubMed] [Google Scholar]
- Robinson CR, Sauer RT 1998. Optimizing the stability of single-chain proteins by linker length and composition mutagenesis. Proc Natl Acad Sci 95: 5929–5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoyama D, Yamashita M, Sagata N 2007. Mechanism of degradation of CPEB during Xenopus oocyte maturation. Proc Natl Acad Sci 104: 18001–18006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER 2003. A neuronal isoform of the Aplysia CPEB has prion-like properties. Cell 115: 879–891 [DOI] [PubMed] [Google Scholar]
- Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER 2010. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell 140: 421–435 [DOI] [PubMed] [Google Scholar]
- Sohn SY, Bae WJ, Kim JJ, Yeom KH, Kim VN, Cho Y 2007. Crystal structure of human DGCR8 core. Nat Struct Mol Biol 14: 847–853 [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JD 1999. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol Cell 4: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol 20: 1515–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba G, White K 2008. The third RNA recognition motif of Drosophila ELAV protein has a role in multimerization. Nucleic Acids Res 36: 1390–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne N, Bolon DN 2007. Dimerization of Hsp90 is required for in vivo function. Design and analysis of monomers and dimers. J Biol Chem 282: 35386–35395 [DOI] [PubMed] [Google Scholar]
- Wayne N, Lai Y, Pullen L, Bolon DN 2010. Modular control of cross-oligomerization: Analysis of superstabilized Hsp90 homodimers in vivo. J Biol Chem 285: 234–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wormington M, Peltz SW 2001. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol 2: 237–246 [DOI] [PubMed] [Google Scholar]
- Yao G, Chiang YC, Zhang C, Lee DJ, Laue TM, Denis CL 2007. PAB1 self-association precludes its binding to poly(A), thereby accelerating CCR4 deadenylation in vivo. Mol Cell Biol 27: 6243–6253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeger SL, Liang KY 1986. Longitudinal data analysis for discrete and continuous outcomes. Biometrics 42: 121–130 [PubMed] [Google Scholar]