This paper presents a novel approach for the detection of microRNA (miRNA) activity in different cell types. This is based on the use of a retroviral vector combined with a dual luciferase reporter that allows detection of miRNA activity in primary cells.
Keywords: UTR, microRNA, retrovirus, primary cells, dual luciferase
Abstract
MicroRNA–mRNA interactions are commonly validated and deconstructed in cell lines transfected with luciferase reporters. However, due to cell type-specific variations in microRNA or RNA-binding protein abundance, such assays may not reliably reflect microRNA activity in other cell types that are less easily transfected. In order to measure miRNA activity in primary cells, we constructed miR-Sens, a MSCV-based retroviral vector that encodes both a Renilla luciferase reporter gene controlled by microRNA binding sites in its 3′ UTR and a Firefly luciferase normalization gene. miR-Sens sensors can be efficiently transduced in primary cells such as human fibroblasts and mammary epithelial cells, and allow the detection of overexpressed and, more importantly, endogenous microRNAs. Notably, we find that the relative luciferase activity is correlated to the miRNA expression, allowing quantitative measurement of microRNA activity. We have subsequently validated the miR-Sens 3′ UTR vectors with known human miRNA-372, miRNA-373, and miRNA-31 targets (LATS2 and TXNIP). Overall, we observe that miR-Sens-based assays are highly reproducible, allowing detection of the independent contribution of multiple microRNAs to 3′ UTR–mediated translational control of LATS2. In conclusion, miR-Sens is a new tool for the efficient study of microRNA activity in primary cells or panels of cell lines. This vector will not only be useful for studies on microRNA biology, but also more broadly on other factors influencing the translation of mRNAs.
INTRODUCTION
MicroRNAs (miRNAs) are short single-stranded RNAs binding to mRNAs with partial complementarity in miRNA-binding sites, resulting in translational repression and/or mRNA destabilization. Most well-recognized targets are located in 3′ UnTranslated Regions (UTRs), which mediate miRNA-dependent translational repression and mRNA destabilization (Bartel 2009). Several computational approaches have been developed to predict miRNA targets based on seed (nucleotides 2–8 of the miRNA) sequence complementarity, combined with various additional criteria such as thermodynamic stability of the miRNA:mRNA pair, conservation of the target sequence, G:U pairing, and calculated accessibility of the site (Mazière and Enright 2007; Bartel 2009; Thomas et al. 2010). Unfortunately, these prediction algorithms are not always consistent with each other, and any given method has a significant rate of false-positive predictions (Mazière and Enright 2007; Voorhoeve 2010; Zhang and Verbeek 2010). Furthermore, all of these predictions ultimately require functional validation in a cell-based assay to prove their validity (Thomas et al. 2010).
One of the methods to show a direct interaction between miRNA and mRNA target site is using constructs containing wild-type or mutant 3′ UTRs fused behind a reporter, among which luciferase is the most convenient. Usually, the miRNA–mRNA interaction is tested in highly transfectable cell lines such as 293/T or MCF-7. The miRNA-mediated translational repression is measured by changes in activity of the 3′ UTR–containing luciferase reporter in response to cotransfection with miRNAs. Correction for cell viability, transfection efficiency, and nonspecific effects on translation is done through a second luciferase gene that lacks the 3′ UTR, which can be present on a separate plasmid or on the same plasmid under a separate promoter. Notably, both luciferase activities can be tested sequentially on the same sample, which greatly reduces variations due to cell number, viability, or reporter delivery (so-called dual-luciferase system) (Thomas et al. 2010).
However, miRNA–target interactions are not just dependent on the presence of the binding site and the miRNA, but are also influenced by the presence of other miRNAs or RNA-Binding Proteins (RBP) (van Kouwenhove et al. 2011). RBPs can be present in a cell type-specific fashion and inhibit (Kedde et al. 2007), or enhance the activity of a miRNA toward a target mRNA (Kedde et al. 2010). Additionally, RBPs such as hnRNP-L can be activated by specific conditions to neutralize miRNA activity (Jafarifar et al. 2011). Moreover, it is becoming apparent that the activity of a given miRNA toward its target is affected by the presence of competing endogenous mRNAs, whose presence will be specific to each particular cell type (Salmena et al. 2011). These and other findings indicate that the functional validation of miRNA–mRNA interactions should not be agnostic of the cell type in which the miRNA is thought to exert its translational control on the presumed target.
Unfortunately, the dual-luciferase assay is presently limited to cells that can be transfected with a reasonable efficiency, and thus cannot be easily used in primary cells or in cell lines with low-transfection efficiency. Efficient transient transfection also has the disadvantage of producing relatively high levels of reporter mRNA, obscuring competitive effects with endogenous mRNAs. These observations justify the development of a retroviral vector for the testing of miRNA-mediated translational repression, which can be used in primary cells or other cells that cannot be (efficiently) transfected.
To measure miRNA activity in human primary cells, we prepared a retroviral vector (miR-Sens) expressing both a reporter luciferase (Renilla) and a control luciferase (Firefly). We found that efficient retroviral transduction requires the deletion of two strong internal Poly Adenylation Signals (PAS) from the original dual-luciferase construct. Although this extends the 3′ UTR of the reporter luciferase, we could show that this vector still sensitively detects miRNA activities. Since the control luciferase is now expressed from the same integration sites as the reporter, this system is more robust than when using two independent luciferase constructs. Consequently, the reproducibility of the retroviral dual-luciferase system is such that it is possible to detect the independent contribution of multiple miRNAs on a target 3′ UTR, and we show that the relative luciferase activity is correlated to the miRNA expression, allowing quantitative measurement of miRNA activity. A combination of a retroviral vector and a dual-luciferase reporter therefore provides reproducibility in the detection of miRNA activity in primary cells or in panels of non-easily transfectable cell lines.
RESULTS
Construction of the miR-Sens vector
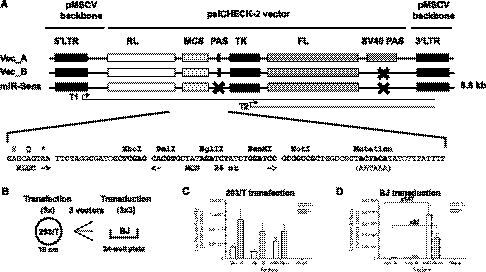
We set out to construct a retroviral vector that allows easy detection of miRNA–mRNA interactions in primary cells (Fig. 1A). We therefore modified a MSCV-based vector to contain two luciferase reporters inserted between the MSCV packaging sequence and the 3′ LTR (Vec_A) (Fig. 1A). In this vector, the Renilla luciferase expression—the reporter—is driven by the viral 5′ LTR promoter, while the Firefly luciferase expression depends on the Thymidine Kinase promoter and is included as a normalization reference (Fig. 1A).
FIGURE 1.
miR-Sens vector construction and validation. (A) Structure of the miR-Sens vector and sequence of the multiple cloning site (MCS). The MSCV packaging sequence between the 5′ LTR and the RL is not shown on this graph. The vectors are not to scale. (B) Flowchart for the validation of the miR-Sens vector. Three independent 293/T transfections with the three vectors (Vec_A, Vec_B, and miR-Sens) were done on three different days. Supernatants were collected on the third day post-transfection, and flash frozen. On the same day, the 293/T cells were harvested and equal amounts of cells were flash-frozen in liquid nitrogen. (C) Results of the 293/T cell transfection. The 293/T transfected cell pellets were tested in triplicate on the same day. (D) Results of the BJ cell transduction. BJ cells were transduced in triplicate on the same day with the three independent viral supernatants collected from the three 293/T cell supernatants. BJ cells were assayed 3 d after transduction as described in the Materials and Methods. Results correspond to the Mean ± SEM. (LTR) Long Terminal Repeat; (RL) Renilla luciferase; (PAS) synthetic PolyAdenylation Sequence; (T1) Expected Renilla luciferase transcript; (T2) Expected Firefly luciferase transcript; (TK) Thymidine Kinase promoter; (FL) Firefly luciferase; (SV40 PAS) Simian Virus 40 late PolyAdenylation Signal.
Efficient retroviral production requires the transcription of a long RNA that should not be abrogated by any polyadenylation sequence before reaching the 3′ Long Terminal Repeat (LTR) (Yang et al. 1999). The 3′ LTR is required for integration of the virus and itself contains a viral polyadenylation sequence (Jern and Coffin 2008). As expected, we found that cloning the two luciferase reporter genes, including their two polyadenylation sequences (PASs) that are used in the plasmid-based expression constructs to separate the two luciferase transcripts, severely reduced viral RNA titers (Fig. 1C,D). To maintain the independence of the two luciferase reporter transcripts while attempting to increase viral titers, we first deleted only the SV40 late polyadenylation sequence (Vec_B). Moreover, the synthetic polyadenylation signal (AATAAA), which terminates the Renilla transcript before the TK promoter and firefly-encoding sequences, was changed into an inactive polyadenylation signal (ACTACA) (Retelska et al. 2006; Proudfoot 2011) as shown in Figure 1A (miR-Sens). This later modification results in the inclusion of the TK promoter and Firefly luciferase-encoding sequences into the 3′ UTR of the Renilla reporter construct (predicted transcript T1) (Fig. 1A). Since the Firefly luciferase ORF is the second in this transcript, we expect it not to be translated, and Firefly luciferase activity to still be derived solely from the predicted transcript T2, which does not contain the miRNA target sequences to be tested (Fig. 1A).
Removal of both internal polyadenylation sequences is required for efficient virus production
To test the effect of these changes on viral production, we have compared the Renilla and Firefly luciferase activities from these vectors after transfection of 293/T packaging cells or after subsequent transduction of primary BJ fibroblasts with equivalent amounts of packaging cell supernatant containing the respective retroviruses (Fig. 1B). After transient transfection, Renilla and Firefly luciferase activities did not differ by more than twofold between the different constructs, indicating that all plasmids were transfected at similar levels, and that all mRNAs transcribed from the three constructs could be translated efficiently (Fig. 1C). In contrast, we found that, as expected, the vectors containing one or two internal polyadenylation sequences resulted in an average Renilla and Firefly luciferase activity after transduction that was >10-fold lower than for the miR-Sens vector (Fig. 1D; Blø et al. 2008). Having thus established a vector that produces sufficient titer to allow reliable measurement of the ratio of the reporter over the control luciferase activity (RL/FL), we set out to prove that the Renilla reporter is still sensitive to miRNA-mediated repression despite the necessary elongation of its 3′ UTR.
The extended Renilla reporter transcript can still function as an Ago-2-dependent microRNA sensor
To establish that the amount of Firefly luciferase translated from transcript 2 (Fig. 1A) is not influenced by the fate of the partially overlapping transcript 1, we first inserted sequences complementary to miRNAs in the multiple cloning site of the miR-Sens vector behind the Renilla luciferase ORF (Fig. 1A). When a miRNA targets an Ago2-containing RISC to this sensor, the Renilla luciferase encoding mRNA is cleaved. The transcript that encodes for the Firefly luciferase (transcript T2) (Fig. 1A) will not contain the miRNA sensor, and should thus be unaffected by the presence or absence of the miRNA, which is important if the Firefly luciferase activity is to be used as normalization control.
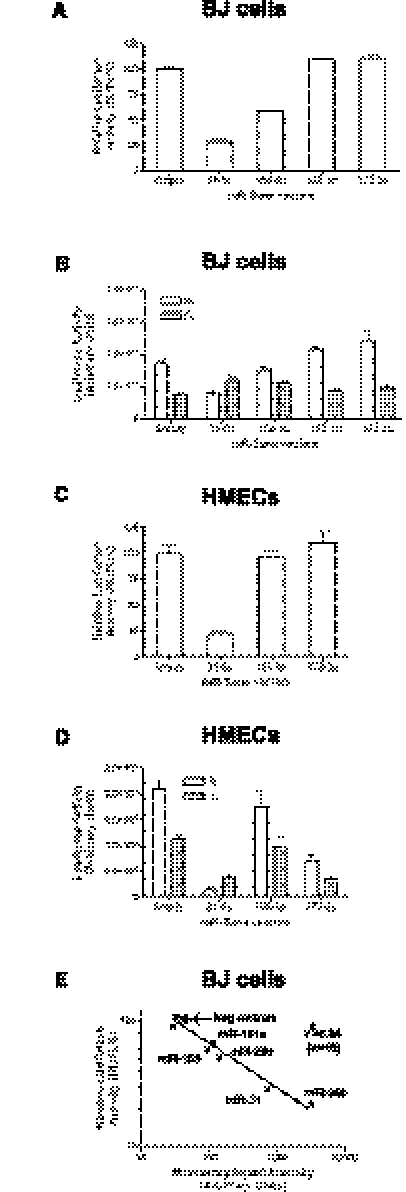
Indeed, Firefly luciferase activity was similar between constructs containing sensors for miR-31, miR-155 (both detectable in primary BJ cells by microarray), and miR-372 and miR-373 (both undetectable), indicating comparable titers and transduction efficiencies for the generated retroviral sensors (Fig. 2B,D). In contrast, Renilla luciferase activity declined according to the presence of the corresponding miRNAs in the primary cells (Fig. 2A). Further corroboration that the miR-Sens vector could functionally detect miRNAs in primary cells was obtained by transducing these retroviral sensors in primary human epithelial cells (HMECs) (Fig. 2C), which express miR-31, but not miR-373 or miR-155 (Ma et al. 2007; Valastyan et al. 2009). The relative reduction in RL activity due to the presence of a microRNA is even apparent when using a virus batch with a lower titer (e.g., miR-Sens 31-5p and 373-3) (Fig. 2D) or after dilution of the virus (data not shown).
FIGURE 2.
Efficient detection of miR expression with miR-Sens sensors. (A) Relative luciferase activities for miRNA sensors in BJ cells. (B) Absolute levels of Firefly and Renilla luciferase activities from A, indicating similar retroviral titer and transduction efficiency for various miR-Sens vectors. (C) Relative luciferase activities for miRNA sensors in HMECs. (D) Absolute levels of Firefly and Renilla luciferase activities from C, indicating that a change in the viral titer does not compromise the specificity of the miR-Sens vector. (E) Correlation between constitutive miRNAs expression and the relative luciferase activity of miR-Sens sensors in BJ cells (n = 10). Five nonexpressed miRNAs are used as negative controls (hsa-miR-29b-2-5p, hsa-miR-29c-5p, 302c-3p, 372-3p, and 373-3p).
Interestingly, when testing sensors against several microRNAs, we observed that the log-transformed relative luciferase activity of the respective sensor and miRNA levels as measured by microRNA array (n = 10 different miRNAs) correlated linearly in BJ cells (Fig. 2E). This illustrates that the sensor, when expressed from the integrated virus, is responsive over a wide range of microRNA levels. These findings indicate that the miR-Sens sensors are suitable for quantitative measurement of miRNA expression in primary cells.
Elongation of the reporter 3′ UTR does not compromise the detection of miRNA activity on 3′ UTRs in BJ cells
Unlike Ago2-mediated cleavage of completely complementary targets, microRNA targeted RISC represses translation and stability of mRNAs containing partially complementary target sequences through a variety of mechanisms (Wu and Belasco 2008) that can be influenced by the presence of RNA-binding proteins (van Kouwenhove et al. 2011) and RNA structural features such as 3′ UTR length (Lewis et al. 2003; Filipowicz et al. 2008; Mayr and Bartel 2009). Efficient retroviral production requires the deletion of the strong PAS that separates the reporter and normalization transcripts in the plasmid-based dual-luciferase system, resulting in a significant and constitutive extension of the 3′ UTR of the Renilla firefly encoding transcript (Fig. 1A). Thus, while the internal polyadenylation sequences of the miR-Sens are dispensable for Ago2-mediated cleavage of miRNA sensors, an extended heterologous 3′ UTR downstream from the 3′ UTR of interest could possibly interfere with efficient miRNA activity.
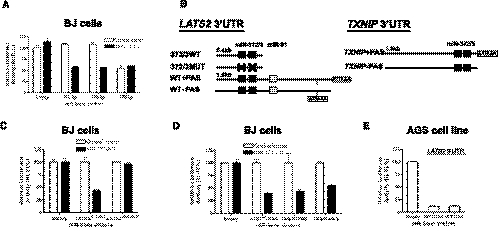
To be able to measure microRNA activity on 3′ UTRs of interest and verify target predictions in primary cells, we had to first address this possibility. We therefore prepared miR-Sens constructs, in which we introduced behind the reporter luciferase the 3′ UTR of LATS2, a known miR-372 and miR-373 target whose repression by these microRNAs in primary BJ cells has been shown to contribute to the oncogenic transformation of these cells (Fig. 3A; Voorhoeve et al. 2006).
FIGURE 3.
Validation of miRNA activity in LATS2 and TXNIP 3′ UTRs in the absence of PAS. (A) Relative luciferase activities in BJ control or miR-371-2-3 cluster expressing cells for miR-372-3p and -373-3p sensors. (B) LATS2 (ENST00000382592) and TXNIP (ENST00000369317) 3′ UTRs. (Black rectangles) miR-372 and miR-373 binding sites; (gray rectangle) miR-31 site; (black cross) miRNA-binding site mutation of miR-372 and miR-373 (Voorhoeve et al. 2006); (373/3WT and 372/3MUT) wild-type and double miR-372 and miR-373 mutant binding sites in LATS2 3′ UTR; (31MUT) miR-31 mutant binding site in LATS2 3′ UTR; (AATAAA) alternative PAS for LATS2 3′ UTR (EST ACC#BQ009415). We have confirmed that only the most 3′ LATS2 PAS was used in BJ cells by 3′ RACE PCR (data not shown). (C) Relative luciferase activities in BJ control or miR-371-2-3 cluster expressing cells for LATS2 short wild-type and double-mutant 3′ UTRs. (D) Relative luciferase activities in BJ control or miR-371-2-3 cluster expressing cells for LATS2wt and TXNIP 3′ UTRs in the absence of the most 3′ A(A/T)AAA sequence (Table 2). Comparable decrease of the relative luciferase activity was obtained when including the most 3′ A(A/T)AAA sequence of LATS2 and TXNIP 3′ UTRs in BJ cells and (E) AGS cell line. In these experiments, the relative luciferase activity of the miR-Sens vectors in control cells is normalized to 100% (double-normalization).
Primary BJ cells, transduced with miR-Vec 371-2-3, which expresses the oncogenic microRNAs 371, 372, and 373 under a strong promoter, were validated to express miR-372 and miR-373 using a mir-Sens vector with the relevant complementary sensors (Fig. 3A). miR-Sens vectors containing part of the wild-type LATS2 3′ UTR or a mutant in which the two miR-372 and miR-373 binding sites have been inactivated (Voorhoeve et al. 2006) were then introduced (Fig. 3B). Indeed, the miRNA cluster overexpression decreased the relative luciferase activity, which was dependent on miR-372 and miR-373 binding sites in the 3′ UTR (Fig. 3C), indicating that, despite the elongation of the reporter 3′ UTR, miR-Sens can be used to detect miRNA activity on 3′ UTRs of relevant target genes.
Many mRNAs contain more than one polyadenylation signal in their 3′ UTR (Mayr and Bartel 2009; Mangone et al. 2010), e.g., LATS2 has been found to have an internal polyadenylation site (AATAAA, EST BQ009415) (Fig. 3A) in addition to the most 3′ site. We wanted to determine whether inclusion of these internal polyadenylation signals, like the strong SV40 PAS, would reduce the titer to a point where the readout becomes unreliable. This information would help to determine whether internal PASs need to be mutated when cloning a longer version of a 3′ UTR in this vector. Therefore, we cloned and tested full-length versions of the LATS2 3′ UTR (ENST00000382592) containing an internal AATAAA PAS, with and without the most 3′ ATTAAA PAS into the miR-Sens vector as shown in Figure 3B.
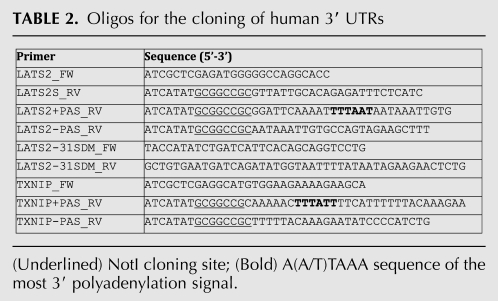
Inclusion of the endogenous upstream sequence element and A(A/T)TAAA signal of the PAS in the LATS2 and TXNIP (another miRNA-372 and miRNA-373 target) (Yan et al. 2011) 3′ UTRs decreased retroviral titer by about twofold as estimated by Firefly luciferase activity after transduction (Table 2, below; data not shown). However, the remaining signal was sufficiently strong, and the experimental variation sufficiently small, to accurately measure miRNA cluster activity against the LATS2 and TXNIP 3′ UTRs in BJ cells or AGS, a gastric cancer cell line that expresses these microRNAs (Fig. 3D,E; Cho et al. 2009; data not shown). This observation indicates that these PASs do not strongly affect viral titer through premature RNA termination in the packaging 293T cells, suggesting that full-length 3′ UTRs cloned, e.g., using oligo-dT anchored primers, would be suitable for functional tests.
TABLE 2.
Oligos for the cloning of human 3′ UTRs
miR-sens can detect the contribution of multiple miRs to translational repression of the same target
One important advantage of incorporating the two reporters in one retrovirus, in contrast to using a normalization method such as an independent retroviral vector expressing Firefly luciferase (Wu et al. 2010), is illustrated by the improved precision of the assay when measuring the ratio Renilla over Firefly luciferase activity. Due to unavoidable differences in number and location of retroviral integrations, we observe that the luciferase values vary by ∼10% between independent transductions (Fig. 4A, RL, FL). However, since these two reporters are coexpressed from the same construct, the variability of their ratio is reduced by more than threefold (Fig. 4A, RL/FL), allowing an improved precision when assessing subtle effects of miRNAs on their targets.
FIGURE 4.
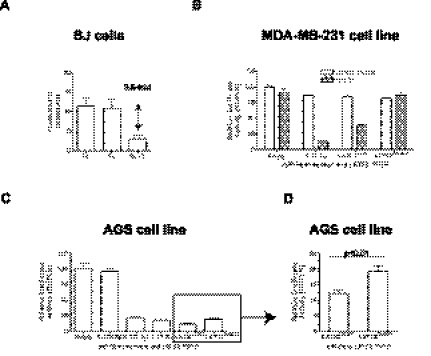
Multiple miRs contribute to LATS2 3′ UTR mediated translational repression in BJ cells and AGS cell line. (A) Average precision of the relative luciferase assay using miR-Sens reporter in BJ cells. The precision of the measure for the Renilla, the Firefly luciferase, and the ratio of RL/FL luciferase signal was calculated based on three independent transductions for each miR-Sens vectors from Figure 2A and data not shown (n = 6 constructs). The average precision and its standard deviation were subsequently determined. (B) Effect of the miR-31 site mutation in LATS2 3′ UTR on the relative luciferase activity in MDA-MB-231 cells. miR-31-5p sensor was used as positive control for miR-31 expression. miR-31 was overexpressed with miR-Vec blast in MDA-MB-231 cell line. (C) Same as in B in AGS cell line. miR-373-3p was used as a positive control for miR-373 expression. miR-371-2-3 and miR-31 are constitutively expressed in AGS cell line. (D) Enlarged results from C. In these experiments, only the relative luciferase activity of the miR-Sens empty vector in control cells is normalized to 100%.
We and others have identified a miR-31 binding site in the LATS2 3′ UTR (Fig. 3A,B; Liu et al. 2010). To illustrate the reproducibility of the miR-Sens vector and its resulting sensitivity to detect small changes in relative activity of microRNAs, we investigated the contribution of this miR-31 site into the overall miRNA activity on LATS2 3′ UTR; miR-31 was initially overexpressed in the MDA-MB-231 cell line, which does not express miR-31, miR-372, or miR-373 (Valastyan et al. 2009; data not shown). miR-31 expression in MDA-MB-231 cell line was validated with a miR-31 sensor and found to be similar to miR-31 expression in HMECs (Figs. 2C, 4B). The full-length LATS2 3′ UTR was indeed responsive to miR-31 expression, and mutation of the miR-31 site in LATS2 3′ UTR showed that this effect was direct (Fig. 4B).
To test whether the miR-Sens vector could also detect the contribution of miR-31 to LATS2 translational repression in the context of strong repression by two other microRNAs, we transduced the wild-type and miR-31 mutant LATS2 3′ UTR reporters into the AGS cell line, which constitutively express miR-31 as well as miR-372 and miR-373 (Fig. 4C). We found that the mutation of the miR-31 site resulted in a modest but significant increase in the RL/FL ratio (Fig. 4D). The relief of miR-31 mediated repression (as compared with the 3′ UTR with the mutated miR-31 binding site) in AGS cells that also repress the LATS2 3′ UTR through miR-372 and miR-373 was similar to that seen in BJ cells (data not shown), which do not express these miRNAs. This result illustrates that the combination of a retroviral vector and a dual-luciferase reporter vector is appropriate for the study of specific miRNA-mediated translational repression, even in the presence of strong repression by other miRNAs. Taken together, these results indicate that the miR-Sens vector is sensitive and reproducible enough to detect even slight functional changes in the translational repression due to endogenously expressed, overexpressed, and/or multiple miRs in human primary cells and cell lines.
DISCUSSION
We have developed a dual-luciferase reporter retroviral vector for the functional validation of miRNAs in primary cells that are dividing, but not easily transfectable. To our knowledge, there has been no report of a dual-luciferase reporter retroviral vector, although single luciferase reporter lentiviral vectors have been published (Wu et al. 2010) or are commercially available (pLSG_UTR_RenSP, Sigma; pLenti-UTR-Luc, ABM). While we had to remove the internal polyadenylation sequences to rescue the viral titer, we have found that the miR-Sens vector retained its ability to detect miRNA activity on both sensors and 3′ UTRs.
Compared with viral vectors containing a single luciferase reporter, the addition of the second luciferase gene improves the precision of the assays by threefold, and permits the detection of combinatorial miRNA effects (Fig. 4). Importantly, if the Firefly (control) luciferase expression would have been driven by the 5′ LTR promoter while the Renilla (reporter) luciferase was expressed from the TK promoter, then the activity of the control luciferase might have been influenced by the inclusion of the miRNA-binding site into its extended/artificial 3′ UTR. One caveat is that the extended 3′ UTR of the miR-Sens vector might introduce spurious regulatory elements such as microRNA- or RNA-Binding Protein-sites that could interfere with translation of the reporter luciferase. Also, introducing 3′ UTR-encoding sequences in front of the TK promoter could introduce (DNA) regulatory elements that affect transcription of the control luciferase. It is therefore important to normalize data to control miR-Sens vectors (containing the same extended 3′ UTR minus the 3′ UTR under investigation) or control cells not expressing the microRNA of interest.
Beside retroviral transduction-based assays, the miR-Sens vector can be used for transfection-based experiments in appropriate cells. However, given the 10-fold higher luciferase expression that we observed after transient transfection in 293T cells, sponge effects of the reporter could become a source of concern. The miR-Sens vector can also be used to detect sudden decreases in miRNA activity, which should result in a relative increase in the Renilla reporter activity. A rapid increase in miRNA abundance or activity would not be readily apparent due to the long half-life of the Renilla protein (Loening et al. 2010; data not shown). Relevantly, even with 10-fold lower levels of Renilla expression, the miR-Sens system is still reliable, allowing, for instance, the incorporation of destabilized Renilla, which is expressed at lower steady-state levels, but which will show a more rapid response to translational repression after induction of miRNA activity. This modification should offer new perspectives to the study of the tightly regulated miRNA turnover as reported for miR-122 or miR-141 (Katoh et al. 2009; Kim et al. 2011). Likewise, given the reported payload of pMSCV (Baldeschi et al. 2003), we expect that 3′ UTRs as long as 5 kb can be cloned in the miR-Sens vector and tested in target cells without loss of sensitivity. So far, we have prepared more than 40 3′ UTRs with sizes ranging up to 3 kb without significant loss of titer. Notably, the miR-Sens vector can be used in any mammalian dividing primary cells, except for cells in which the 5′ LTR promoter is not active, such as ES cells. Moreover, the dual-luciferase cassette from miR-Sens can be transferred to a lentiviral backbone in order to transduce nondividing cells.
We think that the potential applications of the miR-Sens vector go beyond the miRNA field. By cloning the 3′ UTR of a gene of interest behind the reporter and transducing cells in which this gene might be regulated (for instance after growth factor signaling or during differentiation), it can be established that, indeed, the expression of this gene is regulated through its 3′ UTR. Potential regulation by miRNAs, RBPs, or other factors (Gong and Maquat 2011) can then be deduced by careful examination of the responsive sequences within the 3′ UTR. Indeed, increasing evidence suggests that miRNAs also control mRNA transcription/translation through their interaction with open reading frames (ORFs) or 5′ UTRs (Orom et al. 2008; Bartel 2009; Huang et al. 2010; Shin et al. 2010; Brest et al. 2011). miR-Sens can be easily modified to test the activity of miRNAs in these nonconventional miRNA target sites. Notably, a T2A ribosome-skipping site conveniently inserted between the open reading frame of the gene of interest and the Renilla luciferase should ensure that the half-life of the target ORF does not influence the abundance of the reporter luciferase (Szymczak et al. 2004). Additionally, our protocol allows medium throughput assays with up to dozens of cells types to be transduced in triplicate with a single viral supernatant batch, widening the opportunities of testing hypotheses regarding translational control of a gene in panels of primary or tumor cells.
In conclusion, we have developed a new tool for the functional study of miRNAs in primary cells. We have demonstrated that the miR-Sens vector combines the reproducibility of a dual-luciferase reporter with the ease of use of retroviral transductions, without losing its sensitivity to miRNA-mediated translational control. We think that this vector will not only be useful for laboratories interested in miRNA biology, but also more broadly in the translational regulation of mRNAs.
MATERIALS AND METHODS
Construction of the miR-Sens vector
The miR-Sens vector was constructed by ligating the NheI–BamHI fragment of the psiCHECK2 (Promega) vector to a pMSCV vector at EcoRI/BamHI sites, using oligo adapters (pMSCV_EcoRINheI_FW: AATTCAAGCTTACATG and pMSCV_EcoRINheI_RV: CTAGCATGTAAGCTTG). The SV40 polyadenylation signal was deleted by ligating the XhoI–BamHI fragment containing the pMSCV backbone to a XbaI/XhoI fragment containing the TK promoter and the Firefly luciferase using the oligos pMSCV_XbaIBamHI-FW: CTAGAAAGCTTGATCG and pMSCV_XbaIBamHI-RV: GATCCGATCAAGCTTT creating an additional HindIII site. The multiple cloning site (MCS) of the miR-Sens was modified with the following oligos to delete the EcoRI site and introduce PmlI, BglII, and BamHI sites (XhoI-NotI-Linker_FW: TCGAGCACGTGCTATAGATCTATCTGGATCCGC and XhoI-NotI-Linker_RV: GGCCGCGGATCCAGATAGATCTATAGCACGTGC) between the XhoI and NotI sites. The polyadenylation signal immediately downstream from the Renilla luciferase was mutated by PCR on the psiCHECK-2 vector using the following primers (SICHECK-pASDM_FW: GGATCCGCGGCCGCTGGCCGCTACTACATATCTT and SICHECK-pASDM_RV: CAGGGTCGCTCGGTGTTC). The PCR product was digested with MluI and NotI restriction enzymes and subsequently ligated to the cut vector. The sequence of the miR-Sens construct was confirmed by restriction analysis for the presence/deletion of sites and partial resequencing.
Cloning of miRNA sensors and 3′ UTRs
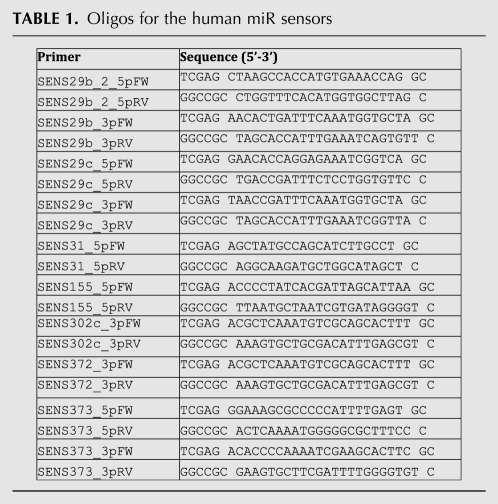
MiRNA sensor vectors were prepared by ligation of annealed oligos at the XhoI–NotI site in the miR-Sens vector (Table 1). The 3′ UTRs were PCR amplified from human BJ cell genomic DNA with the primers from Table 2 and cloned at the same site. miR-31 site mutagenesis was done by PCR on the wild-type LATS2 3′ UTR using primers from Table 2. PCR amplification was carried out with iProof enzyme (Bio-Rad). The wild-type and mutant short LATS2 3′ UTRs have been reported previously (Voorhoeve et al. 2006). Constructs identity and orientation were confirmed by sequencing.
TABLE 1.
Oligos for the human miR sensors
miR-expressing vectors
The miR-Vec Blast retroviral vector was used for stable expression of miRNAs after Blasticidin S selection (Invitrogen) of transduced cells (Voorhoeve et al. 2006). MiR-31 was cloned from BJ cell genomic DNA with the following primers (MIR31S_FW: TAGATCGGATCCCATCTTCAAAAGCGGACACTCTA and MIR31S_RV: GATCGAATTCACAATACATAGCAGGACAGGAAGTAAG) between the BamHI and EcoRI sites. The miRNA-371, miRNA-372, and miRNA-373 (subsequently referred to as miRNA 371-2-3 cluster) expression vector has been reported before (Voorhoeve et al. 2006). The identity and orientation of the PCR products were confirmed by sequencing.
Retroviral production and cell transduction
Retroviruses were prepared by transfection of 293/T cells with DNA precipitated using Calcium Phospate according to standard procedures. Retroviruses were pseudotyped with either an ecotropic (pCL-Eco) or amphotropic (pCL-Ampho, for miR-Sens only) envelope. miR-Sens vector plasmid DNA was cotransfected along with a packaging plasmid and a pmCherry-C1 vector to monitor transfection efficiency under a fluorescence microscope, the efficiency routinely exceeding 50%. Retroviral supernatants were harvested at 48 and 72 h after transfection, pooled on ice, aliquoted and flash-frozen with liquid nitrogen. Cell transduction (20–50,000 cells) was carried out with 150 μL of viral supernant and 350 μL of media in the presence of eight microg/mL of polybrene (Sigma) in triplicate in a 24-well plate overnight. The media was replaced the next morning with 0.5 mL of fresh media. Cells were assayed for luciferase activities after 72 h.
Cell lines
The primary BJ cells (human foreskin fibroblast cells) expressing RasV12ERTAM, GFP-st, and the murine ecotropic receptor, and the AGS cell line (human gastric cancer cell line) have been previously reported (Voorhoeve et al. 2006; Oh et al. 2011). The MDA-MB-231-eco cell line (human breast cancer cell line, kindly provided by Dr. D.M. Virshup [Duke-NUS, Singapore]) and HMECs (human mammary epithelial cells, purchased from ATCC) were engineered to express the murine ecotropic receptor. HMECs were subsequently transduced with a retroviral vector expressing hTert as described previously (Voorhoeve et al. 2006). The 293/T cell line was provided by Dr. B. Reversade (IMB, Singapore). Cells were grown according to standard procedures. BJ and MDA-MB-231-eco cells were transduced with ecotropic miR-Vec blast retroviruses expressing miR-371-2-3, miR-31 or the inactive Telomerase RNA component (TERC RNA nt 1–211) as reported before, and selected with Blasticidin S (Invitrogen) for 4–7 d (Voorhoeve et al. 2006). miR-31 expression was confirmed by quantitative PCR in MDA-MB-231 cells using EXIQON miR-31 assay with U6 RNA as a control.
miRNA microarray data
Total BJ cell RNA from three independent cultures was isolated by combining TRizol with a modified protocol from Invitrogen RNA pureLink kit to allow for the extraction of RNAs smaller than 200 nt. The quality of the total RNA was verified by an Agilent 2100 Bioanalyzer profile. A total of 1000 ng of total RNA from BJ cells and reference samples were labeled with Hy3 and Hy5 fluorescent label, respectively, using the miRCURY LNA Array power labeling kit (Exiqon) following the manufacturer's instructions. The Hy3-labeled sample and a Hy5-labeled reference RNA sample were mixed pairwise and hybridized to the miRCURY LNA Array version fifth Generation (Exiqon), which contains capture probes targeting all registered human miRNAs from miRBASE version 15.0. The hybridization was performed according to the miRCURY LNA array manual using a Tecan HS4800 hybridization station (Tecan). The miRCURY LNA array microarray slides were scanned using the Agilent G2565BA Microarray Scanner System (Agilent Technologies, Inc.) and the image analysis was carried out using the ImaGene 9.0 software (BioDiscovery Inc.). The raw Hy3 fluorescence intensities were used for the linear regression without background substraction.
Luciferase assays
The Dual-Luciferase Reporter kit (Promega) was used for the detection of the luciferase activities according to the manufacturer's protocol. Cells were lysed briefly in 24-well plates for 20 min at room temperature with 100 μL of 1X PLB buffer. Subsequently, 10 μL of the lysate was tested with 50 μL of each reagent in a black 96-well plate (NUNC). The luciferase activities were detected with a Tecan Infinite M200 microplate reader. The results (mean ± SD) from three independent cell transductions are shown and experiments were repeated at least twice with similar results unless otherwise stated. The threshold for a significant change in the relative Renilla over Firefly luciferase activity compared with control vector was calculated based on the standard deviation of the ratio from two independent transductions in triplicate in BJ and AGS cells (four experiments). Values outside of the mean ± 3SD range are considered significantly changed from the control.
Statistics
The statistical analysis was done using Graphpad Prism V4 (Graphpad). We have used the unpaired t-test or the one-way ANOVA test where appropriate for statistical comparison of results. The linearity was tested by linear regression. Precision of a series of replicates was calculated as follows: Precision(%) = SD/mean.
ACKNOWLEDGMENTS
We acknowledge Dr. P. Tan, Dr. D.M. Virshup, and Dr. M. Fivaz (Duke-NUS), Dr. B. Reversade (IMB, Singapore) and Dr. J. Kluiver (UMCG, The Netherlands) for sharing reagents. We thank Ted Chang for critically reviewing of this manuscript. This work was supported by the Association for International Cancer Research (Grant no. 09-0126) and Duke-NUS Graduate Medical School.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.031831.111.
REFERENCES
- Baldeschi C, Gache Y, Rattenholl A, Bouillé P, Danos O, Ortonne J-P, Bruckner-Tuderman L, Meneguzzi G 2003. Genetic correction of canine dystrophic epidermolysis bullosa mediated by retroviral vectors. Hum Mol Genet 12: 1897–1905 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blø M, Micklem DR, Lorens JB 2008. Enhanced gene expression from retroviral vectors. BMC Biotechnol 8: 19 doi: 10.1186/1472-6750-8-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier J-F, Hébuterne X, et al. 2011. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet 43: 242–245 [DOI] [PubMed] [Google Scholar]
- Cho WJ, Shin JM, Kim JS, Lee MR, Hong KS, Lee J-H, Koo KH, Park JW, Kim K-S 2009. miR-372 regulates cell cycle and apoptosis of ags human gastric cancer cell line through direct regulation of LATS2. Mol Cells 28: 521–527 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Gong C, Maquat LE 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Wu S, Ding J, Lin J, Wei L, Gu J, He X 2010. MicroRNA-181a modulates gene expression of zinc finger family members by directly targeting their coding regions. Nucleic Acids Res 38: 7211–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarifar F, Yao P, Eswarappa SM, Fox PL 2011. Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J 30: 1324–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P, Coffin JM 2008. Effects of retroviruses on host genome function. Annu Rev Genet 42: 709–732 [DOI] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S-I, Baba T, Suzuki T 2009. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev 23: 433–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JAF, Slanchev K, le Sage C, Nagel R, Voorhoeve M, van Duijse J, Ørom UA, et al. 2007. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131: 1273–1286 [DOI] [PubMed] [Google Scholar]
- Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JAF, Elkon R, Agami R 2010. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol 12: 1014–1020 [DOI] [PubMed] [Google Scholar]
- Kim Y-K, Yeo J, Ha M, Kim B, Kim VN 2011. Cell adhesion-dependent control of microRNA decay. Mol Cell 43: 1005–1014 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih I, Jones-Rhoades M, Bartel DP, Burge CB 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798 [DOI] [PubMed] [Google Scholar]
- Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, Demidenko E, Korc M, Shi W, Preis M, et al. 2010. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest 120: 1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loening AM, Dragulescu-Andrasi A, Gambhir SS 2010. A red-shifted Renilla luciferase for transient reporter-gene expression. Nat Methods 7: 5–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA 2007. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449: 682–688 [DOI] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. 2010. The landscape of C. elegans 3′UTRs. Science 329: 432–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazière P, Enright AJ 2007. Prediction of microRNA targets. Drug Discov Today 12: 452–458 [DOI] [PubMed] [Google Scholar]
- Oh H-K, Tan AL-K, Das K, Ooi CH, Deng N-T, Tan IB, Beillard E, Lee J, Ramnarayanan K, Rha SY, et al. 2011. Genomic loss of miR-486 regulates tumor progression and the OLFM4 antiapoptotic factor in gastric cancer. Clin Cancer Res 17: 2657–2667 [DOI] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH 2008. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 30: 460–471 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ 2011. Ending the message: Poly(A) signals then and now. Genes Dev 25: 1770–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retelska D, Iseli C, Bucher P, Jongeneel CV, Naef F 2006. Similarities and differences of polyadenylation signals in human and fly. BMC Genomics 7: 176 doi: 10.1186/1471-2164-7-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP 2011. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 146: 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C, Nam J-W, Farh KK-H, Chiang HR, Shkumatava A, Bartel DP 2010. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell 38: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DAA 2004. Correction of multi-gene deficiency in vivo using a single “self-cleaving” 2A peptide-based retroviral vector. Nat Biotechnol 22: 589–594 [DOI] [PubMed] [Google Scholar]
- Thomas M, Lieberman J, Lal A 2010. Desperately seeking microRNA targets. Nat Struct Mol Biol 17: 1169–1174 [DOI] [PubMed] [Google Scholar]
- Valastyan S, Reinhardt F, Benaich N, Wang ZC, Richardson AL, Weinberg RA 2009. A pleiotropically acting MicroRNA, miR-31, inhibits breast cancer metastasis. Cell 137: 1032–1046 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- van Kouwenhove M, Kedde M, Agami R 2011. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer 11: 644–656 [DOI] [PubMed] [Google Scholar]
- Voorhoeve PM 2010. MicroRNAs: Oncogenes, tumor suppressors or master regulators of cancer heterogeneity? Biochim Biophys Acta 1805: 72–86 [DOI] [PubMed] [Google Scholar]
- Voorhoeve PM, le Sage C, Schrier M, Gillis AJM, Stoop H, Nagel R, Liu Y-P, van Duijse J, Drost J, Kulalert W, et al. 2006. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124: 1169–1181 [DOI] [PubMed] [Google Scholar]
- Wu L, Belasco JG 2008. Let me count the ways: Mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell 29: 1–7 [DOI] [PubMed] [Google Scholar]
- Wu S, Huang S, Ding J, Zhao Y, Liang L, Liu T, Zhan R, He X 2010. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene 29: 7211–7218 [DOI] [PubMed] [Google Scholar]
- Yan G-R, Xu S-H, Tan Z-L, Liu L, He Q-Y 2011. Global identification of miR-373-regulated genes in breast cancer by quantitative proteomics. Proteomics 11: 912–920 [DOI] [PubMed] [Google Scholar]
- Yang S, Delgado R, King SR, Woffendin C, Barker CS, Yang ZY, Xu L, Nolan GP, Nabel GJ 1999. Generation of retroviral vector for clinical studies using transient transfection. Hum Gene Ther 10: 123–132 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Verbeek FJ 2010. Comparison and integration of target prediction algorithms for microRNA studies. J Integr Bioinform 7: 127 doi: 10.2390/biecoll-jib-2010-127 [DOI] [PubMed] [Google Scholar]