The tRNAHis guanylyltransferase (Thg1) family of enzymes comprises members from all three domains of life (Eucarya, Bacteria, Archaea). Although the initial activity associated with Thg1 enzymes was a single 3′-to-5′ nucleotide addition reaction that specifies tRNAHis identity in eukaryotes, the discovery of a generalized base pair–dependent 3′-to-5′ polymerase reaction greatly expanded the scope of Thg1 family-catalyzed reactions to include tRNA repair and editing activities in bacteria, archaea, and organelles. Here the authors review recent advances toward understanding diverse Thg1 family enzyme functions and mechanisms. They also discuss possible evolutionary origins of Thg1 family–catalyzed 3′-to-5′ addition activities and their implications for the currently observed phylogenetic distribution of Thg1-related enzymes in biology.
Keywords: 3′-to-5′ polymerase, Thg1-like protein, tRNA editing, tRNAHis guanylyltransferase, tRNAHis recognition
Abstract
The tRNAHis guanylyltransferase (Thg1) family of enzymes comprises members from all three domains of life (Eucarya, Bacteria, Archaea). Although the initial activity associated with Thg1 enzymes was a single 3′-to-5′ nucleotide addition reaction that specifies tRNAHis identity in eukaryotes, the discovery of a generalized base pair–dependent 3′-to-5′ polymerase reaction greatly expanded the scope of Thg1 family–catalyzed reactions to include tRNA repair and editing activities in bacteria, archaea, and organelles. While the identification of the 3′-to-5′ polymerase activity associated with Thg1 enzymes is relatively recent, the roots of this discovery and its likely physiological relevance were described ∼30 yr ago. Here we review recent advances toward understanding diverse Thg1 family enzyme functions and mechanisms. We also discuss possible evolutionary origins of Thg1 family–catalyzed 3′-to-5′ addition activities and their implications for the currently observed phylogenetic distribution of Thg1-related enzymes in biology.
INTRODUCTION
Virtually all known nucleic acid polymerases catalyze the addition of nucleotides with a 5′-to-3′ polarity. This is the case for both template-dependent polymerases (e.g., DNA polymerases, RNA polymerases, reverse transcriptases) (Joyce and Steitz 1995) and template-independent ones (e.g., polyA polymerase, terminal transferases, CCA-adding enzymes) (Ratliff 1981; Martin and Keller 1996; Yue et al. 1996), as well as enzymes containing an internal template (e.g., telomerase) (Blackburn 1992). These reactions are mechanistically equivalent, involving the attack of the 3′-OH of the polynucleotide chain on the triphosphate moiety of the incoming nucleotide, liberating pyrophosphate (Fig. 1A). However, similar reaction chemistry can synthesize nucleic acids in the 3′-to-5′ direction. In this scenario, the 3′-OH of an incoming nucleotide attacks the 5′ end of a polynucleotide having a 5′-tri (or di)-phosphate (Fig. 1B).
FIGURE 1.
Chemically equivalent mechanisms of 5′-to-3′ and 3′-to-5′ nucleic acid synthesis. (A) 5′-to-3′ pathway of nucleotide addition catalyzed by canonical DNA/RNA polymerases. The reaction involves attack of the 3′-hydroxyl of the growing polynucleotide chain on the 5′-triphosphate of the incoming NTP, with release of pyrophosphate (PPi). (B) Alternative 3′-to-5′ pathway for polynucleotide synthesis catalyzed by Thg1 family enzymes. The reversal of functional groups with respect to the nucleic acid and NTP substrates results in the net extension of the nucleic acid chain in the opposite direction to extension by other DNA/RNA polymerases.
The preponderance of 5′-to-3′ polymerization has been suggested to be due to the advantage conferred during proofreading. Removal of a mismatched nucleotide by the 3′-to-5′ exonuclease activity of DNA polymerases (Lehman and Richardson 1964), as well as backtracking and cleavage of nascent RNA chains by RNA polymerases (Uptain et al. 1997), regenerates a 3′-OH end that is competent to participate in the addition of the next nucleotide without further activation. In contrast, removal of nucleotides from the 5′ end of a polynucleotide results in a 5′ monophosphate (Deutscher and Kornberg 1969), which requires either the addition of phosphates or activation (e.g., via an adenylylated intermediate) prior to subsequent reactions (Fig. 1). However, the ability to add ≥1 nucleotides to the 5′ ends of RNAs could clearly be advantageous in some situations. Examples of 3′-to-5′ polymerization are rare, but recent work has uncovered an entire family of reverse polymerases that add nucleotides to the 5′ ends of RNAs (Price and Gray 1999b; Gu et al. 2003; Abad et al. 2010, 2011; Heinemann et al. 2010, 2011; Rao et al. 2011). Unlike the capping reaction that attaches a 5′ G residue to RNA polymerase II transcripts via a 5′-to-5′ linkage (Banerjee 1980), this emerging family of polymerases creates standard 3′-to-5′ phosphodiester bonds (Price and Gray 1999b; Gu et al. 2003; Abad et al. 2010, 2011). Moreover, the ability of Thg1 polymerases to also catalyze 5′-phosphate activation casts some doubt on the “proofreading” rationale for 5′-to-3′ polymerase evolution, since even the additional consumption of an ATP molecule that would occur during this type of proofreading step is likely outweighed by the substantial number of ATP molecules needed to ligate Okazaki fragments synthesized in the 5′-to-3′ direction during lagging strand synthesis. The unusual protein superfamily that carries out 3′-to-5′ polymerization is the focus of this review.
The founding member of the reverse polymerase family of enzymes is yeast (Saccharomyces cerevisiae) tRNAHis guanylyltransferase (Thg1), an essential protein responsible for the addition of an extra G to the 5′ end of tRNAHis (Gu et al. 2003). Early studies by Söll and colleagues demonstrated that the addition of the 5′ guanylate occurs post-transcriptionally via a 3′-to-5′ linkage (Cooley et al. 1982). G addition by Thg1 is a nontemplated reaction, with the G inserted opposite the conserved A73 of tRNAHis (Nameki et al. 1995). Surprisingly, however, when the A73 discriminator nucleotide is changed to C73, yeast Thg1 catalyzes the sequential 3′-to-5′ addition of multiple nucleotides to the 5′ end of the tRNA, using the 3′ portion of the acceptor stem as a template (Jackman and Phizicky 2006).
The 3′-to-5′ polymerase reaction carried out by Thg1 is reminiscent of another activity that modifies the 5′ end of tRNAs, namely, the editing reaction that occurs in the mitochondria of certain eukaryotic microbes (Price and Gray 1998). Editing corrects acceptor stem mismatches encoded within mitochondrial tRNA genes through a process that includes removal of the mismatched nucleotides from the 5′ end of the tRNA and 3′-to-5′ re-synthesis using the 3′ portion of the acceptor stem as template (Price and Gray 1999b; Laforest et al. 2004; Bullerwell and Gray 2005). The striking parallels between the tRNA 5′ editing and Thg1 activities suggested that the enzymes catalyzing these reactions were likely to be evolutionarily related (Price and Gray 1999b; Gu et al. 2003).
Genes with sequence similarity to Thg1 have now been identified in a range of organisms, encompassing the domains Archaea and Bacteria, as well as Eucarya (Gu et al. 2003; Heinemann et al. 2009, 2010, 2011; Abad et al. 2010, 2011; Rao et al. 2011). As discussed below, characterization of representative bacterial and archaeal Thg1-like proteins (TLPs) revealed that these enzymes display robust 3′-to-5′ templated synthesis using various 5′-truncated tRNAs as templates (Abad et al. 2010; Rao et al. 2011). In addition, recent evidence suggests a role for TLPs in 5′ editing of mitochondrial tRNAs in the amoeboid protist, Dictyostelium discoideum (Abad et al. 2011). Thus, TLP family members appear to play roles in tRNA 5′ editing and general tRNA repair. It seems likely, therefore, that such quality control activities are more prevalent than generally appreciated.
Remarkably, although the sequence of Thg1 shows no obvious similarities to canonical polymerases, the recently solved crystal structure of the human Thg1 (hTHG1) enzyme exhibits striking structural homology with 5′-to-3′ DNA polymerases, including the placement of residues critical for catalysis (Hyde et al. 2010). These findings suggest that 5′-to-3′ and 3′-to-5′ polymerization activities may be evolutionarily related.
YEAST tRNAHisGUANYLYLTRANSFERASE: FOUNDING MEMBER OF THE Thg1 SUPERFAMILY
An additional 5′-guanylate residue is a hallmark of virtually all tRNAHis species (Sprinzl et al. 1998). The presence of G at the −1 position (according to standard tRNA numbering) serves as an important identity element for histidylation of tRNAHis by its cognate aminoacyl-tRNA synthetase, HisRS (Himeno et al. 1989; Nameki et al. 1995; Giegé et al. 1998; Rosen and Musier-Forsyth 2003). Notable exceptions to the rule are approximately 20 α-proteobacterial species, whose tRNAHis lacks an additional G−1 residue and which contain a simultaneous variation in HisRS that allows efficient aminoacylation of the G−1-deficient tRNA (Fig. 2A; Ardell and Andersson 2006; Wang et al. 2007; Yuan et al. 2011). In Escherichia coli and chloroplasts, the G−1 nucleotide is encoded in the genome, present in the precursor-tRNA transcript, and retained in the final mature tRNA species after removal of the rest of the 5′ leader sequence by ribonuclease P (RNase P) (Fig. 2B; Burkard and Söll 1988; Burkard et al. 1988). A similarly encoded G−1 residue is found throughout Bacteria (with the exception of α-Proteobacteria) and in many, but not all, archaeal genomes for which sequences are available. A different origin for the essential G−1 residue in eukaryotes was first described in Schizosaccharomyces pombe and Drosophila melanogaster (Cooley et al. 1982; Williams et al. 1990). In these species, G is not encoded at the −1 position of cytoplasmic tRNAHis genes; instead, a requirement for post-transcriptional enzymatic addition of the G−1 residue was demonstrated (Fig. 2C). Despite the importance of the G−1 nucleotide for tRNAHis identity and the unique nature of this requirement for a single additional nucleotide among well-studied tRNAs, the identity of the gene encoding the enzyme that catalyzes the G−1 addition reaction remained a mystery for >20 yr.
FIGURE 2.
Multiple mechanisms for specifying tRNAHis identity. (A) tRNAHis identity in several groups of α-proteobacteria, including Rhodobacterales and Caulobacterales. In these species, tRNAHis genes lack an encoded G−1, and the precursor tRNA (5′ leader sequence indicated in blue) is cleaved by RNase P to leave the tRNA without the usual G−1 residue. The HisRS in these organisms is atypical and recognizes alternative identity elements in the α-proteobacterial tRNAHis for G−1-independent aminoacylation. (B) tRNAHis identity in E. coli and chloroplasts. tRNAHis genes in these cases are encoded with a G−1 residue in the 5′ leader sequence (indicated in blue), which is retained following 5′ end processing by RNase P to yield the G−1-containing tRNA that is the substrate for aminoacylation by HisRS. Many archaea and all bacteria, with the exception of the few α-proteobacterial species described in A, encode G−1 in their tRNAHis genes and therefore presumably obtain the G−1 identity element by this mechanism. (C) tRNAHis identity in eukaryotes. In eukaryotes, tRNAHis genes do not contain the G−1 identity element in the precursor sequence (indicated in blue), and the residue is added post-transcriptionally by Thg1, after 5′ end processing by RNase P. A small number of eukaryotic species lack an identifiable Thg1 enzyme in their genomes; the G−1-status and its requirement for tRNAHis identity have not been investigated in these species.
Using a biochemical genomics approach, in which a library of all 6000 predicted open reading frames in yeast can be assayed simultaneously for a given biochemical activity, an essential gene was identified (YGR024c), the product of which incorporated [α-32P]-GTP into unlabeled yeast tRNAHis transcripts lacking the additional G−1 residue (Gu et al. 2003). TheYGR024c gene encodes a 24-kD polypeptide, and the purified protein exhibits ATP-dependent guanylyltransferase activity with tRNAHis transcripts carrying a 5′-monophosphate. Conditional depletion of the YGR024c (now THG1) ORF in yeast led to complete loss of G−1-containing tRNAHis and concomitant loss of aminoacylation, confirming the participation of the YGR024c gene product in G−1-addition in vivo (Gu et al. 2005). When expressed and purified from E. coli, this protein exhibited the expected activities of the tRNAHis guanylyltransferase enzyme and was named Thg1 (tRNAHis guanylyltransferase) (Gu et al. 2003). This designation is used in place of the earlier TGT (Jahn and Pande 1991) to avoid confusion with another well-characterized tRNA modification enzyme, the tRNA guanine transglycosylase. Interestingly, a second ORF (YDL076c) was also identified in the screen for G−1 addition activity in yeast and the N-terminal GST-tagged YDL076c gene product copurified with tRNAHis guanylyltransferase activity. The role, if any, of this nonessential gene in tRNAHis maturation remains unknown.
AN UNUSUAL 3′-TO-5′ NUCLEOTIDE ADDITION REACTION
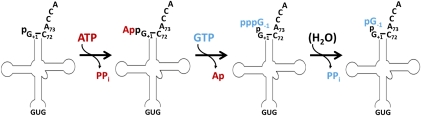
Biochemical characterization of purified yeast Thg1 confirmed several features of the proposed 3′-to-5′ addition mechanism for enzymatic G−1 addition to tRNAHis (Fig. 3), particularly the requirement for ATP to activate the 5′ end of the monophosphorylated tRNA in the first step of the reaction, via creation of a 5′-5′ phosphoanhydride with AMP (Jahn and Pande 1991; Gu et al. 2003). In the second step, Thg1 uses the 3′-hydroxyl of GTP to attack the activated App-tRNA intermediate, yielding G−1-containing tRNAHis, with the additional nucleotide linked by a standard 3′-to-5′ phosphodiester bond. In a third step, Thg1 removes a pyrophosphate moiety from the 5′ end of the added G−1 residue, producing mature tRNAHis in the form that is preferred for HisRS recognition. Importantly, despite the similarity of the first two chemical steps to reactions catalyzed by members of the DNA/RNA ligase family of enzymes, no sequence similarity was identified between Thg1 and ligases that might suggest a similar molecular mechanism for Thg1 catalysis. Additionally, while the ligase reaction is well known to proceed via formation of a covalent adenylylated-enzyme intermediate (involving a highly conserved lysine residue) (Shuman and Hurwitz 1981; Tomkinson et al. 1991; Shuman and Lima 2004), no adenylylated-Thg1 was detected with purified enzyme, even after prolonged incubation with labeled [α-32P]-ATP.
FIGURE 3.
Three-step mechanism for G−1 addition to tRNAHis by yeast Thg1. First, the 5′-monophosphorylated tRNAHis is activated by adenylylation using ATP. In the second step, the G−1 nucleotide (in the form of GTP) is transferred to the activated 5′ end, releasing AMP. In the third step, the 5′-pyrophosphate is removed from the G−1 nucleotide to yield the monophosphorylated G−1-containing tRNAHis that is the optimal substrate for HisRS.
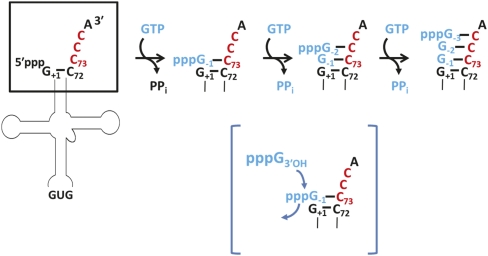
In (most) bacteria, the additional G−1 residue on tRNAHis is involved in a Watson-Crick base pair with an invariant bacterial C73 discriminator nucleotide, creating an unusual 8-bp aminoacyl acceptor stem (Fig. 2B). In contrast, in eukaryotes, the discriminator nucleotide is universally an A73, and the addition of G−1 does not form an additional Watson-Crick base pair at the beginning of the aminoacyl acceptor stem (Fig. 2C). Alteration of the universally conserved A73 in yeast tRNAHis to the bacterial C73 led to a surprising observation. Instead of adding a single G−1 residue to the 5′ end of the C73-tRNAHis variant, wild-type yeast Thg1 added a series of three G residues to this substrate, creating Watson-Crick base pairs with C73, C74, and C75 at the 3′ end of the tRNA (Fig. 4; Jackman and Phizicky 2006). The number of added G residues depended on the number of sequential C residues acting as a template at the 3′ end of the tRNA, since introduction of an A residue at either position 74 or 75 terminated the multiple nucleotide addition reaction. Up to six G residues could be added to a variant tRNAHis containing an extended run of six C residues at its 3′ end. The template-dependent polymerization reaction was not limited to G, since Thg1 also added multiple C residues to a tRNAHis variant containing G73, G74, and G75 (Jackman and Phizicky 2006). Multiple nucleotide addition only occurred in the presence of the correct Watson-Crick base-pairing NTP. Thus, yeast Thg1 catalyzes a second biochemical activity, polymerizing Watson-Crick base-paired nucleotides in the opposite direction (3′-to-5′) to all studied DNA and RNA polymerases: the first example of an enzyme able to carry out this activity. Thg1-catalyzed 3′-to-5′ polymerization has also been observed in vivo in yeast using tRNAHis variant substrates (Preston and Phizicky 2010), but a physiological function for the polymerization reaction in yeast remains unknown. Nonetheless, the unexpected ability of Thg1 to recognize and use Watson-Crick base pairs suggests the possibility of a biological function for the reverse polymerase activity.
FIGURE 4.
Yeast Thg1 catalyzes template-dependent 3′-to-5′ polymerase activity. Utilizing tRNAHis variant substrates that contain C73 instead of the wild-type A73, yeast Thg1 catalyzes sequential addition of up to three G-residues to the 5′ end of the tRNA. The reverse polymerase reaction only occurs in the presence of the correct Watson-Crick base-pairing nucleotide (GTP in the case of the C73-tRNA). For multiple nucleotide additions, the 5′-triphosphorylated end resulting from the previous nucleotide addition is the activated end for attack by the 3′-hydroxyl of the subsequent nucleotide, as shown in detail in brackets.
Interestingly, in addition to Thg1-catalyzed RNA-dependent RNA reverse polymerization (RDRrevP), RNA-dependent DNA reverse polymerase (RDDrevP) activity has also been observed with yeast Thg1 (Jackman and Phizicky 2006), suggesting the possibility of Thg1 involvement in either DNA or RNA metabolism, in addition to its demonstrated role with tRNAHis. The ability of the single Thg1 enzyme to catalyze RDRrevP and RDDrevP activities suggests a resemblance between Thg1 and repair-type polymerases, which also exhibit more relaxed specificities for sugar-nucleotide substrates.
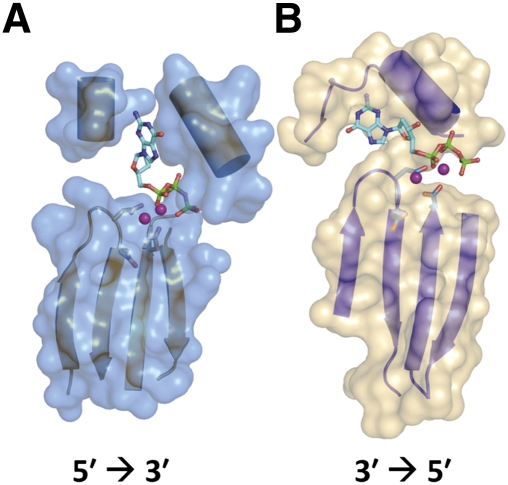
When tRNA substrates containing activated 5′ ends are employed, the reverse polymerization reaction does not require inclusion of ATP, suggesting a mechanism of reverse polymerization in which each successive nucleotide is added to the 5′-triphosphorylated end created by the preceding nucleotide addition (Fig. 4; Jackman and Phizicky 2006). The chemistry of the 3′-to-5′ polymerization reaction, involving attack of NTP 3′-OH groups on 5′-triphosphorylated polynucleotide substrates, is strikingly reminiscent of the chemistry of canonical 5′-to-3′ polymerization (Fig. 1). The 2.95 Å crystal structure of hTHG1 provided important insight, revealing that hTHG1 shares unexpected structural similarity with canonical 5′-to-3′ nucleotide polymerases, particularly to A-family polymerases such as T7 DNA polymerase (Fig. 5; Hyde et al. 2010). The enzyme crystallized with two bound dGTP nucleotides in the active site, and orientation of the bound dGTP combined with kinetic investigation of selected hTHG1 variants suggests that the captured structure represents the enzyme poised for the first step of the reaction, adenylylation (Jackman and Phizicky 2008; Hyde et al. 2010; Smith and Jackman 2011). Although the structural and biochemical data suggest that, as with all known 5′-to-3′ DNA/RNA polymerases, a two-metal-ion mechanism of catalysis is used for 3′-to-5′ polymerization, many aspects of the molecular mechanism of Thg1 catalysis remain to be elucidated. In particular, yet to be addressed is the basis for tRNA substrate binding and recognition, as well as the basis for selection of Watson-Crick base-paired nucleotides for the polymerization reaction and for the selection of the non-Watson-Crick-paired G−1 residue for addition to tRNAHis in eukaryotes. Nonetheless, the structure provides the first glimpse into the active site of this unique enzyme family. The observation that the active site that catalyzes canonical 5′-to-3′ polymerization is comparable to that used to catalyze 3′-to-5′ polymerization raises important evolutionary questions regarding the selection of the 5′-to-3′ polymerase reaction as the dominant enzyme activity observed in nature.
FIGURE 5.
Thg1 and canonical 5′-to-3′ polymerases share significant structural similarity. A model of the active site of T7 DNA polymerase (A; Protein Data Bank identification 1T7P) and human Thg1 (B; Protein Data Bank 3OTB), each co-crystallized with bound dNTP, reveals that both enzymes share a similar secondary structure characteristic of the polymerase palm domain, including two metal ions that are critical for catalysis by each enzyme. The bound nucleotide visible in the T7 DNA polymerase structure reveals the position of the incoming dNTP, poised for attack by the 3′-hydroxyl of the elongating primer (data not shown). The bound nucleotide observed in the hThg1 structure is believed, based on kinetic and mutational data, to mimic the position of the nucleotide used to activate the 5′ end of the tRNA, consistent with the reversed orientation of the nucleotide substrates for 5′-to-3′ and 3′-to-5′ polymerization.
TLPS IN BACTERIA AND ARCHAEA
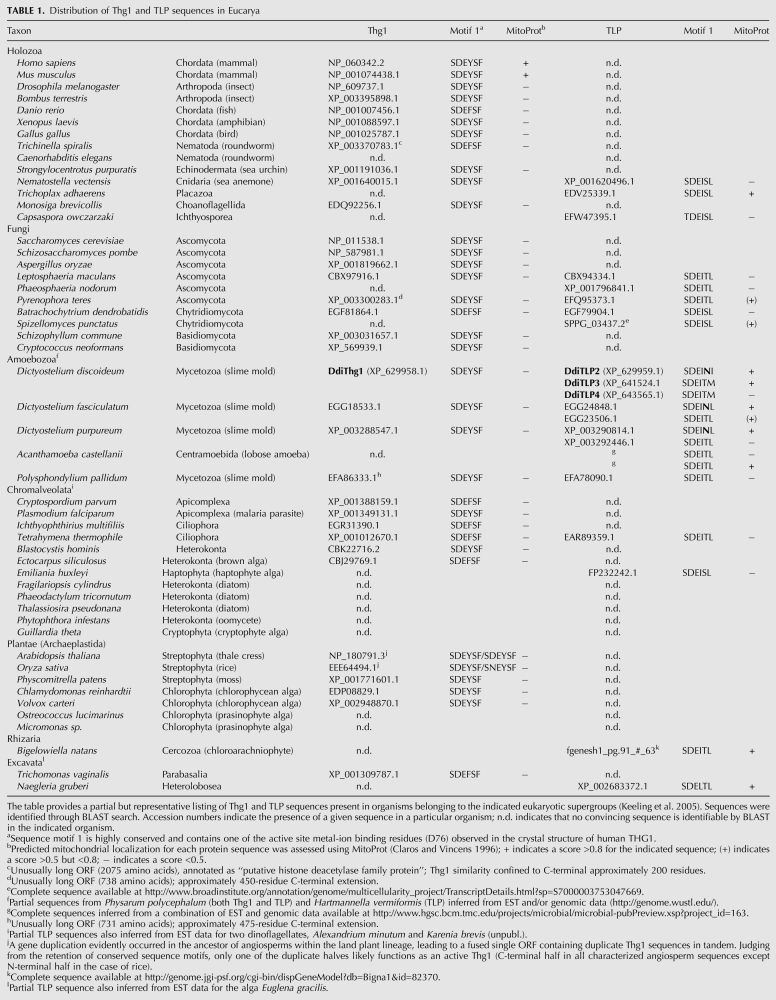
The discovery of the unusual template-dependent 3′-to-5′ polymerization activity catalyzed by yeast Thg1 prompted a search for alternative functions for 3′-to-5′ addition. Genes with sequence similarity to yeast Thg1 had been identified in a number of archaeal and bacterial species through BLAST searches. Some of the archaeal genes, and all of the bacterial genes, reside in organisms that already encode a G−1 residue at the 5′ end of their tRNAHis genes (see Fig. 2B), raising the question of whether the Thg1-related gene is present in these species for another purpose. Sequence comparison between eucaryal Thg1 family enzymes and the bacterial and archaeal genes pinpointed domain-specific differences in regions that are of demonstrated importance for the G−1 addition activity of yeast Thg1, most notably the absence of a highly conserved, eukaryote-specific HINNLYN sequence (motif 2) in the archaeal and bacterial genes (Jackman and Phizicky 2008) and variations in active site motif 1 (Table 1). We proposed a classification system based on these sequence differences, such that the designation “Thg1” is reserved for bona fide yeast Thg1 orthologs that contain the HINNLYN sequence (so far only observed in eukaryotes), while the designation “Thg1-like protein” (TLP) refers to all homologs that do not cluster with yeast Thg1 in phylogenetic reconstructions (Fig. 6). Although the majority of TLPs identified on the basis of these sequence differences are found in the domains Bacteria and Archaea, several TLPs have been identified in the genomes of eukaryotic microbes, as described below (Table 1). Classification based on sequence similarity is preferred because of the likely functional importance of the domain-specific sequence differences that have been identified (see below) and the potential for multiple functions for some or all of the Thg1-related genes.
TABLE 1.
Distribution of Thg1 and TLP sequences in Eucarya
FIGURE 6.
Phylogenetic reconstruction depicting the evolutionary relationships among members of the Thg1 superfamily. Numbers in parentheses refer to the number of sequences used in the construction of the maximum likelihood (ML) phylogenetic tree (Supplemental Fig. S1) upon which the figure is based. Numbers along edges are bootstrap values (see below). Names of phyla or supergroups in each clade are listed. The branching positions of the four Dictyostelium discoideum TLPs (DdiTLP1 to 4; see text) are indicated by arrows. Methodology: Thg1 and TLP sequences were retrieved in December 2011 using the BLASTP program (Altschul et al. 1997) from the nr database at the NCBI (http://www.ncbi.nlm.nih.gov/). BLAST outputs were checked manually to identify homologs (no arbitrary cut-off e-value was used). The corresponding sequences were aligned using MAFFT v6.857b (Katoh et al. 2002). The resulting alignments were then inspected visually and refined manually using the ED program from the MUST package (Philippe 1993). Prior to phylogenetic analyses, regions of doubtful homology were removed manually from the alignments using NET from the MUST package (data sets are available on request). ML phylogenetic trees were computed with PHYML 3.0 (Guindon and Gascuel 2003) using the LG model (Le and Gascuel 2008) and a γ correction to take into account the heterogeneity of the evolutionary rates across sites (using four discrete classes of sites and an estimated α parameter). Branch robustness of the resulting trees was estimated using the nonparametric bootstrap procedure implemented in PHYML using 100 replicates of the original data set and the same parameters as for tree reconstruction. In order to obtain further insights into the evolutionary history of these proteins, an accurate phylogenetic analysis was performed using the same methods as outlined above with a subset of Thg1/TLP homologs reflecting the taxonomic diversities of this gene family.
TLPs from several archaeal species (Methanosarcina acetivorans, Methanosarcina barkeri, Methanopyrus kandleri, Methanothermobacter thermoautotrophicus, Methanosaeta thermophile, and Pyrobaculum aerophilum) and two bacteria (Bacillus thuringiensis and Myxococcus xanthus) have been biochemically characterized (Heinemann et al. 2009, 2010; Abad et al. 2010, 2011; Rao et al. 2011). All exhibit tRNAHis guanylyltransferase activity, using 3′-to-5′ addition to add a G−1 residue to a variety of histidyl-tRNAs (derived from yeast, bacteria, and archaea). However, tRNAHis species from several of these organisms (M. acetivorans, M. barkeri, B. thuringiensis, and M. xanthus) do not predictably require post-transcriptional addition of G−1, although the possibility of a role in tRNAHis maturation, owing to loss of the encoded G−1 from the transcript during maturation by RNase P, as has been observed in the case of tRNAHis in some plants (Placido et al. 2010), cannot be excluded. As with yeast Thg1, TLPs catalyze efficient Watson-Crick template-dependent 3′-to-5′ nucleotide addition, attaching G−1 to C73-containing tRNAHis in vitro and in vivo in yeast (Abad et al. 2010; Heinemann et al. 2010; Rao et al. 2011). However, in contrast to bona fide Thg1 enzymes (exemplified by yeast or human Thg1), TLPs catalyze inefficient addition of non-Watson-Crick-paired G−1 to A73-tRNAHis, consistent with the absence of this substrate in the domains Bacteria and Archaea (Abad et al. 2010; Rao et al. 2011). Interestingly, B. thuringiensis TLP (BtTLP) adds a −1 residue to A73-tRNAHis in vivo in yeast, thus supporting growth of a yeast thg1Δ strain, although Watson-Crick-dependent U−1 addition to tRNAHis may contribute to the observed complementation in vivo (Heinemann et al. 2010; Rao et al. 2011). The ability of Thg1/TLP enzymes from all three domains of life to catalyze Watson-Crick-templated, but not nontemplated 3′-to-5′ nucleotide addition, suggests that the templated activity was likely a property of the earliest Thg1 ancestors (Abad et al. 2010). The non-Watson-Crick-dependent G−1 addition reaction appears to be a unique derived trait of the eucaryal Thg1 enzymes.
ALTERNATIVE FUNCTIONS FOR Thg1/TLP ENZYMES
Additional functions for Thg1/TLP enzymes might reasonably be expected to make use of their common ability to catalyze template-dependent 3′-to-5′ nucleotide addition. Indeed, TLPs from bacteria and archaea exhibit a strong biochemical preference for repair of 5′-truncated tRNAs, using templated 3′-to-5′ addition to restore the missing nucleotides, recreating a fully base-paired aminoacyl acceptor stem (Rao et al. 2011). The 5′-tRNA repair reactions were observed with tRNA substrates missing each of the four NTPs (G, A, U, or C), were evident with tRNAs other than tRNAHis, and in all cases were catalyzed with greater efficiency (from sixfold to 300-fold as reflected by kcat/KM) than identical Watson-Crick base-paired additions occurring at the −1 position of the tRNA. Addition of up to four missing nucleotides has been observed (F Mohammad and JE Jackman, unpubl.), suggesting a role for RDRrevP activity in maintaining 5′ ends of tRNA. The use of the 5′-tRNA repair pathway in vivo in Bacteria and/or Archaea has not yet been demonstrated, but substrates for 5′-tRNA repair could conceivably arise through 5′ end truncation by exonucleases, improper 5′ end processing, or even imprecision in initiation of transcription of “leaderless” tRNAs. In this way, TLP-catalyzed 5′ end repair might complement the well-known systems that maintain high-quality tRNAs by protection of 3′ ends (Zhu and Deutscher 1987; Schürer et al. 2001).
Moreover, alternative functions for Thg1 family enzymes may not be limited to TLPs. Although addition of G−1 to tRNAHis is the only essential function of yeast Thg1 in rapidly growing cells (Preston and Phizicky 2010), several phenotypes that have been associated with yeast and human Thg1 remain unexplained. Yeast Thg1 interacts with the Orc2 component of the origin recognition complex (Rice et al. 2005); decreased expression of yeast and human Thg1 leads to severe cell-cycle progression defects (Guo et al. 2004); and high level Thg1 expression has been associated with renal disease in humans (Murphy et al. 2008). With no obvious connection to tRNAHis metabolism, it is tempting to speculate that some or all of these phenotypes may be related to the template-dependent 3′-to-5′ addition activity of these enzymes.
5′ EDITING OF MITOCHONDRIAL tRNAS
Another example of reverse polymerization was provided by the discovery, initially in the amoeboid protist, Acanthamoeba castellanii (eukaryotic supergroup Amoebozoa), of an activity that repairs terminal mismatches in the acceptor stems of mitochondrial tRNAs (Fig. 7; Lonergan and Gray 1993a). In A. castellanii, the mitochondrial genome encodes 15 tRNAs, 12 of which are predicted to have mismatches in one or more of the first 3 bp of the acceptor stem (Lonergan and Gray 1993a,b; Burger et al. 1995). Investigation of the sequences of mature mitochondrial tRNAs demonstrated that in all cases, predicted terminal mismatches are corrected by replacement of the nucleotide on the 5′ side of the mismatch to create a standard Watson-Crick base pair with the corresponding nucleotide on the 3′ side of the stem (e.g., UxC→G:C) (Fig. 7; Lonergan and Gray 1993a; Price and Gray 1999a). As well, G:U and U:G pairs occurring within the first three acceptor stem positions (but not elsewhere) are edited in the same way to canonical base pairs (Price and Gray 1999a). In several other amoeboid protists—the cellular slime molds, D. discoideum, Dictyostelium fasciculatam, Dictyostelium purpureum, and Polysphondylium pallidum—multiple acceptor stem mismatches in mtDNA-encoded tRNA genes predict the existence of a similar tRNA editing activity; indeed, repair of mismatches has been directly demonstrated in the case of P. pallidum mitochondrial tRNAs (Schindel 2004). In Physarum polycephalum, a plasmodial slime mold, the mtDNA encodes only five tRNAs, two of which have recently been shown to undergo 5′ editing (Gott et al. 2010). These results suggest that 5′ editing of mitochondrial tRNAs is widespread within Amoebozoa.
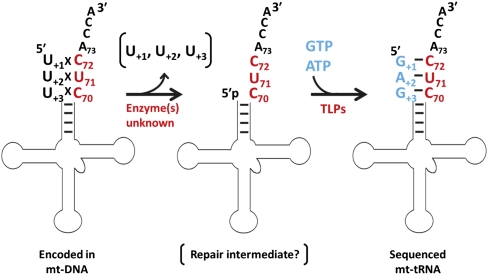
FIGURE 7.
tRNA 5′ editing repairs encoded mismatches found in some mitochondrial tRNAs. The mitochondrial tRNA 5′ editing reaction comprises at least two activities. First, up to three mismatched nucleotides are removed from the 5′ ends of tRNA species, such as the mt-tRNAAsp from A. castellanii shown here, by (an) unidentified enzyme(s). A 5′-monophosphate-containing repair intermediate species is generated that lacks the mismatches, although the precise biochemical structure of this intermediate has not been determined. In the second step, the aminoacyl acceptor stem is repaired using 3′-to-5′ polymerase activity, with 3′ end nucleotides (shown in red) serving as the template for addition of the correct Watson-Crick pairing NTPs. The biochemical activities catalyzed by two TLPs from D. discoideum are consistent with a role for TLPs in this step of the editing reaction.
A similar situation has been described in the distantly related chytridiomycete fungus, Spizellomyces punctatus (supergroup Opisthokonta), whose mitochondrial genome encodes eight tRNAs, all of which are predicted to have mismatches in one or more of the first 3 bp of the acceptor stem (Laforest et al. 1997). Again, these mismatches were shown to be corrected in the mature mitochondrial tRNAs by replacement of the nucleotide on the 5′ side of the mismatch by one generating a standard base pair with the nucleotide on the 3′ side of the mismatch (Laforest et al. 2004). Modeling of mtDNA-encoded tRNA secondary structures is highly suggestive of this type of tRNA editing in other protist lineages (e.g., in the chlorarachniophyte alga Bigelowiella natans, supergroup Rhiizaria, and the heterolobosean protist Naegleria gruberi, supergroup Excavata), although it should be recognized that in some protists, acceptor stem mismatches have been reported to be repaired through 3′ rather than 5′ tRNA editing (Leigh and Lang 2004).
A tRNA 5′ editing activity capable of removing the first three 5′-terminal nucleotides of mature tRNA substrates and replacing these nucleotides in a template-directed reaction (using the 3′ side of the acceptor stem as guide) has been partially purified from A. castellanii mitochondria (Price and Gray 1999b). This activity includes a 3′-to-5′ nucleotidyltransferase that restores missing 5′ nucleotides sequentially, stopping at the normally unpaired discriminator nucleotide, the fourth nucleotide from the 3′ end. With synthetic tRNA substrates carrying a 5′ monophosphate terminus, the polymerization reaction is strictly ATP-dependent, presumably reflecting a requirement for 5′ end activation (likely through adenylylation, as in the case of Thg1). However, this ATP requirement is abrogated if the substrate carries a 5′ triphosphate group; moreover, editing proceeds whether or not substrates carry a 3′-CCA. A mitochondrial tRNA 5′ editing activity that is virtually indistinguishable biochemically from the system described for A. castellanii has been partially purified and characterized from S. punctatus mitochondria (Bullerwell and Gray 2005). The existence of apparently identical activities in distantly related organisms that are separated phylogenetically by apparently nonediting taxa raises intriguing questions about the evolutionary origin of this editing system (discussed below). Moreover, the lack of conservation of editing sites among orthologous tRNAs from related organisms suggests that these activities are well suited to function as generalized quality control mechanisms in biological systems.
Thg1 AND THE CONNECTION TO tRNA 5′ EDITING
Early on, biochemical similarities between G−1 addition to tRNAHis and mitochondrial tRNA 5′ editing were noted (Price and Gray 1999b; Gu et al. 2003), although an evolutionary and/or functional connection between the two was not immediately apparent. The first indication of such a connection came from annotation of the D. discoideum genome sequence, which revealed four Thg1 homologs, subsequently designated DdiTLP1–4 (Abad et al. 2011), thereby defining the Thg1 superfamily, cl01644 (pfam04446, “Thg1”; COG4021, “uncharacterized conserved protein [function unknown]”). On the basis of BLAST similarity and active site motif, DdiTLP1(now DdiThg1) appeared to be a Thg1 ortholog, whereas DdiTLP2–4 are more similar to bacterial/archaeal TLP sequences, with which they cluster in phylogenetic trees (Fig. 6) and similarly lack the eucaryal-specific SDE(Y/F)SF and HINNLYN sequence motifs (Table 1). Notably, DdiTLP2 and DdiTLP3 are predicted to contain N-terminal mitochondrial targeting sequences, suggesting a role for one or both of these enzymes in mitochondrial tRNA 5′ editing in D. discoideum.
To test these predictions, recombinant DdiTLP1–4 proteins were expressed in and purified from E. coli, and their biochemical activities were individually characterized (Abad et al. 2011). As anticipated, DdiTLP1(DdiThg1) proved to be a bona fide Thg1 ortholog, a G−1 addition enzyme likely responsible for cytoplasmic tRNAHis maturation in D. discoideum. On the other hand, DdiTLP3 and DdiTLP4 exhibited biochemical activities consistent with a role for these enzymes in tRNA 5′ editing, based on their ability to efficiently and accurately repair the 5′ ends of synthetic tRNA substrates lacking one or more 5′ nucleotides. Although a role for DdiTLP2 was not established in this initial study, more recent results (Y Long and JE Jackman, unpubl.) suggest that this predicted mitochondrion-targeted protein may function in G−1 addition during processing and maturation of the mtDNA-encoded tRNAHis, whose gene sequence specifies U rather than G at the −1 position.
The availability of the D. discoideum TLP2–4 sequences led to the rapid identification of TLP homologs in other eukaryotes. Initially, it was puzzling that neither the authentic yeast Thg1 nor the orthologous D. discoideum sequence (DdiThg1) was able to retrieve a Thg1 homolog when used as a TBLASTN query in searches of A. castellanii genomic or EST sequence data; however, searches using DdiTLP2, -3, or -4 sequences as query readily identified two A. castellanii homologs, one of which contains a predicted mitochondrial targeting sequence (MW Gray, unpubl.) (Table 1). These A. castellanii TLPs are currently being cloned and expressed in order to investigate their biochemical properties. Curiously, it appears that A. castellanii does not encode a bona fide Thg1 ortholog, raising the question of whether its cytoplasmic tRNAHis acquires a G−1 residue. In fact, as discussed in the next section, the phylogenetic distribution among eukaryotes of Thg1 (a protein highly conserved at the sequence level) and TLPs (more rapidly diverging in sequence) is quite puzzling.
PHYLOGENETIC DISTRIBUTION AND EVOLUTION OF Thg1 AND ITS HOMOLOGS
Comprehensive database searches show that the phylogenetic distribution of Thg1 homologs falls into four distinct categories: Thg1 only (most eukaryotes), TLP(s) only (Archaea, Bacteria, and a small number of eukaryotes, notably including several fungi), both Thg1 and TLP(s) (mainly Amoebozoa, but also a few other examples), and neither Thg1 nor TLP(s) (a limited number among sequenced eukaryotes, but several examples from multiple kingdoms) (Table 1). Authentic Thg1 orthologs are limited to eukaryotes but, surprisingly, are not universally present within the domain Eucarya. Conversely, many (but by no means all) bacterial and archaeal taxa contain TLP genes, which are also present in some (but by no means all) eukaryotic species (Fig. 6). Finally, TLP genes are present in the genomes of certain bacteriophages (ABY63144.1, AEO93637.1) and giant viruses infecting Acanthamoeba species (YP_003986757.1, AEQ32982.1).
The apparent absence of Thg1 and/or TLP genes in eukaryotes might be ascribed to lack of completeness of available sequence information, except that the results in Table 1 derive for the most part from deeply sequenced genomic and/or ESTs data sets. In the case of A. castellanii, for example, BLAST searches of a deeply sequenced EST database readily retrieved multiple ESTs encompassing virtually the entire coding region of the two TLPs listed in Table 1, but not a single EST corresponding to authentic Thg1. The presence of both Thg1 and TLP(s) in certain eukaryotes (e.g., D. discoideum) can be rationalized in terms of distinct functions (G−1 addition to cytoplasmic tRNAHis vs. mitochondrial tRNA 5′ editing). However, the apparent absence of Thg1 in other eukaryotes (like A. castellanii) is more difficult to explain, unless in these cases both functions are carried out by one or more TLPs. In cases where no Thg1 homolog at all can be identified, maturation or aminoacylation of cytoplasmic tRNAHis could conceivably proceed via a different mechanism.
As discussed above, a function in generalized repair of tRNA 5′ ends has been argued in the case of TLP-containing bacteria and archaea (Rao et al. 2011), but what do we make of those prokaryotic species (e.g., E. coli) whose completely sequenced genomes contain no evidence of a TLP gene? Considering that the primary structure of yeast Thg1 provided no clue by way of conserved domains as to its function(s) (Gu et al. 2003), is it conceivable that organisms lacking a Thg1 homolog might rely on a different enzyme family and/or employ a different mechanism to carry out generalized repair of tRNA 5′ ends, if in fact such repair is an essential/important function?
In phylogenetic reconstructions (Fig. 6; Heinemann et al. 2010, 2011), Thg1 sequences are clearly monophyletic. In many of our analyses, archaeal homologs also form a monophyletic group, although statistical support is relatively low and the long-branch chrenarchaeote subgroup does cluster as a separate clade in some analyses (see, e.g., Fig. 6; Heinemann et al. 2010, 2011). TLPs from crenarchaeotes are clearly distinguishable from euryarchaeote ones, consistent with notable differences in conserved sequence blocks within the two groups. Bacterial TLPs are robustly distributed into two distinct groups, designated Group 1 and Group 2 by Heinemann et al. (2010). Eukaryotic TLPs branch as a sister clade of Group 1 bacterial TLPs (a single exception is the starlet sea anemone, Nematostella vectensis, whose TLP consistently branches within bacterial TLP Group 2) (Fig. 6). The two bacteriophage TLPs are individually most closely related to TLPs from the bacterial genus they infect, whereas the two eukaryotic viral TLPs are each other's closest relative, together branching within the eukaryotic TLP clade.
Phylogenetic analysis also demonstrates that DdiTLP3 and DdiTLP4 are paralogs, with single orthologs present in other Dictyostelium species (Dictyostelium fasciculatum, D. purpureum) as well as P. pallidum. On the other hand, DdiTLP2, which has a single ortholog in D. fasciculatum and D. purpureum, is not closely related to DdiTLP3/4. The relatively divergent Ddi/Dfa/Dpu TLP2 clade usually branches deeply within the eukaryotic TLP clade; indeed, in some analyses it branches outside the latter grouping, often together with or close to the crenarchaeote TLP clade (Fig. 6; Heinemann et al. 2010), a result likely due to the effect of long-branch attraction. We suggest that DdiTLP2 may have had a separate evolutionary origin from DdiTLP3/4 (notably, motif 1 is SDEINL in DdiTLP2 orthologs, as in a number of archaeal TLPs, as opposed to the canonical motif 1—SDEIT(M/L) in DdiTLP3/4 orthologs—that characterizes bacterial and other eukaryotic TLPs) (Table 1).
The broad distribution of Thg1 orthologs in eukaryotes and TLPs in archaea and their phylogenetic congruence with the overall phylogeny of the organisms containing them suggest a pattern of vertical inheritance within these two groups from a common ancestral sequence (Heinemann et al. 2010). Considering that Thg1 orthologs are clearly monophyletic and are found only in eukaryotes, whereas TLP-type enzymes are found in all three domains of life, we conclude that Thg1 is a eukaryote-specific invention, evolving from a TLP-like ancestral sequence at an early stage in eukaryotic evolution. In contrast, as noted by Heinemann et al. (2010), the sparse distribution of TLPs within bacteria and their distribution into two distinct phylogenetic groups (Groups 1 and 2) (Fig. 6) are best explained by at least two horizontal transfers of archaeal-type TLPs into bacteria, followed by additional TLP gene transfers within bacteria: an inference with which we concur. The sister-group relationship between bacterial Group 1 TLPs and eukaryotic TLPs (Fig. 6) might suggest that the latter have evolved from the former via one or more bacteria-to-eukaryote horizontal gene transfers; however, the actual direction of the transfer cannot be established definitively from existing data.
Although a number of eukaryotic TLPs are evidently targeted to and function in mitochondria, an origin of eukaryotic TLPs from the α-proteobacterial progenitor of mitochondria seems unlikely, given the wholesale absence of TLP genes in sequenced α-proteobacterial genomes and the curious lack of G−1 in tRNAHis of these organisms (Wang et al. 2007). In any event, convincing evidence of monophyly of Thg1 and probably also of eukaryotic TLPs argues that their punctate distribution within eukaryotes is likely due to an early emergence of these genes within Eucarya followed by selective loss of Thg1 and/or TLP genes within particular lineages. Nevertheless, the evolutionary pathway of TLP genes within eukaryotes remains enigmatic at present.
EVOLUTIONARY ORIGIN OF MITOCHONDRIAL tRNA EDITING
To explain the evolutionary emergence of mitochondrial tRNA 5′ editing, a model invoking “constructive neutral evolution” (Covello and Gray 1993; Stoltzfus 1999) has been elaborated (Price and Gray 1998; Gray 2001). This multistep model invokes the appearance of a potential RNA editing activity via an alteration in a pre-existing enzyme activity through a process of gene duplication and divergence. In this case, we imagine the appearance in mitochondria of an activity that has the capacity to remove the first three 5′ nucleotides from a tRNA and replace them with nucleotides that form standard Watson-Crick base pairs with the nucleotide on the 3′ side of the acceptor stem (Fig. 7). The existence of such an activity is proposed to relax evolutionary constraints on mtDNA-encoded tRNA genes, such that first three positions on the 5′ side of the acceptor stem are free to mutate without regard to whether or not they are able to form a base pair with their 3′ partner. Any mispairing is thus able to be corrected at the RNA level. As the number of such mismatches increases, negative selection operating through an evolutionary ratchet ensures that the editing system is effectively “locked in” as another step in the genetic information pathway.
TLP genes, specifying a generalized 5′ tRNA repair activity that is widespread among eukaryotes, provide a simple explanation for the emergence of at least the 3′-to-5′ polymerase component of a mitochondrial tRNA 5′ editing system, since all that is required is that a nucleus-encoded TLP gene product acquires targeting information that allows it to be imported into mitochondria. Such a scenario might happen independently in different eukaryotic lineages, resulting in a punctate distribution of mitochondrial tRNA 5′ editing systems that have virtually indistinguishable biochemical properties: as, e.g., in the amoeboid protozoon A. castellanii (Price and Gray 1999b) and the chytid fungus S. punctatus (Bullerwell and Gray 2005). Our recent demonstration that D. discoideum TLPs possess the requisite 3′-to-5′ polymerase activity with 5′-truncated tRNA substrates (Abad et al. 2011) strongly supports this evolutionary model.
CONCLUSIONS AND FUTURE PROSPECTS
With the discovery of the tRNAHis guanylyltransferase enzyme family, a new type of enzyme chemistry—3′-to-5′ polymerization of nucleic acids—has been added to the panoply of catalyzed reactions in biology. The initial association of yeast THG1 with the maturation of tRNAHis removed this enzyme family from the ranks of “conserved proteins of unknown function” and solved the long-standing mystery surrounding the identity of the G−1 addition enzyme in eukaryotes. However, investigations of Thg1/TLP orthologs in other organisms have since revealed that the use of 3′-to-5′ polymerase activity is likely more widespread than has hitherto been appreciated, extending beyond just a simple role in eukayotic tRNAHis maturation.
Given the similarity in the actual chemical steps catalyzed by Thg1/TLPs during 3′-to-5′ polymerization to the chemistry used by 5′-to-3′ polymerases, and the recently observed similarity in active site structure between these two types of nucleic acid polymerases, these findings also raise evolutionary questions regarding the predominance of 5′-to-3′ polymerases in extant species. The difficulties for the cell attendant with limitation of nucleic acid synthesis to the 5′-to-3′ direction, such as shortening of 5′ ends of linear chromosomes and the constraints of lagging strand synthesis during bidirectional DNA replication, could presumably have been precluded by a resort to 3′-to-5′ synthesis in biology, perhaps even in a supporting role to canonical 5′-to-3′ polymerases. Why this did not happen in the course of polymerase evolution is a fundamental and currently unanswered question. A complete understanding of the molecular mechanism of the 3′-to-5′ polymerase enzyme family will be critical in order to fully understand the molecular basis for the biochemical differences between 3′-to-5′ and 5′-to-3′ polymerase enzymes and their implications for modern biological systems.
The continually increasing numbers and quality of available genome sequences will likely reveal additional members of the Thg1/TLP superfamily and possibly additional functions for some of these gene products. However, even among existing well-studied genomes, some curious exceptions remain to be investigated. The apparent lack of a Thg1 or TLP homolog in some eukaryotes (such as C. elegans) raises the question of whether a G−1-containing tRNAHis is present in these particular organisms and, if so, whether G−1 addition is accomplished via a different mechanism in these species. Among the possibilities, an encoded G−1 could be generated by processing, as in bacteria, or a separate, non-Thg1 reverse polymerase could substitute for Thg1 in these species. Notably, in plant mitochondria, a tRNAHis guanylyltransferase activity has been found, but neither of the two Thg1 orthologs that have been identified in plants appears to localize to this organelle (Placido et al. 2010). Alternatively, G−1 may be dispensable in some cases, as it is in α-proteobacteria. Characterization of the tRNAHis and requirements for HisRS recognition in such eukaryotes is likely to be informative.
Similarly, the reasons for the punctate distribution of TLPs among bacteria, archaea, and eukaryotic microbes compared with the more widespread occurrence of Thg1 in eukaryotes are unknown. The predicted need for 5′-editing to produce functional mitochondrial tRNAs correlates well with the presence of TLPs in many eukaryotic microbes, but the lack of tRNA substrates with predicted mismatched nucleotides in bacteria and archaea suggests that the presence of TLPs in these species is not due to a role in 5′-editing. Thus, bona fide physiological substrates for bacterial and archaeal TLPs remain to be identified. Despite the biochemical similarities between bacterial/archaeal TLPs and the putative mitochondrial tRNA editing enzymes, the possibility of both tRNA and non-tRNA substrates for 5′ end polymerase activities remains an exciting possibility. We note that the ability of TLPs to catalyze addition without the strong tRNAHis preference exhibited by eukaryal enzymes lends support to a role for TLPs in repairing 5′ ends of a broad distribution of substrates.
Compared with the situation in archaea/bacteria, a role for TLPs in eukaryotic mitochondria is more readily apparent, based on the demonstrated requirement for 3′-to-5′ polymerase activity to repair tRNA 5′ ends in the organelle. However, many biochemical questions regarding the mitochondrial tRNA editing reaction must be further explored. Ideally, biochemical purification of the activity would lead to direct identification of a TLP as the active reverse polymerase. Attempts to identify TLPs in highly purified preparations of the editing complex from A. castellanii mitochondria have so far not been successful; however, improved mass spectrometry techniques to deal with limited samples along with the possibility of generating antibodies against purified recombinant TLPs may prove useful in this respect. The question of the activity that removes mismatched nucleotides from the 5′ end of tRNAs so that the reverse polymerase can operate also remains to be answered. Again, by analogy with canonical polymerases, it is possible that 5′-to-3′ exonuclease activity is an intrinsic activity of TLPs; in this regard, we note that the putative nuclease activity consistently copurifies with the 3′-to-5′ polymerase activity during isolation of the editing complex from A. castellanii (A Lohan and MW Gray, unpubl.). Alternatively, separate enzyme(s) may be responsible for catalyzing this step. Further biochemical characterization using synthetic tRNA substrates may prove informative in demonstrating the type (endo- vs. exo-) and identity of this nuclease.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
M.W.G. acknowledges support of research in this area from the Canadian Institutes of Health Research (operating grant MOP-4124), and work on Thg1 family enzymes in the laboratory of J.E.J. is supported by NIH R01-GM087543. We thank Dr. Laura Eme (Department of Biochemistry and Molecular Biology, Dalhousie University) for valuable discussions relating to evolution of the Thg1 superfamily, and for conducting the particular ML phylogenetic analysis depicted in Figure 6, as well as for generating the figure. We also thank Dr. Brian Hyde and Dr. Sylvie Doublié (Department of Cell and Molecular Biology, University of Vermont) for generously providing the structures shown in Figure 5.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.032300.112.
REFERENCES
- Abad MG, Rao BS, Jackman JE 2010. Template-dependent 3′–5′ nucleotide addition is a shared feature of tRNAHis guanylyltransferase enzymes from multiple domains of life. Proc Natl Acad Sci 107: 674–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abad MG, Long Y, Willcox A, Gott JM, Gray MW, Jackman JE 2011. A role for tRNAHis guanylyltransferase (Thg1)-like proteins from Dictyostelium discoideum in mitochondrial 5′-tRNA editing. RNA 17: 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardell DH, Andersson SGE 2006. TFAM detects co-evolution of tRNA identity rules with lateral transfer of histidyl-tRNA synthetase. Nucleic Acids Res 34: 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK 1980. 5′-Terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol Rev 44: 175–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH 1992. Telomerases. Annu Rev Biochem 61: 113–129 [DOI] [PubMed] [Google Scholar]
- Bullerwell CE, Gray MW 2005. In vitro characterization of a tRNA editing activity in the mitochondria of Spizellomyces punctatus, a chytridiomycete fungus. J Biol Chem 280: 2463–2470 [DOI] [PubMed] [Google Scholar]
- Burger G, Plante I, Lonergan KM, Gray MW 1995. The mitochondrial DNA of the amoeboid protozoon, Acanthamoeba castellanii: complete sequence, gene content and genome organization. J Mol Biol 245: 522–537 [DOI] [PubMed] [Google Scholar]
- Burkard U, Söll D 1988. The 5′-terminal guanylate of chloroplast histidine tRNA is encoded in its gene. J Biol Chem 263: 9578–9581 [PubMed] [Google Scholar]
- Burkard U, Willis I, Söll D 1988. Processing of histidine transfer RNA precursors. Abnormal cleavage site for RNase P. J Biol Chem 263: 2447–2451 [PubMed] [Google Scholar]
- Claros M, Vincens P 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241: 779–786 [DOI] [PubMed] [Google Scholar]
- Cooley L, Appel B, Söll D 1982. Post-transcriptional nucleotide addition is responsible for the formation of the 5′ terminus of histidine tRNA. Proc Natl Acad Sci 79: 6475–6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello PS, Gray MW 1993. On the evolution of RNA editing. Trends Genet 9: 265–268 [DOI] [PubMed] [Google Scholar]
- Deutscher MP, Kornberg A 1969. Enzymatic synthesis of deoxyribonucleic acid. XXIX. Hydrolysis of deoxyribonucleic acid from the 5′ terminus by an exonuclease function of deoxyribonucleic acid polymerase. J Biol Chem 244: 3029–3037 [PubMed] [Google Scholar]
- Giegé R, Sissler M, Florentz C 1998. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res 26: 5017–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gott JM, Somerlot BH, Gray MW 2010. Two forms of RNA editing are required for tRNA maturation in Physarum mitochondria. RNA 16: 482–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. 2001 Speculations on the origin and evolution of editing. In RNA editing (ed. BL Bass), pp. 160–184. Oxford University Press, Oxford, UK. [Google Scholar]
- Gu W, Jackman JE, Lohan AJ, Gray MW, Phizicky EM 2003. tRNAHis maturation: An essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes Dev 17: 2889–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Hurto RL, Hopper AK, Grayhack EJ, Phizicky EM 2005. Depletion of Saccharomyces cerevisiae tRNAHis guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m5C. Mol Cell Biol 25: 8191–8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Guo D, Hu K, Lei Y, Wang Y, Ma T, He D 2004. Identification and characterization of a novel cytoplasm protein ICF45 that is involved in cell cycle regulation. J Biol Chem 279: 53498–53505 [DOI] [PubMed] [Google Scholar]
- Heinemann IU, O'Donoghue P, Madinger C, Benner J, Randau L, Noren CJ, Söll D 2009. The appearance of pyrrolysine in tRNAHis guanylyltransferase by neutral evolution. Proc Natl Acad Sci 106: 21103–21108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann IU, Randau L, Tomko RJ Jr, Söll D 2010. 3′-5′ tRNAHis guanylyltransferase in bacteria. FEBS Lett 584: 3567–3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann IU, Nakamura A, O'Donoghue P, Eiler D, Söll D 2011. tRNAHis-guanylyltransferase establishes tRNAHis identity. Nucleic Acids Res 40: 333–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H, Hasegawa T, Ueda T, Watanabe K, Miura K-i, Shimizu M 1989. Role of the extra G-C pair at the end of the acceptor stem of tRNAHis in aminoacylation. Nucleic Acids Res 17: 7855–7863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde SJ, Eckenroth BE, Smith BA, Eberley WA, Heintz NH, Jackman JE, Doublié S 2010. tRNAHis guanylyltransferase (THG1), a unique 3′-5′ nucleotidyl transferase, shares unexpected structural homology with canonical 5′-3′ DNA polymerases. Proc Natl Acad Sci 107: 20305–20310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Phizicky EM 2006. tRNAHis guanylyltransferase catalyzes a 3′-5′ polymerization reaction that is distinct from G−1 addition. Proc Natl Acad Sci 103: 8640–8645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Phizicky EM 2008. Identification of critical residues for G−1 addition and substrate recognition by tRNAHis guanylyltransferase. Biochemistry 47: 4817–4825 [DOI] [PubMed] [Google Scholar]
- Jahn D, Pande S 1991. Histidine tRNA guanylyltransferase from Saccharomyces cerevisiae. II. Catalytic mechanism. J Biol Chem 266: 22832–22836 [PubMed] [Google Scholar]
- Joyce CM, Steitz TA 1995. Polymerase structures and function: variations on a theme? J Bacteriol 177: 6321–6329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K , Miyata T 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW 2005. The tree of eukaryotes. Trends Ecol Evol 20: 670–676 [DOI] [PubMed] [Google Scholar]
- Laforest MJ, Roewer I, Lang BF 1997. Mitochondrial tRNAs in the lower fungus Spizellomyces punctatus: tRNA editing and UAG ‘stop’ codons recognized as leucine. Nucleic Acids Res 25: 626–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforest M-J, Bullerwell CE, Forget L, Lang BF 2004. Origin, evolution, and mechanism of 5′ tRNA editing in chytridiomycete fungi. RNA 10: 1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25: 1307–1320 [DOI] [PubMed] [Google Scholar]
- Lehman IR, Richardson CC 1964. The deoxyribonucleases of Escherichia coli. IV. An exonuclease activity present in purified preparations of deoxyribonucleic acid polymerase. J Biol Chem 239: 233–241 [PubMed] [Google Scholar]
- Leigh J, Lang BF 2004. Mitochondrial 3′ tRNA editing in the jakobid Seculamonas ecuadoriensis: a novel mechanism and implications for tRNA processing. RNA 10: 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonergan KM, Gray MW 1993a. Editing of transfer RNAs in Acanthamoeba castellanii mitochondria. Science 259: 812–816 [DOI] [PubMed] [Google Scholar]
- Lonergan KM, Gray MW 1993b. Predicted editing of additional transfer RNAs in Acanthamoeba castellanii mitochondria. Nucleic Acids Res 21: 4402 doi: 10.1093/nar/21.18.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Keller W 1996. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J 15: 2593–2603 [PMC free article] [PubMed] [Google Scholar]
- Murphy M, Docherty NG, Griffin B, Howlin J, McArdle E, McMahon R, Schmid H, Kretzler M, Droguett A, Mezzano S, et al. 2008. IHG-1 amplifies TGF-β1 signaling and is increased in renal fibrosis. J Am Soc Nephrol 19: 1672–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nameki N, Asahara H, Shimizu M, Okada N, Himeno H 1995. Identity elements of Saccharomyces cerevisiae tRNAHis. Nucleic Acids Res 23: 389–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H 1993. MUST, a computer package of Management Utilities for Sequences and Trees. Nucleic Acids Res 21: 5264–5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placido A, Sieber F, Gobert A, Gallerani R, Giegé P, Maréchal-Drouard L 2010. Plant mitochondria use two pathways for the biogenesis of tRNAHis. Nucleic Acids Res 38: 7711–7717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston MA, Phizicky EM 2010. The requirement for the highly conserved G−1 residue of Saccharomyces cerevisiae tRNAHis can be circumvented by overexpression of tRNAHis and its synthetase. RNA 16: 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DH, Gray MW 1998. Editing of tRNA. In Modification and editing of RNA (ed. H Grosjean, R Benne), pp. 289–305. ASM Press, Washington, DC [Google Scholar]
- Price DH, Gray MW 1999a. Confirmation of predicted edits and demonstration of unpredicted edits in Acanthamoeba castellanii mitochondrial tRNAs. Curr Genet 35: 23–29 [DOI] [PubMed] [Google Scholar]
- Price DH, Gray MW 1999b. A novel nucleotide incorporation activity implicated in the editing of mitochondrial transfer RNAs in Acanthamoeba castellanii. RNA 5: 302–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao BS, Maris EL, Jackman JE 2011. tRNA 5′-end repair activities of tRNAHis guanylyltransferase (Thg1)-like proteins from Bacteria and Archaea. Nucleic Acids Res 39: 1833–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratliff RL. 1981 Terminal deoxynucleotidyltransferase. In The Enzymes (ed. PD Boyer), pp. 105–118. Academic Press, Inc., New York. [Google Scholar]
- Rice TS, Ding M, Pederson DS, Heintz NH 2005. The highly conserved tRNAHis guanylyltransferase Thg1p interacts with the origin recognition complex and is required for the G2/M phase transition in the yeast Saccharomyces cerevisiae. Eukaryot Cell 4: 832–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen AE, Musier-Forsyth K 2003. Recognition of G-1:C73 atomic groups by Escherichia coli histidyl-tRNA synthetase. J Am Chem Soc 126: 64–65 [DOI] [PubMed] [Google Scholar]
- Schindel ET. 2004 “Editing of mitochondrial tRNAs in Polysphondylium pallidum.” MS thesis, Dalhousie University, Halifax, Nova Scotia. [Google Scholar]
- Schürer H, Schiffer S, Marchfelder A, Mörl M 2001. This is the end: processing, editing and repair at the tRNA 3′-terminus. Biol Chem 382: 1147–1156 [DOI] [PubMed] [Google Scholar]
- Shuman S, Hurwitz J 1981. Mechanism of mRNA capping by vaccinia virus guanylyltransferase: characterization of an enzyme–guanylate intermediate. Proc Natl Acad Sci 78: 187–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S, Lima CD 2004. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr Opin Struct Biol 14: 757–764 [DOI] [PubMed] [Google Scholar]
- Smith BA, Jackman JE 2011. Kinetic analysis of 3′–5′ nucleotide addition catalyzed by eukaryotic tRNAHis guanylyltransferase. Biochemistry 51: 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 26: 148–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus A 1999. On the possibility of constructive neutral evolution. J Mol Evol 49: 169–181 [DOI] [PubMed] [Google Scholar]
- Tomkinson AE, Totty NF, Ginsburg M, Lindahl T 1991. Location of the active site for enzyme-adenylate formation in DNA ligases. Proc Natl Acad Sci 88: 400–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uptain SM, Kane CM, Chamberlin MJ 1997. Basic mechanisms of transcript elongation and regulation. Annu Rev Biochem 66: 117–172 [DOI] [PubMed] [Google Scholar]
- Wang C, Sobral BW, Williams KP 2007. Loss of a universal tRNA feature. J Bacteriol 189: 1954–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JB, Cooley L, Söll D 1990. Enzymatic addition of guanylate to histidine transfer RNA. Methods Enzymol 181: 451–462 [DOI] [PubMed] [Google Scholar]
- Yuan J, Gogakos T, Babina AM, Söll D, Randau L 2011. Change of tRNA identity leads to a divergent orthogonal histidyl-tRNA synthetase/tRNAHis pair. Nucleic Acids Res 39: 2286–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue D, Maizels N, Weiner AM 1996. CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyltransferase superfamily: characterization of the CCA-adding enzyme from the archaeal hyperthermophile Sulfolobus shibatae. RNA 2: 895–908 [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Deutscher MP 1987. tRNA nucleotidyltransferase is not essential for Escherichia coli viability. EMBO J 6: 2473–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]