The authors utilized next-generation sequencing to analyze small noncoding RNA expression patterns during different stages of early zebrafish development. Their main focus is on miRNAs and piRNAs. This work defines the expression patterns for 198 miRNAs during early vertebrate development and also shows significant 3′ end variation that suggests a novel form of targeted degradation. Their analysis also resulted in the identification of eight new miRNAs, as well as the existence of tRNA-derived small RNAs in zebrafish. Finally, the authors identify a large set of piRNAs during early development.
Keywords: microRNA, zebrafish, piRNA, development
Abstract
During early vertebrate development, a large number of noncoding RNAs are maternally inherited or expressed upon activation of zygotic transcription. The exact identity, expression levels, and function for most of these noncoding RNAs remain largely unknown. miRNAs (microRNAs) and piRNAs (piwi-interacting RNAs) are two classes of small noncoding RNAs that play important roles in gene regulation during early embryonic development. Here, we utilized next-generation sequencing technology to determine temporal expression patterns for both miRNAs and piRNAs during four distinct stages of early vertebrate development using zebrafish as a model system. For miRNAs, the expression patterns for 198 known miRNAs within 122 different miRNA families and eight novel miRNAs were determined. Significant sequence variation was observed at the 5′ and 3′ends of miRNAs, with most extra nucleotides added at the 3′ end in a nontemplate directed manner. For the miR-430 family, the addition of adenosine and uracil residues is developmentally regulated and may play a role in miRNA stability during the maternal zygotic transition. Similar modification at the 3′ ends of a large number of miRNAs suggests widespread regulation of stability during early development. Beside miRNAs, we also identified a large and unexpectedly diverse set of piRNAs expressed during early development.
INTRODUCTION
The importance of small RNA-mediated gene regulation has been increasingly recognized in recent years, playing multiple roles during development (Pauli et al. 2011). miRNAs and endogenous siRNAs have been shown to regulate gene expression by silencing specific genes, whereas piRNAs have been implicated mainly in genome protection and/or maintenance in germ cells via silencing of transposable elements (Bartel and Chen 2004; Aravin et al. 2007; Brennecke et al. 2007; Houwing et al. 2007; Czech et al. 2008; Flynt and Lai 2008; Tam et al. 2008). miRNAs have been identified in organisms as diverse as viruses, unicellular algae, plants, worms, flies, fish, and mammals (Bartel 2004; Zhao et al. 2007; Skalsky and Cullen 2010). For those miRNAs thus far analyzed, expression patterns are highly regulated, both temporally and spatially (Wienholds et al. 2005; Landgraf et al. 2007; Ruby et al. 2007). miRNAs mainly exert their effects by blocking translation and/or destabilizing mRNAs (Lim et al. 2005; Giraldez et al. 2006; Baek et al. 2008; Guo et al. 2010). piRNA expression is thought to be mostly restricted to germ cells and some somatic cells in flies (Halic and Moazed 2009; Malone et al. 2009; Haase et al. 2010).
Mature miRNAs are 22–23 nucleotides (nt) in length. They are derived from longer primary transcripts (pri-miRNAs) that contain multiple stem–loop structures that undergo two sequential cleavages by the enzymes Drosha and Dicer to produce mature small RNA duplexes (Hutvagner et al. 2001; Ketting et al. 2001; Lee et al. 2002, 2003; Cai et al. 2004). Generally, one of the two strands is then incorporated into an RNA Induced Silencing Complex (RISC) with one or more Argonaute proteins (Schwarz et al. 2003). In contrast to miRNAs, piRNAs are slightly larger (25–30 nt) and, at least in flies and fish, are derived from a reciprocal amplification loop in which primary piRNAs associate with Piwi proteins to direct cleavage of transposable element mRNAs (Brennecke et al. 2007; Houwing et al. 2008). The initial cleavage products then serve to promote production of sense-strand secondary piRNAs which, in turn, generate more primary piRNAs by pairing with and directing cleavage of antisense RNAs derived from discrete repetitive genomic loci, referred to as piRNA clusters (Brennecke et al. 2007; Malone et al. 2009). This model of biogenesis is referred to as the Ping-Pong model, because the production of piRNAs from one strand drives the generation of piRNAs from the other strand (Brennecke et al. 2007).
Zebrafish is widely used as a model system to study early vertebrate development (Kimmel et al. 1995). Previous work has shown that miRNAs play important functional roles during cell specification and differentiation (Giraldez et al. 2005; Flynt et al. 2007; Mishima et al. 2009). In zebrafish, miRNA expression patterns have been extensively examined using direct cloning and microarray analyses (Giraldez et al. 2005; Wienholds et al. 2005; Thatcher et al. 2007). These studies have shown that most miRNAs are not highly expressed during the first 12 h of zebrafish development, but that the overall pattern becomes increasingly diverse and complex as development proceeds (Chen et al. 2005; Wienholds et al. 2005; Thatcher et al. 2007). Despite the seeming lack of diversity during the earliest stages of development, miRNA function is clearly essential, as maternal-zygotic Dicer mutant fish show severe defects and die by 7 dpf (Giraldez et al. 2005). One of the most abundant early expressed families of miRNAs is the miR-430 family, which functions to induce deadenylation, degradation, and clearance of maternal mRNAs, facilitating the maternal-zygotic transition (Giraldez et al. 2006). A similar phenomenon occurs in Xenopus laevis through the action of the miR-427 family (Lund et al. 2009).
In zebrafish, piRNAs are mainly expressed in ovaries and testes (Houwing et al. 2007, 2008). Ziwi-mutant fish, which lack the zebrafish PIWI protein, exhibit extensive apoptosis in adult germ cells and are sterile (Houwing et al. 2007). This suggests an essential role in genome maintenance, but the exact role and mechanism of action for these small RNAs remains mostly unknown.
Recently, the utilization of next-generation sequencing technologies has revolutionized our ability to dissect transcriptomes, even for RNAs expressed at low levels. Here, we used RNA-Seq to examine the expression of miRNAs and piRNAs across four stages of early zebrafish development. We discovered the presence of large numbers of miRNAs, both maternally deposited and zygotically transcribed. Many miRNAs show widespread variation at their 3′ ends, mostly by the addition of nontemplate-directed nucleotides that are added in a developmental stage-specific manner. We also show that piRNAs are extensively expressed throughout early embryonic development, suggesting a broad role during the earliest cell divisions.
RESULTS
Small RNA sequencing
We isolated total RNA from zebrafish embryos at four different stages of early development (256-cell, sphere, shield, and 1 dpf) representing 2.5, 4, 6, and 24 h post fertilization (hpf), respectively. We chose these stages because they coincide with key gene expression changes during early vertebrate development. At the 256-cell stage, the majority of RNAs are maternally deposited. The sphere stage represents the time during which the embryo shifts from utilization of many maternally deposited RNAs and initiates zygotic transcription. During the shield stage, gastrulation continues to generate the three germ layers. By 1 dpf, the major organ systems have formed and hearts are beating. To examine gene expression patterns during these specific stages, small RNA libraries (15–30 nt) were prepared for high-throughput sequencing using the Illumina platform (Hafner et al. 2008). We constructed independent libraries for each stage, and generated a total of 29,963,921 sequence reads. In order to validate the reliability of the different high-throughput sequencing runs, experimental duplicates of small RNA libraries were prepared from sphere stage RNA, and independently sequenced. A significant correlation was observed between the results from the two independent libraries (R2 = 0.89; Supplemental Fig. 1). In addition, the miRNA profiles revealed by our sequencing data across all four stages were largely consistent with prior miRNA expression analyses (Chen et al. 2005; Wienholds et al. 2005; Thatcher et al. 2007).
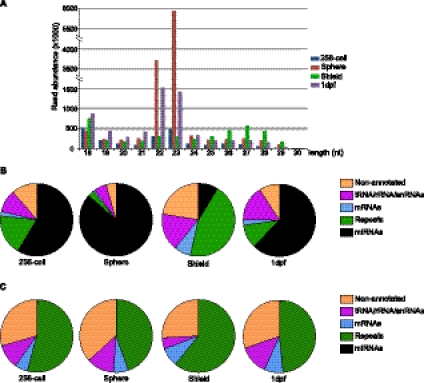
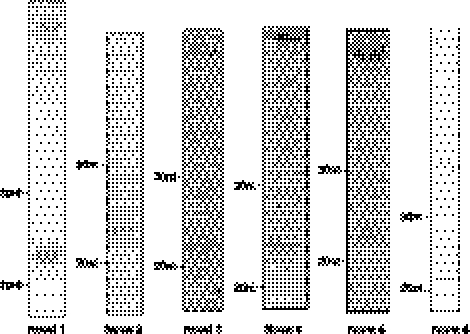
Analysis of the size distribution of all reads within each library revealed at least three classes between 18 and 30 nt. The major size class peaked at 22–23 nt (Fig. 1A). Based on the sequences and genomic positions of these reads, this class represents miRNAs. From all four stages, ∼55% of the total reads were identified as miRNAs, representing 198 distinct miRNAs (based on sequence alignment with miRBase Release 16) (Griffiths-Jones 2004; Griffiths-Jones et al. 2006, 2008). At the sphere stage, 99% of the miRNA reads were derived from just one family, the miR-430 family. To better examine the diversity of noncoding reads, we grouped identical sequences, which showed that a wide diversity of small RNAs were recovered (Fig. 1B,C). Examination of the reads after such a grouping revealed a second size class peaking at 26–28 nt (Fig. 1A; Fig. 5A,B, below). Based on RNA sequences and genomic mapping, these RNAs are derived primarily from repetitive elements. The smallest size class peaked at 18 nt and consists almost entirely of tRNA-derived small RNAs (see below) and rRNA-related small RNAs (data not shown).
FIGURE 1.
Sequencing summary. (A) Size distribution of all sequencing reads between 18 and 30 nt. RNA reads derived from four developmental stages are indicated in different colors. The size distribution and abundance of the reads from each stage are as indicated. (B) Read frequency for all sequences. The identity and frequency of small RNAs reads from different developmental time points are as indicated. (C) Read frequency for unique sequences. In contrast to B, where the total read frequencies were charted, small RNA reads derived from the same miRNA were grouped together as a single subset. This analysis shows that a large number of unique reads are derived from distinct genomic elements, mostly repetitive elements.
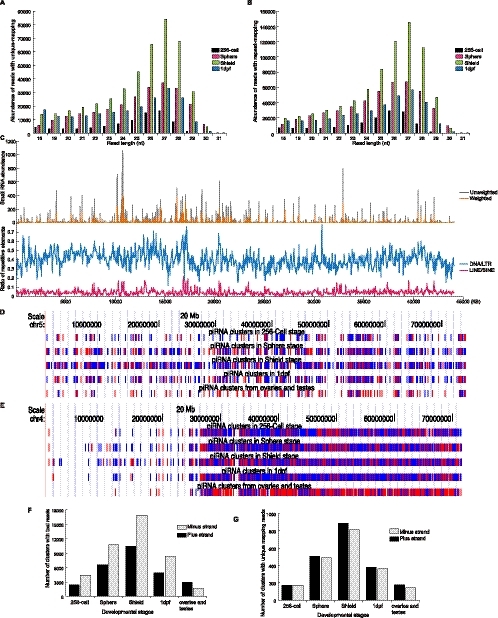
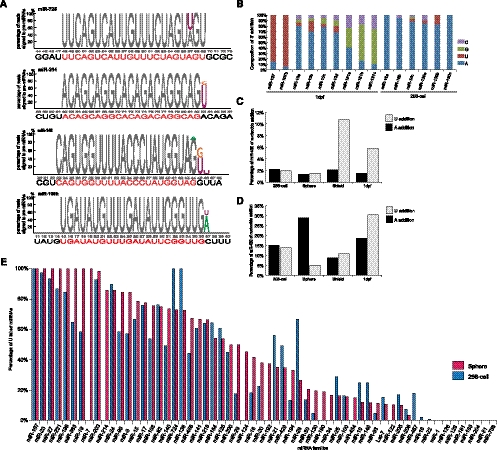
FIGURE 5.
Zebrafish piRNA expression. (A,B) After filtering out miRNA reads, the size distribution and abundance of reads mapping to unique (A) or repetitive (B) genomic loci is shown. Different colors represent reads from different developmental stages. (C) Correlation between the abundance of reads derived from shield stage small RNAs and the density of transposable elements along chromosome 11. At the top, small RNAs that map to chromosome 11 are indicated either as unweighted (gray), meaning the total number of reads irrespective of how many positions (copies) along a chromosome that might encode this RNA, or weighted (orange line), meaning the total number of reads divided by the number of positions or copies within the zebrafish genome. Transposable elements and transposons were divided into DNA/LTR (blue) and LINE/SINE (red) based on the origin of the small RNA reads. The density of repetitive elements (ratio) was determined by the percentage of nucleotides mapping to transposable elements per 50 kb. (D,E) Genomic localization of piRNA clusters. Vertical lines represent piRNA clusters from four developmental time points and also from data generated from adult ovaries and testes across either chromosome 4 or chromosome 5. Red lines indicate plus strands, while blue lines indicate minus strands. (F,G) Quantification of expression of piRNA clusters from total reads (F) or reads derived from unique genomic loci (G). Black and gray columns represent the strands from which the reads originated.
miRNA expression analysis
Analysis of the sequencing reads showed that 198 known miRNAs were detected that could be grouped into 122 families. Most of the miRNA reads were derived from just one arm of the hairpin structures that constitute each precursor miRNA. Short reads corresponding to the loop and the other precursor arm (star sequence arm) were also detected, but at much lower frequencies, consistent with proposed miRNA biogenesis models (Kim et al. 2009). However, we also found exceptions in which the star reads were much more prevalent than the mature reads, as previously reported (Soares et al. 2009; Jagadeeswaran et al. 2010; Yang et al. 2011) (e.g., miR-735 and miR-135b; see Supplemental Table 1 for full list). An extreme example of diverse read lengths with different 5′ and 3′ ends was detected for reads derived from miR-2190, one of the most recently annotated zebrafish miRNAs (Soares et al. 2009). We found multiple small RNAs derived from this locus with different ends, suggesting random cleavage at multiple sites across the proposed precursor hairpin (see Supplemental Table 1). The miR-2190 putative hairpin overlaps with two rRNA genes in the zebrafish genome, and that fact, together with the observed sequence heterogeneity, suggests that miR-2190 is more likely to be a product of rRNA degradation rather than an authentic miRNA.
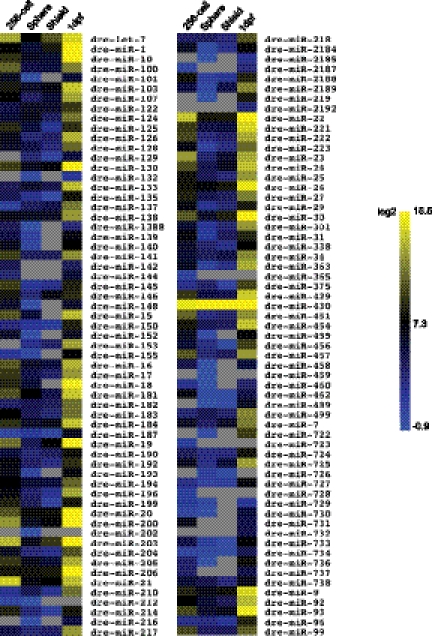
Quantitation of miRNA expression patterns was determined based on the read frequency for each mature miRNA. Previous work demonstrated a significant correlation between read numbers and miRNA levels, with the caveat that bias cannot be completely eliminated due to secondary structures or other variables (Landgraf et al. 2007). Nevertheless, normalization between developmental stages has usually been performed using miRNA read numbers divided by the total number of genome matching reads in each library (Chen et al. 2005; Ameres et al. 2010). We initially analyzed expression patterns in this manner (Supplemental Fig. 3A). We found that this method was not appropriate, as overall transcriptional activity significantly increases as development proceeds, resulting in a large increase in the size of libraries dervived from small RNA. Also, there is a large increase in the total number of piRNAs and miRNA reads mostly derived from the miR-430 family, which increased from 25% of the total miRNA reads at the 256-cell stage to 99% at the sphere stage. If the libraries are normalized using individual miRNA read numbers, divided by the total number of genome matching reads in each library, changes in the expression of individual miRNAs can become obscured by increased overall transcription as development proceeds, as well as by the extreme abundance of reads derived from just the miR-430 family. Thus, we normalized the values from each library based on the levels of two miRNAs that are present at moderate levels in all libraries (let-7 and miR-9). For this, we performed RT–qPCR to determine the levels of these miRNAs relative to U6 snRNA (Supplemental Fig. 2). We then used the relative values of let-7 and miR-9 to normalize the read frequencies for each library (Fig. 2; Supplemental Fig. 3B; Friedlander et al. 2009). Reassuringly, the expression patterns were quite similar, whether normalized to miR-9 levels or let-7 levels, even though the RT–qPCR levels for these miRNAs differ by an order of magnitude, and despite the fact that let-7a levels might be biased using deep-sequencing approaches (Linsen et al. 2009). Normalization based on miR-9 or let-7 resulted in very different heat maps compared with that obtained when total genome matching reads of each library were used for normalization (Supplemental Fig. 3).
FIGURE 2.
miRNA family expression profiles. miRNA family read frequency was normalized (see Materials and Methods) and compared across four developmental stages. Yellow indicates high expression and blue indicates low expression. Gray indicates undetectable levels of expression.
Quantitation and normalization of sequencing data to analyze gene expression patterns is not trivial and is subject to ongoing research and debate (Meyer et al. 2010). Thus, as a further test of whether normalization based on miR-9 or let-7 is valid, we used the weighted trimmed mean of the log expression ratios (the trimmed mean of M values [TMM]) to normalize the data (Robinson and Oshlack 2010). TMM uses raw data to estimate appropriate scaling factors that facilitates analysis of differential expression patterns. When we used TMM normalization, we reassuringly found that the derived heat maps were very similar to those obtained after normalization with either miR-9 or let-7 (Supplemental Fig. 3C). Finally, we used Northern blots on six miRNAs with differing raw read numbers to confirm our normalization method, and the results completely supported our heat maps (Supplemental Fig. 4).
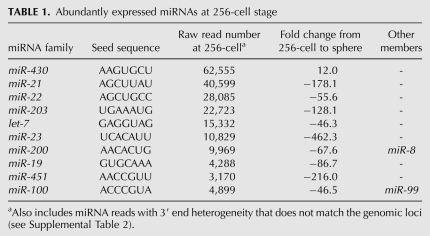
Developmentally, we found that 180 miRNAs belonging to 109 different families are present at the 256-cell stage. Because this is before the onset of zygotic transcription, these miRNAs are maternally deposited and, interestingly, are present at relatively high levels, suggesting an important role during the earliest stages of development (Newport and Kirschner 1982; Schier 2007). A total of 88 out of the 109 miRNA families that are expressed at the 256-cell stage are down-regulated at least twofold as development and zygotic transcription proceeds. The 10 most abundant miRNAs expressed at this stage and their subsequent changes in expression are listed in Table 1. For the sphere state, a total of 182 different miRNAs were detected. Many of the maternally inherited miRNAs that were detected at the 256-cell stage were not observed by the sphere stage. From the sphere stage onward, miRNAs can be roughly divided into two distinct groups based on their expression patterns during early development. The first group is composed of 60 miRNA families that are expressed at low levels across all four stages, while the second group is composed of 48 miRNAs that show decreased expression at the sphere stage, followed by at least a twofold increase in expression as development proceeds (Fig. 2). The miRNAs in the second group appear to be more broadly expressed.
TABLE 1.
Abundantly expressed miRNAs at 256-cell stage
Identification of novel miRNAs
Analysis of all sequencing reads during early zebrafish development resulted in the identification of eight miRNAs that had not been previously reported in the zebrafish genome (Table 2). For this, we analyzed all small RNA reads from the four developmental stages using the miRDeep algorithm (Friedlander et al. 2008). Following this pipeline, predicted miRNAs were filtered for novel miRNA identification by comparison to sequences deposited in the miRNA Registry. To increase accuracy, miRDeep-predicted novel miRNAs that genomically overlap with tRNA, rRNA, or transposable elements, were eliminated, and our analyses were restricted to only those that were identified in at least two different developmental stages and/or two independent libraries. With this restriction, five novel candidate miRNAs were identified, two of which are conserved across vertebrates, whereas the other three appear to be zebrafish specific (Supplemental Fig. 5). The remaining three were discovered independent of miRDeep and match miRNA sequences reported for other species. For these three, we aligned small RNAs to known miRNA hairpin sequences of other species in conjunction with secondary structure analyses of the corresponding zebrafish genomic loci. Of the three, two show conservation of the mature miRNA strand, while one shows conservation of the passenger strand. For all eight new zebrafish miRNAs, we validated their presence by examining and ensuring that their flanking sequences and corresponding genomic loci fit existing models of miRNA biogenesis (see Supplemental Material, Novel miRNA Sequences). To independently confirm the expression of these miRNAs, we performed Northern blots on total RNA from the same four stages of development as used for deep sequencing. As expected, these miRNAs were expressed at low levels, but we were still able to detect bands of the appropriate size for the mature sequences for six out the eight novel miRNAs and bands corresponding to pre-miRNAs for five of the eight (Fig. 3). We could not detect signals for the two remaining novel miRNAs, presumably due to expression at levels below the limits of detection. Nevertheless, based on the criteria used to identify these miRNAs, we believe they constitute newly reported miRNAs.
TABLE 2.
Novel miRNAs
FIGURE 3.
Northern blot of novel miRNAs. Northern blots were performed to verify the expression of predicted novel miRNAs. Mature miRNAs were detected for six of the eight predicted miRNAs, and miRNA precursors were detected for five of the six miRNAs.
miRNA sequence variation
Sequence variation at the 5′ and 3′ ends of mature miRNA sequences has been reported following deep-sequencing analyses (Landgraf et al. 2007; Morin et al. 2008; Lehrbach et al. 2009; Burroughs et al. 2010; Fernandez-Valverde et al. 2010; Kamminga et al. 2010). Such variation is due to mismatches between the reads and their corresponding genomic loci, with most mismatches detected at the 3′ ends. We also observed significant sequence variation, mostly nucleotide additions at the 3′ ends (Fig. 4A; Supplemental Table 2). Within the mature miRNA sequences, <1% of the reads differed from their genomic loci. In contrast, ∼40% of the total reads were 1–2 nt longer at the 3′ end than the mature sequence reported in miRBase, whereas much less 5′ end variation was detected. In some cases, the additional nucleotides matched the sequence of the pre-miRNA, suggesting that cleavage events during miRNA processing are not always precise. In the case of miR-2190, sequence heterogeneity was observed at both the 5′ and 3′ ends, with no clear preference for specific precursor cleavage sites, again arguing against this being a bona fide miRNA.
FIGURE 4.
miRNA sequence heterogeneity. (A) Significant sequence variation was detected among mature miRNA reads, especially at 3′ ends. Shown are compilations from four different miRNAs with the extent of variation at any particular nucleotide indicated by the size of the font. (Below) In red is the mature miRNA sequence with adjacent genomic sequence in black. Nontemplate-directed 3′ additions are shown in color. (B) Similar composition of 3′ addition among different miRNAs of the same family. The extent and base composition of miRNA tailing is indicated for a representative subset of miRNA families at the indicated stages of development. The percentage of reads with different nucleotide 3′ ends is shown in different colors. (C) Modification of mature miR-430 reads. The ratio of both A- or U-tailed mature miR-430 family reads to total mature miR-430 reads is shown across four developmental stages. (D) Modification of miR-430 reads subject to trimming. A large number of miR-430 reads did not contain the normal terminal guanosine. For this subset, the ratio of A- or U-tailed RNAs miR-430 reads is shown across four developmental stages. (E) The percentage of U-tailed miRNA reads for all tailed miRNA reads in both 256-cell and sphere stage embryos. All miRNAs with 3′ additions of nontemplate-directed U residues are shown for the 256-cell and sphere stage. The percentage of U addition increased from the 256-cell stage to the sphere stage.
We also detected significant 3′ sequence variation (∼10% of reads) due to the addition of nongenomically encoded nucleotides, mostly adenine, uracil, or both (Supplemental Fig. 6A). Rarely did we detect addition of cytosine or guanosine, although miR-181 had reads with guanosine addition and miR-738 had reads with cytosine addition. Among miRNA families, the composition and extent of sequence variation was nearly identical between family members (Fig. 4B). Additions at the 3′ end in this manner (tailing) have been reported previously, with proposals that the extra adenine nucleotides might stabilize miRNA half-life (Katoh et al. 2009; Fernandez-Valverde et al. 2010; Burns et al. 2011). Our data are consistent with this idea, but also support the hypothesis that tailing with U residues may be a mechanism to target miRNAs for degradation. Support for this hypothesis is based on variation at the 3′ end of miR-430 family members. As above, this family is the most abundant miRNA at the sphere stage, where it constitutes 97% of the reads and functions to target maternal mRNAs for degradation (Giraldez et al. 2005, 2006). We found that 3′ nontemplate-directed uracil addition among miR-430 family members is relatively low until the sphere stage, after which time increasing addition of U residues was observed. In contrast, the extent of modification by the addition of A residues was not significantly different across these stages. Increasing modification by the addition of U residues coincided with declining levels of detectable miR-430 from the sphere stage onward (Fig. 4C).
To further examine the effects of nucleotide addition on the levels of miR-430, we also analyzed the 3′ ends of miR-430 reads that lacked the last guanosine from the mature miRNA. These reads are likely to be derived from mature miR-430 RNAs that have undergone trimming and subsequent tailing at the time of library preparation. For these RNAs, we found a significant increase in adenosine addition at the sphere stage (Fig. 4D). In contrast, uracil addition for these reads was lowest at the sphere stage. Together with the modifications described above for mature miR-430 family members, the sequencing data are consistent with the hypothesis that U and A additions are associated with miRNA destabilization and stabilization, respectively.
To more broadly assess the dynamics of nucleotide addition between stages, we next examined the subset of modified miRNAs whose expression overlaps both the 256-cell stage and the sphere stage. For these miRNAs, we calculated the percentage of reads that showed U addition compared with all tailed reads. For each miRNA shown in Figure 4E, we show the extent of U modification at the 256 cell stage (blue) compared with U modification at the sphere stage (red). Increased levels of U modification were observed for the majority of miRNAs as the embryo proceeds from the 256-cell stage through the sphere stage. Interestingly, if all modifications on all miRNAs are examined across stages of development, we see a similar phenomenon with the majority of modifications detected at the 256-cell stage, followed by the sphere stage, and then decreasing levels of 3′ modification thereafter. This is shown by the extent and amounts of black lines above the x axis in Supplemental Figure 6B. The data are consistent with the idea that there is active stabilization/destabilization prior to and through the beginning of the maternal zygotic transition, after which multiple mechanisms are in play to control the expression and stability of a given miRNA.
Zebrafish piRNA expression
When we examined the total reads from all four developmental time points, a distinct subset of small RNAs was detected that derive from genomic regions that do not encode miRNA genes (Fig. 1). These RNAs represent a diverse array of sequences with a peak size distribution of ∼26–28 nt, and they map to either repetitive or unique genomic sequences (Fig. 1). Their overall detection was clearly not saturated, as >80% were detected only once. Based on size, they most likely represent piRNAs (Fig. 5A,B). Three additional criteria suggest that these RNAs are bona fide piRNAs. First, 10,892 reads matched sequences previously reported to be piRNAs in zebrafish (Houwing et al. 2007). Second, in genome wide, a much, much larger number of reads (674,777 sequences from 1,068,353 reads from shield stage) mapped to transposable elements with a strong bias toward the antisense strand (Fig. 5C; Supplemental Fig. 7). Finally, for those reads that mapped to the sense strands of transposable elements, there was a strong preference for an A at position 10, while those derived from antisense strands showed a preference for a U at position 1. This is completely consistent with the Ping-Pong model of piRNA biogenesis (Supplemental Fig. 8).
Over 65% of the piRNA reads that originated from repetitive elements were derived from the LTRs of transposable elements (Supplemental Fig. 9). A different subset mapped back to unique genomic loci, primarily intergenic regions of the zebrafish genome. For both, the reads showed a periodicity and strand preference. Based on the genomic loci of both types of piRNAs, it appears that the majority of the piRNA reads are derived from long transcripts that are then subsequently processed into smaller RNAs. The genomic regions that give rise to these long transcripts are referred to as piRNA clusters (Brennecke et al. 2007; Houwing et al. 2007). We defined piRNA clusters as genomic fragments encoding at least 10 unique piRNAs with gaps no greater than 1 kb (Supplemental Table 3). By grouping reads into defined piRNA clusters, we were able to quantify expression patterns based on the abundance of clusters instead of absolute piRNA reads, allowing examination of the expression of piRNAs during early zebrafish development. Previous work reported piRNA expression in adult ovaries and testes (Houwing et al. 2007), but we detected widespread embryonic piRNA expression across all four developmental stages (Fig. 5D,E). Beginning at the sphere stage and becoming much more prevalent at the shield stage, we detected increased piRNA expression (Fig. 5D–G). The increase was not due to a decrease in other small RNAs, because the sequencing runs were not saturated and, for example, we detected a robust increase in piRNA reads between 256-cell and sphere stage embryos, coincident with the large increase in miR-430 reads. This suggests enhanced transcriptional activity of piRNA clusters and transposable elements, with a resultant increase in piRNA production that likely functions to maintain genome integrity during early cell divisions and as germ cells are set aside. Compared with piRNA expression data from adult ovaries and testes (GEO: GSE7131; http://www.ncbi.nlm.nih.gov/geo/ under series number GSE7131) (Houwing et al. 2007), there is a significant reduction in piRNA expression in adult tissues compared with the early developmental stages we examined. At least for ovaries and testes, the expression patterns in adult tissues most closely resemble those observed at the 256-cell stage. This is consistent with maternally contributed piRNAs being most similar in pattern to those detected in ovaries and testes.
Previous analysis of piRNAs in zebrafish showed that in adult ovaries and testes, the majority of piRNAs are derived from the plus strands of genome (Houwing et al. 2007). In contrast, during early development, we observed a preference for piRNAs from minus strands (Fig. 5F,G). Given the fact that zebrafish piRNAs have a strong bias toward the antisense strand of transposons, this suggests that the expressed transposable or repetitive elements that are associated with piRNAs at very early developmental stages are, at least partially, different from those expressed in adult ovaries and testes. It is not clear why such strand preferences exist at the different times of development. Regardless of strand of origin, the data suggest that piRNAs have important functions not only in adult ovaries and testes, but also at multiple stages of early development.
tRNA-derived small RNAs in zebrafish
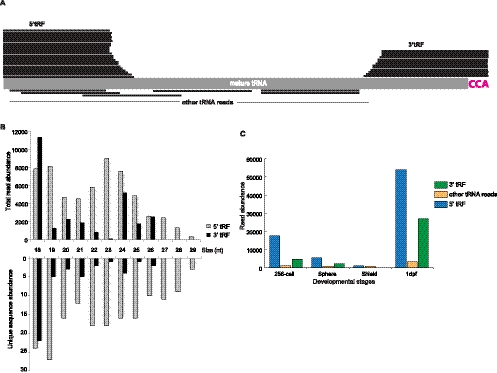
In addition to miRNAs and piRNAs, we also detected a large number of other small noncoding RNAs. One particularly interesting subset (∼225 different sequences; 56,311 total reads) was derived from tRNAs (Fig. 6). These RNAs matched the 5′ and 3′ ends of tRNAs with a size range between 18 and 28 nt (Fig. 6A,B; Supplemental Table 4). The 3′ end reads contained the universal 3′ CCA sequence that is added post-transcriptionally, indicating that these RNAs are derived from mature tRNAs, consistent with previous reports (Cole et al. 2009; Lee et al. 2009; Haussecker et al. 2010). Interestingly, the sizes of the 3′ tRNA reads are mainly 18 nt, while the 5′ tRNA reads are distributed more widely, suggesting that they might be under different selection mechanisms or have different functions (Fig. 6B). Given the nonrandom accumulation of these small RNAs from just the 5′ and 3′ ends, it is hard to argue that these RNAs are degradation products. Indeed, other reads across the entire mature tRNA sequence were observed at much, much lower frequencies (17-fold less at 256-cell and 1 dpf) (Fig. 6C). We note that the tRNA-derived fragments we detect are likely different from angiogenin-mediated tRNA fragments found in stress-induced cells, because the cleavage sites we detect are different from those reported (Thompson et al. 2008; Yamasaki et al. 2009; Emara et al. 2010).
FIGURE 6.
Zebrafish tRNA-derived Fragments (tRF). (A) Graphic representation of tRF alignments to zebrafish tRNAs. Most reads mapped to the 5′ and 3′ ends or mature tRNAs. (B) Size distribution of all identified tRF reads from four developmental stages in terms of either unique read sequence abundance or total read abundance. (C) The raw abundance of the 5′ tRF and 3′ tRF reads at four developmental stages, as indicated.
DISCUSSION
miRNA expression
miRNAs are postulated to play important roles during the maternal-zygotic transition, when embryos reshape and reprogram the transcriptional landscape (Giraldez 2010). Using deep sequencing, we determined the expression patterns for 198 distinct miRNAs, and we identified five novel miRNAs and three new conserved zebrafish miRNAs expressed during early development. Maternally inherited miRNAs and a subset of miRNAs that are expressed very early in development are important for zebrafish embryogenesis (Giraldez et al. 2005, 2006). Compared with prior approaches involving direct cloning or microarrays, the increased sensitivity of miRNA detection with deep sequencing affords the opportunity to more completely define the expression patterns and, ultimately, the function of small noncoding RNAs during early vertebrate development, including those expressed in single cells or specific tissues (Wienholds et al. 2005; Thatcher et al. 2007). The 198 miRNAs that we identified and profiled can be classified into 122 miRNA families. A total of 70% of these families are expressed at or below the limits of detection using microarrays. Fortunately, for those miRNAs expressed at detectable levels using microarrays, there was a strong correlation between the data sets, lending confidence that data generated from our sequencing libraries are accurate (Wienholds et al. 2005; Thatcher et al. 2007).
Previous studies showed that zebrafish miRNA expression patterns become increasingly complex as development proceeds (Wienholds et al. 2005; Thatcher et al. 2007). Our results revealed more widespread miRNA expression at earlier stages of development than previously observed. Beginning at the 256-cell stage, a large number (178 out of 216 currently known zebrafish miRNAs) of miRNAs are detectable, with several expressed at relatively high levels. These miRNAs are likely maternally deposited and may either regulate initial development or function similar to the miR-430 family to target maternally inherited mRNAs for degradation during the maternal-zygotic transition (Newport and Kirschner 1982; Giraldez et al. 2005; Schier 2007). We found that fully 99% of the miRNA reads detected during the sphere stage are derived from miR-430 family members. Interestingly, maternally inherited miRNAs appear to be rapidly down-regulated during the maternal-zygotic transition, raising the possibility that they are subject to similar forms of regulation as maternally inherited mRNAs. Beyond maternally inherited miRNAs, our analyses are also consistent with important early roles for a number of miRNAs, including miR-203, miR-1, and miR-133, which function to balance proliferation and differentiation during early development (Yi et al. 2008; Mishima et al. 2009). In contrast to previous results, we detected significant levels of let-7, raising the question as to its exact role in early development (Kloosterman et al. 2004; Wienholds and Plasterk 2005).
Normalization of miRNA sequencing data
Typically, total genome matching reads are used to account for differences in the sequencing depth of individual libraries when normalizing small RNA sequencing data. This certainly applies when biological replicates are being sequenced; for example, when comparing two independent libraries from the same developmental stage. However, if the overall transcriptome changes dramatically between samples, normalization based solely on total genome matching reads can obscure important biology (Robinson and Oshlack 2010). During early development, cell numbers are increasing dramatically, concomitant with extraordinary changes in gene expression as differentiation proceeds. This creates a unique set of problems when normalizing sequencing data. We chose to normalize based on the relative expression levels of either miR-9 or let-7. These miRNAs were chosen because they are expressed at moderate levels across the four stages of early development that we examined. The resulting heat maps were very similar, whether using miR-9 or let-7, and Northern blots of six miRNAs were completely consistent with the derived heat maps. Interestingly, when we normalized using TMM, the resulting heat maps were also very similar to those obtained with either miR-9 or let-7. Thus, we believe that the strategy we adopted to normalize our small RNA sequencing data during early development is appropriate. Nevertheless, we realize that under conditions where differential gene expression patterns are changing dramatically, every available method has certain limitations, and other methods can certainly be used to quantitatively analyze small RNA sequencing data, e.g., TMM (Robinson and Oshlack 2010) and DESeq package from http://www-huber.embl.de/users/anders/DESeq/ (Anders and Huber 2010).
Sequence variation
miRNA 3′ tailing and trimming were previously reported in worms, flies, mouse, and human cells (Landgraf et al. 2007; Katoh et al. 2009; Lehrbach et al. 2009; Ameres et al. 2010; Burroughs et al. 2010; Fernandez-Valverde et al. 2010). We detected many tailed miRNAs containing additional nontemplate-directed nucleotides, mostly A and U, primarily at 3′ ends (Fig. 4A). The extent of 3′ variation may actually be higher than that reported here, since in some cases (e.g., let-7) the extra A and U residues match the pre-miRNA flanking sequence, so their origin is not clear. The temporal pattern of A versus U tailing suggests that the addition of A residues might stabilize miRNA half-life, whereas U addition may promote degradation. This seems to hold true for the miR-430 family, but may apply more broadly, since we detected a large percentage of tailed miRNAs during very early development, with decreasing modification at the later stages. It is possible that different mechanisms regulate miRNA half-life during the maternal zygotic transition compared with later stages, but our data suggest a common mechanism.
piRNA expression
piRNAs have been reported to be specifically expressed in the germline to maintain genome integrity (Halic and Moazed 2009). We identified many piRNAs in 256-cell stage embryos, consistent with maternal inheritance. We also found a peak of piRNA expression at the shield stage, with readily detectable levels out to 1 dpf. Except for the shield stage, the pattern is similar to that reported in mouse embryos (Ohnishi et al. 2010). However, the robust expression levels at the shield stage, when the germ layers are forming, may indicate an important function for piRNAs during early development. Previous work has reported that piRNAs can induce deadenylation and decay of nanos, a maternal mRNA in Drosophila embryos (Rouget et al. 2010). In zebrafish, primordial germ cells (PGCs) are first specified during early embryogenesis and give rise to germ cells after proper migration and further differentiation. Zebrafish PGCs show maximal migration activity amongst a far larger number of somatic cells at the beginning of gastrulation (5.5–6 hpf) (Raz 2003; Blaser et al. 2005). The shield stage coincides with the onset of gastrulation, suggesting that the widespread expression of piRNAs that we detect at this time may play an important role in the maintenance and proper migration of PGCs. Only a very limited number of PGCs have formed or are forming during this time, raising the possibility of piRNAs originating from other cells beside PGCs in embryos. Consistent with a requirement for piRNAs during these early stages, morpholino-mediated knockdown of Ziwi protein inhibited and/or blocked proper germ-cell migration (H Dai, C Wei, and JG Patton, unpubl.). Future studies are needed to understand the temporal and spatial manner of piRNA expression and their specific functions during early embryonic development.
TRNA-derived small RNAs
Small tRNA-derived fragments were previously reported in different cell lines (Cole et al. 2009; Lee et al. 2009; Haussecker et al. 2010). Our sequencing analyses expand the number of species that express this class of small RNA, indicating that the biogenesis and function of tRNA-derived small RNAs is conserved, suggesting an important role. Several lines of evidence raise the possibility that these small RNAs function in gene silencing in a Dicer-independent manner (Cole et al. 2009; Haussecker et al. 2010). It will be important to determine how or whether these RNAs regulate gene expression, especially in light of the fact that these RNAs are themselves temporally regulated during development.
MATERIALS AND METHODS
Small RNA purification, cloning, and sequencing
Zebrafish embryos were collected at the 256-cell stage, sphere stage, shield stage, and 1 d post fertilization (dpf). Total RNA was isolated from embryos using Trizol. RNAs were fractionated on 15% denaturing polyacrylamide gels, and small RNAs between 15 and 30 nt were excised and purified. cDNA libraries were generated using specific linkers and RT/PCR, as previously described (Hafner et al. 2008). Libraries were sequenced in the Genome Technology Core of Vanderbilt University using the Illumina sequencing platform. The numbers of sequencing reads from the four developmental stages are listed in Supplemental Table 5.
3′ ligation adaptor: AMP-5′-pCTGTAGGCACCATCAATdideoxyC-3′.
5′ ligation adaptor: 5′-ACACUCUUUCCCUACACGACGCUCUUCCGAUC-3′.
RT primer: ATTGATGGTGCCTACAG.
PCR forward primer: 5′-AATGATACGGCGACCACCGAACACTCTTTCCCTACACGACG-3′.
PCR reverse primer: 5′-CAAGCAGAAGACGGCATACGATTGATGGTGCCTACAG-3′.
Small RNA identification
Initial reads were processed to remove the linker sequences using an in-house dynamic alignment algorithm, which allows one mismatch and a minimal of five starting nucleotides in the linker sequences. Small RNAs matching the zebrafish genome (Zv8) from Ensembl (http://www.ensembl.org) were retrieved with Bowtie, a short read alignment algorithm widely used for aligning short DNA sequences, using the default parameters which allow, at most, two mismatches in the “seed of 28 nt” with maximum quality values of 70 (Langmead et al. 2009). After elimination of reads derived from known mRNAs, tRNAs, rRNAs, and snRNAs, miRNAs were next annotated based on a perfect match to miRNA hairpin sequences as reported in miRbase (http://www.miRbase.org) with further filtering to remove reads derived from miRNA passenger strand and loops. To identify miRNA reads with nontemplate-directed nucleotide additions, reads with exactly one mismatch at the very 3′ end were specifically retrieved from the original data set. For analysis of known miRNAs, sequencing frequencies of less than two reads per library were discarded. For quantitation of expression levels, miRNA reads with nontemplated addition of 3′ nucleotides were included in the counts.
To identify piRNAs, consensus sequences from zebrafish repetitive elements were retrieved from Repbase (Kohany et al. 2006) (http://www.girinst.org/repbase/index.html) and RepeatMaskers using the UCSC genome browser (http://genome.ucsc.edu). Small RNAs perfectly mapping to these consensus sequences and their genomic flanking regions were sorted into piRNA libraries. piRNAs with unique mapping positions in the genome were isolated and analyzed independently. For piRNAs with multiple mapping positions, the abundance of each piRNA was weighted by the number of the piRNA reads divided by the number of its genomic mapping positions, assuming all genomic loci contribute equally to overall read numbers. piRNA clusters were manually defined as genomic loci when at least 10 different piRNAs mapped to positions with <1 kb between the reads, as previously described (Brennecke et al. 2007; Houwing et al. 2007).
miRNA prediction
We prepared libraries from RNAs between 15 and 30 nt, but only analyzed genome-derived small RNAs of 18 nt or longer that aligned to known hairpin sequences from other species listed in miRbase (http://www.mirbase.org/ftp.shtml). Small reads either perfectly matching the stem regions of these hairpins or overlapping with known mature or passenger miRNAs were then retrieved using Bowtie with default parameters. The dynamic stability of secondary structures of flanking genomic sequences for all retrieved RNAs and their relative localization in the zebrafish genome were further analyzed using RNAfold (Hofacker 2003) (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). Predicted hairpin sequences sharing high similarity with known hairpins from other species were further screened, focusing on the small RNAs on the stem regions to identify putative zebrafish miRNA homologs. For prediction analyses, RNAs whose sequencing frequency was less than five reads per library were discarded. The 5′ ends of novel miRNAs were determined by the starting position of the most abundant small RNA reads mapping to the stem of putative hairpin sequences. Predicted miRNAs were further validated in Ensembl using RFAM and miRBase. Also, miRDeep was independently utilized to predict novel miRNAs from raw sequence data (Friedlander et al. 2008). To identify novel miRNAs, samples from the four different stages were subjected to miRDeep analysis with a cutoff score of 2. Predicted miRNAs by miRDeep were further filtered by aligning to the databases of known miRNAs (miRBase), transposable elements, rRNAs, tRNAs, snRNAs, and other known small RNAs (UCSC genome browsers). To ensure the authenticity of these predicted miRNAs, only small RNAs that appeared in at least two developmental stages were retained. From among the 327 novel miRNAs predicted by miRDeep, five were finally recognized as authentic novel miRNAs.
qRT–PCR
qRT–PCR assays (Taqman RT kit and Taqman miRNA custom assays, ABI) were performed to measure the expression levels of let-7a and miR-9. Relative levels were normalized to U6 snRNA. cDNAs were synthesized from 10 ng of total RNA extracted from zebrafish embryos. Products of RT reactions without reverse transcriptase served as a negative control, and each measurement was derived from three biological replicates. Expression levels were determined based on the threshold cycle values (Ct) of each miRNA relative to that of U6 and assigned as 2-ΔΔCt.
Northern blots
A total of 18–23 μg of total RNA (10 μg for miR-430) was extracted from zebrafish embryos at different developmental stages, separated on 15%–20% denaturing polyacrylamide gels, and transferred to membranes for Northern blot analysis of known miRNAs following procedures as previously described (Thatcher et al. 2007). For novel miRNA detection, 13–15 μg of small RNAs (<200 nt) were enriched from 220–250 μg of total RNA extracted from 1 dpf zebrafish embryos using mirVana miRNA isolation kits (Applied Biosystems). The sequences of miRNA Northern probes (miRNA StarFire probes, IDT) were complementary to the mature sequences for let-7a, miR-455, miR-21, miR-22a, miR-200b, and miR-430b. For the novel miRNAs, the probes were as follows:
novel miRNA 1: GGTAACCATTGGCTTCCATTGT;
novel miRNA 2: AACTGGTAACCATTGACCTTCA;
novel miRNA 3: ACTGGCTACCAGCATGGTACA;
novel miRNA 4: CAGTATCAGCGAGTATTTCCAT;
novel miRNA 5: CCTTCCAGTGTAGGATTCACA;
novel miRNA 8: GGAACCCTTCTCCACTTC.
miRNA expression profiles
Absolute read numbers were collected and sorted into miRNA families based on sequence records in miRbase. We defined miRNA families as those miRNAs sharing the same seed sequence along with high-sequence similarity toward the 3′ end. We also normalized expression levels across four developmental stages relative to let-7a and miR-9 levels, as determined using qRT–PCR. Read numbers were transformed into log2 values and displayed with a MultiExperiment Viewer (Saeed et al. 2003, 2006) (http://www.tm4.org/mev/). The raw log2 value of all single miRNAs and miRNA families throughout development are listed in Supplemental Table 6.
DATA DEPOSITION
Sequencing data have been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under the series number GSE27722. Public data with series number GSE7131 are also available at http://www.ncbi.nlm.nih.gov/geo/.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Elizabeth Thatcher, Nan Li, and Stacey Lawrence for their assistance in preparing the small RNA libraries. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University and was supported by grants from the National Institutes of Health to J.G.P. (GM 075790 and EY019759), the Searle Scholars Program (A.R.), and the National Science Foundation (DEB-0844968 to A.R.). C.W. was supported in part by a Discovery Grant from Vanderbilt University.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.029090.111.
REFERENCES
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106 doi: 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ 2004. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 5: 396–400 [DOI] [PubMed] [Google Scholar]
- Blaser H, Eisenbeiss S, Neumann M, Reichman-Fried M, Thisse B, Thisse C, Raz E 2005. Transition from non-motile behaviour to directed migration during early PGC development in zebrafish. J Cell Sci 118: 4027–4038 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Burns DM, D'Ambrogio A, Nottrott S, Richter JD 2011. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature 473: 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, et al. 2010. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res 20: 1398–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Manninga H, Slanchev K, Chien M, Russo JJ, Ju J, Sheridan R, John B, Marks DS, Gaidatzis D, et al. 2005. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev 19: 1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G 2009. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P 2010. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem 285: 10959–10968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Valverde SL, Taft RJ, Mattick JS 2010. Dynamic isomiR regulation in Drosophila development. RNA 16: 1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Lai EC 2008. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet 9: 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG 2007. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet 39: 259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26: 407–415 [DOI] [PubMed] [Google Scholar]
- Friedlander MR, Adamidi C, Han T, Lebedeva S, Isenbarger TA, Hirst M, Marra M, Nusbaum C, Lee WL, Jenkin JC, et al. 2009. High-resolution profiling and discovery of planarian small RNAs. Proc Natl Acad Sci 106: 11546–11551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ 2010. microRNAs, the cell's Nepenthe: clearing the past during the maternal-to-zygotic transition and cellular reprogramming. Curr Opin Genet Dev 20: 369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF 2005. MicroRNAs regulate brain morphogenesis in Zebrafish. Science 308: 833–838 [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S 2004. The microRNA Registry. Nucleic Acids Res 32: D109–D111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ 2010. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev 24: 2499–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T 2008. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Moazed D 2009. Transposon silencing by piRNAs. Cell 138: 1058–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA 2010. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16: 673–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL 2003. Vienna RNA secondary structure server. Nucleic Acids Res 31: 3429–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129: 69–82 [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF 2008. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J 27: 2702–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran G, Zheng Y, Sumathipala N, Jiang H, Arrese EL, Soulages JL, Zhang W, Sunkar R 2010. Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genomics 11: 52 doi: 10.1186/1471-2164-11-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJ, Roovers EF, Ladurner P, Berezikov E, Ketting RF 2010. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J 29: 3688–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T 2009. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev 23: 433–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310 [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH 2004. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res 32: 6284–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J 2006. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 7: 474 doi: 10.1186/1471-2105-7-474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-2-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN 2002. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21: 4663–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23: 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA 2009. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol 16: 1016–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- Linsen SE, de Wit E, Janssens G, Heater S, Chapman L, Parkin RK, Fritz B, Wyman SK, de Bruijn E, Voest EE, et al. 2009. Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods 6: 474–476 [DOI] [PubMed] [Google Scholar]
- Lund E, Liu M, Hartley RS, Sheets MD, Dahlberg JE 2009. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA 15: 2351–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer SU, Pfaffl MW, Ulbrich SE 2010. Normalization strategies for microRNA profiling experiments: a ‘normal’ way to a hidden layer of complexity? Biotechnol Lett 32: 1777–1788 [DOI] [PubMed] [Google Scholar]
- Mishima Y, Abreu-Goodger C, Staton AA, Stahlhut C, Shou C, Cheng C, Gerstein M, Enright AJ, Giraldez AJ 2009. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev 23: 619–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18: 610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Kirschner M 1982. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell 30: 675–686 [DOI] [PubMed] [Google Scholar]
- Ohnishi Y, Totoki Y, Toyoda A, Watanabe T, Yamamoto Y, Tokunaga K, Sakaki Y, Sasaki H, Hohjoh H 2010. Small RNA class transition from siRNA/piRNA to miRNA during pre-implantation mouse development. Nucleic Acids Res 38: 5141–5151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Rinn JL, Schier AF 2011. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 12: 136–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz E 2003. Primordial germ-cell development: the zebrafish perspective. Nat Rev Genet 4: 690–700 [DOI] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25 doi: 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M 2010. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467: 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17: 1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J 2006. TM4 microarray software suite. Methods Enzymol 411: 134–193 [DOI] [PubMed] [Google Scholar]
- Schier AF 2007. The maternal-zygotic transition: death and birth of RNAs. Science 316: 406–407 [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Skalsky RL, Cullen BR 2010. Viruses, microRNAs, and host interactions. Annu Rev Microbiol 64: 123–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares AR, Pereira PM, Santos B, Egas C, Gomes AC, Arrais J, Oliveira JL, Moura GR, Santos MA 2009. Parallel DNA pyrosequencing unveils new zebrafish microRNAs. BMC Genomics 10: 195 doi: 10.1186/1471-2164-10-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher EJ, Flynt AS, Li N, Patton JR, Patton JG 2007. MiRNA expression analysis during normal zebrafish development and following inhibition of the Hedgehog and Notch signaling pathways. Dev Dyn 236: 2172–2180 [DOI] [PubMed] [Google Scholar]
- Thompson DM, Lu C, Green PJ, Parker R 2008. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14: 2095–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RH 2005. MicroRNA function in animal development. FEBS Lett 579: 5911–5922 [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH 2005. MicroRNA expression in zebrafish embryonic development. Science 309: 310–311 [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC 2011. Widespread regulatory activity of vertebrate microRNA* species. RNA 17: 312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Poy MN, Stoffel M, Fuchs E 2008. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature 452: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y 2007. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev 21: 1190–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]