Small nucleolar and small Cajal body RNAs (snoRNAs and scaRNAs) are generated by exonucleolytic shortening of longer precursors. Removal of the last few nucleotides at the 3′ end is known to be distinct from the removal of the bulk of excess 3′ sequence. This manuscript shows that, in human cells, the noncanonical poly(A) polymerase PAPD5 adds oligo(A) tails to the short intronic extensions remaining beyond the mature 3′ end of H/ACA box snoRNAs. The poly(A) specific ribonuclease (PARN) then removes these tails and also the remaining intronic nucleotides. In agreement with a role of PARN in snoRNA and scaRNA processing, the enzyme is concentrated in nucleoli and Cajal bodies.
Keywords: deadenylation, 3′ end processing, poly(A) polymerase, poly(A)-specific ribonuclease, small nucleolar RNA
Abstract
Small nucleolar and small Cajal body RNAs (snoRNAs and scaRNAs) of the H/ACA box and C/D box type are generated by exonucleolytic shortening of longer precursors. Removal of the last few nucleotides at the 3′ end is known to be a distinct step. We report that, in human cells, knock-down of the poly(A) specific ribonuclease (PARN), previously implicated only in mRNA metabolism, causes the accumulation of oligoadenylated processing intermediates of H/ACA box but not C/D box RNAs. In agreement with a role of PARN in snoRNA and scaRNA processing, the enzyme is concentrated in nucleoli and Cajal bodies. Oligo(A) tails are attached to a short stub of intron sequence remaining beyond the mature 3′ end of the snoRNAs. The noncanonical poly(A) polymerase PAPD5 is responsible for addition of the oligo(A) tails. We suggest that deadenylation is coupled to clean 3′ end trimming, which might serve to enhance snoRNA stability.
INTRODUCTION
Eukaryotic cells contain many different small nucleolar RNAs (snoRNAs), which function in the site-specific modification of ribosomal RNAs: C/D box snoRNAs direct ribose 2′-O-methylation, and H/ACA box snoRNAs are essential for the isomerization of uridine to pseudouridine. The two types of snoRNAs are distinguished by unique structural and sequence elements, and each is associated with a distinct set of four different core proteins. Among these, fibrillarin (Nop1p), complexed with C/D box RNAs, carries out 2′-O-methylation, and dyskerin (Cbf5p), complexed with H/ACA box snoRNAs, catalyzes pseudouridylation. The snoRNAs direct fibrillarin and dyskerin to specific sites by base-pairing with the substrate RNA. A smaller number of related molecules, the small Cajal body RNAs (scaRNAs), play comparable roles in the modification of spliceosomal snRNAs, a process taking place in Cajal bodies (Bachellerie et al. 2002; Filipowicz and Pogacic 2002; Kiss 2002; Reichow et al. 2007; Kiss et al. 2010).
All snoRNAs are processed by exonucleases from precursors, which can be produced in different ways. In Saccharomyces cerevisiae, most snoRNAs are encoded in individual transcription units. Their 3′ ends are generated by 3′ exonuclease digestion from an entry point created by cotranscriptional processing coupled to termination (Fatica et al. 2000; Steinmetz et al. 2001; Grzechnik and Kufel 2008). Some snoRNAs are synthesized as polycistronic transcripts, which are dissected into precursors to individual snoRNAs by the endonuclease Rnt1p (Chanfreau et al. 1998a,b). Almost all vertebrate and some yeast snoRNAs are encoded in introns (Liu and Maxwell 1990; Leverette et al. 1992; Fragapane et al. 1993; Kiss and Filipowicz 1993; Tycowski et al. 1993). The pre-snoRNA sequences are liberated from the primary transcript by endonuclease cleavage (Caffarelli et al. 1996; Villa et al. 1998) or, predominantly, by splicing and intron debranching; exonucleolytic processing ensues (Cecconi et al. 1995; Kiss and Filipowicz 1995; Ooi et al. 1998; Petfalski et al. 1998). In budding yeast, 3′ exonucleolytic shortening is catalyzed by the exosome (Allmang et al. 1999; van Hoof et al. 2000). Interestingly, final trimming of the last ∼3 nt is a distinct step and requires the exosome-associated nuclease, Rrp6p (Allmang et al. 1999; van Hoof et al. 2000; Grzechnik and Kufel 2008). In mammals, the exonucleases responsible for 3′ processing of pre-snoRNAs have not, to our knowledge, been examined experimentally. However, in an in vitro processing system, trimming of the final 5–8 nt at the 3′ ends of snoRNAs is distinct: It is slow in the case of H/ACA box RNAs and, under similar conditions, does not take place for C/D box RNAs (Kiss and Filipowicz 1993, 1995; Tycowski et al. 1993). Mature snoRNA sequences are thought to be protected from exonuclease digestion by binding of the snoRNP proteins and, if present, by the 5′ cap (Terns et al. 1995; Balakin et al. 1996; Caffarelli et al. 1996; Watkins et al. 1996; Ganot et al. 1997b; Bortolin et al. 1999). In fact, crystal structure analysis showed binding of the so-called PUA domain of Cbf5/dyskerin to the ACA box very close to the RNA 3′ end (Li and Ye 2006).
In S. cerevisiae cells with mutations affecting the nuclear exosome and in a Schizosaccharomyces pombe pab2 mutant, lacking the nuclear poly(A) binding protein, polyadenylation of precursors and intermediates of snoRNA maturation becomes apparent (Allmang et al. 1999; van Hoof et al. 2000; Wyers et al. 2005; Carneiro et al. 2007; Grzechnik and Kufel 2008; Lemay et al. 2010). 3′ cleavage and transcription termination of pre-snoRNAs is coupled to polyadenylation (Grzechnik and Kufel 2008), and 3′ ends produced by Rnt1p as well as intermediates of exosome digestion can also receive poly(A) tails (van Hoof et al. 2000; LaCava et al. 2005; Grzechnik and Kufel 2008). Much of this polyadenylation is catalyzed by the “canonical” poly(A) polymerase, Pap1p, but the enzyme may depend on oligo(A) primers provided by the noncanonical poly(A) polymerase, Trf4p (Carneiro et al. 2007; Grzechnik and Kufel 2008). Trf4p and the related enzyme, Trf5p, polyadenylate a great variety of nuclear RNAs to facilitate their degradation by the exosome or their processing to mature, functional molecules (Kadaba et al. 2004; LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005; Egecioglu et al. 2006; Houseley and Tollervey 2006; San Paolo et al. 2009; Lemay et al. 2010; Wlotzka et al. 2011).
The poly(A)-specific ribonuclease, PARN, is a homodimeric 3′ exonuclease that prefers poly(A) as a substrate and is moderately stimulated by a 5′ cap on the RNA (Körner and Wahle 1997; Körner et al. 1998; Dehlin et al. 2000; Gao et al. 2000; Martinez et al. 2001; Wu et al. 2005, 2009). The enzyme is widely, but not universally, conserved. Its substrate preferences suggest roles in mRNA metabolism. In fact, Xenopus PARN is responsible for the so-called default deadenylation of maternal mRNA during oocyte maturation (Körner et al. 1998; Dehlin et al. 2000; Copeland and Wormington 2001), and a role in mRNA degradation has also been suggested in mammalian somatic cells (e.g., Cevher et al. 2010). However, a comprehensive picture of the enzyme's function in RNA metabolism is lacking.
This paper defines a new function for mammalian PARN in the processing of H/ACA box snoRNAs: The noncanonical poly(A) polymerase PAPD5 adds oligo(A) tails to the last few nucleotides remaining after exonucleolytic degradation of the 3′ flanking intron; these oligoadenylated processing intermediates are then trimmed by PARN.
RESULTS
PARN is localized in nucleoli and Cajal bodies
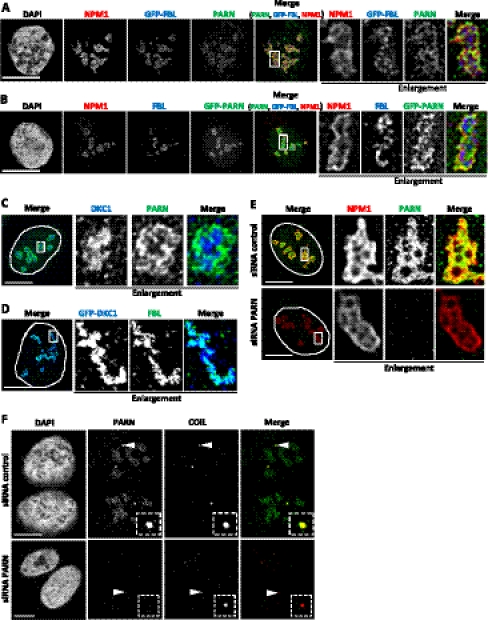
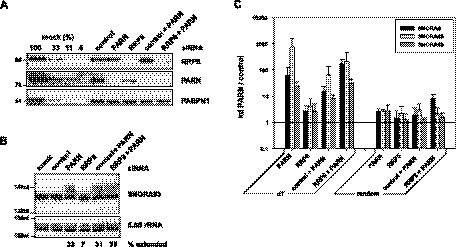
Immunostaining of U2OS cells with a PARN-specific antibody (Fig. 1; Supplemental Fig. S1A) revealed a nuclear localization in agreement with earlier data (Yamashita et al. 2005). The intensely stained structures were identified as nucleoli by costaining with antibodies against the markers nucleophosmin (NPM1) or fibrillarin (FBL) as well as by transfection of GFP-fibrillarin. Quantitation of the fluorescence signal in the plane of the confocal section indicated that the concentration of PARN in nucleoli was four times higher than in the nucleoplasm; the cytoplasmic level was indistinguishable from background. Whereas fibrillarin was localized mainly to the “interior” of nucleoli, PARN predominantly colocalized with nucleophosmin at the outer rim (Fig. 1A). This localization was confirmed by the expression of GFP-PARN, which colocalized with nucleophosmin but barely with fibrillarin (Fig. 1B). GFP-DKC1 colocalized with fibrillarin but, congruently, Dyskerin (DKC1) did not colocalize with PARN (Fig. 1C,D). Specificity of nucleolar localization was confirmed by loss of PARN immunostaining in nucleoli upon knock-down (Fig. 1E). Nucleolar morphology was largely unaffected by PARN knock-down, as indicated by the localization of nucleophosmin.
FIGURE 1.
PARN is localized in nucleoli and Cajal bodies. (A) Localization of endogenous PARN and nucleophosmin (NPM1) was analyzed by immunostaining in U2OS cells transfected with GFP-fibrillarin (GFP-FBL). All three proteins are localized in nucleoli. Significant colocalization of PARN with NPM1, but not with GFP-FBL, is observed within the nucleoli upon enlargement (2.5-fold magnification) of boxed region. (B) Localization of GFP-PARN was analyzed in U2OS cells counterstained for nucleophosmin (NPM1) and fibrillarin (FBL). Significant colocalization of GFP-PARN with NPM1, and to a much lesser extent with FBL, is observed in nucleoli upon enlargement of boxed region. (C,D) Localization of PARN, endogenous dyskerin (DKC1), GFP-dyskerin, and FBL was analyzed in U2OS cells. DKC1 colocalizes with FBL but not PARN at the interior of nucleoli, as observed upon enlargement of regions boxed in the left panel. (E) Localization of PARN in nucleoli is lost upon PARN knock-down, as indicated by counterstaining for nucleophosmin (NPM1). Enlargements (2.5-fold magnification) of boxed regions in left panel are shown in the right panels. (F) Localization of PARN was analyzed in cells counterstained for coilin (COIL). Both proteins colocalize in Cajal bodies in U2OS cells transfected with control siRNA. Upon PARN knock-down, the signal in nucleoli and some Cajal bodies is lost or severely diminished. Enlargements (2.5-fold magnification) of Cajal bodies indicated by arrow heads are shown as boxed insets. Note that PARN knock-down appears to affect Cajal body number and size; so far, this effect has not been further analyzed. In C–E, nuclei are indicated by dashed lines. Bar indicates 10 μm.
In addition to nucleoli, Cajal bodies, identified by coilin staining, also reacted strongly with PARN antiserum (Fig. 1F). Although neither GFP-PARN nor Flag-PARN was observed in Cajal bodies, severely diminished Cajal body staining upon PARN knock-down confirmed the specificity of antibody labeling. A localization of PARN in nucleoli and Cajal bodies was also observed in HEK293 cells (Supplemental Fig. S1B,C).
Identification of SNORA68 as a potential substrate of PARN
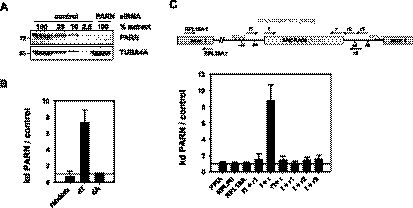
Microarray analysis was used to identify candidate PARN substrate RNAs by their increased abundance upon siRNA-mediated knock-down of the enzyme (Fig. 2A). Potential mRNA substrates of PARN identified by these analyses will be treated elsewhere. The array contained probe sets for 16 H/ACA box snoRNAs. Even though oligo(dT) priming was used for probe preparation, two such RNAs, SNORA61 and 68, were consistently detectable, and both were enriched upon PARN knock-down (average 1.6-fold and 8.5-fold in two independent experiments). The array also contained probe sets for seven C/D box snoRNAs, two of which (SNORD104, SNORD123) were consistently detectable, but neither was enriched upon knock-down of PARN.
FIGURE 2.
Oligoadenylated SNORA68 is enriched in PARN knock-down cells. (A) U2OS cells were transfected with PARN siRNA 1 or control siRNA. PARN levels were determined by Western blotting. A fixed amount of the PARN knock-down extract was compared with a dilution series of control extract. Tubulin (TUBA4A) served as a loading control. This is one representative example of many knock-down experiments. (B) The level of SNORA68 was analyzed by qRT-PCR. Total RNA was reverse-transcribed with random, oligo(dT) or oligo(dA) primers as indicated. SNORA68 sequences were quantified by qPCR and cross-normalized to PPIA and RPLP0. The qPCR of the oligo(dA)-primed sample was normalized to PPIA only, which is amplified due to an internal oligo(U) stretch. Data are plotted as abundance of SNORA68 in knock-down over control cells. (C) Oligo(dT)-primed cDNA as in B was analyzed by qPCR with primer combinations covering different parts of the RPL18A pre-mRNA as indicated at the top. Data were cross-normalized to PPIA and RPLP0 and are plotted as abundance of each RNA species in knock-down cells over control cells. In B, error bars indicate the standard deviation of five independent biological experiments, except the oligo(dA)-primed sample (average of two experiments). In C, the standard deviation is based on two independent biological experiments with two technical replicates each.
Quantitative RT-PCR confirmed a ∼10-fold enrichment of SNORA68 in PARN knock-down cells when the cDNA was prepared by oligo(dT) priming but not with random or oligo(dA) priming (Fig. 2B). Human SNORA68 is encoded in intron 3 of the RPL18A gene. Quantitative RT-PCR with different primer combinations was used to determine whether the increased SNORA68 signal reflected the abundance of the mature snoRNA or its processing intermediates or precursor. Whereas the abundance of an adenylated form of the mature snoRNA was increased upon PARN knock-down, RNA species detected by primers placed in flanking intron sequences of the host gene were not affected. Similarly, abundance of exon 1, reflecting the sum of mature mRNA, precursor, and processing intermediates, was not changed (Fig. 2C).
Many adenylated H/ACA box snoRNAs are substrates of PARN
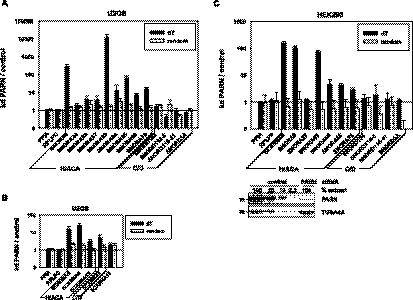
Another type of microarray, comprising probe sets for miRNAs and other small RNAs, was used to determine which and how many small RNAs are potential substrates of PARN. Again, hybridization probes were generated by oligo(dT) priming. The array contained probes for 274 C/D box RNAs. Roughly one-third gave reproducible signals in two biological replicates, but only one of them, SNORD3A, was weakly enriched in the knock-down sample. Probes for 96 H/ACA box snoRNAs were present on the array. Eighty percent were consistently detectable, and one-third of these were enriched after PARN knock-down (Table 1; Supplemental Table S1). Similar data were obtained for the scaRNAs; six out of nine detectable H/ACA box scaRNAs were enriched in the PARN knock-down, but only one out of four detectable C/D box scaRNAs (U90 = SCARNA7). One out of two detectable combined C/D and H/ACA box scaRNAs was also more abundant after depletion of PARN (U85 = SCARNA10). MiRNAs were not detectably affected (Table 1; Supplemental Table S1).
TABLE 1.
H/ACA box RNAs are substrates of PARN
Five H/ACA box snoRNAs that were increased upon PARN knock-down (SNORA9, 63, 65, and 76, and SNORA68 as a positive control) were validated by qRT-PCR. The abundance of all was increased (up to 10,000-fold) when oligo(dT) priming was used but not with random priming of RT-reactions (Fig. 3A). Four additional H/ACA box snoRNAs (SNORA24, 33, 37, and 64) that were not enriched in microarray analysis were also tested by oligo(dT)-primed qRT-PCR. Increases between two- and 16-fold suggested that all four are potential PARN substrates (Fig. 3A). In contrast, analysis of four C/D box snoRNAs revealed no significant change. This included SNORD3A, the only C/D box snoRNA suggested as a possible PARN substrate by microarray analysis. Quantitative RT-PCR also confirmed three adenylated H/ACA box scaRNAs to be increased by PARN depletion (Fig. 3B). The combined C/D and H/ACA box RNA SCARNA10 was twofold enriched, but this was also the case when random priming was used. SCARNA7 was the only C/D box RNA analyzed for which an increase in knock-down cells was found both by microarray and qRT-PCR.
FIGURE 3.
H/ACA, but not C/D box, RNAs are enriched upon PARN knock-down. (A) Levels of snoRNAs were analyzed by reverse-transcription of total RNA prepared from control and PARN knock-down U2OS cells using random or oligo(dT) priming and gene-specific qPCR. Data were cross-normalized to PPIA and RPLP0 messages and are plotted as RNA level in knock-down over control cells. (B) Relative scaRNA levels were determined in U2OS cells by qRT-PCR as described in A. (C) Relative snoRNA levels in HEK293 cells were analyzed by qRT-PCR as in A. The Western blot control for the knock-down efficiency shown below the histogram is a representative example. In A–C, data represent the average of at least three independent biological experiments; bars indicate standard deviations.
Knock-down of PARN in HEK293 cells also resulted in increased levels of five out of six H/ACA box snoRNAs and the single H/ACA box scaRNA examined; SNORA37 was the only exception. In contrast, the levels of four C/D box snoRNAs analyzed did not increase significantly (Fig. 3C). The effect of PARN depletion on adenylated H/ACA box RNAs in HEK293 cells was smaller than in U2OS cells. This might be due to an about fivefold higher expression of PARN in HEK293 cells, as determined by Western blots (Supplemental Fig. S2).
In summary, the microarray data show that many adenylated H/ACA box RNAs accumulate when PARN levels drop. Quantitative RT-PCR demonstrated increased levels of all H/ACA box RNAs tested; therefore, many, possibly all, H/ACA box RNAs are substrates of PARN. With a single exception, C/D box RNAs were not detectably affected by PARN knock-down.
PARN catalyzes deadenylation of H/ACA box snoRNAs
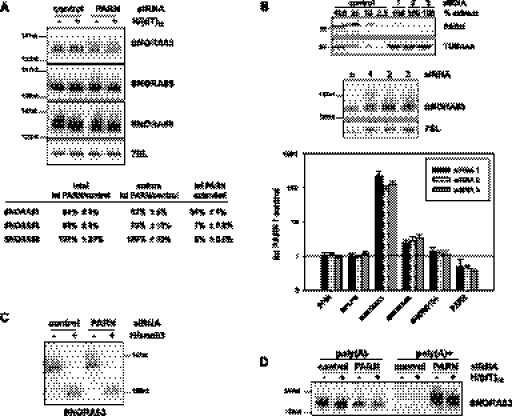
Among all RNAs examined, SNORA63 (= E3) was most strongly affected by the PARN knock-down, with an up to 10,000-fold increase of its adenylated form, as determined by qRT-PCR. Northern blot analysis showed that, upon depletion of PARN, approximately one-third of this RNA was extended by up to ∼15 nt. Extended forms of SNORA65 and 68 were also detectable at ∼7% and 5% of the total population, respectively (Fig. 4A). Knock-down of PARN with two other siRNAs also led to the accumulation of extended H/ACA box RNAs, as detected by Northern blotting (SNORA63) and qRT-PCR (SNORA63 and 68) (Fig. 4B).
FIGURE 4.
Extended H/ACA box RNAs are oligoadenylated. (A) U2OS cells were treated with siRNA against PARN or control siRNA. Nuclear RNA was hybridized to dT12 and treated with RNase H or not. RNAs were analyzed by Northern blotting, with 7SL RNA serving as a loading control. Size markers (in nucleotides) are indicated. Quantifications reported in the bottom panel (see Materials and Methods) are based on normalization to the 7SL RNA. For SNORA63 and 68, n = 4; for SNORA65, n = 3. (B) U2OS cells were transfected with PARN siRNA 1, 2, or 3, or control siRNA. PARN levels were determined by Western blotting (top panel). Total RNA was analyzed by Northern blotting with a SNORA63 probe (middle panel). In the same RNA preparations, levels of snoRNAs were analyzed by oligo(dT)-primed reverse-transcription and gene-specific qPCR (bottom panel). Data were cross-normalized to PPIA and RPLP0 messages and are plotted as RNA level in knock-down over control cells. Error bars indicate the standard deviation of two independent biological experiments, each with two technical repeats. (C) The same RNA preparations as in A were hybridized with a DNA oligonucleotide matching the 3′ end of SNORA63 and treated with RNAse H or not. RNAs were analyzed by Northern blotting with a probe directed against SNORA63. The experiment also served as a positive control for the RNase H digestion in A and D. (D) Total RNA was prepared from control and PARN knock-down U2OS cells and separated by oligo(dT) cellulose chromatography. 2 μg of poly(A)− and 0.25 μg of poly(A)+ RNA were hybridized to oligo(dT)12 and treated with RNase H or not. The RNA was analyzed by Northern blotting and hybridization with a SNORA63 probe.
Quantification of Northern blots revealed that, upon PARN knock-down, the mature form of SNORA63 was reduced to 62% of the amount in control cells, and the extended form (34% of total SNORA63) accumulated at the expense of the mature form. Similarly, mature SNORA65 was decreased to 79% of the level in control cells (Fig. 4A). This is strong evidence that 3′ trimming by PARN is on the pathway leading to mature snoRNA and does not contribute to snoRNA decay. Mature SNORA68 was not detectably decreased.
Cleavage of SNORA63 with RNase H and a DNA oligonucleotide hybridizing at the 3′ end of the mature RNA resulted in a 5′ fragment that had the same size irrespective of the PARN knock-down. Thus, the additional nucleotides of the extended form must be at the 3′ end (Fig. 4C). Oligo(dT) selection enriched the extended form of SNORA63 (Fig. 4D). In agreement with the qRT-PCR data and the substrate specificity of PARN, this shows that this RNA is oligoadenylated. Surprisingly, RNase H cleavage in the presence of oligo(dT) had only a minor effect on the extended forms of SNORA63, SNORA65, and SNORA68 (Fig. 4A,D), suggesting that the oligo(A) tails are short and additional nucleotides might contribute to the size difference detected by Northern blotting.
Oligo(A) tails are attached to 3′ trimming intermediates
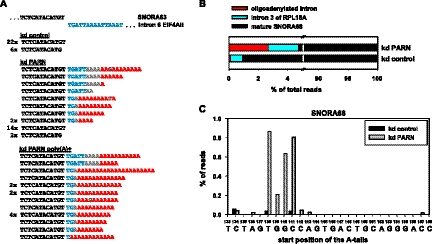
More detailed information concerning the nature of the extended SNORA63 species was obtained from sequencing of cDNA clones generated by adapter ligation and RT-PCR (Fig. 5A). All clones derived from control cells either had the mature 3′ end or were shortened by one nucleotide; extended molecules were not found. In contrast, all clones derived from oligo(dT)-selected RNA from PARN knock-down cells had oligo(A) tails between 5 and 21 nt, one of which was disrupted by a single G residue. None of the oligo(A) tails was attached to the mature snoRNA end; instead, up to 6, possibly 10, nt of 3′ flanking intron sequence were present between the mature sequence and the oligo(A) tail. (In many clones, the first 1–4 nt of the oligo(A) tail could have been either derived from the primary transcript or added post-transcriptionally.) Of 25 sequences obtained from nonselected nuclear RNA from PARN knock-down cells, six unique clones had nonencoded oligo(A) tails similar to those observed in the poly(A)+ sample: The length was between 1 and 11 nt, there were two non-A residues in the population, and the oligo(A) tails were attached to the same intron nucleotides as in the poly(A)+ population. One additional clone ended in an oligo(A) tract that could have been encoded or added post-transcriptionally. The remaining clones either had the mature 3′ end or were 1 nt short. The fraction of oligoadenylated molecules (36%) was in good agreement with the extended fraction present in the Northern blot (Fig. 4A).
FIGURE 5.
3′ extended H/ACA box RNAs carry intron nucleotides plus oligo(A) tails. (A) Nuclear RNA from control and PARN knock-down U2OS cells and poly(A)+ RNA from knock-down cells were the same preparations as in Figure 4. SNORA63 clones were prepared by linker ligation, cDNA synthesis, nested PCR, and cloning (Materials and Methods). Sequences of individual clones derived from each of the three RNA preparations are compiled. Mature snoRNA sequences are in black, remaining intron nucleotides are blue, post-transcriptionally added sequences are in red. Nucleotides of ambiguous origin are gray. Numbers on the left indicate how often each sequence was found. Unique sequences have no number. (B) Nuclear RNA from control and PARN knock-down U2OS cells was used to analyze SNORA68 by linker ligation, cDNA synthesis, PCR, and deep sequencing. The percentage of reads ending with the mature end (black), containing additional intron nucleotides (blue) or nonencoded oligo(A) sequences (red) are shown. 100% is the number of reads that matched the SNORA68 sequence and could be grouped as described in Materials and Methods. (C) The positions of nonencoded oligo(A) sequences found by deep sequencing of SNORA68 are indicated above the intron sequence. Position 133 corresponds to the last nucleotide of mature SNORA68. Oligo(A) sequences shown at, for example, position 138 were attached to nucleotide 137. Sequences from the PARN knock-down and the control sample are gray and black as indicated.
Deep sequencing was employed for the analysis of SNORA68. There were 626,838 Illumina sequencing reads of 30 nt, representing nonselected RNA from PARN knock-down cells, that matched the terminal part of SNORA68 and/or the beginning of the 3′ flanking intron, and 617,104 of those could be grouped into three classes (see Materials and Methods): 95.27% ended either at nt 133, the predicted 3′ end of mature SNORA68, or 1 nt upstream. There were 12,496 reads (2.02%) representing intermediates of 3′ exonuclease processing, possessing intron sequences extending past nt 133 but lacking nonencoded nucleotides. In addition, 16,688 reads (2.70%) had nonencoded oligo(A) sequences at their 3′ ends (Fig. 5B). A very small number of reads showed an oligo(A) sequence attached to the mature SNORA68 3′ end; the vast majority of A tails was attached 4–7 nt downstream, and very few were found >9 nt downstream (Fig. 5C). Their average length was 4.5 nt (Supplemental Fig. S3), and they contained a small percentage of non-A residues (1.82% G, 0.49% T, 0.21% C; this exceeded the observed frequency of sequencing errors in the sample). That the 3′ extensions of both SNORA63 and 68 were composed of intron sequence followed by mostly very short A tracts explains their poor sensitivity to RNase H/oligo(dT) digestion.
An RNA sample derived from control cells resulted in 51,845 sequences matching SNORA68, of which 49,903 could be grouped as above. Here, the percentage of mature 3′ ends, including those 1 nt short, was 99.06%. The fraction of reads containing nonencoded A tails was very low, as expected (72; 0.14%). Interestingly, the fraction of reads representing 3′-extended RNAs without nonencoded nucleotides was also lower (395; 0.79%) than in the PARN knock-down sample (Fig. 5B).
RRP6 plays a minor role in H/ACA box snoRNA processing
The data presented so far indicate that mammalian PARN has taken over a role played by Rrp6p in S. cerevisiae. Whether mammalian RRP6 plays a role in H/ACA box processing was tested directly by depletion of RRP6, either by itself or in combination with a PARN knock-down. In the latter case, combination of PARN siRNA with an irrelevant siRNA was used as a control. Under these conditions, the expected increase in the abundance of oligoadenylated snoRNAs was seen, but the effect was weaker than in a single knock-down (Fig. 6C). Although depletion of RRP6 was efficient (Fig. 6A), Northern blotting showed a marginal accumulation of extended SNORA63 precursors of uncertain significance (Fig. 6B). Quantitative RT-PCR analysis of SNORA9, 63, and 65 confirmed a minor accumulation of oligoadenylated species upon RRP6 knock-down or, more prominently, in the double knock-down, compared to the single knock-down of PARN (Fig. 6C). The effect of RRP6 depletion was seen more convincingly in the analysis of individual experiments as opposed to averages (Supplemental Fig. S4). The data suggest that RRP6 plays a secondary role in 3′ trimming of H/ACA box snoRNAs that becomes more visible at low PARN levels.
FIGURE 6.
RRP6 plays a minor role in H/ACA snoRNA processing. U2OS cells were treated with control siRNA and siRNAs directed against PARN or RRP6 in combinations as indicated. (A) Western blot of nuclear extracts shows knock-down efficiency of PARN and RRP6. PABPN1 served as loading control. Size markers are given in kD. (B) Nuclear RNA was analyzed by Northern blotting with a SNORA63 probe. 5.8S rRNA served as a loading control. Size markers are given in nt. Numbers at the bottom indicate the fraction of extended SNORA63 (see Materials and Methods). (C) Total RNA was isolated, and relative levels of snoRNAs were determined by qRT-PCR, as in Figure 3A. Data represent the average of three independent biological experiments (except “random priming, control + PARN;” only two replicates); error bars correspond to the standard deviation. Some of these data are presented in greater detail in Supplemental Figure S4.
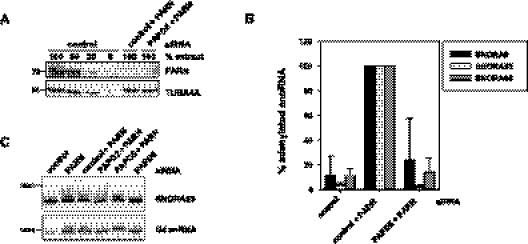
Oligo(A) tails of SNORA63 are added by PAPD5
The character of the snoRNA oligo(A) tails suggests their addition by a noncanonical poly(A) polymerase. The human genome contains seven predicted members of this family. When one of these enzymes, PAPD5 (hTRF4-2), was depleted simultaneously with PARN, the increased abundance of oligoadenylated SNORA9, 63, and 65 was strongly reduced or abolished, as determined by qRT-PCR (Fig. 7A,B). Northern blots confirmed the enrichment of extended SNORA63 in the PARN knock-down and its absence in a PARN-PAPD5 double knock-down (Fig. 7C). SiRNAs directed against PAPD2 did not detectably revert the phenotype caused by PARN depletion, even though the knock-down efficiency was comparable to that of PAPD5 (Fig. 7C and data not shown). Preliminary experiments also failed to show effects for any of the remaining five noncanonical poly(A) polymerases. Although we cannot exclude contributions of other enzymes, the strong effect of the PADP5 knock-down indicates that the H/ACA box snoRNAs are oligoadenylated primarily by this enzyme.
FIGURE 7.
PAPD5 is responsible for SNORA63 oligoadenylation. U2OS cells were treated with control siRNA and/or siRNAs directed against PARN, PAPD2, or PAPD5 in combinations as indicated. (A) A representative Western blot shows the knock-down efficiency of PARN. (B) Total RNA was isolated, and relative levels of adenylated snoRNAs were determined by qRT-PCR, as in Figure 3. SnoRNA levels in cells treated with a combination of PARN and control siRNA were set to 100%. Data represent the average of five independent experiments with two different siRNAs against PAPD5 (two and three experiments, respectively); error bars correspond to the standard deviation. As determined by qRT-PCR, 17 ± 12% of PAPD5 mRNA remained in the double knock-down. (C) Nuclear RNA was analyzed by Northern blotting with a SNORA63 probe. Remaining mRNA levels were 23% for PAPD2, 20% for PADP5 in the double knock-down, and 15% for PADP5 in the single knock-down, as determined by qRT-PCR. U4 snRNA served as a loading control. Size markers (in nt) are indicated.
When PAPD5 alone was knocked down, no accumulation of extended SNORA63 molecules was visible in a Northern blot (Fig. 7C), and sequencing of 28 cDNA clones revealed only mature 3′ ends.
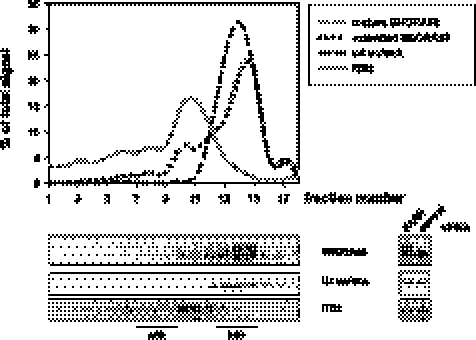
Oligoadenylated SNORA63 is associated with preribosomal particles
SNORA63 is responsible for pseudouridylation at position 4390 of 28S rRNA (Ganot et al. 1997a). The association of SNORA63 with nascent ribosomal particles was probed by sucrose gradient centrifugation of nuclear extract from PARN knock-down cells. Free snoRNPs have a sedimentation constant of 10–15S, but varying proportions can be found in association with rapidly sedimenting preribosomal particles (Maxwell and Fournier 1995). Indeed, whereas the majority of SNORA63 sedimented more slowly than the 25S marker, the U4/U6 snRNP (Behrens et al. 1993), ∼20% formed a rapidly sedimenting shoulder at the same position as the precursor to 28S rRNA. Importantly, the proportions of mature and 3′ extended forms of SNORA63 were constant throughout the gradient (Fig. 8). This suggests that the incompletely processed RNAs may be functional.
FIGURE 8.
Oligoadenylated SNORA63 is associated with preribosomal particles. Nuclear extract was prepared from U2OS cells after PARN knock-down and fractionated by sucrose gradient centrifugation. The distribution of SNORA63, U4 snRNA, and ribosomal precursor RNA was analyzed by Northern blotting. The probe for the ribosomal precursor hybridized with the internal transcribed spacer (ITS2) 5′ of the mature 28S rRNA sequence. Signals were quantified and are given as line diagrams (upper panel). RNA from PARN knock-down cells was compared to RNA from a control knock-down (right panel).
DISCUSSION
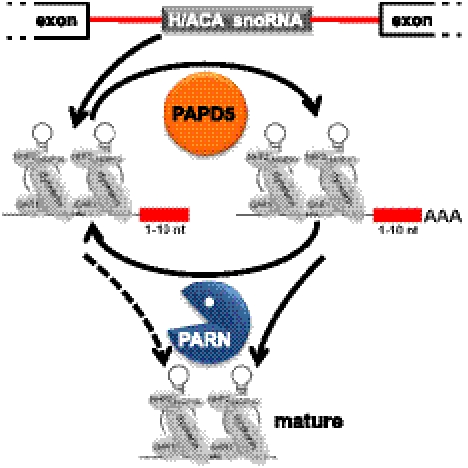
This paper reports a novel step in the maturation of mammalian H/ACA box snoRNAs and the unanticipated involvement of two enzymes, PAPD5 and PARN. PAPD5 attaches oligo(A) tails to late intermediates of 3′ shortening of H/ACA box snoRNAs. PARN removes these oligo(A) tails and, presumably, the remaining intron nucleotides, leading to precisely trimmed snoRNA 3′ ends (Fig. 9). We suggest that this may serve to stabilize mature snoRNAs against further 3′ shortening.
FIGURE 9.
A model for the roles of PAPD5 and PARN in the maturation of H/ACA box snoRNAs. A late intermediate in the 3′ shortening of the snoRNA precursors, retaining the last few nucleotides of the intron, is the substrate for adenylate addition by PAPD5. The oligo(A) tails are removed by PARN. We speculate that the RNAs may go through several cycles of oligoadenylation/deadenylation. At some point, or in several steps, PARN may also remove the remaining intron stub.
Most of the intron sequence flanking the 3′ side of mammalian snoRNAs is likely degraded by the exosome, as in yeast. PARN does not seem to be involved in this process, as depletion of the enzyme did not cause any detectable accumulation of longer 3′ processing intermediates. Thus, if addition of oligo(A) tails plays any role at earlier stages of 3′ shortening, as in yeast, a nuclease different from PARN is likely to be responsible for their removal. Trimming of the last few intron nucleotides is a distinct step. In yeast, accumulation of the corresponding intermediates in an rrp6 mutant suggests that Rrp6p is responsible for 3′ trimming (Allmang et al. 1999; van Hoof et al. 2000; Grzechnik and Kufel 2008). In mammalian cells, removal of the last 5–9 nt of the 3′ flanking intron stood out as a slow step during in vitro processing of H/ACA box snoRNA precursors (Kiss and Filipowicz 1993, 1995), and PARN appears to have taken over the trimming function for these RNAs. This conclusion is based both on a clearly detectable phenotype of PARN depletion and on a marginal effect of RRP6 knock-down that seems to be enlarged upon simultaneous knock-down of both enzymes. Recently it has been reported that pull-down of human RRP6 coprecipitated several C/D box snoRNP-specific proteins, whereas components of H/ACA box snoRNPs were not found (Lubas et al. 2011). This is consistent with our results suggesting that RRP6 does not play a major role in 3′ trimming of H/ACA box snoRNAs and also suggests a possible reason why we do not find C/D box snoRNAs among PARN substrates.
Oligoadenylation is known to promote RNA shortening by the exosome in yeast (see Introduction). This is easily explained in mechanistic terms: While the exosome has no specificity for poly(A), the oligo(A) tail provides an unstructured entry point for the enzyme, which can then digest further into the RNA due to its processive activity and the assistance of the RNA-dependent ATPase Mtr4p (Kadaba et al. 2004; LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005). In contrast, PARN has a preference for poly(A) and, on uncapped substrates such as intron-encoded snoRNAs, a very low processivity. As a consequence, the enzyme deadenylates an RNA but does not advance efficiently into non-poly(A) sequences in vitro (Körner and Wahle 1997; Dehlin et al. 2000; Martinez et al. 2000). Thus, an oligo(A) tail would not be expected to facilitate digestion of additional sequences by the same enzyme: Once an RNA has been deadenylated, PARN, due to its distributive activity, would not remain bound to the RNA to shorten it further. In this view, the role of PARN would be restricted to removing the oligo(A) tails added by PAPD5 to permit the subsequent removal of the remaining intron nucleotides by a different exonuclease. However, analysis of SNORA68 under conditions of PARN knock-down revealed not only an accumulation of oligoadenylated species but also of 3′ trimming intermediates lacking nonencoded nucleotides (Fig. 5B). Thus, PARN promotes removal of the remaining intron nucleotides; in the simplest case, PARN itself catalyzes the reaction. This would also be consistent with the fact that oligo(A) tails are mostly attached to G residues (Fig. 5): G is the nucleotide least efficiently removed by PARN (Martinez et al. 2000) and may thus block the advance of the enzyme, permitting oligo(A) addition. Mechanistically, removal of the intron stub by PARN might be explained by a (hypothetical) interaction of the enzyme with one of the snoRNP proteins, which would increase its processivity and facilitate the digestion of non-A sequences, just like an interaction with the 5′ cap does (Dehlin et al. 2000; Martinez et al. 2001). In a similar manner, 3′ trimming of yeast snoRNA precursors by Rrp6p is facilitated by Rrp47p, which appears to mediate an interaction with snoRNP proteins (Mitchell et al. 2003; Costello et al. 2011), in agreement with earlier suggestions that snoRNP proteins play an active role in 3′ processing (Kufel et al. 2000; Lafontaine and Tollervey 2000).
The length of the 3′ extensions transiently accumulating during in vitro processing of H/ACA box RNAs, 5–9 nt (Kiss and Filipowicz 1993, 1995), matches quite well the lengths of the oligoadenylated intron stubs observed here. Even though the in vitro system contained ATP, analysis of the 3′ extended processing intermediate did not reveal any signs of oligoadenylation. This agrees with the deep sequencing analysis of SNORA68 from “wild-type” cells, in which unadulterated intron reads were fivefold more abundant than oligoadenylated intron reads (Fig. 5B). One reasonable model to explain these results would be that the processing intermediate undergoes repeated cycles of oligoadenylation/deadenylation, with the equilibrium favoring the deadenylated state. Eventually, the intron stub will be taken off together with the oligo(A) tail (Fig. 9). Among the limited number of sequenced SNORA63 clones, no intron stubs lacking oligo(A) were found. Thus, intron stub removal may be more tightly coupled to deadenylation in this case.
Surprisingly, depletion of PAPD5 did not affect 3′ end maturation of snoRNAs. We can only speculate about possible explanations. The methods used for analysis may have been too blunt to detect a low level of extended snoRNA species. Our data do not exclude roles of other noncanonical poly(A) polymerases in snoRNA processing. Finally, oligoadenylation may be favorable but not essential for 3′ trimming. For example, oligoadenylation of yeast snoRNAs has been speculated to be limited to those RNAs that have problems in snoRNP assembly (Costello et al. 2011). Oligoadenylation of snoRNAs by PAPD5 is consistent with the enzyme's nuclear localization (Rammelt et al. 2011) with some nucleolar accumulation (Lubas et al. 2011) and with a proteomic analysis that detected both C/D box- and H/ACA box-specific proteins in a PAPD5 immunoprecipitate (Lubas et al. 2011).
Quantitative RT-PCR showed that PARN depletion resulted in increased levels of all H/ACA box RNAs examined, including those that did not display a response in microarray analysis. With the particular type of microarray used, levels of background hybridization were difficult to judge, and, consequently, the correlation between microarray and qRT-PCR data was only qualitative. The latter method appeared more sensitive and consistently yielded higher enrichment factors. Based on the limited number of RNAs examined by qRT-PCR, it seems likely that most, if not all, H/ACA box RNAs are deadenylated by PARN. That only a fraction of each RNA species accumulated in an extended form upon PARN knock-down may have several reasons: metabolic stability of mature snoRNAs, the incomplete elimination of PARN, and a possible redundancy of PARN with other 3′ exonucleases, e.g., RRP6. The majority of C/D box snoRNAs do not appear to be PARN substrates. Differences between C/D box and H/ACA box RNAs with respect to 3′ end formation have been seen before. For example, in contrast to H/ACA box RNAs, C/D box RNAs did not undergo final 3′ trimming in an in vitro processing system (Tycowski et al. 1993). 3′ extended processing intermediates of C/D box snoRNAs have been observed in vivo and appear to pass through the coiled body as part of their maturation pathway (Verheggen et al. 2002); this has not been reported for H/ACA box RNAs.
Both types of snoRNAs are targeted to nucleoli (Samarsky et al. 1998; Narayanan et al. 1999), where they modify rRNAs. Likewise, scaRNAs are targeted to Cajal bodies for snRNA modification (Richard et al. 2003). The localization of PARN suggests that these subcompartments are also the sites where 3′ trimming as the final step in H/ACA box RNA maturation takes place. However, PARN is associated predominantly with the outer rim of the nucleoli and does not colocalize with dyskerin and fibrillarin; thus, it is not stably associated with snoRNAs. Nucleolar localization of PARN is in agreement with proteomic analyses of purified organelles (Scherl et al. 2002; Andersen et al. 2005).
As the amount of mature SNORA63 is but moderately reduced upon PARN knock-down, testing for a defect in SNORA63 function is not straightforward. Nevertheless, the sedimentation pattern of 3′ extended SNORA63 molecules suggested their association with preribosomal particles; thus they are likely to be functional. In yeast, even the lariat precursor of an intron-encoded snoRNA is functional in rRNA methylation (Ooi et al. 1998). Why, then, are snoRNAs trimmed to a precise 3′ end? We speculate that this serves to stabilize the RNAs: Any dangling RNA 3′ overhang is likely to be a substrate for enzymes like PAPD5 or Trf4p. Oligoadenylation can attract the exosome, which, with the help of the associated ATPase Mtr4p, may be able to invade even structured RNPs. Neat trimming of the 3′ end prevents oligoadenylation and consequential degradation. Removal of the last few nucleotides of the intron by PARN, inefficient due to its preference for adenylate and its low processivity, may ensure that digestion does not proceed into essential sequences. The moderately reduced amounts of total SNORA63 and SNORA65 upon PARN knock-down (Fig. 4A) could reflect such a decreased stability of 3′ extended snoRNAs.
MATERIALS AND METHODS
Cell culture, siRNA treatment, and microscopy
U2OS and HEK293 cells were grown in DMEM GlutaMax (Invitrogen) supplemented with 10% FBS at 37°C with 5% CO2 and 90% humidity. Cells were transfected with siRNAs (Eurofins MWG Operon) by Lipofectamine RNAiMAX (Invitrogen; 1 μL per 20 pmol siRNA). In double knock-downs, a 1:1 mixture of both siRNAs was used; total siRNA concentration was kept constant. Analyses were carried out 72 h after transfection. In all experiments, knock-down efficiencies were checked by Western blotting or, if no antibodies were available, by qRT-PCR. Transfection with plasmids, immunostaining, and imaging procedures have been described (Stöhr et al. 2006). Sequential Z-stack images were acquired on a Leica SP5 LSM microscope equipped with a white light laser and deconvolved with the 3D-deconvolution tool provided by Leica. Images were processed with ImageJ. The relative fluorescence intensity and analyzed area size of nuclei, nucleoli, and Cajal bodies was determined using ImageJ. Fluorescence intensity of the nucleoplasm was determined by subtracting the total fluorescence intensity of Cajal bodies and nucleoli from total fluorescence intensity determined per nucleus.
Reagents
The PARN antibody was raised in rabbits against recombinant protein (Eurogentec). For immunolocalization, an affinity-purified preparation was used which recognized a single band in Western blots. For controlling knock-down efficiencies, the first bleeding of the serum was used without affinity purification. Other antibodies were: Anti-tubulin (mouse monoclonal DM1A, Sigma-Aldrich); anti-fibrillarin (rabbit polyclonal, Abcam); anti-dyskerin (goat polyclonal; Santa Cruz); anti-coilin (mouse monoclonal, BD); anti-nucleophosmin (mouse monoclonal, Abcam), anti-RRP6 (rabbit, affinity-purified; Sigma-Aldrich), and anti-PABPN1 (Krause et al. 1994). Secondary antibodies for Western blotting were horseradish peroxidase-conjugated anti-mouse or anti-rabbit from DAKO. The plasmid expressing the GFP-fibrillarin fusion was a gift of Yaron Shav-Tal. The GFP-PARN fusion was generated by amplification of the PARN coding sequence and insertion into EGFP-C2 via Bgl II/Sal I. The plasmid expressing GFP-dyskerin was a gift of Tamas Kiss.
RNA and DNA oligonucleotides are listed in Supplemental Table S2.
RNA analysis
Total RNA was extracted by the TRIZOL method. Nuclear RNA was obtained from nuclear extracts by the same method (Trask et al. 2009). Polyadenylated RNA was isolated from 1 mg total RNA with an mRNA Purification Kit (GE Healthcare). Two purification steps were carried out at room temperature.
For qRT-PCR, total RNA was treated with RNasefree DNase I (Roche). 2 μg RNA was reverse-transcribed with random, (dT)12 or (dA)12 primers and MMLV RT RNase H Minus (Promega). qRT-PCR was performed using QuantiTect SYBR green master mix (Qiagen) in light cycler 1.5 or 480 SYBR Green Master I in light cycler 480 II (both Roche Diagnostics). For all primer pairs, annealing temperature was 60°C. Relative changes of mRNA amounts were calculated by the ΔCt method (Livak and Schmittgen 2001). Some adenylated snoRNAs were very rare in control cells, resulting in high Ct values. As these were measured with limited precision, the enrichment factors are also approximate.
Probes for Northern blots were synthetic DNA oligonucleotides or T7 transcripts generated from templates obtained by RT-PCR. T7 RNA polymerase (Stratagene) was used with [α-32P]-UTP (Hartmann). RNA was separated on agarose-formaldehyde (rRNA precursor) or polyacrylamide-8.3 M urea (all other RNAs) gels, transferred to Hybond-N (GE Healthcare) by capillary or semidry electro-blotting and, after UV irradiation, hybridized in Church buffer at the appropriate temperature. Hybridization signals were detected with a PhosphorImager and analyzed by ImageQuant. For quantitation of extended snoRNAs, the corresponding area in the RNA from control cells was subtracted as background. This was between 3% and 11% of the total snoRNA signal and probably mostly due to smearing of the mature RNA. For quantitation of mature or total snoRNA, equally sized areas from an “empty” part of the same blot were subtracted as background.
For RNase H digestion, 10 μg of RNA were incubated with 700 pmol oligonucleotide SNORA63 r or (dT)12 with RNase H (Promega) in MMLV RT buffer (Promega) for 45 min at 37°C. The reaction was stopped by addition of formamide loading buffer and analyzed by Northern blotting.
Microarrays
For the analysis of small RNAs, total RNA from two independent knock-down experiments was further purified by phenol/chloroform extraction and EtOH precipitation, subjected to quality control using an Agilent Bioanalyzer, and used to prepare double-stranded cDNA (Superscript II, Life Technologies) primed with oligo-dT containing an T7 RNA polymerase promoter (Genset SA). Biotinylated cRNA was synthesized (3′IVT Express Kit, Affymetrix), fragmented, and hybridized to an Affymetrix GeneChip miRNA array according to the manufacturer's protocols. Microarray analyses were carried out by Knut Krohn (Core Unit DNA Technologies, IZKF Leipzig). Data were normalized using the RMA and PLIER protocol with R software. The resulting signal intensities using the RMA algorithm ranged between 20 and 3000. Probe sets with signal intensities higher than 100 were considered significant. The average of signal intensities of the control and PARN knock-down replicates was compared.
Sequencing
0.7 μg total RNA or 0.09 μg oligo(dT)-selected RNA was ligated to 100 pmol adaptor oligonucleotide with 30 U T4 RNA Ligase 1 (NEB) in 20 μl for 48 h at 16°C. After phenol/chloroform extraction and ethanol precipitation, cDNA was synthesized with 200 U MMLV RT RNase H Minus (Promega) and 200 pmol of oligonucleotides complementary to the adaptor sequences. Nested PCR was carried out with Pfu polymerase (Fermentas) and primers as described in Supplemental Table S2, and products were purified with the High Pure PCR Product Purification Kit (Roche). For SNORA63, PCR products were phosphorylated, ligated into the EcoR V site of pBlueScript II SK, and transformed into E. coli XL1 blue. Individual clones were sequenced with the BigDye Terminator v 1.1 Ready Reaction Cycle Sequencing Kit (Applied Biosystems) and T7 promotor primer on an ABI Prism 310 Genetic Analyzer.
For Illumina sequencing of SNORA68, Illumina GX1 and GX2 sequences were introduced by another round of PCR. Thirty-six sequencing cycles were carried out in two single flow cell lanes on the Illumina Genome Analyzer IIx. Image analysis and base calling were performed using Illumina pipeline v1.7. Each lane contained 12 samples distinguished by different index sequences (corresponding to the 5′ end of the adaptor oligonucleotide), of which one index per lane indicated SNORA68 samples (control cells and PARN knock-down cells). Reads from the two samples exactly matching the corresponding index sequence were extracted, and those reads passing the Illumina chastity filter and having quality values of ≥20 assigned to at least 12 bases within the first half of the read were kept for further analysis. Filtering criteria were as described (Minoche et al. 2011). Reads containing uncalled bases were removed. The index was trimmed off, and the reverse complement was built.
The filtered reads were mapped against the reference sequence of human SNORA68 (133 bases) and the flanking 3′ intron sequence (214 bases) (corresponding to chromosome 19, positions 17973397-17973743 of the human genome assembly GRCh37/hg19). One mismatch was allowed within the first 10 bases of a read and any number of mismatches in the following 20 bases using the mapping program Bowtie (Langmead et al. 2009) (Bowtie version 0.12.6 parameters: -S -l 10 -n 1 -e 800–nomaqround–norc). This selection criterion allowed for the presence of up to 21 nonencoded nucleotides at the 3′ end of a read. The majority of reads (626,838 from the PARN knock-down sample and 51,845 from the control sample) matched the region between positions 103 and 158 of the reference sequence; these were sorted into four groups as follows: The first group contained reads matching positions 103–132 or 104–133 with, at most, one mismatch in the entire sequence; these were considered mature 3′ ends of SNORA68. The second group contained reads extending beyond the mature 3′ end (i.e., covering positions 105–134 to 129–158) and matching the reference intron sequence with, at most, one mismatch after position 133. The third group contained reads with nonencoded A residues at the 3′ end, either directly following position 133 or following intron sequence with, at most, one internal mismatch. For inclusion in this third group, no more than one interruption by G, C, or T in a row was allowed in the 3′ part when the read was screened for A residues starting from the last base. In this way, A tails of different lengths could be detected which were interspersed with non-A residues to a low extent. To account for sequencing errors, the number of reads with single nonencoded A residues at the 3′ end was corrected for the expected number of A-substitution errors. Such cases occurred only in the sample prepared from PARN knock-down cells for which an error rate of <1% was estimated according to the average quality values of the reads. As a control, no comparable number of homopolymeric tails of any of the three other nucleotides was detectable in the data set when corresponding criteria were applied. The fourth group comprised reads not falling into any of the first three groups (1942 = 3.71%, control; 9734 = 1.55%, PARN knock-down). The sums of the first three groups from each sample (49,903 reads, control; 617,104 reads, PARN knock-down) were set as 100% for the purpose of comparing the two RNA populations. Analyses were performed with scripts written in Perl v5.8.9 and using Unix shell commands.
Western blotting
Cells were harvested, washed with PBS, and lysed in 50 mM Tris/HCl, pH 7.2–7.6, 200 mM NaCl, 1% Triton-×100, 0.25% deoxycholate, 1 mM EGTA, 1 mM EDTA. Proteins were resolved by SDS-10% polyacrylamide gel electrophoresis and electro-blotted onto nitrocellulose membranes, which were then blocked with 5% milk in TN-Tween and incubated with the primary antibody. Secondary horseradish peroxidase-conjugated antibodies and Super Signal West-Pico Stable-Western blotting substrate (Thermo Scientific) were used for detection.
Gradient analysis
Nuclear extract (Trask et al. 2009) from siRNA-treated U2OS cells was loaded on a linear 10%–45% sucrose gradient (150 mM NaCl, 10 mM HEPES pH 7.6, 1.5 mM MgCl2). After centrifugation in a SW40Ti rotor (Beckman) at 40,000 rpm and 4°C for 4 h, 18 fractions were collected. RNA was isolated by phenol/chloroform extraction and isopropanol precipitation and analyzed by Northern blot. Parallel sedimentation of cytoplasmic extract was used as a standard for determination of the 80S ribosome peak.
DATA DEPOSITION
The microarray data have been submitted to the ArrayExpress database (www.ebi.ac.uk/arrayexpress; accession number E-MEXP-3344).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Knut Krohn and the IZKF Leipzig DNA Core Facility at the Faculty of Medicine of the University of Leipzig (Projekt Z03) for carrying out the microarray analyses; Yaron Shav-Tal (Bar-Ilan University, Israel) and Tamas Kiss (Toulouse University, France, and Hungarian Academy of Sciences, Szeged, Hungary) for reagents; Edouard Bertrand and Utz Fischer for comments on the manuscript; and Maria Carmo-Fonseca for support. Funded by the DFG (GRK 1591 and RA 1892/1-1; S.H., C.R., and E.W.) and Kultusministerium des Landes Sachsen-Anhalt (S.H. and E.W.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.032292.112.
REFERENCES
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D 1999. Functions of the exosome in rRNA, snoRNA, and snRNA synthesis. EMBO J 18: 5399–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Lam YW, Leung AKL, Ong S-E, Lyon CE, Lamond AI, Mann M 2005. Nucleolar proteome dynamics. Nature 433: 77–83 [DOI] [PubMed] [Google Scholar]
- Bachellerie J-P, Cavaillé J, Hüttenhofer A 2002. The expanding snoRNA world. Biochimie 84: 775–790 [DOI] [PubMed] [Google Scholar]
- Balakin AG, Smith L, Fournier MJ 1996. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86: 823–834 [DOI] [PubMed] [Google Scholar]
- Behrens S-E, Tyc K, Kastner B, Reichelt J, Lührmann R 1993. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol Cell Biol 13: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolin M-L, Ganot P, Kiss T 1999. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J 18: 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarelli E, Fatica A, Prislei S, De Gregorio E, Fragapane P, Bozzoni I 1996. Processing of the intron-encoded U16 and U18 snoRNAs: The conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J 15: 1121–1131 [PMC free article] [PubMed] [Google Scholar]
- Carneiro T, Carvalho C, Braga J, Rino J, Milligan L, Tollervey D, Carmo-Fonseca M 2007. Depletion of the yeast nuclear exosome subunit Rrp6 results in accumulation of polyadenylated RNAs in a discrete domain within the nucleolus. Mol Cell Biol 27: 4157–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi F, Mariottini P, Amaldi F 1995. The Xenopus intron-encoded U17 snoRNA is produced by exonucleolytic processing of its precursor in oocytes. Nucleic Acids Res 23: 4670–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevher MA, Zhang X, Fernandez S, Kim S, Baquero J, Nilsson P, Lee S, Virtanen A, Kleiman FE 2010. Nuclear deadenylation/polyadenylation factors regulate 3′ processing in response to DNA damage. EMBO J 29: 1674–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G, Legrain P, Jacquier A 1998a. Yeast RNAse III as a key processing enzyme in small nucleolar RNAs metabolism. J Mol Biol 284: 975–988 [DOI] [PubMed] [Google Scholar]
- Chanfreau G, Rotondo G, Legrain P, Jacquier A 1998b. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J 17: 3726–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland PR, Wormington M 2001. The mechanism and regulation of deadenylation: Identification and characterization of Xenopus PARN. RNA 7: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello JL, Stead JA, Feigenbutz M, Jones RM, Mitchell P 2011. The C-terminal region of the exosome-associated protein Rrp47 is specifically required for box C/D snoRNA 3′ maturation. J Biol Chem 286: 4535–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehlin E, Wormington M, Körner CG, Wahle E 2000. Cap-dependent deadenylation of mRNA. EMBO J 19: 1079–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu DE, Henras AK, Chanfreau G 2006. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA 12: 26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Morlando M, Bozzoni I 2000. Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J 19: 6218–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Pogacic V 2002. Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol 14: 319–327 [DOI] [PubMed] [Google Scholar]
- Fragapane P, Prislei S, Michienzi A, Caffarelli E, Bozzoni I 1993. A novel small nucleolar RNA (U16) is encoded inside a ribosomal protein intron and originates by processing of the pre-mRNA. EMBO J 12: 2921–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Bortolin M-L, Kiss T 1997a. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809 [DOI] [PubMed] [Google Scholar]
- Ganot P, Caizergues-Ferrer M, Kiss T 1997b. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev 11: 941–956 [DOI] [PubMed] [Google Scholar]
- Gao M, Fritz DT, Ford LP, Wilusz J 2000. Interaction between a poly(A)-specific ribonuclease and the 5′ cap influences mRNA deadenylation rates in vitro. Mol Cell 5: 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzechnik P, Kufel J 2008. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol Cell 32: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2006. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep 7: 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T 2002. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148 [DOI] [PubMed] [Google Scholar]
- Kiss T, Filipowicz W 1993. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J 12: 2913–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Filipowicz W 1995. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev 9: 1411–1424 [DOI] [PubMed] [Google Scholar]
- Kiss T, Fayet-Lebaron E, Jady BE 2010. Box H/ACA small ribonucleoproteins. Mol Cell 37: 597–606 [DOI] [PubMed] [Google Scholar]
- Körner C, Wahle E 1997. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J Biol Chem 272: 10448–10456 [DOI] [PubMed] [Google Scholar]
- Körner CG, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E 1998. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J 17: 5427–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause S, Fakan S, Weis K, Wahle E 1994. Immunodetection of poly(A) binding protein II in the cell nucleus. Exp Cell Res 214: 75–82 [DOI] [PubMed] [Google Scholar]
- Kufel J, Allmang C, Chanfreau G, Petfalski E, Lafontaine DLJ, Tollervey D 2000. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol Cell Biol 20: 5415–5424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Lafontaine DLJ, Tollervey D 2000. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol Cell Biol 20: 2650–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay J-F, D'Amours A, Lemieux C, Lackner DH, St-Sauveur VG, Bähler J, Bachand F 2010. The nuclear poly(A)-binding protein interacts with the exosome to promote synthesis of noncoding small nucleolar RNAs. Mol Cell 37: 34–45 [DOI] [PubMed] [Google Scholar]
- Leverette RD, Andrews MT, Maxwell ES 1992. Mouse U14 snRNA is a processed intron of the cognate hsc70 heat shock pre-messenger RNA. Cell 71: 1215–1221 [DOI] [PubMed] [Google Scholar]
- Li L, Ye K 2006. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 443: 302–307 [DOI] [PubMed] [Google Scholar]
- Liu J, Maxwell ES 1990. Mouse U14 snRNA is encoded in an intron of the mouse cognate hsc70 heat shock gene. Nucleic Acids Res 18: 6565–6571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell 43: 624–637 [DOI] [PubMed] [Google Scholar]
- Martinez J, Ren Y-G, Thuresson A-C, Hellman U, Astrom J, Virtanen A 2000. A 54-kDa fragment of the poly(A)-specific ribonuclease is an oligomeric, processive, and cap-interacting poly(A)-specific 3′ exonuclease. J Biol Chem 275: 24222–24230 [DOI] [PubMed] [Google Scholar]
- Martinez J, Ren Y-G, Nilsson P, Ehrenberg M, Virtanen A 2001. The mRNA cap structure stimulates rate of poly(A) removal and amplifies processivity of degradation. J Biol Chem 276: 27923–27929 [DOI] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ 1995. The small nucleolar RNAs. Annu Rev Biochem 35: 897–934 [DOI] [PubMed] [Google Scholar]
- Minoche AE, Dohm JC, Himmelbauer H 2011. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and Genome Analyzer systems. Genome Biol 12: R112 doi: 10.1186/gb-2011-12-11-r112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D 2003. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol Cell Biol 23: 6982–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A, Lukowiak A, Jady BE, Dragon F, Kiss T, Terns R, Terns MP 1999. Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J 18: 5120–5130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SL, Samarsky DA, Fournier MJ, Boeke JD 1998. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: Intron length effects and activity of a precursor snoRNA. RNA 4: 1096–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petfalski E, Dandekar T, Henry Y, Tollervey D 1998. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol 18: 1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rammelt C, Bilen B, Zavolan M, Keller W 2011. PAPD5, a noncanonical poly(A) polymerase with an unusual RNA-binding motif. RNA 17: 1737–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichow SL, Hamma T, Ferré-D'Amaré AR, Varani G 2007. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res 35: 1452–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Darzaq X, Bertrand E, Jady BE, Verheggen C, Kiss T 2003. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J 22: 4283–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky DA, Fournier MJ, Singer RH, Bertrand E 1998. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J 17: 3747–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Paolo S, Vanacova S, Schenk L, Scherrer T, Blank D, Keller W, Gerber AP 2009. Distinct roles of non-canonical poly(A) polymerases in RNA metabolism. PLoS Genet 5: e1000555 doi: 10.1371/journal.pgen.1000555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherl A, Couté Y, Déon C, Callé A, Kindbeiter K, Sanchez J-C, Greco A, Hochstrasser D, Diaz J-J 2002. Functional proteomic analysis of human nucleolus. Mol Biol Cell 13: 4100–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Conrad NK, Brow DA, Corden JL 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413: 327–331 [DOI] [PubMed] [Google Scholar]
- Stöhr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Hüttelmaier S 2006. ZBP1 regulates mRNA stability during cellular stress. J Cell Biol 175: 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns MP, Grimm C, Lund E, Dahlberg JE 1995. A common maturation pathway for small nucleolar RNAs. EMBO J 14: 4860–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask HW, Cowper-Sal-Lari R, Sartor MA, Gui J, Heath CV, Renuka J, Higgins A-J, Andrews P, Korc M, Moore JH, et al. 2009. Microarray analysis of cytoplasmic versus whole cell RNA reveals a considerable number of missed and false positive mRNAs. RNA 15: 1917–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu M-D, Steitz JA 1993. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev 7: 1176–1190 [DOI] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol 20: 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189 doi: 10.1371/journal.pbio.0030189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C, Lafontaine DLJ, Samarsky DA, Mouaikel J, Blanchard J-M, Bordonné R, Bertrand E 2002. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J 21: 2736–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T, Ceradini F, Presutti C, Bozzoni I 1998. Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol Cell Biol 18: 3376–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Leverette RD, Xia L, Andrews MT, Maxwell ES 1996. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA 2: 118–133 [PMC free article] [PubMed] [Google Scholar]
- Wlotzka W, Kudla G, Grannemann S, Tollervey D 2011. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J 30: 1790–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Reuter M, Lilie H, Liu Y, Wahle E, Song H 2005. Structural insight into poly(A) binding and catalytic mechanism of human PARN. EMBO J 24: 4082–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Nilsson P, Henrikson N, Niedzwiecka A, Lim MK, Cheng Z, Kokkoris K, Virtanen A, Song H 2009. Structural basis of m7GpppG binding to poly(A)-specific ribonuclease. Structure 17: 276–286 [DOI] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle J-C, Dufour ME, Boulay J, Régnault B, Devaux F, Namane A, Seraphin B, et al. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Chang T-C, Yamashita Y, Zhu W, Zhong Z, Chen C-YA, Shyu A-B 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 12: 1054–1063 [DOI] [PubMed] [Google Scholar]