The pseudoknot of the telomerase RNA of Saccharomyces cerevisiae is characterized biophysically in vitro in the absence of protein cofactors. Under those conditions, it is shown that a conformational equilibrium exists between states with a pseudoknot and without.
Keywords: telomerase, pseudoknot, RNA thermodynamics, conformational equilibrium, TLC1
Abstract
Recent structural and functional characterization of the pseudoknot in the Saccharomyces cerevisiae telomerase RNA (TLC1) has demonstrated that tertiary structure is present, similar to that previously described for the human and Kluyveromyces lactis telomerase RNAs. In order to biophysically characterize the identified pseudoknot secondary and tertiary structures, UV-monitored thermal denaturation experiments, nuclear magnetic resonance spectroscopy, and native gel electrophoresis were used to investigate various potential conformations in the pseudoknot domain in vitro, in the absence of the telomerase protein. Here, we demonstrate that alternative secondary structures are not mutually exclusive in the S. cerevisiae telomerase RNA, tertiary structure contributes 1.5 kcal mol−1 to the stability of the pseudoknot (∼half the stability observed for the human telomerase pseudoknot), and identify additional base pairs in the 3′ pseudoknot stem near the helical junction. In addition, sequence conservation in an adjacent overlapping hairpin appears to prevent dimerization and alternative conformations in the context of the entire pseudoknot-containing region. Thus, this work provides a detailed in vitro characterization of the thermodynamic features of the S. cerevisiae TLC1 pseudoknot region for comparison with other telomerase RNA pseudoknots.
INTRODUCTION
Telomerase is the ribonucleoprotein (RNP) complex responsible for the maintenance of telomeric DNA at the ends of linear chromosomes. Telomere shortening occurs during successive rounds of chromosomal replication and can lead to end-to-end fusions, chromosomal degradation, and, eventually, to the loss of genetic information (Greider 1996). The incorporation of incorrect telomeric repeats at the ends of chromosomes has been found to cause chromosomal instability and to compromise cellular viability in vertebrates, ciliates, and yeast (McEachern and Blackburn 1995; Kirk et al. 1997; Guiducci et al. 2001). The telomerase enzyme is composed of the telomeric template-containing telomerase RNA (TER; TLC1 in Saccharomyces cerevisiae), the telomerase reverse transcriptase protein (TERT; Est2p in Saccharomyces cerevisiae), and a variety of other accessory proteins (Kelleher et al. 2002; Harrington 2003; Collins 2006). In addition to containing the template sequence, the telomerase RNA also provides a scaffold for accessory protein subunits and contributes to catalysis and repeat addition processivity (Tzfati et al. 2000; Chen and Greider 2003; Lai et al. 2003; Mason et al. 2003; Zappulla and Cech 2004; Zappulla et al. 2005).
Telomerase RNAs vary widely among different species (even closely related species) in both size and sequence composition: ∼160 nt in ciliates (Romero and Blackburn 1991; Lingner et al. 1994), ∼500 nt in vertebrates (Chen et al. 2000), and ∼930–1600 nt in yeast (Singer and Gottschling 1994; Webb and Zakian 2008). Due to the large size of yeast telomerase RNAs and their sequence diversity, a single structural model of the yeast TER has yet to be irrefutably proven. Nevertheless, based on the various proposed secondary structures of telomerase RNAs from S. cerevisiae (Chappell and Lundblad 2004; Dandjinou et al. 2004; Lin et al. 2004; Zappulla and Cech 2004), Kluyveromyces lactis (Brown et al. 2007; Shefer et al. 2007), Candida albicans (Gunisova et al. 2009), Candida glabrata (Kachouri-Lafond et al. 2009), and Schizosaccharomyces pombe (which is relatively distant from the budding yeasts) (Webb and Zakian 2008), several predicted conserved secondary structural elements (Chen and Greider 2004) have been validated, including the template, a template boundary element, and a large loop that includes the template, a pseudoknot, a critical three-way junction motif (Brown et al. 2007), and a loop-closing helical region.
In 2007, Shefer et al. proposed a three-dimensional model of the pseudoknot in this region for the K. lactis telomerase RNA in which a triple helix is formed by Hoogsteen base-pairing between the uridine-rich 5′ loop (loop 1) of the pseudoknot and Watson-Crick A-U base pairs in the pseudoknot 3′ stem (stem 2) (Shefer et al. 2007), similar to the triple helix identified in the human telomerase RNA (Theimer et al. 2005). This prediction for K. lactis was consistent with the observation that triple helix-disrupting mutations abolished telomerase function as well as the formation of a stable pseudoknot structure. Compensatory mutations that formed pH-dependent Hoogsteen C-G●C+ triples restored the pseudoknot structure in a pH-dependent manner and partially restored telomerase function in vivo (Shefer et al. 2007), as was previously observed for the human telomerase RNA Hoogsteen triple base pairs (Theimer et al. 2005). It has been reported that a similar triple helix in the S. cerevisiae TLC1 contributes to telomerase activity but not by binding the TERT protein (Qiao and Cech 2008). Previous studies had suggested that the pseudoknot region of the telomerase RNA provides part of the binding region for the telomerase reverse transcriptase catalytic protein subunit (Chappell and Lundblad 2004; Lin et al. 2004). However, since mutations in the triple-helix region did not appear to affect binding between the various RNA mutants and the Est2 RNA binding domain, it has been proposed that the triple-helix structure is involved in the catalytic cycle, rather than being responsible for binding the reverse transcriptase protein subunit (Qiao and Cech 2008). Recently, targeted 2′-O methylation at single nucleotide positions within the putative pseudoknot region of S. cerevisiae TER demonstrated that mutations adjacent to but not within the previously proposed triple-helix structure (Qiao and Cech 2008) resulted in lower telomerase activity in vivo (Huang and Yu 2010).
In order to investigate the thermodynamic and structural features of the S. cerevisiae telomerase RNA pseudoknot-containing region, we have used mainly UV-monitored thermal denaturation experiments and native gel electrophoresis experiments, supported by 1D and 2D NMR spectroscopy, to examine a series of RNA pseudoknot and hairpin constructs in vitro. Here, we have confirmed that the alternative hairpin secondary structures proposed in 2004 (Fig. 1A; Chappell and Lundblad 2004; Dandjinou et al. 2004; Lin et al. 2004; Zappulla and Cech 2004; for review, see Chen and Greider 2004) are in conformational equilibrium with the predominant pseudoknot conformation in vitro and that the presence of the conserved “alternative” H5 hairpin sequence is most likely maintained to prevent dimerization and alternate conformations from forming in the wild-type pseudoknot sequence. In addition, correlations were made between the thermodynamic stabilities of mutations in the pseudoknot domain and in vitro telomerase activity (Qiao and Cech 2008). This study thoroughly characterizes the thermodynamic properties of the S. cerevisiae telomerase RNA pseudoknot and upstream 5′ sequences in vitro, allowing for comparison to the structurally and thermodynamically well-characterized human telomerase RNA pseudoknot (Theimer et al. 2003, 2005) and other telomerase RNAs, as they are more completely characterized.
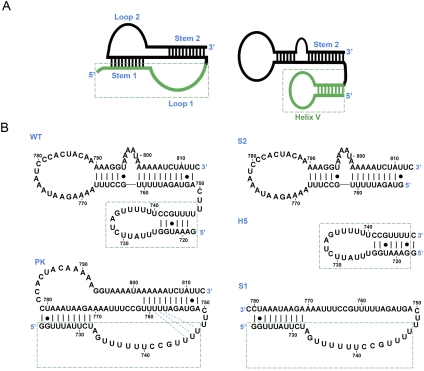
FIGURE 1.
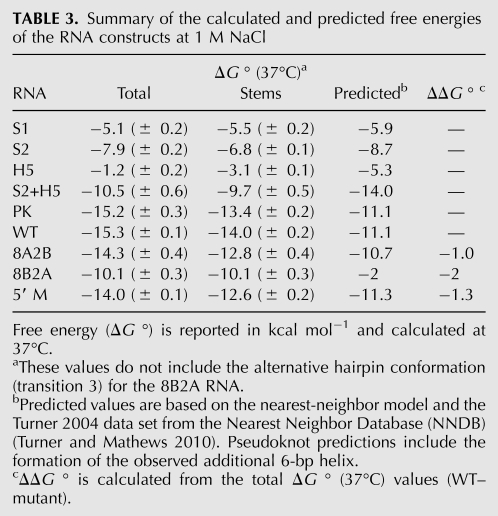
RNA construct design for the pseudoknot region of the S. cerevisiae TLC1. (A) Secondary structural representations of the pseudoknot region based on the original structural models (Chappell and Lundblad 2004; Dandjinou et al. 2004; Lin et al. 2004; Zappulla and Cech 2004). The helix V-forming sequence is indicated in green. (B) RNA constructs for the S. cerevisiae TLC1 pseduoknot region. Proposed Hoogsteen base triples (Shefer et al. 2007; Qiao and Cech 2008) are indicated on the PK sequence with blue dashed lines between loop 1 and stem 2. In both A and B, the sequences proposed to form the helix V hairpin are boxed in green. Nucleotides are numbered based on the complete S. cerevisiae TLC1 sequence in all figures.
RESULTS
RNA hairpin and pseudoknot constructs
As originally described in 2004, the potential pseudoknot-containing region of the S. cerevisiae TLC1 had two conflicting proposed structures, one containing a pseudoknot structure only (Lin et al. 2004), while the others consisted of three minor variations on hairpin structures (Fig. 1A; Chappell and Lundblad 2004; Dandjinou et al. 2004; Zappulla and Cech 2004). Although later studies demonstrated the presence of the pseudoknot conformation in this sequence (Qiao and Cech 2008), these studies did not preclude the possibility of a hairpin conformation in equilibrium with the pseudoknot. Therefore, five RNA constructs were designed to include these possible conformations, as well as to thoroughly characterize the secondary structure and thermodynamic properties of the RNA pseudoknot (Fig. 1B). The longest construct, WT, consisted of nt 718–813 from the S. cerevisiae TLC1 and contains all of the nucleotides involved in the proposed secondary structures for this region. The PK construct (nt 723–813) eliminated the five nucleotides from the 5′ end of the WT sequence which were originally proposed to form helix V stem base pairs (Lin et al. 2004), eliminating the possibility of forming this competing hairpin. The S1 construct (nt 723–781) was designed to simplify the assignment of melting transitions in the PK and WT optical melting profiles and contains the 5′ helical stem of the pseudoknot conformation (stem 1). The last two constructs, S2 and H5, contain the individual helical stems predicted by all three hairpin-based structural models (Fig. 1A; Chappell and Lundblad 2004; Dandjinou et al. 2004; Zappulla and Cech 2004). The S2 construct (nt 751–813) consists of the 3′ helical stem of the pseudoknot conformation (stem 2) and was synthesized with a single G nucleotide added at the 5′ end to promote efficient in vitro transcription by T7 RNA polymerase. Finally, the H5 construct, which forms only the helix V hairpin (nt 718–747), was designed to include one additional Watson-Crick G-C base pair at the end of the helical stem for additional stability and increased in vitro transcription efficiency.
In order to identify secondary structural elements in the PK and WT RNAs, optical transitions observed in the thermal melting profiles must first be assigned to the melting of helical stems. In the simplest case, the PK melting profile should appear to be a summation of the S1 and S2 melting profiles, with predictable variations in melting temperature and enthalpy based on pseudoknot loop entropic penalties (Gultyaev et al. 1999) and minor alterations in base-pairing. In addition, if only a pseudoknot conformation exists, the WT melting profile should duplicate the PK melting profile under similar solution conditions. Complete melting profiles (plotted as ∂A/∂T at 260 and 280 nm versus T) for all RNAs investigated in this study were collected under a variety of monovalent (50, 100, 200, 500 mM, and 1 M NaCl) and divalent (10, 50, 100, 500 μM, and 1 mM MgCl2) salt conditions. Fitting of these data to an unfolding model consisting of a set of sequential, independent two-state unfolding transitions returns the thermodynamic parameters tm and ΔH, as well as the amplitudes of the hyperchromicity at each wavelength (A260 and A280, respectively) for each transition. All optical melting profiles which were fit to more than one transition are presented in figures with two panels. The left panel contains the raw data and the final calculated curves, based on the unfolding model, which fit the entire data set. The right panel contains the calculated individual transitions (at both wavelengths) which combine to form the total calculated curve and are provided to facilitate comparisons of optical melting profiles.
Characterization of the S1 and S2 hairpins
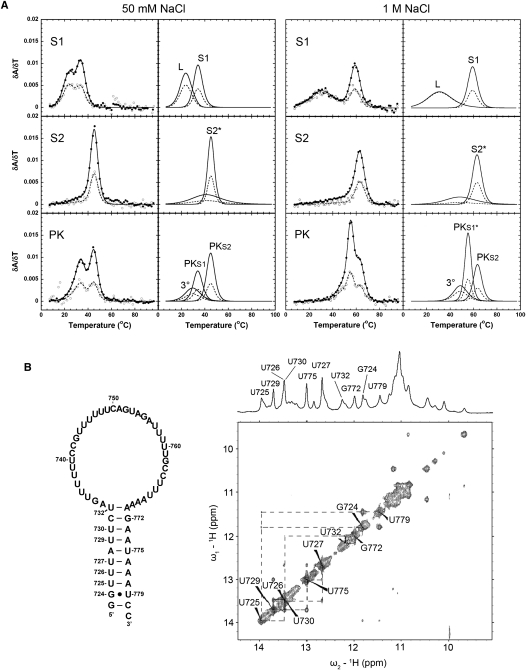
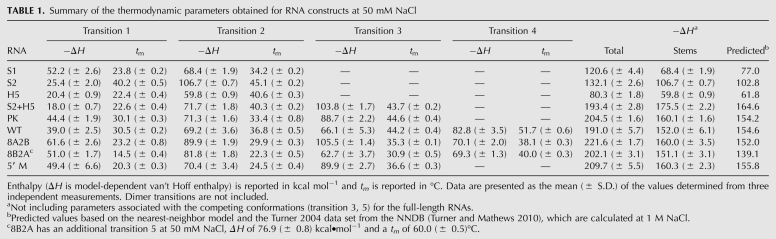
The S1 RNA includes the base-pairing sequences that make up the 5′ stem (stem 1) of the pseudoknot with a large hairpin loop (Fig. 1B). Two optical unfolding transitions are clearly observed for the S1 melting profile under all measured conditions. At 50 mM NaCl, both transitions are characterized by relatively low melting temperatures (23.8 ± 0.2°C and 34.2 ± 0.2°C, respectively) (Fig. 2A; Table 1). Based on the stem 1 sequence, the high A260/280 ratio of the second transition (A260/A280 = 2.55 ± 0.16) (Supplemental Table S1) is consistent with the hyperchromic signature expected for an A-U rich base-paired region (Fresco et al. 1963). The enthalpy and free energy of the second transition, ΔH = −80.0 ± 2.0 kcal mol−1 (Table 2) and ΔG ° (37°C) = −5.5 ± 0.2 kcal mol−1 (Table 3), at 1 M NaCl, are in good agreement with the enthalpy and free energy predicted for this 10-bp stem from Turner rules (ΔH and ΔG ° [37°C] of −77.0 kcal mol−1 and −5.9 kcal mol−1, respectively, at 1 M NaCl) (Turner and Mathews 2010). Therefore, the second unfolding transition was assigned to the unfolding of the expected helical stem 1 region (S1, Fig. 2A). Additional base-pairing in the loop might explain the additional transition in the thermal melting profile (L, Fig. 2A), since a similar transition was observed for the stem 1 hairpin of the human telomerase RNA pseudoknot, which was assigned to a series of noncanonical U●U and U●C base pairs in the hairpin loop region (Theimer et al. 2003).
FIGURE 2.
Comparison of the melting profiles for the S1, S2, and PK RNAs. (A) Experimental optical melting profiles at 50 mM NaCl (left) and 1M NaCl (right). Profiles on the left show every fifth data point obtained at 260 nm (●) and 280 nm (○) and the calculated fits to the 260-nm (solid lines) and 280-nm data (dashed lines), with the individual transitions at 260 nm (solid lines) and 280 nm (dashed lines), which combine to generate the total calculated curves, shown separately to the right. The individual transition fits are labeled according to the correlated structural elements: (L) loop structure, (S1) stem 1, (S2*) stem 2 including 6-bp helix, (3°) tertiary structure, (PKS1) pseudoknot stem 1, (PKS1*) pseudoknot stem 1 and 6-bp helix, (PKS2) pseudoknot stem 2, respectively. (B) Hairpin schematic and 500-MHz 1D and 2D imino proton spectra of the S1 RNA at 10°C. The hairpin stem imino protons assigned in the NMR spectrum are numbered on the hairpin schematic.
TABLE 1.
Summary of the thermodynamic parameters obtained for RNA constructs at 50 mM NaCl
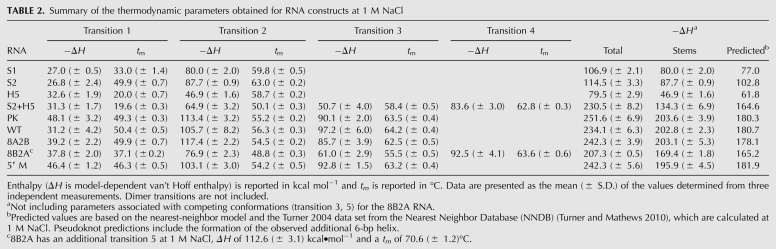
TABLE 2.
Summary of the thermodynamic parameters obtained for RNA constructs at 1 M NaCl
TABLE 3.
Summary of the calculated and predicted free energies of the RNA constructs at 1 M NaCl
Based on the high content of U nucleotides on both the 5′ and 3′ sides of the loop in the S1 sequence, a series of noncanonical U●U base pairs could also be responsible for the first low temperature unfolding transition in the S1 hairpin. Assignment of the imino proton resonances of the S1 RNA using 2D NMR spectroscopy (Fig. 2B) indicated that, in addition to the formation of the base pairs expected for the 10-bp stem, there was an additional group of imino protons resonances identified between 9 and 11.5 ppm with detectable NOE connectivities. The chemical shifts, NOEs, salt dependence, and intensities of the additional peaks (Fig. 2B; data not shown) are consistent with the behavior observed for noncanonical U●U and U●C base pairs in the human telomerase RNA hairpin loop (Theimer et al. 2003). Consistent with this assignment, these peaks are not observed in a modified S1 RNA, which has the same base-pairing sequence as stem 1 except for an additional GC base pair and a UUCG tetraloop in place of the S1 hairpin loop sequence (Fig. 3A,C). The melting profile of S1-mod is also simplified, with the complete disappearance of the first S1 transition under all measured salt conditions and, as expected, only imino proton peaks for the predicted Watson-Crick base pairs in the helical stem in the imino region of the 2D H2O NOESY spectrum (Fig. 3B,C). Therefore, the additional transition in the thermal melting profile of the S1 RNA was assigned to the unfolding of noncanonical base pairs in the loop region of the S1 hairpin (L, Fig. 2A).
FIGURE 3.
Spectral properties of the S1-mod RNA. (A) RNA construct for the S1-mod RNA. The added G-C base pair and UUCG tetraloop are indicated in gray. (B) Experimental optical melting profiles at 50 mM (top) and 1M NaCl (bottom). Profiles show every fifth data point obtained at 260 nm (●) and 280 nm (○), and the calculated fits to the 260-nm (solid lines) and 280-nm data (dashed lines) indicated. (C) 500-MHz 1D and 2D NOESY imino proton spectra of the S1-mod RNA at 10°C. The hairpin stem imino protons assigned in the NMR spectrum are numbered on the hairpin schematic in A.
The S2 RNA includes base-pairing sequences that make up the 3′ stem (stem 2) of the pseudoknot, which is also the second hairpin of the two-hairpin model (Fig. 1A,B; Chappell and Lundblad 2004; Dandjinou et al. 2004; Zappulla and Cech 2004). The melting profile of the S2 RNA was fit to two unfolding transitions, one broad, lower tm, low-amplitude transition, and one very sharp, higher amplitude transition (Fig. 2A; Table 1). The high A260/A280 ratio and enthalpy of the second transition are generally consistent with expectations for the unfolding of the A-U rich 5′ region of the hairpin stem (Fresco et al. 1963), and the total enthalpy of this transition matches predictions for the unfolding of both the 10-bp and 6-bp helices (Fig. 1B) simultaneously. Addition of the enthalpy of the first, lower amplitude transition results in significantly higher enthalpy than expected, which suggests loop structure, but, based on the hairpin loop sequence (adenine rich with very little canonical base-pairing potential and no complementary pyrimidine tracts), it is unclear what type of hairpin loop structure might form. When the S2 RNA is unfolded at higher salt conditions, we see an unusual effect (Fig. 2A). While the melting temperatures increase for both transitions, and the enthalpy of the first transition remains approximately the same, as expected with increasing monovalent salt concentrations, the enthalpy of the second transition drops by 20 kcal mol−1 (Table 2), approximating the value expected for the 10-bp stem alone. A similar effect was observed in the presence of increasing concentrations of magnesium (see broadening of melt profile with increasing magnesium concentrations) (Supplemental Fig. S1). However, despite the unusual behavior of the stem enthalpy, the predicted ΔH and ΔG ° (37°C) of −102.8 and −8.7 kcal mol−1, respectively, based on the presence of both helices, more closely matches the experimentally determined values (−114.5 ± 3.3 and −7.9 ± 0.2 kcal mol−1, respectively) (Tables 2, 3) than the predictions for the 10-bp stem alone (−69.5 and −4.2 kcal mol−1, respectively). Therefore, based on the enthalpies of these transitions and comparisons with additional data described below, we concluded that, under low-salt conditions, the S2 hairpin forms both helices, but alternative conformations in the upper stem and loop appear to occur under higher salt conditions, competing with the 6-bp helix. We were unable to assign the initial low amplitude transition specifically, although it could be an alternative conformation in the upper helical region in competition with the expected 6-bp helix, loop stacking interactions, non-two-state unfolding behavior caused by the 6-nt bulge loop between stems, or a combination of these processes.
Assignment of the pseudoknot unfolding pathway
Hairpin-type pseudoknots typically unfold through a multistep pathway, either via a stem 1 (5′) or stem 2 (3′) hairpin intermediate before becoming completely unfolded. Assignment of the S1 (5′ hairpin) and S2 (3′ hairpin) unfolding transitions and comparison of these profiles to the PK pseudoknot melting profile were essential for accurate assignment of the PK unfolding pathway. The RNA construct, PK, was designed to form only the pseudoknot conformation, which contains the two helical stems and the tertiary structural interactions within the pseudoknot (Fig. 1B). Three transitions were observed for the PK melting profile, consistent with three sequential unfolding steps (Fig. 2A; Table 1). Based on comparison of the melting profiles and thermodynamic parameters of the PK, S1, and S2 RNA constructs under low-salt conditions (Table 1), the last two transitions appear to correspond to the unfolding of stem 1 (S1) followed by stem 2 (S2), respectively, although the enthalpy associated with the 6-bp helix does not appear to be present in the pseudoknot under low-salt conditions (PKS1 and PKS2, Fig. 2A). Consistent with this assignment, the enthalpy of these two transitions together is −160.1 ± 1.9 kcal mol−1, compared to the predicted value of −154.2 kcal mol−1 for the pseudoknot stems (stem 1 and the 10-bp stem 2 only) (Fig. 1B; Table 1).
The first melting transition of PK was assigned to tertiary structure unfolding in the PK RNA (3°, Fig. 2A), consistent with the Hoogsteen base-pairing between the uridine-rich loop (loop 1) of the pseudoknot and Watson-Crick A-U base pairs in the pseudoknot 3′ stem (stem 2) proposed for the S. cerevisiae telomerase RNA pseudoknot (Shefer et al. 2007; Qiao and Cech 2008). Based on these data alone, we cannot rule out the possibility of additional or alternative tertiary interactions; we can only identify the magnitude of the observed tertiary structure-based stability contribution. The tertiary structural transition in the PK pseudoknot contributes −44.4 ± 1.9 kcal mol−1 to the stability of the pseudoknot structure, under low-salt conditions (Table 1). A similar unfolding transition was observed and assigned to tertiary structure unfolding in melting profiles of the human telomerase RNA pseudoknot, which contains the same type of Hoogsteen base-pairing interactions in the same topological region of the pseudoknot (Theimer et al. 2003, 2005).
Unlike the melting profiles of the simpler hairpin molecules, the PK melting profile changes appearance significantly with increasing monovalent and divalent salt concentrations (Fig. 2A; Supplemental Fig. S2), with all three transitions becoming more tightly coupled on the temperature coordinate. In fact, the high-salt profile of PK is very similar in shape to that observed for the human telomerase RNA pseudoknot (hTR ΔU177), with the expected variations in A260/A280 ratio, based on stem base-pair content (Theimer et al. 2003, 2005). Interestingly, although the enthalpies associated with tertiary structure formation and the third unfolding transition (assigned to stem 2) remained the same, the enthalpy of the second transition (assigned to stem 1) increased by ∼40 kcal mol−1 (Table 2). Based on comparisons to the S1 and S2 hairpin profiles under similar solution conditions and the predicted enthalpy of base-pairing for the stem 2 6-bp helix, the additional enthalpy was assigned to the formation of the 6-bp helix observed in the S2 construct (PKS1*, Fig. 2A). In the context of the PK pseudoknot, formation of this helix results in a bulge of 6 nt in stem 2 (as seen in the stem of the S2 construct, Fig. 1B) in a similar location to the observed conserved bulge in the human telomerase RNA pseudoknot (Chen et al. 2000; Theimer et al. 2005), as well as a 3-nt (AAA) loop at the helical junction. Inclusion of these base pairs and the appropriate penalty for the bulge loop in stem 2 result in predicted ΔH and ΔG ° (37°C) values of −180.3 and −11.1 kcal mol−1, respectively, which better match the experimentally determined values of −203.6 ± 3.9 and −13.4 ± 0.2 kcal mol−1 for the stems (not including the tertiary structural transition) than predictions which do not include the additional 6-bp helix (Tables 1–3). We, therefore, conclude that, while this helix is not formed under low-salt conditions, this region of the PK pseudoknot becomes structured under higher monovalent salt concentrations and in the presence of magnesium (see similarities in melt profiles between high monovalent and divalent salt concentrations) (Supplemental Fig. S2), which may be more representative of the conformation present in the cell in the presence or absence of the telomerase reverse transcriptase protein. In addition, based on the melting data, it is most likely that this helix stacks with stem 1 at the helical junction, causing them to unfold simultaneously. The PK unfolding pathway, therefore, proceeds as 3°structure → stem 1 (and 6-bp stem 2 helix, at high salt) → stem 2 → unfolded RNA, which is different than the human telomerase RNA pseudoknot unfolding pathway (3°structure → stem 2 → stem 1 → unfolded) (Theimer et al. 2003).
A comparison of the proposed triple helix in the S. cerevisiae telomerase RNA (Qiao and Cech 2008) and the solution structure of the human telomerase RNA pseudoknot (Theimer et al. 2005) may explain this discrepancy. In the case of the human telomerase RNA, the pseudoknot contains a unique triple helix composed of three Hoogsteen U-A●U triple base pairs, Hoogsteen loop 1-loop 2-bp interactions which span the pseudoknot helical junction and two stem 1-loop 2 minor groove triple base pairs, with a large number of base stacking interactions to contribute to the enthalpy, the melting temperature, and to the hyperchromic signature of the tertiary structural transition (Theimer et al. 2003, 2005). The secondary structural model of the triple helix in the S. cerevisiae pseudoknot or any other yeast pseudoknot, in contrast, currently provides no evidence for additional stacking interactions between the Hoogsteen base triples and additional structural elements between loop 2 and stem 1. In addition, the nucleotide sequences near the helical junction do not appear to be base-paired under low-salt conditions, suggesting that the tertiary structural network in the S. cerevisiae TLC1 is either shorter or discontinuous, since the junction region of this pseudoknot does not appear to be as stable or as structured as that of the human telomerase pseudoknot under similar conditions.
Additional structure in the H5 hairpin loop
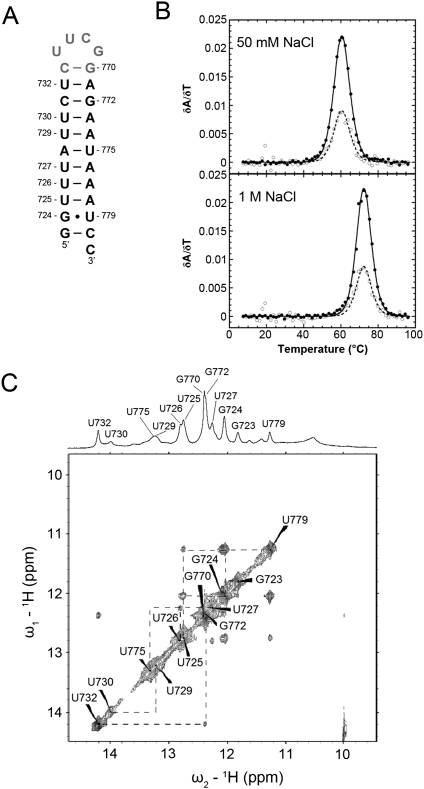
The H5 hairpin consists of the base-pairing sequence for the proposed helix V structure from the two-hairpin secondary structural model (Fig. 1B; Chappell and Lundblad 2004; Dandjinou et al. 2004; Zappulla and Cech 2004). Two optical melting transitions are observed in the H5 melting profiles under low-salt conditions (L, H5, Fig. 4A; Table 1). The second transition observed under low-salt conditions, with the higher melting temperature, was assigned to the unfolding of the monomeric helix V stem (Fig. 1B), based on consistency with stability predictions and between the experimental A260/A280 ratio and the specific stem sequence. The helix V stem has three G-C, two G●U, and three A-U base pairs in the helix V stem, which is reasonably consistent with the observed hyperchromicity at both 260 nm and 280 nm (Supplemental Table S1; Fresco et al. 1963). While the experimentally determined ΔH of −46.9 ± 1.6 kcal mol−1 and ΔG ° (37°C) of −3.1 ± 0.1 kcal mol−1 for this transition (at 1M NaCl) are smaller than the predicted values (ΔH of −61.8 kcal mol−1 and ΔG ° of −5.3 kcal mol−1), this does not include the enthalpy and free energy associated with the first transition (Tables 2, 3). An additional transition is present under high-salt conditions (D, Fig. 4A). Based on the high enthalpy of this transition (ΔH of −131.0 ± 4.5 kcal mol−1 at 1M NaCl) and the strong salt dependence of the intensity of this transition, this transition was identified as the unfolding of an H5 dimer species. Native gel electrophoresis of varying concentrations of the H5 RNA confirms the presence of dimerized H5 at higher RNA sample concentrations (Fig. 4B), based on the appearance of an H5 RNA band which migrates slower than the S2 RNA, which is nearly twice the size of the H5 monomer.
FIGURE 4.
Comparison of the melting profiles for the H5, H5+S2, and WT RNAs. (A) Profiles on the left show every fifth data point obtained at 260 nm (●) and 280 nm (○), and the calculated fits to the 260-nm (solid lines) and 280-nm data (dashed lines), with the individual transitions at 260 nm (solid lines) and 280 nm (dashed lines), which combine to generate the total calculated curves, shown separately to the right. The individual transition fits are labeled according to the correlated structural elements: (L) loop structure, (H5) helix V, (D) dimer, (S2*) stem 2 including 6-bp helix, (3°) tertiary structure, (HPs) the overlapped H5 and S2* transitions, (PKS1) pseudoknot stem 1, (PKS1*) pseudoknot stem 1 and 6-bp helix, (PKS2) pseudoknot stem 2, respectively. (B) Native polyacrylamide gel and H5 hairpin schematic. Monomer (M) and dimer (D) species are indicated to the left of the gel, and the S2 RNA was loaded as a size marker (61 nt) for comparison to the H5 dimer (64 nt). The H5 RNA sample concentrations were 0.5, 1.0, 1.75, 2.5, and 3.5 μM, respectively. The hairpin stem imino protons assigned in the NMR spectrum are numbered on the hairpin schematic. (C) 500-MHz 1D and 2D NOESY imino proton spectra of the H5 RNA at 10°C.
Finally, the first unfolding transition has a very low amplitude and modest enthalpy (L, Fig. 4A; Table 1). Based on the sequence of the H5 loop, particularly the high content of U nucleotides on both the 5′ and 3′ sides of the loop, this transition was predicted to result from a series of noncanonical U●U pairs in the hairpin loop region, similar to the melting transition assignment above for the S1 RNA. In the imino proton region of the 2D H2O NOESY spectrum (Fig. 4C), the H5 RNA shows imino proton peaks between 9 and 11 ppm with observable NOE connectivities, in addition to the ten imino proton resonances predicted for the Watson-Crick and G●U wobble base pairs in the helix V stem, even under low-salt conditions. Although peaks from the noncanonical U●U base pairs tend to have higher intensities with stronger NOE connectivities at higher salt concentrations, the H5 RNA had to be investigated in the absence of monovalent salt to prevent dimer formation. Despite the unfavorable low-salt conditions, the NMR experiments do indicate the presence of multiple additional base pairs in the H5 loop region, which confirmed the assignment of the first low melting temperature transition to H5 loop structure unfolding (L, Fig. 4A). It is interesting to note that both the helix 5 and stem 1 hairpins of the yeast telomerase RNAs have loop structures including potential sequential noncanonical U●U base pairs, similar to what was previously observed in the stem 1 hairpin of the human telomerase RNA pseudoknot (Theimer et al. 2003).
The H5 and S2 sequences do not interact to form a bimolecular pseudoknot
In the absence of pseudoknot formation, one would expect that the melting profile of the entire sequence (nt 718–813) would simply be the linear addition of the two individual melting profiles for the H5 and S2 hairpins. An equimolar mixture of the H5 and S2 constructs was used to simulate the appearance of the competing two-hairpin conformation melting profile. The melting profile of the H5-S2 equimolar mixture at 50 mM NaCl is not clearly additive upon initial visual inspection (Fig. 4A). The melting temperature, enthalpy, and A260/A280 ratio of the first low-amplitude transition are a very good match for the loop structure transition of H5 alone. However, the remaining three expected transitions (two for S2 and one from H5) are overlapped and combined into the second, higher melting temperature transition. This is due to the similar melting temperatures of the H5 and S2 stems, 40.6 ± 0.3°C and 45.1 ± 0.2°C, respectively, as well as the H5 stem and the S2 additional transition (40.2 ± 0.5°C) (Table 1). It was possible to fit the higher amplitude second peak to two transitions (H5, S2*, Fig. 4A) with melting temperatures of 40.3 ± 0.2°C and 43.7 ± 0.2°C, respectively, although it was not feasible to also include the S2 low-amplitude transition. Thus, the transition at 40.3°C most likely includes both the overlapped H5 stem and S2 low-temperature transitions, while the transition at 43.7°C belongs to the S2 stem (including the 6-bp helix).
Under high-salt conditions (1M NaCl), the H5 and S2 stem unfolding transitions are slightly better separated on the temperature coordinate and, as a result, all four expected peaks are observable (L, H5, S2*, and the unlabeled S2 low-amplitude transition, Fig. 4A). In addition, the H5 dimer transition (D) observed for the H5 RNA alone at high-salt conditions also reappears in the H5-S2 mixture under high-salt conditions. There is no evidence in the thermal melting profiles of additional secondary structure formed in the H5-S2 mixture or the presence of the hypothetical tertiary contact between the two loops proposed in Dandjinou et al. (2004). However, this does not preclude the possibility that such interactions do occur in a unimolecular construct, in the absence of the nucleation penalties associated with bimolecular interactions.
The equimolar mixture of H5 and S2 was also examined using native PAGE experiments, and no alternative conformations were observed (data not shown). This indicates that, even when annealed together at high concentrations, the H5 and S2 hairpins do not interact to form a bimolecular pseudoknot construct with a discontinuity in the loop 1 sequence. Bimolecular pseudoknot formation has been observed for a functional S. cerevisiae TER pseudoknot domain in the past (Qiao and Cech 2008), as well as the human telomerase RNA pseudoknot (Theimer et al. 2003), where the discontinuity occurs in the loop 2 region, far from any critical structural features. In this case, it is not clear if the bimolecular pseudoknot does not form because the discontinuity is in the loop 1 region, which has been shown to be involved in tertiary contacts with stem 2 (Shefer et al. 2007; Qiao and Cech 2008), or because of the stability of the H5 helix, which would compete with bimolecular structure formation. Based on the S2 melting profiles in the presence and absence of the H5 hairpin, the experimental data best agree with the two-hairpin model proposed by Zappulla and Cech (2004) and Dandjinou et al. (2004), as the most likely hairpin conformation in competition with the pseudoknot conformation. The pseudoknot domain in the context of the entire sequence could, therefore, exist either in the pseudoknot alone (Lin et al. 2004) or in a structural equilibrium between the two competing conformations.
WT exists in equilibrium between the pseudoknot and two-hairpin conformations
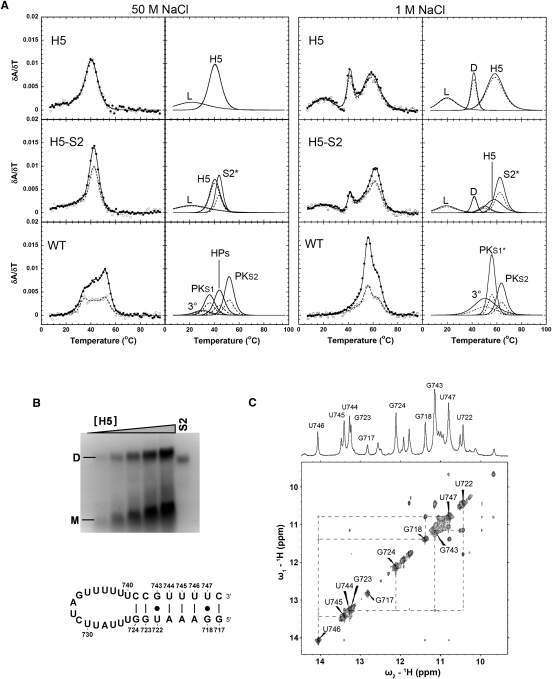
The RNA construct WT, contains all of the nucleotides (nt 718–813) involved in the secondary structural models of the pseudoknot region (Fig. 1A). The only difference between this construct and the PK construct is five additional nucleotides at the 5′ end of the RNA, which are not required for the formation of the pseudoknot but are required for the formation of the competing H5 stem. Four transitions are observed in the melting profiles of WT RNA under low-salt conditions (3°, PKS1, HPs, PKS2, Fig. 4A). Visual inspection of the melting profile makes it clear that, under low-salt conditions, the WT RNA does not simply form a two-hairpin structure similar to that observed for the H5-S2 equimolar mixture (the two-hairpin model mimic) (Fig. 4A), nor does it precisely match the pseudoknot melting profile (Fig. 2A).
Since pseudoknot unfolding pathways involve a hairpin unfolding intermediate, we investigated the possibility that the WT melting profile was a combination of the PK and H5+S2 melting profiles due to equilibrium between the two conformations. Examining the melting profile on this basis, it was possible to assign the second transition to unfolding of stem 1 in the pseudoknot conformation (PKS1, transition 2 in the PK melting profile), and the fourth transition to unfolding of stem 2 of the pseudoknot conformation (PKS2, transition 3 in the PK melting profile), although both transitions occur at somewhat higher melting temperatures in WT (Figs. 2A, 4A; Table 1). The third transition of WT bears a strong resemblance to the second transition in the H5+S2 melting profile, with only minor perturbations of the melting temperature, enthalpy, and A260/A280 ratio, so it was assigned to the unfolding of two helical stems (HPs, the helix V and the stem 2 hairpins). Finally, the first peak in the WT melting profile is assigned to the unfolding of the tertiary structure in the pseudoknot conformation (3°) with some contribution from the H5 hairpin loop structure unfolding. Therefore, at low monovalent salt concentrations, the pseudoknot region of the S. cerevisiae TLC1 exists in a conformational equilibrium between the pseudoknot (Lin et al. 2004) and the two-hairpin (H5-S2) (Dandjinou et al. 2004) conformations, with both states significantly populated.
Under high-salt conditions (monovalent and divalent), the H5+S2 transition disappears, and the melting curves of the WT and PK RNAs are very similar, consistent with our assignments of the unfolding transitions (high monovalent PK, WT, Figs. 2A, 4A, respectively; increasing magnesium concentration, Supplemental Figs. S2, S3, respectively). In fact, within the standard deviation of our measurements, the WT and PK RNA constructs form pseudoknots with essentially the same stability (Table 3), including the appearance of the 6-bp helix in stem 2 stacked across the helical junction onto stem 1 and unfolding, simultaneously (PKS1*, high-salt transition, Fig. 4A). Therefore, the conformational equilibrium in the WT RNA is shifted to favor exclusively the pseudoknot conformation at higher monovalent or divalent salt concentrations.
Compensatory base-pair-substituted WT constructs dimerize in solution
Previously, in vitro telomerase activity assays were used in conjunction with compensatory base-pair mutational analysis, to map the sequences in the pseudoknot region which are essential for telomerase activity (Qiao and Cech 2008). Helix-disrupting mutants (2A, 2B, 8A, and 8B) and helix-restoring double mutants (8A/2B and 8B/2A) (Fig. 5A) demonstrated that disrupting base-pairing in stem 1 significantly reduced telomerase activity and that this activity was completely restored by the helix-restoring complementary mutation (Qiao and Cech 2008). These results further suggested that the exact base-pair orientation of the stem 1 helix was not essential for telomerase activity, since these mutant RNAs had ∼20% more telomerase activity than the wild-type sequence (Qiao and Cech 2008). It was previously demonstrated that single nucleotide, base-pair, or base triple substitutions in tertiary structural regions of the human telomerase RNA pseudoknot resulted in decreases in telomerase activity that correlated closely with changes in the thermodynamic stability of the RNA pseudoknot (Theimer et al. 2005). Therefore, we investigated the thermodynamic stabilities of the S. cerevisiae TLC1 helix-restoring mutations in stem 1 to see if there was a similar correlation between thermodynamic stability and telomerase activity.
FIGURE 5.
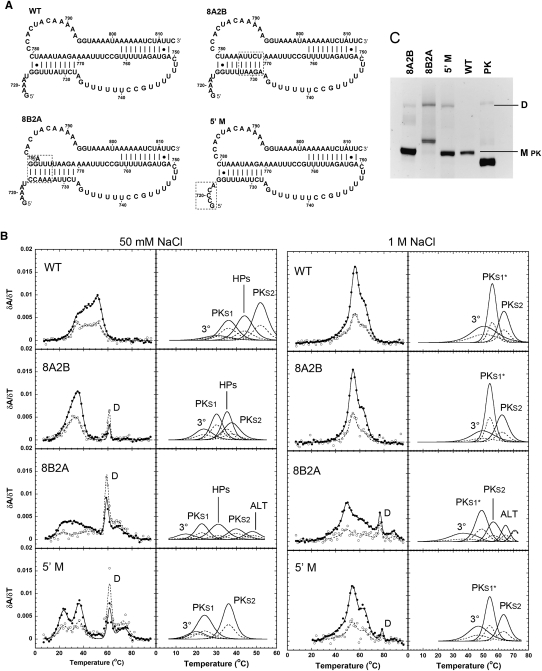
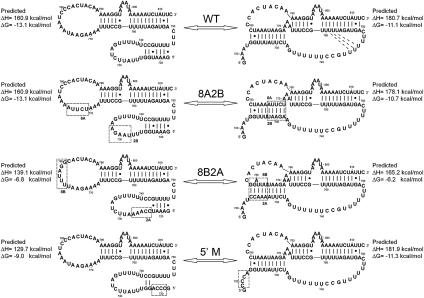
Comparisons between the WT (WT) pseudoknot domain construct and stem 1 and 5′-terminal mutant RNAs. (A) RNA constructs for the wild-type pseudoknot region (WT) and the 8A2B, 8B2A, and Δ5′ mutations. Dashed boxes enclose the mutated regions. (B) Profiles on the left show every fifth data point obtained at 260 nm (●) and 280 nm (○), and the calculated fits to the 260-nm (solid lines) and 280-nm data (dashed lines), with the individual transitions at 260 nm (solid lines) and 280 nm (dashed lines), which combine to generate the total calculated curves, shown separately to the right. The individual transition fits are labeled according to the correlated structural elements: (3°) tertiary structure, (PKS1) pseudoknot stem 1, (HPs) the overlapped H5 and S2* transitions, (PKS1*) pseudoknot stem 1 and 6-bp helix, (PKS2) pseudoknot stem 2, (ALT) alternative structure, respectively. (C) Native polyacrylamide gel. Monomer (M pk) and dimer (D) species are indicated to the right of the gel, and the PK RNA was loaded for a size comparison to the monomeric species.
Two RNA constructs were designed to examine the effects of stem 1 compensatory base-pair substitutions on the stability of the pseudoknot (Fig. 5A). The RNA construct 8A2B is a combination of the two stem 1 disrupting mutants, 8A (771AGAAU775 → UCUUA) and 2B (728AUUCU732 → UAAGA), swapping the two strands of the stem. A similar pair of compensatory mutations was made to obtain the RNA construct 8B2A, with six consecutive nucleotides mutated in the 8B region (776AAAUCC781 → UUUAGG) and five consecutive nucleotides mutated in the 2A region (723GGUUU727 → CCAAA). However, due to the G●U wobble base pair in this region of the wild-type stem, this construct has a 1-nt bulge (A779) in stem 1, which was present in the originally designed 8B2A construct (Qiao and Cech 2008). This bulge would result in destabilization of stem 1 base-pairing in the region closest to the 5′ end and be predicted to form a much less stable pseudoknot structure than 8A2B or WT.
Under low-salt conditions, the 8A2B RNA melting profile contained a set of overlapping peaks at lower temperatures and a single sharp peak at higher temperatures (D, Fig. 5B). The single unfolding transition at ∼60°C is extremely sharp, with a very high enthalpy, consistent with the formation of a dimeric species. In order to confirm that this transition corresponds to dimerization of the 8A2B RNA, native gel electrophoresis was performed on the wild-type and mutant RNA constructs (Fig. 5C). The 8A2B RNA migrates in three bands on the gel, while WT has only one band visible on the native gel. The lower band of 8A2B is by far the most intense and comigrates with the single band observed for WT. The top band in 8A2B has a much slower mobility in the gel and is significantly weaker than the main band, consistent with the smaller amplitude of the proposed dimer peak in the thermal melting profile. There is also a broad band between the monomer and dimer band, which corresponds to a weakly populated alternative conformation. Based on these results, we focused on the low-temperature transitions and their comparison to the WT profiles.
In addition to the expected effects on the pseudoknot conformation, the 8A2B substitution also alters the sequences of the helix V and stem 2 loops of the two-hairpin conformation (Fig. 6; Chappell and Lundblad 2004; Dandjinou et al. 2004; Lin et al. 2004; Zappulla and Cech 2004). Since the mutations only occur in the loop regions of the two hairpins, the identified base-pairing in the stems are still maintained in the 8A2B construct, and thus the unfolding transitions associated with those stems and the overall stability of the potential two-hairpin conformation was not expected to be greatly affected. However, the 728AUUCU732 → UAAGA (2B) mutation does substitute in four purine nucleotides on the 5′ side of the loop which could possibly form one G●U wobble and three Watson-Crick A-U base pairs with the tract of uridine nucleotides on the 3′ side of the H5 loop, which could affect the overall stability of the H5 hairpin stem and loop.
FIGURE 6.
Predicted structures and stabilities for the competing two-hairpin and pseudoknot conformations for WT pseudoknot domain constructs. The mutated sequences are indicated by dashed boxes. Predicted stabilities are based on the combination of the nearest-neighbor model and Turner 2004 data set from the Nearest Neighbor Database (NNDB) (Turner and Mathews 2010) and pseudoknot entropic penalties (Gultyaev et al. 1999), as appropriate. The predicted 3° structure between stem 2 and loop 1 triples (Shefer et al. 2007; Qiao and Cech 2008) is indicated on the WT pseudoknot structure with dashed black lines.
The lower-temperature region (5–50°C) contains a set of four overlapping transitions, which are similar in shape and A260/A280 ratios to those of the WT RNA, although the tms of these transitions are lower than wild type (3°, PKS1, HPs, PKS2, Fig. 5B; Table 2). Based on similarity to the WT melting profile, the first peak in the 8A2B melting profile is assigned to unfolding of secondary structure in the H5 loop region and tertiary interactions in the pseudoknot (3°). The increased enthalpy of this transition is consistent with the possibility of increased Watson-Crick base-pairing in the H5 hairpin loop. The second peak corresponds to the unfolding of stem 1 in the pseudoknot conformation (PKS1) and the fourth peak to the unfolding of stem 2 in the pseudoknot conformation (PKS2). The third peak is assigned, as in WT, to the unfolding of the helix V stem and the S2 hairpin of the competing two-hairpin conformation (HPs). Proportionally, this peak is larger in the 8A2B melting profile than in the WT profile, suggesting that the two-hairpin conformation is more populated in the 8A2B RNA than in the wild-type sequence. The decreased melting temperatures in the mutant 8A2B profile most likely come from the combination of the expected modest decrease in the stability of stem 1 based on less stable nearest-neighbor interactions compared to the wild-type sequence (Fig. 6; Turner et al. 1988), a possible minor decrease in the stem 2 hairpin stability based on changes in the loop sequence stability, as well as altered tertiary structural contacts in the pseudoknot conformation.
Under high-salt conditions (monovalent and divalent), the melting profile of 8A2B is nearly identical to the WT melting profile (3°, PKS1*, HPs, PKS2, Figs. 4A, 5B; Supplemental Figs. S3, S4). The 7–10°C difference in melting temperature between 8A2B and WT under low-salt conditions decreases to a 2–3°C difference under high-salt conditions, the dimer peak almost completely vanishes, and the two-hairpin conformation transition disappears, as observed for the WT profile. This indicates that increasing the salt concentration preferentially stabilizes the monomer pseudoknot conformation over the two-hairpin (HPs) and dimer (D) conformations. Under high-salt conditions, the 8A2B RNA pseudoknot is only ∼1.0 kcal mol−1 less stable than the WT RNA pseudoknot (Table 3), consistent with the expected decrease in stability of the 8A2B helix (∼0.4 kcal mol−1). In summary, optical melting data and native gel electrophoresis suggest that the mutant 8A2B folds into a pseudoknot with very similar properties to the WT sequence, with only a small difference in thermodynamic stability, consistent with its wild-type telomerase activity in vitro (Qiao and Cech 2008).
At 50 mM NaCl, the optical melting profile of the 8B2A RNA also can be divided into two regions: 5–50°C and 50–95°C (3°, PKS1*, HPs, PKS2, ALT, Fig. 5B). The 60°C transition again bears the characteristic hallmarks of some type of dimerization (D), and the dimer peak is more intense than in the 8A2B melting profile. The 8B2A RNA migrates in three bands by native gel electrophoresis (Fig. 5C). The top band has the same mobility as the proposed 8A2B dimer band and a greater intensity, consistent with the increased amplitude of the dimer transition observed in the optical melting profile. The lowest band, which comigrates with WT and 8A2B, is very faint, although it was reproducibly present. Unlike WT and 8A2B, there is another much darker band above this band, suggesting that this sequence forms a significant amount of an alternative conformation. Although the Mfold program (Zuker 2003) cannot predict pseudoknots, it does predict the two-hairpin conformation of the WT region for both WT and 8A2B. However, the Mfold program also predicts three variations of a three-helix junction-containing structure for the 8B2A sequence (Supplemental Fig. S5), consistent with the possible alternative conformation suggested by the optical melting data (ALT) and native gel electrophoresis. The Kinefold pseudoknot prediction program (Xayaphoummine et al. 2005) predicted both the two-hairpin and pseudoknot (including the 6-bp helix) conformations for the WT and 8B2A sequences but not the three-helix junction conformation for 8B2A.
In addition to the expected effects on the pseudoknot conformation, the 2A substitution, 723GGUUU727 → CUAAA, also alters the sequences of the helix V stem and loop and the stem 2 loop of the two-hairpin conformation (Fig. 6; Chappell and Lundblad 2004; Dandjinou et al. 2004; Lin et al. 2004; Zappulla and Cech 2004). In the helix V loop region, this mutation results in the loss of two of the G-C base pairs in the stem and replaces three potential U●U base pairs in the loop with three Watson-Crick A-U base pairs. Since this mutation significantly perturbs the base-pairing of the H5 hairpin, the overall stability of the potential two-hairpin conformation was expected to be significantly affected. In the low-temperature region (5–50°C) of the 8B2A melting profile, the profile can best be fit to five unfolding transitions. Like the 8A2B mutation, the second peak appears to correspond to the unfolding of stem 1 in the pseudoknot conformation (PKS1) and the fourth peak to the unfolding of stem 2 in the pseudoknot conformation (PKS2), with a similar decrease in melting temperatures. The first unfolding transition most likely corresponds to the unfolding of tertiary structure (3°) and overlapping H5 loop unfolding effects. However, the third (HPs or ALT) and fifth (ALT) transitions bear little to no resemblance to any of the previously assigned transitions, suggesting that these transitions are the result of the possible alternative three-helix conformation of this sequence predicted based on the native gel electrophoresis (Fig. 5C) and by the Mfold prediction program (Zuker 2003). Under high-salt conditions (monovalent and divalent), the melting profile of 8B2A more closely approximates the WT melting profile (3°, PKS1*, PKS2, Fig. 5B; Supplemental Fig. S6), although the alternative conformation (ALT) is still highly populated.
The 8B2A RNA pseudoknot is 5.2 kcal mol−1 less stable than the WT RNA pseudoknot, which would be expected to result in a significant decrease in telomerase activity, based on results from the human telomerase RNA investigations. However, our calculations indicate that the stability of the competing two-hairpin conformation would also decrease by ∼6.3 kcal mol−1 compared to the calculated stability of the WT two-hairpin conformation (Fig. 6). This may explain why the 8B2A pseudoknot retains wild-type in vitro telomerase activity (Qiao and Cech 2008), when pseudoknot stability differences of this magnitude in the human telomerase RNA system resulted in <10% activity in vitro (Theimer et al. 2005).
Abolishing the H5 helix results in the formation of only the pseudoknot conformation
The last RNA construct in this study, 5′ M (Fig. 5A), was generated by mutating the five consecutive nucleotides at the 5′ end of WT before the pseudoknot stem 1–forming sequence to prevent the formation of a stable helix V stem (with a single G718 nucleotide maintained at the 5′ end to promote efficient in vitro transcription by T7 RNA polymerase). The mutation made in 5′ M was designed to disrupt four of the helix V stem base pairs, with only a slight perturbation of the stability of the pseudoknot conformation, since the modified sequence is not expected to be involved in pseudoknot formation (Figs. 5A, 6). The expected differences in pseudoknot stability for this RNA are based on examination of the thermodynamic parameters associated with contributions from the 5′ dangling single-stranded nucleotides (U in WT [WT] and A in M, respectively) (Turner and Mathews 2010).
At 50 mM NaCl, the 5′ M RNA optical melting profile, like the other full-length mutant melting profiles, has a set of melting transitions for the monomeric conformations at the low temperature region (5–50°C) and a sharp dimer peak (D) at ∼60°C (Fig. 5B). Native gel electrophoresis (Fig. 5C) confirmed the presence of one species with the same mobility as the WT sequence and one lower mobility species with the same mobility as the previously identified dimeric species. As predicted, the 5′ M monomer transitions bear a very strong resemblance to the PK optical melting transitions (3°, PKS1, PKS2, Figs. 2A, 5B), with no evidence of the two-hairpin transition seen in the WT melting profiles. In fact, the enthalpies and A260/A280 ratios for the PK, WT, and 5′ M RNAs are completely consistent with formation of the same basic pseudoknot structure (Tables 1–3; Supplemental Table S1). It is interesting to note, however, that under low-salt conditions, the WT RNA melting temperatures are uniformly higher than those of the PK and 5′ M RNAs (Table 1). The 5′ M melting temperatures for all three transitions are between 10–15°C lower than WT (WT), while the melting temperatures for PK are closer to WT melting temperatures.
At 1M NaCl, the melting profile of 5′ M changes shape, similar to the PK and WT profiles (Figs. 2B, 5B). As seen with all other modified WT sequences, the dimer transition (D) shrinks significantly, indicating a decrease in the population of the dimeric species as the pseudoknot is stabilized by increasing monovalent salt concentrations (or divalent salt concentrations) (Supplemental Fig. S6). At 1M NaCl, the melting temperatures for the equivalent transitions in WT, PK, and 5′ M under high-salt conditions are all within ∼1–2°C of each other, with the 5′ M melting temperatures consistently the lowest of the three (Table 2). This indicates that whatever causes the differences in tm under low-salt conditions is alleviated, for the most part, at higher salt concentrations. Overall, these results indicate that the presence and sequence of the 5′ single-stranded nucleotides do have a small but measureable effect on the stability of the pseudoknot, which is not entirely consistent with the predictions based on a 5′ single-stranded nucleotide stacking on a regular Watson-Crick helix. This is most likely due to proximity and potential transient interactions with the nearby loop 2 nucleotides.
DISCUSSION
In this study, the pseudoknot domain of TLC1 was characterized as an isolated construct using in vitro biophysical methods. Previous studies have demonstrated that TLC1 can be considered a flexible scaffold for protein binding and that minimized secondary structural elements are predicted to fold like the wild-type 1.2-kb sequence (Zappulla et al. 2005; Qiao and Cech 2008). All of the designed minimized TLC1 RNAs (mini-T RNAs, 384–500 nt) retained essential telomerase function in vivo and in vitro (Zappulla et al. 2005), and the micro-T RNA (170 nt), which consists of only the core pseudoknot domain, the template sequence, and a loop-closing helical region, exhibits similar activity to the mini-T (500 nt) RNA in vitro, in the presence of the telomerase protein (Qiao and Cech 2008). In addition, hybrid constructs where the pseudoknot domain of TLC1 was replaced by the Oxytricha nova and human telomerase RNA pseudoknots were also demonstrated to generate functional telomerase enzymes in vivo (Chappell and Lundblad 2004). Therefore, the TLC1 pseudoknot domain was identified as an independently folding RNA domain, and the folding properties of this individual domain most likely remain the same in isolation and in the context of the entire TLC1 RNA, making this an ideal candidate for a reductionist approach.
Similar in vitro investigations of the isolated hTR pseudoknot indicated that the pseudoknot thermodynamic properties correlated closely with in vitro telomerase activity assays involving the full-length RNA and the human telomerase reverse transcriptase protein (Theimer et al. 2005). In vitro studies have also been performed on isolated pseudoknot domains from K. lactis and ciliate telomerase RNAs (Shefer et al. 2007; Ulyanov et al. 2007). While it is expected that interactions with the telomerase reverse transcriptase and other accessory proteins in the context of the entire TLC1 sequence, can and probably do affect the conformation of the pseudoknot domain, understanding the fundamental properties of the isolated RNA structural element provides a groundwork for elucidating the pseudoknot properties and function in the more complicated in vivo environment.
A structural role for the H5 stem forming sequence
Recent work demonstrating both the existence of the pseudoknot conformation and validating the presence of tertiary structure in the pseudoknot (Shefer et al. 2007; Qiao and Cech 2008) in the S. cerevisiae TLC1 eliminated much of the structural ambiguity in this region based on previous phylogenetic studies (Chen and Greider 2004). However, in this study, we have demonstrated that, while the pseudoknot conformation is absolutely present in RNA constructs of this region, the two-hairpin models are not invalidated by the presence of the pseudoknot conformation. Our data demonstrate that the pseudoknot is in equilibrium with a two-hairpin unfolding intermediate, that is essentially identical to the one predicted by two of the models (Fig. 1A; Dandjinou et al. 2004; Zappulla and Cech 2004), under low-salt conditions. This result potentially explains the observed conservation of sequences in the 5′ portion of the helix V pairing region (Chappell and Lundblad 2004; Dandjinou et al. 2004; Zappulla and Cech 2004) which would not be predicted to be important based on the pseudoknot model alone (Lin et al. 2004).
Examination of the 62 Saccharomyces telomerase RNA sequences currently available in the Rfam database (Daub et al. 2008; Gardner et al. 2009, 2011) reveals that the only complete base-pair covariation (GC → AU or AU → GC) in the pseudoknot region occurs in the H5 stem. The G724-C741 base pair is converted in 14 sequences to A724-U741, with 10 additional sequences containing the wobble G724●U741 pair (base-pair covariation mapped on the hairpin and pseudoknot structures) (Supplemental Fig. S8). In the pseudoknot conformation, G724 is located in the 5′-terminal single-stranded region and C741 in loop 1 outside of the tertiary structural region and would have no structural reason to covary. In fact, nearly all of the nucleotide substitutions found in the H5 helix-forming sequences (718–724 and 741–747) maintain base-pairing, typically by converting an A-U or G-C pair to a G●U wobble base pair, or by converting a G●U wobble to an A-U base pair. These mutations both maintain the integrity of the H5 helix and the thermodynamic equilibrium between the pseudoknot and hairpin conformations, since the thermodynamically weaker G●U wobble and A-U base pairs in the H5 stem are not converted to the more stable G-C base pair, which would potentially shift the equilibrium more toward the hairpin conformation.
Our experiments indicate that altering the conserved helix V sequences which are not involved in pseudoknot structure tends to result in dimerization, modest decreases in pseudoknot stability, and alternative conformations, suggesting that preventing alternative structure formation may be the primary reason for phylogenetic conservation of relatively weak base pairs in the helix V-specific sequences. Therefore, the potential to form the two-hairpin conformation, a pseudoknot-folding pathway intermediate, is likely to be maintained in this system to prevent the formation of different, potentially thermodynamically or kinetically trapped structures in the context of the entire RNA, which would not easily interconvert with the functionally relevant pseudoknot conformation.
Tertiary structure in the S. cerevisiae TLC1 pseudoknot
Our studies have determined that tertiary structure contributes, on average, −1.5 kcal mol−1 to the free energy (ΔG ° (37°C), 1 M NaCl) of the pseudoknot conformation, approximately half the amount of stabilization due to tertiary structure in the human telomerase RNA pseudoknot (Theimer et al. 2005). This difference is consistent with the different properties of the two pseudoknots at their respective helical junctions. In the case of the human telomerase RNA, the tertiary structure spans a highly structured, uninterrupted, coaxially stacked helical junction, generating a network of six continuous tertiary structural contacts across the helical junction, along stem 2, down to the bulge U177 nucleotide in stem 2 (Theimer et al. 2005; Kim et al. 2008). The structure near the helical junction in stem 2 of the S. cerevisiae TER pseudoknot does not appear to be base-paired under low-salt conditions. Even when base-pairing does occur at higher monovalent salt concentrations, the expected 3-nt bulge loop on the continuous strand between stem 1 and stem 2 and the predicted 6-nt bulge loop further down in stem 2 (in approximately the same position as the U177 bulge nucleotide in the human telomerase RNA [Chen et al. 2000]) would likely prevent a continuous stretch of tertiary structural contacts from forming across the helical junction. In addition, the Hoogsteen base triples identified for the S. cerevisiae TER pseudoknot occur below the proposed 6-bulge loop (Qiao and Cech 2008), further separating this tertiary structural element from stacking with possible tertiary structural elements near the helical junction. While the tertiary structural thermodynamic stability identified in this study is consistent with the known interactions, it does not preclude the formation of additional, as yet unidentified, tertiary structural interactions in the S. cerevisiae TER pseudoknot, since the thermodynamic parameters associated with tertiary structure formation are not well-established.
Additional tertiary contacts closer to the helical junction, similar to those seen in the human telomerase RNA pseudoknot (Theimer et al. 2005; Kim et al. 2008) and proposed for the K. lactis TER pseudoknot (Brown et al. 2007; Shefer et al. 2007), can be proposed to form in the S. cerevisiae pseudoknot between the three consecutive A-U base pairs in the 6-bp helix (close to the helix junction) and a run of three U nucleotides (U738–740) at the 5′ end of loop 1 (Fig. 6; Supplemental Fig. S8). Mutation of the A790–U795 sequence in S. cerevisiae does not have a large impact on telomerase activity (Qiao and Cech 2008). However, a similar magnitude effect was seen in this region of the K. lactis pseudoknot, which was still proposed to be base-paired (Shefer et al. 2007). The phylogenetic conservation of the U738–740 nucleotides is 100% based on the 62 Saccharomyces telomerase RNA sequences in the Rfam database (Daub et al. 2008; Gardner et al. 2009, 2011), similar to the three U nucleotides (745–747) demonstrated to be involved in tertiary structure (Qiao and Cech 2008). Consistent with the possibility of forming loop 2-loop1 Hoogsteen base pairs and/or stem 2-loop 1 Hoogsteen base triples, A789-A790-A791-A792 (loop 2 and stem 2), U765-U766 (stem 2), and U738-U739-U740 (loop 1) are absolutely conserved (Supplemental Fig. S8), as are the known pseudoknot stem-forming and tertiary structure-forming sequences in the S. cerevisiae telomerase RNA pseudoknot (Qiao and Cech 2008). Nearly all of the other loop nucleotides in the pseudoknot vary in sequence in 10%–50% of the sequences in the Rfam database (Supplemental Fig. S8). Thus, thermodynamic identification of the 6-bp helix in stem 2 and examination of the phylogenetic conservation of pseudoknot sequences suggest the possibility of additional tertiary structures in the S. cerevisiae telomerase RNA pseudoknot still to be characterized.
Correlation of pseudoknot thermodynamic stability and telomerase activity
Previous studies included in vitro telomerase activity assays of stem 1 mutations in the S. cerevisiae TLC1 pseudoknot domain (Qiao and Cech 2008). We investigated the possibility that there would be a correlation between thermodynamic stability and telomerase activity for these mutations in the S. cerevisiae TLC1 pseudoknot, similar to previous studies of human telomerase RNA pseudoknot tertiary structural mutations (Theimer et al. 2005). The S. cerevisiae mutations swap 5 bp at a time in stem 1 of the pseudoknot, the activity of the mutations was nearly 20% greater than wild type (Qiao and Cech 2008), and it is not known if these specific base pairs are involved in tertiary structural contacts in the S. cerevisiae TLC1 pseudoknot structure. In both cases (8A2B and 8B2A), we observed that the tertiary structural transitions were still present and fairly similar to the wild-type tertiary structural transition, consistent with their robust telomerase activities, and that base-pair orientation in stem 1 is not critical to telomerase activity.
The 8A2B pseudoknot behaves very similarly to the WT pseudoknot under high salt and in the presence of magnesium, with a modest decrease in stability (ΔG ° = 1.0 kcal mol−1) for the pseudoknot and no significant predicted effect on hairpin stability. In the case of the 8B2A pseudoknot, both the pseudoknot and hairpin conformations of the 8B8A mutations decrease in stability by 5.4 (experimentally determined) and 6.3 (predicted) kcal mol−1, respectively. Without determining the actual stabilities of the mutated hairpin elements, it is not possible to definitively calculate the changes in the pseudoknot-hairpin equilibrium. However, predictions should be reasonably accurate for hairpin structures in the absence of noncanonical and tertiary structural interactions. This suggests that wild-type telomerase activity is maintained by the 8A2B pseudoknot because the change in the conformational equilibrium between the pseudoknot and hairpin conformations is not large, and therefore the dominant conformation under physiological conditions remains the pseudoknot with essential wild-type tertiary structure.
Comparisons with other yeast telomerase RNA pseudoknot regions
The pseudoknot triple helix and the three-helix junction (p6.1 in the human telomerase RNA) have been identified as tertiary structural motifs that are conserved across the highly divergent yeast, ciliate, and vertebrate telomerase RNAs (Lin et al. 2004; Theimer et al. 2005; Brown et al. 2007; Shefer et al. 2007; Qiao and Cech 2008), based on examination of the available data in the literature and in the Rfam database (Daub et al. 2008; Gardner et al. 2009, 2011). Structural similarities between the Kluveromyces and Saccharomyces pseudoknot families include the separation of the stem 2 A-U base pairs and loop 1 nucleotides which form the triple helix from the helical junction, a region of nucleotides at the helical junction which may or may not be base-paired, similar conservation patterns, and a greater emphasis on conservation of stem 2 sequences for function than on stem 1 sequences (Shefer et al. 2007; Qiao and Cech 2008). Recently characterized telomerase RNA pseudoknots from Candida species (Gunisova et al. 2009) also have similar characteristics, including proposed triple-helix interactions. Intriguingly, several of the proposed Candida pseudoknot structures have two potential triple-helix regions in stem 2 which are separated by a bulge, as we propose for the S. cerevisiae pseudoknot sequences. Unlike our model, the stem 2 bulge in the Candida sequences is small, typically 1 nt. However, the computer algorithm used in that work specifically included a parameter which dictated that the bulge loop in stem 2 be no more than 2 nt (Gunisova et al. 2009), thus pseudoknot structures which included a larger bulge would not have been predicted. The Schizosaccharomyces pombe telomerase RNA has also been identified and predicted (using Kinefold software) to have a pseudoknot structure (Webb and Zakian 2008). Without a larger number of related sequences for comparison, the pseudoknot in S. pombe has not yet been characterized; however, it does appear to contain the appropriate stretch of uridine nucleotides in the loop 1 region for base triple formation.
CONCLUSIONS
Overall, this work characterizes the structural and thermodynamic properties of the S. cerevisiae TLC1 pseudoknot in vitro and the contributions of tertiary structure to pseudoknot stability, merges the existing structural models of the pseudoknot region into a coherent and internally consistent model of this domain, provides a more detailed description of the pseudoknot secondary structure, and compares these properties to the human telomerase RNA pseudoknot and other known yeast telomerase RNA sequences. The potential for additional tertiary structure in the Saccharomyces telomerase pseudoknot has been suggested, and a role for the conserved H5 sequence has been proposed, based on the experimental data. Further experimentation in the presence of the telomerase protein in vitro and in vivo (in the presence of additional TLC1 structural elements) will be necessary to demonstrate the functional relevance of these observations and to confirm the presence or absence of the proposed additional tertiary structural contacts.
MATERIALS AND METHODS
RNA synthesis and purification
RNA oligonucleotides were prepared by large scale in vitro transcription using purified His6-tagged T7 RNA polymerase and partially double-stranded DNA templates as described previously (Milligan et al. 1987; Du et al. 1996; Qiu et al. 1996). DNA template sequences for RNA transcription were ordered from Integrated DNA Technologies (IDT) at 1-μmol scale. Transcribed RNAs were purified by denaturing polyacrylamide gel electrophoresis (PAGE, 19:1 acrylamide: Bis-acrylamide 7M urea) in 1× TBE (90 mM Tris-borate, 2 mM EDTA), visualized by UV shadowing, and electroeluted using a Whatman electroeluter. The recovered RNA was loaded onto a HiTrap Q column (GE Healthcare), eluted with 1.5 M NaCl, and ethanol precipitated. The purified RNAs were resuspended in H2O followed by desalting and concentration by ultrafiltration using an Amicon stirred cell (Millipore) to a concentration of 40∼500 μM and stored at −20°C.
Thermal denaturation
RNA samples for thermal denaturation experiments (0.13–1.63 μM) were prepared in 10 mM MOPS (Sigma) buffer, pH 7.0 and varying concentrations of NaCl (50, 100, 200, 500 mM, and 1 M) and/or MgCl2 (50, 100, 200, 500 μM, and 1 mM MgCl2), annealed at 65°C for 5 min, slow-cooled to room temperature, and equilibrated in the spectrophotometer at 5°C for 20 min prior to data collection.
Thermal melting experiments were collected on a Varian Cary 300 UV-VIS spectrophotometer equipped with a Peltier heating accessory and temperature probe in double-beam mode. Absorbance was recorded as a function of probe temperature at 260 and 280 nm simultaneously. The temperature was increased at a rate of 0.3°C per minute from 5–95°C, with data collected at 0.3°C intervals. Melting profiles were obtained by taking the first derivative of the absorbance with respect to temperature (∂A/∂T) and subjecting it to nonlinear least-squares parameter estimation of tm,i (melting temperature), ΔH,i (van't Hoff enthalpy), A260i and A280i (∂A/∂T amplitude at 260 and 280 nm) for each ith transition on application of a sequential independent two-state unfolding model by using the tmelt program (Theimer et al. 1998). The tmelt program provides a total calculated curve for the entire set of data at each wavelength, as well as individual curves for each transition that was used in the calculation of the final total fit to the data, at both wavelengths.
Nuclear magnetic resonance spectroscopy
Purified RNAs were concentrated and exchanged into NMR buffer by ultrafiltration (10 mM potassium phosphate buffer, pH 6.0, 50 mM potassium chloride for S1/S1-mod RNAs, and 10 mM potassium phosphate buffer, pH 6.0 for H5 RNA) using an Amicon stirred cell (Millipore). NMR sample concentrations were ∼0.5 mM. No salt was added to the H5 NMR sample to prevent the dimer formation observed in UV denaturation studies. All NMR spectra were acquired using an Avance Bruker spectrometer operating at a 1H frequency of 700 MHz equipped with a single Z-axis gradient cryoprobe. Exchangeable proton spectra were measured in 90% H2O/10% D2O at 283K. Exchangeable protons in the Watson-Crick, as well as non-Watson-Crick, base pairs were examined and assigned based on 1H 2D NOESY spectra. The H2O resonance was suppressed in 90% H2O samples using a 1-1 spin-echo water suppression scheme (Connor et al. 1987). Spectra were processed using TOPSPIN 2.1 (Bruker, Inc.), and assignments were made using SPARKY 3 (University of California, San Francisco, CA).
Native polyacrylamide gel electrophoresis
RNA samples for native PAGE were annealed in nanopure H2O at 65°C for 5 min and slow-cooled to room temperature. Each RNA sample was then mixed with 6× loading buffer (0.25% [w/v] bromophenol blue, 0.25% [w/v] xylene cyanol, and 30% [v/v] glycerol in H2O [Sigma]) and diluted with nanopure H2O. The final concentrations of these RNA samples ranged from 50 nM–1.5 μM. Samples were loaded onto native polyacrylamide gels; 20% 19:1 acrylamide:bisacrylamide, 1× TBM (90 mM Tris/Borate, 10 mM magnesium chloride) for H5 and S2 RNAs, and 12% 19:1 acrylamide:bisacrylamide, 1× TBE (90 mM Tris/Borate, 10 mM EDTA) for WT and WT mutants. Gels were run at 4°C and 20 W for ∼4 h. After electrophoresis, gels were stained with SYBR Gold stain (Invitrogen) and photographed using the ChemiDoc XRS system from Bio-Rad.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
Work was funded through the University at Albany, State University of New York start-up funds.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.030924.111.
REFERENCES
- Brown Y, Abraham M, Pearl S, Kabaha MM, Elboher E, Tzfati Y 2007. A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res 35: 6280–6289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell AS, Lundblad V 2004. Structural elements required for association of the Saccharomyces cerevisiae telomerase RNA with the Est2 reverse transcriptase. Mol Cell Biol 24: 7720–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-L, Greider CW 2003. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J 22: 304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-L, Greider CW 2004. An emerging consensus for telomerase RNA structure. Proc Natl Acad Sci 101: 14683–14684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-L, Blasco MA, Greider CW 2000. Secondary structure of vertebrate telomerase RNA. Cell 100: 503–514 [DOI] [PubMed] [Google Scholar]
- Collins K 2006. The biogenesis and regulation of telomerase holoenzymes. Nat Rev Mol Cell Biol 7: 484–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor S, Everett J, Nicholson JK 1987. Spin-echo proton NMR spectroscopy of urine samples. Water suppression via a urea-dependent T2 relaxation process. Magn Reson Med 4: 461–470 [DOI] [PubMed] [Google Scholar]
- Dandjinou AT, Lévesque N, Larose S, Lucier J-F, Elela SA, Wellinger RJ 2004. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr Biol 14: 1148–1158 [DOI] [PubMed] [Google Scholar]
- Daub J, Gardner PP, Tate J, Ramsköld D, Manske M, Scott WG, Weinberg Z, Griffiths-Jones S, Bateman A 2008. The RNA WikiProject: Community annotation of RNA families. RNA 14: 2462–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Giedroc DP, Hoffman DW 1996. Structure of the autoregulatory pseudoknot within the gene 32 messenger RNA of bacteriophages T2 and T6: A model for a possible family of structurally related RNA pseudoknots. Biochemistry 35: 4187–4198 [DOI] [PubMed] [Google Scholar]
- Fresco J, Klotz L, Richards E 1963. A new spectroscopic approach to the determination of helical secondary structure in ribonucleic acids. Cold Spring Harb Symp Quant Biol 28: 83–90 [Google Scholar]
- Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. 2009. Rfam: Updates to the RNA families database. Nucleic Acids Res 37: D136–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. 2011. Rfam: Wikipedia, clans, and the “decimal” release. Nucleic Acids Res 39: D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW 1996. Telomere length regulation. Annu Rev Biochem 65: 337–365 [DOI] [PubMed] [Google Scholar]
- Guiducci C, Cerone M, Bacchetti S 2001. Expression of mutant telomerase in immortal telomerase-negative human cells results in cell cycle deregulation, nuclear and chromosomal abnormalities, and rapid loss of viability. Oncogene 20: 714–725 [DOI] [PubMed] [Google Scholar]
- Gultyaev AP, van Batenburg FH, Pleij CW 1999. An approximation of loop free energy values of RNA H-pseudoknots. RNA 5: 609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunisova S, Elboher E, Nosek J, Gorkovoy V, Brown Y, Lucier J-F, Laterreur N, Wellinger RJ, Tzfati Y, Tomaska L 2009. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA 15: 546–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L 2003. Biochemical aspects of telomerase function. Cancer Lett 194: 139–154 [DOI] [PubMed] [Google Scholar]
- Huang C, Yu Y-T 2010. Targeted 2′-O methylation at a nucleotide within the pseudoknot of telomerase RNA reduces telomerase activity in vivo. Mol Cell Biol 30: 4368–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachouri-Lafond R, Dujon B, Gilson E, Westhof E, Fairhead C, Teixeira MT 2009. Large telomerase RNA, telomere length heterogeneity, and escape from senescence in Candida glabrata. FEBS Lett 583: 3605–3610 [DOI] [PubMed] [Google Scholar]
- Kelleher C, Teixeira MT, Forstemann K, Lingner J 2002. Telomerase: Biochemical considerations for enzyme and substrate. Trends Biochem Sci 27: 572–579 [DOI] [PubMed] [Google Scholar]
- Kim N-K, Zhang Q, Zhou J, Theimer CA, Peterson RD, Feigon J 2008. Solution structure and dynamics of the wild-type pseudoknot of human telomerase RNA. J Mol Biol 384: 1249–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk KE, Harmon BP, Reichardt IK, Sedat JW, Blackburn EH 1997. Block in anaphase chromosome separation caused by a telomerase template mutation. Science 275: 1478–1481 [DOI] [PubMed] [Google Scholar]
- Lai CK, Miller MC, Collins K 2003. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol Cell 11: 1673–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Ly H, Hussain A, Abraham M, Pearl S, Tzfati Y, Parslow TG, Blackburn EH 2004. A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proc Natl Acad Sci 101: 14713–14718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hendrick LL, Cech TR 1994. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev 8: 1984–1998 [DOI] [PubMed] [Google Scholar]
- Mason DX, Goneska E, Greider CW 2003. Stem-loop IV of Tetrahymena telomerase RNA stimulates processivity in trans. Mol Cell Biol 23: 5606–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Blackburn EH 1995. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376: 403–409 [DOI] [PubMed] [Google Scholar]
- Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res 15: 8783–8798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F, Cech TR 2008. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol 15: 634–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Kaluarachchi K, Du Z, Hoffman DW, Giedroc DP 1996. Thermodynamics of folding of the RNA pseudoknot of the T4 gene 32 autoregulatory messenger RNA. Biochemistry 35: 4176–4186 [DOI] [PubMed] [Google Scholar]
- Romero DP, Blackburn EH 1991. A conserved secondary structure for telomerase RNA. Cell 67: 343–353 [DOI] [PubMed] [Google Scholar]
- Shefer K, Brown Y, Gorkovoy V, Nussbaum T, Ulyanov NB, Tzfati Y 2007. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol Cell Biol 27: 2130–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE 1994. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409 [DOI] [PubMed] [Google Scholar]
- Theimer CA, Wang Y, Hoffman DW, Krisch HM, Giedroc DP 1998. Non-nearest neighbor effects on the thermodynamics of unfolding of a model mRNA pseudoknot. J Mol Biol 279: 545–564 [DOI] [PubMed] [Google Scholar]
- Theimer CA, Finger LD, Trantirek L, Feigon J 2003. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc Natl Acad Sci 100: 449–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer CA, Blois CA, Feigon J 2005. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell 17: 671–682 [DOI] [PubMed] [Google Scholar]
- Turner DH, Mathews DH 2010. NNDB: The nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res 38: D280–D282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D, Sugimoto N, Freier S 1988. RNA structure prediction. Annu Rev Biophys Biophys Chem 17: 167–192 [DOI] [PubMed] [Google Scholar]
- Tzfati Y, Fulton TB, Roy J, Blackburn EH 2000. Template boundary in a yeast telomerase specified by RNA structure. Science 288: 863–867 [DOI] [PubMed] [Google Scholar]
- Ulyanov NB, Shefer K, James TL, Tzfati Y 2007. Pseudoknot structures with conserved base triples in telomerase RNAs of ciliates. Nucleic Acids Res 35: 6150–6160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CJ, Zakian VA 2008. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat Struct Mol Biol 15: 34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xayaphoummine A, Bucher T, Isambert H 2005. Kinefold web server for RNA/DNA folding path and structure prediction including pseudoknots and knots. Nucleic Acids Res 33: W605–W610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR 2004. Yeast telomerase RNA: A flexible scaffold for protein subunits. Proc Natl Acad Sci 101: 10024–10029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Goodrich K, Cech TR 2005. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat Struct Mol Biol 12: 1072–1077 [DOI] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]